Abstract
Research on the gut microbiota of free-ranging mammals is offering new insights into dietary ecology. However, for free-ranging primates, little information is available for how microbiomes are influenced by ecological variation through time. Primates inhabiting seasonal tropical dry forests undergo seasonally specific decreases in food abundance and water availability, which have been linked to adverse health effects. Throughout the course of a seasonal transition in 2014, we collected fecal samples from three social groups of free-ranging white-faced capuchin monkeys (Cebus capucinus imitator) in Sector Santa Rosa, Área de Conservación Guanacaste, Costa Rica. 16S rRNA sequencing data reveal that unlike other primates, the white-faced capuchin monkey gut is dominated by Bifidobacterium and Streptococcus. Linear mixed effects models indicate that abundances of these genera are associated with fluctuating availability and consumption of fruit and arthropods, whereas beta diversity clusters by rainfall season. Whole shotgun metagenomics revealed that the capuchin gut is dominated by carbohydrate-binding modules associated with digestion of plant polysaccharides and chitin, matching seasonal dietary patterns. We conclude that rainfall and diet are associated with the diversity, composition, and function of the capuchin gut microbiome. Additionally, microbial fluctuations are likely contributing to nutrient uptake and the health of wild primate populations.
Subject terms: Microbial ecology, Molecular ecology
Introduction
Mammalian gut microbiome research has demonstrated pervasive interactions between the microbiota and both intrinsic and extrinsic factors of the host, including diet, phylogeny, health, and behavior [1–10]. Comparatively little is known about the microbiota of free-ranging mammals, and the majority of wild studies focus on single snapshots in time [11–14]. For free-ranging primates, knowledge of how ecological and social variation influences relationships among health, ecology, and the gut microbial communities of host animals is a growing area of research [15–20].
Given the difficulty of sample acquisition [21], there remains a paucity of longitudinal microbiome research on free-ranging animals. Through replicate sampling of individual study subjects, longitudinal analyses can reveal how subtle shifts in health and diet affect microbial communities. The few longitudinal studies of free-ranging mammals suggest that gut microbial composition is not stable over time, and is influenced by environmental fluctuations [9, 22, 23]. Seasonal variation in climate can influence habitat productivity, and thus dietary composition of resident animals. For example, marked seasonal variation in the gut microbiota of wild wood mice results from a dietary transition between arthropod-based and plant-based diets corresponding with fluctuations in Proteobacteria, and Lactobacillus [9]. Furthermore, food shortage, thermoregulatory challenges, or disease stress due to seasonal variation can strain physiological systems and be reflected in the gut microbiome [24]. For example, gut microbial communities of gorillas and howler monkeys correspond with variation in dietary items and energy intake [22, 23].
Primates inhabiting the tropical dry forest of Sector Santa Rosa (SSR), Área de Conservación Guanacaste, Costa Rica, encounter extreme seasonal variation [25, 26] due to several months of frequent rainfall (typically mid-May to mid-November) followed by extended dry periods without precipitation, near complete defoliation of trees, and soaring temperatures (typically November through early May; Fig. 1). Resident primates alter their diets and behaviors to mitigate the impacts of climate on food availability, water, and shade [27]. Fruit biomass follows a strongly seasonal pattern, typically with fruit scarcity during the early wet season and again during the early dry season; this alters foraging behavior of resident primates, who increase consumption of embedded invertebrates, flowers, and pith when fruit is scarce [25, 28, 29]. Fruit abundance at SSR has been shown to predict creatinine concentration—a reliable proxy for muscle mass—in white-faced capuchin urine, indicating that they suffer physiologically from reduced energy intake during months with lower fruit abundance [30]. Additionally, capuchin monkeys obtain drinking water from different sources in wet and dry seasons, potentially exposing them to different microbial communities. In the wet season, capuchins drink more frequently from tree holes and lick foliage; in the dry season, they drink from standing pools of ground-based water.
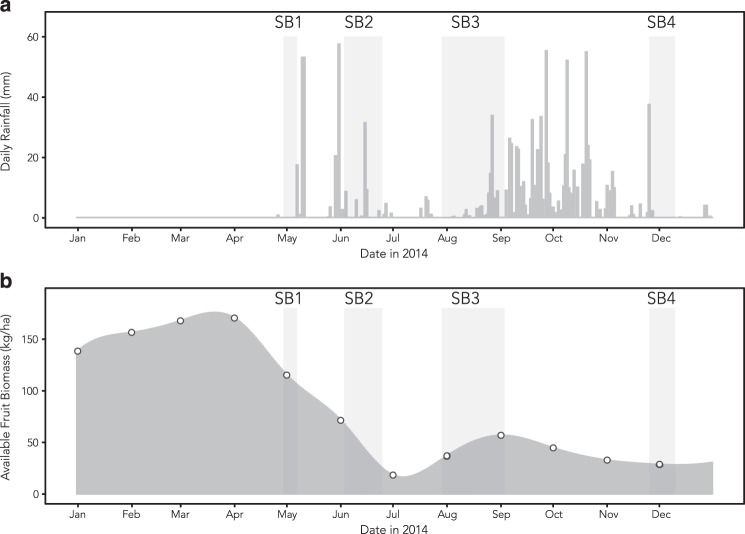
Fig. 1.
a Daily rainfall throughout 2014 at SSR. Note the distinct wet and dry seasons. b Interpolated daily fruit biomass during 2014. The open circles show dates on which biomass was measured. Samples were collected from three groups of white-faced capuchin monkeys during the four shaded sampling bouts (SB1–SB4)
Here, we contribute new data on the impact of environmental seasonality on the gut of white-faced capuchin monkeys. We study the gut microbiota from 24 individuals inhabiting the tropical dry forest of SSR over several months spanning the dry to wet seasonal transition. We hypothesize that bacterial community composition will correspond to seasonal differences in fruit biomass and/or rainfall. Specifically, we hypothesize that (1) if water sources are important drivers of microbial communities, microbial beta diversity will differ between the wet and dry seasons and be more similar within seasonal classifications; (2) if fruit biomass in the habitat affects diet and gut microbial responses, the behavioral activity budget, microbial taxonomic composition, and metagenomic indicators of enzyme activity associated with the degradation of carbohydrates versus chitin and protein will differ during periods of fruit abundance and scarcity; (3) bacteria previously associated with healthy gut function in the comparative literature will be more abundant during periods of high fruit availability versus periods of fruit scarcity when animals are more nutritionally stressed. We integrate environmental, dietary, and behavioral data with 16S amplicons and functional metagenomics in a longitudinal approach that provides a holistic advance in the study of Neotropical primate microbial ecology.
Methods
Seasonality in rainfall and fruit biomass
Sector Santa Rosa, part of the larger Área de Conservación Guanacaste in northwestern Costa Rica (10°45′ to 11°00′ N and 85°30′ to 85°45′ W), is a tropical dry forest ecosystem [31]. This habitat is strongly seasonal, with the majority of the precipitation (900–2400 mm annually) falling mid-May through mid-November [25, 27]. SSR contains both primary forests and secondary forests in various stages of regeneration, following the protection of this area in the late 1970s and subsequent sequestering and reforestation of ranchland [31].
We calculated fruit biomass by combining monthly phenological data with plant species abundances collected from 273 100 × 4 m transects running north-south, and 151 100 × 2 m transects running east-west throughout the study group home ranges (Figure S1). For food species, we included in our fruit biomass calculations all trees of a given species the same size or larger than any tree of that species that capuchins have been observed to feed in from a dataset of ca. 11,500 fruit patch visit records (see Table S1). Fruit biomass calculations follow methods described elsewhere [30, 32, 33] and are detailed in Supplemental Methods. We collected rainfall data with a custom rain gauge checked daily at 7 pm.
Behavioral data and fecal sample collection
White-faced capuchin monkeys (Cebus capucinus imitator) are medium-sized (ca. 5 kg) arboreal monkeys with moderate sexual dimorphism [34]. They are omnivorous, consuming over 120 species of plants and a diet rich in invertebrate and vertebrate prey [25, 35, 36]. In SSR, capuchins have been studied continuously since 1983 and are well-habituated to human observers [37]. We followed three neighboring, habituated groups of capuchin monkeys (GN, LV, RM) 3 to 4 days per month each. Every 30 min, we recorded the activity of the majority of the group using the ethogram developed for long-term data collection in SSR (Table S2). Fruit foraging behaviors were recorded separately from foraging focused on invertebrates, flowers, bromeliad leaves, or pith. Additionally, we recorded travel, resting, social, and other behaviors (e.g., intergroup or predator-focused interactions).
We collected fresh fecal samples from the same monkeys between April 29th and December 10th, 2014. Samples were collected from 8 members per group, totaling 24 individuals, all of whom were identified from facial characteristics and body scars. Sampling occurred in four bouts, once in the late dry season, twice during the rainy season, and once during the following early dry season (Fig. 1, Table S3). Samples from each group were collected in a rotating cycle during the course of 1 to 5 days. Female white-faced capuchin monkeys are philopatric (remaining in natal groups), so we prioritized the collection of samples from females (six females, two males per group) to more robustly sample the natal sex, improving control for potential impacts of group membership on microbiota composition [15]. We collected fecal samples from the forest floor immediately following defecation, and before 12 pm to decrease external changes in microbiota composition. Researchers wore latex gloves and placed fecal samples into empty, sterile 2 ml tubes, which were stored with ice packs in portable coolers. Presence or absence of arthropod exoskeletons was noted and samples were frozen in liquid nitrogen upon returning to the field station, typically between 12 and 3 pm. All samples were stored and shipped frozen to the primate molecular ecology lab at Washington University in St. Louis and stored at −80 °C.
Molecular lab work, variant identification, and functional metagenomics
We extracted DNA from 86 fecal samples in a UV sterilized biosafety cabinet using a standard phenol–chloroform extraction with bead beating [38]. The V4 region of the 16S rRNA gene from each of the 86 samples was amplified in triplicate with standard primers (515F, 806R) [39]. Samples were pooled, tagged with dual indexes, and sequenced on an Illumina MiSeq with V2 chemistry. Whole metagenomic fragment libraries were targeted for 20 samples with adequate DNA concentrations (five individuals spanning the four sampling bouts). Samples were tagged with dual indexes and pooled to run on one lane of an Illumina HiSeq 2500, generating 2 × 125 paired-end reads. 16S reads were checked for quality with FASTQC [40] and trimmed of adaptors and primers with cutadapt [41]. Sample composition was inferred using DADA2, and taxonomy was assigned from the RDP database [42, 43]. Unlike OTU approaches, which rely on arbitrary clustering thresholds, DADA2 identifies unique 16S sequences at the nucleotide level (amplicon sequence variants (ASVs)) using an error controlling algorithm, resulting in improved precision and fewer spurious sequences [43, 44].
Quality control of whole metagenomic shotgun reads was performed with KneedData (https://bitbucket.org/biobakery/kneaddata). We identified coding sequence open reading frames with FragGeneScan+ [45] and searched for sequence homologs in the dbCAN database [46] of carbohydrate-active enzymes (CAZymes) with hidden Markov models using hmmscan in HMMER 3.1 [47]. Amino acid alignment tables were parsed with an E-value of 1e–3. Abundances of individual CAZymes were normalized against the per sample total number of significant CAZyme alignments. Carbohydrate-binding module (CBM) functional classifications were identified by searching the Carbohydrate-Active enZYmes Database (www.cazypedia.org) and clustered by carbohydrate interaction (Table S4). For comparative clarity, we grouped CBMs interacting with cellulose, xylan, fructans, and other plant cell wall components as “Plant” CBMs. Differences in CBM relative abundances across sampling bout and arthropod consumption were identified with Wilcoxon rank sum tests in R.
Prevalence/abundance/diversity metrics
Prior to analysis, we pruned reads belonging to mitochondria, cyanobacteria, chloroplasts, and unknown phyla from the dataset to minimize sequences derived from residual foods in the gut. To assess bacterial richness and diversity within each sampling bout, we calculated Shannon Alpha diversity from the pruned ASV tables with Phyloseq in R [48, 49]. Ribosomal sequence variant abundances were then normalized with the DESeq2 variance stabilizing transformation in Phyloseq. To reduce the effect of rare taxa on beta diversity, we filtered amplicon sequence variants (ASVs) found in fewer than 10% of samples. Weighted Unifrac distance Beta diversity was ordinated with canonical correspondence analysis and non-metric multidimensional scaling (NMDS). We tested group differences in beta diversity with PERMANOVA using the adonis function from vegan in R with 10,000 permutations, while accounting for individual identity as a covariate. ASVs were then agglomerated at the genus level, and differences in relative abundance of the ten most abundant genera across sampling bouts were tested with ANOVA and Tukey’s honest significant difference test in R.
Linear mixed effects models of bacterial abundance
In order to detect differentially abundant ASVs between seasons (wet/dry, abundant/scarce fruit) and diets (presence/absence of arthropod exoskeletons in feces), we measured changes in ASV log2 fold abundance between these binary categories using linear mixed models built in phyloseq and DESeq2. We identified significantly over/under-represented ASVs using Wald tests with Benjamin–Hochberg adjusted p-values of 0.001.
We used the human and mouse microbiome literature to identify genera potentially associated with digestive health (Bacteroides, Bifidobacterium, Clostridium XIVa, Lactobacillus, Lactococcus) or enteric pathogenicity (Enterococcus, Escherichia/Shigella, Haemophilus, Helicobacter, Campylobacter, Pseudomonas, and Streptococcus) [8, 9, 50, 51]. To determine ecological and dietary effects on the abundances of these genera, agglomerations of ASVs in this case, we built linear mixed models in R with lme4, nesting host animal identity within social group membership as a random effect. We selected daily fruit biomass, average daily rainfall within 30 days before sample collection, presence/absence of arthropod exoskeletons in feces, age, and sex as fixed effects. To account for the effects of sex and age, all models included these fixed effects, but alternative models were built with each combination of the remaining three variables. We ran a variance inflation factor test in R, which indicated no collinearity among fixed effects (VIF between 1.0 and 1.4). To assess the appropriateness of model fit, we examined plots of residuals versus fits and qq normality to identify outliers, removing up to three points with substantial leverage based on visual inspection. We ranked models by ∆AICc, and determined directionality of predictors by their slopes. We calculated the importance of each predictor by the summing the weights of the models in which it appeared in R with MuMIn. Additionally, we ran models to predict Shannon Alpha diversity and Weighted Unifrac Beta diversity (position along NMDS axes one and two) using the same approach.
Results
Seasonality in rainfall and fruit biomass was prominent during the study period (Fig. 1). We measured 1112.9 mm of rain, concentrated between late May and early November. We recorded 48,799 trees along the forest transects, cumulatively covering 9.06 ha of forest. Of the species included in our phenology surveys, 10,466 were large enough to be fed in by capuchins, which we included in our biomass calculations. Fruit biomass was highest in the late dry season, spanning Sampling Bout 1. During this period, monkeys spent significantly more of their foraging activity budget foraging on ripe fruit, and overall time spent in foraging versus other behaviors was lower (Fig. 2). In subsequent sampling periods, monkeys spent a greater amount of time foraging outside ripe-fruit patches, almost entirely on invertebrates (Table S2).
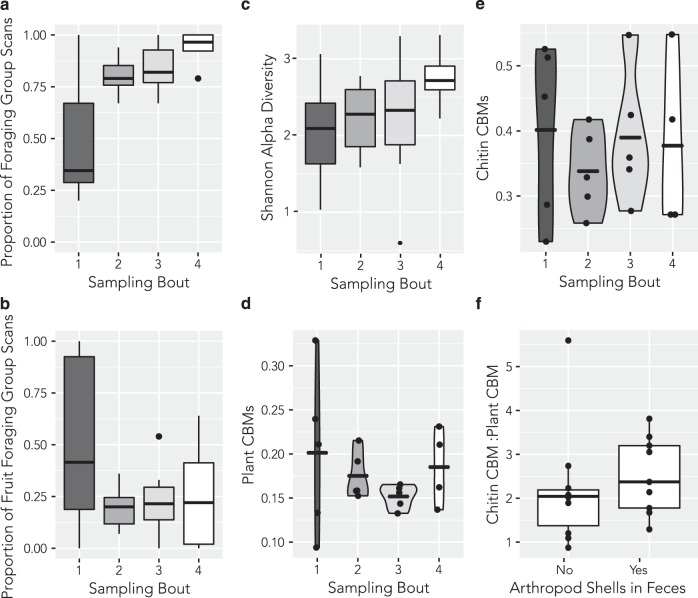
Fig. 2.
a Proportion of behavioral group scans in which white-faced capuchin monkeys were observed foraging. b Proportion of foraging group scans in which the monkeys were observed foraging for fruit. c Shannon Alpha diversities for each sampling bout. Alpha diversity is highest during the early dry season (SB4). d Plant CBMs across sampling bouts. e Chitin CBMs across sampling bouts. f Ratio of chitin CBMs to plant CBMs depending on the presence or absence of arthropod fragments in feces. See Fig. 1 for timing of sampling bouts
Biodiversity of gut microbiota from 16S amplicon sequencing
After DADA2 filtration, merger, and chimera removal, we used 5,527,759 2 × 250 PE reads to identify 3371 unique taxa from 86 capuchin fecal samples with a median of 61,684 reads per sample. After we filtered out low abundance reads, 418 taxa remained, which we agglomerated into 150 genera.
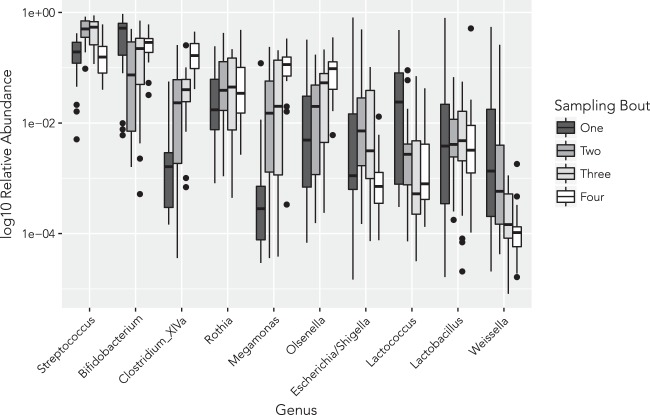
The guts of white-faced capuchins at SSR are dominated by Firmicutes (45.3%, 884 taxa), Actinobacteria (28.8%, 583 taxa), Proteobacteria (9.1%, 823 taxa), and Bacteroidetes (2.8%) (Figure S2). Trace amounts of Acidobacteria (<0.1%), Planctomycetes (<0.1%), Verrucomicrobia (<0.1%), and others are also present (Table S5). The ten most abundant genera accounted for 92% of bacterial abundance in the overall dataset. Bacterial composition was dominated by the genera Streptococcus (29.8%) and Bifidobacterium (23.8%), with lesser contributions from Clostridium XIVa (6.2%), Rothia (6.0%), Megamonas (4.8%), Olsenella (4.8%), Escherichia/Shigella (4.3%), Lactococcus (2.6%), Lactobacillus (2.2%), and Weissella (1.5%), fluctuating across sampling bouts (Fig. 3). Among the ten most abundant genera, significant differences in relative abundances across sampling bouts were observed for Streptococcus, Bifidobacterium, Olsenella, Clostridium XIVa, Megamonas, and Lactococcus (Table S6).
Fig. 3.
Relative abundance (log10 transformed) of the ten most abundant genera across sampling bouts (Dry season: SB one, four; Wet season: SB two, three)
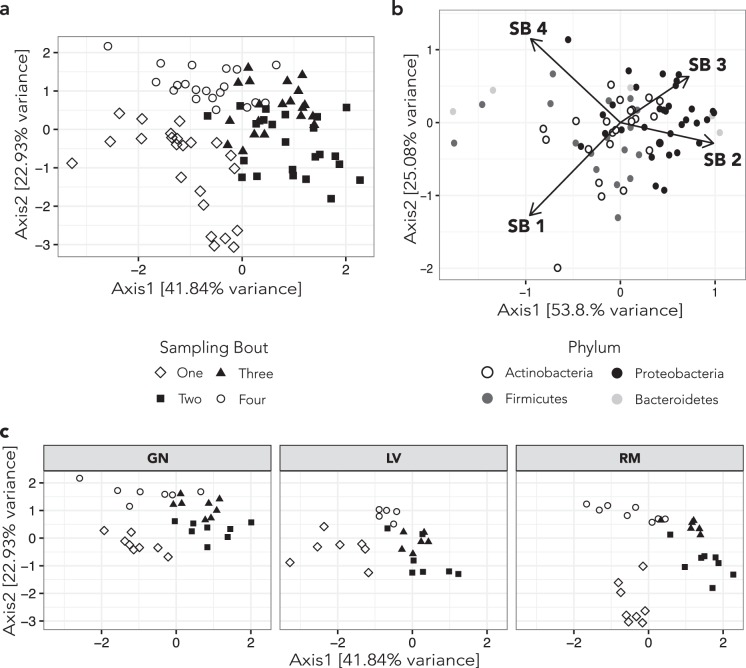
Gut microbial beta diversity among white-faced capuchin monkeys displays a significant seasonal pattern between samples collected during the dry (Sampling Bouts 1 and 4) and wet seasons (Sampling Bouts 2 and 3) (Fig. 4). A PERMANOVA test of Weighted Unifrac distances indicates that seasonal differences hold both across (p = <0.0001; R2 = 0.14) and within (GN: p < 0.001, R2 = 0.17; LV p < 0.001, R2 = 0.20; RM: p < 0.0001, R2 = 0.21) social groups. Shannon Alpha diversity differed significantly across seasons (Fig. 2c; Table 1), with diversity increasing from the late dry season, across the wet season, and into the early stage of the subsequent dry season (ANOVA; p < 0.001; F = 7.9). Pairwise comparisons between sampling bouts were significant (Tukeys’s HSD) for Bouts 4 versus 2 (p < 0.001) and 4 versus 3 (p < 0.01).
Fig. 4.
Weighted unifrac beta diversity ordinated with canonical correspondence analysis. In a (all capuchin samples) and c (capuchin samples facetted by social group), solid black points are from the wet season and hollow black points are from the dry season. Samples vary by rainfall season on Axis 1 and then by sampling bout on Axis 2. b Bacterial ASVs agglomerated by family, with phylum coded by shape, and transformed by five to increase clarity. Arrows represent vectors in the direction of increase during each sample bout. During the wet season (SB2 and SB3) Proteobacteria are more abundant
Table 1.
Results of linear mixed effects models of bacterial abundance, Shannon Alpha diversity, and Weighted Unifrac Beta diversity
| Linear mixed effects model | Importance/Slope | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Response | Fixed effects | K | Log likelihood | AICc | ∆AICc | Weight | Fruit biomass | Arthropod consumption | Rainfall |
| Lactococcus | ~Fruit | 8 | −217.78 | 435.46 | 0.00 | 0.31 | 1.00/+ | 0.39/− | 0.43/+ |
| ~Fruit + Rain | 9 | −216.60 | 453.61 | 0.14 | 0.29 | ||||
| ~Fruit + Arthropods | 9 | −216.72 | 453.83 | 0.37 | 0.26 | ||||
| ~Fruit + Rain + Arthropods | 10 | −216.10 | 455.16 | 1.70 | 0.13 | ||||
| Campylobacter | ~Fruit + Arthropods | 9 | −226.36 | 473.10 | 0.00 | 0.51 | 1.00/− | 0.66/− | 0.27/+ |
| ~Fruit | 8 | −228.44 | 474.75 | 1.65 | 0.22 | ||||
| Clostridium XIVa | ~Fruit + Arthropods | 9 | −168.56 | 357.55 | 0.00 | 0.55 | 1.00/− | 0.72/+ | 0.23/+ |
| ~Fruit | 8 | −170.74 | 359.40 | 1.85 | 0.22 | ||||
| Bifidobacterium | ~Fruit + Rain | 9 | −164.20 | 348.84 | 0.00 | 0.61 | 0.99/+ | 0.33/+ | 0.88/− |
| ~Fruit + Rain + Arthropods | 10 | −163.78 | 350.56 | 1.72 | 0.26 | ||||
| Haemophilus | ~Fruit | 8 | −233.05 | 483.97 | 0.00 | 0.50 | 0.89/− | 0.25/− | 0.27/+ |
| Streptococcus | ~Arthropods | 8 | −181.05 | 379.96 | 0.00 | 0.54 | 0.23/+ | 0.99/− | 0.31/+ |
| ~Rain + Arthropods | 9 | −180.65 | 381.68 | 1.72 | 0.23 | ||||
| Bacteroides | ~Fruit + Arthropods | 9 | −242.56 | 505.49 | 0.00 | 0.59 | 0.86/+ | 0.96/+ | 0.39/− |
| ~Fruit + Rain + Arthropods | 10 | −242.09 | 507.12 | 1.63 | 0.26 | ||||
| Escherischia / Shigella | ~Arthropods | 8 | −244.48 | 506.84 | 0.00 | 0.35 | 0.43/+ | 0.85/− | 0.49/+ |
| ~Rain + Arthropods | 9 | −243.87 | 508.11 | 1.27 | 0.18 | ||||
| ~Fruit + Rain + Arthropods | 10 | −242.59 | 508.12 | 1.28 | 0.18 | ||||
| ~Fruit + Arthropods | 9 | −244.18 | 508.73 | 1.89 | 0.14 | ||||
| Enterococcus | ~Fruit + Arthropods | 9 | −212.15 | 444.71 | 0.00 | 0.71 | 0.98/+ | 0.91/+ | 0.23/+ |
| Shannon Alpha Diverisity | ~Fruit + Arthropods | 9 | −55.32 | 131.05 | 0.00 | 0.44 | 0.72/− | 0.88/+ | 0.29/− |
| ~Arthropods | 8 | −57.36 | 132.61 | 1.56 | 0.20 | ||||
| ~Fruit + Rain + Arthropods | 10 | −54.99 | 132.96 | 1.91 | 0.17 | ||||
| NMDS Axis 2 | ~Rain + Arthropods | 9 | −210.06 | −399.74 | 0.00 | 0.41 | 0.37/+ | 0.78/+ | 0.81/− |
| ~Fruit + Rain + Arthropods | 10 | −210.54 | −398.15 | 1.59 | 0.19 | ||||
| ~Rain | 8 | −207.89 | −397.91 | 1.83 | 0.17 | ||||
| Pseudomonas | ~Rain + Arthropods | 9 | −211.46 | 443.30 | 0.00 | 0.42 | 0.47/+ | 0.71/− | 0.98/+ |
| ~Fruit + Rain + Arthropods | 10 | −210.61 | 444.16 | 0.86 | 0.27 | ||||
| ~Fruit + Rain | 9 | −212.23 | 444.82 | 1.52 | 0.20 | ||||
| Helicobacter | ~Rain | 8 | −204.57 | 427.08 | 0.00 | 0.53 | 0.24/− | 0.26/− | 0.90/+ |
| NMDS Axis 1 | ~Rain | 8 | 165.68 | −313.48 | 0.00 | 0.33 | 0.41/− | 0.52/− | 0.84/+ |
| ~Rain + Arthropods | 9 | 166.61 | −312.85 | 0.63 | 0.24 | ||||
Models with ∆AICc less than two are shown for each response variable. The importance and directionality (slope) of each predictor, as generated from the AICc weights of all models, are shown at right, with strong predictors (≥0.9) in bold and moderate predictors (0.70–0.89) in italics
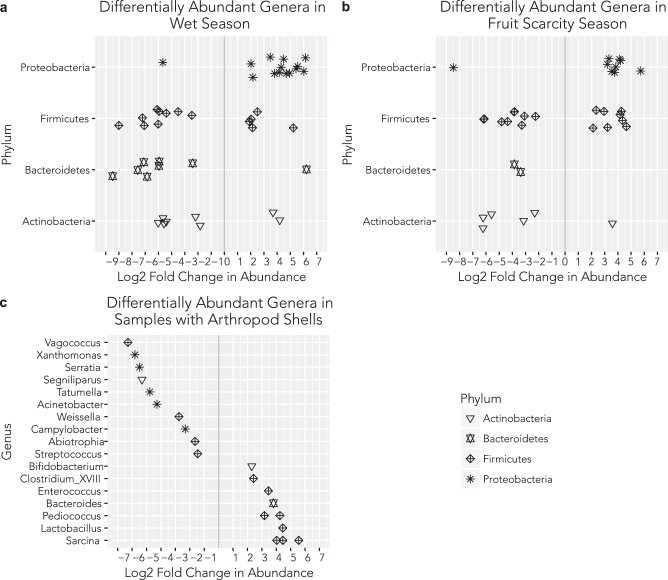
Changes in ASVs showed clear patterns linked to environmental variation (both wet versus dry seasons, and periods of abundant fruit biomass versus fruit scarcity). We identified 78 ASVs from 41 genera and 4 phyla with significantly different log2 fold abundance changes (Fig. 5, Figure S3). During the wet season, when fruit biomass starts to decline, genera of Proteobacteria had significant (adjusted p < 0.001) increases in log2 fold abundance, whereas members of the Actinobacteria and Bacteroidetes were significantly reduced. Finally, we identified nine genera as significantly over- or under-represented in fecal samples that contained arthropod fragments, relative to feces without evidence of insect exoskeletons (Fig. 5c).
Fig. 5.
Log2 fold change in the four dominant phyla in the white-faced capuchin monkey gut microbiome. Each point is a 16S V4 amplicon sequence variant with a significant (adjusted p < 0.001) difference in abundance during a the wet (SB 2 and 3) season and b fruit scarcity (SB 2, 3, and 4) season. Points are displayed with vertical jitter for clarity. During the wet and low fruit seasons, genera from the Proteobacteria are significantly more abundant, whereas those from the Actinobacteria and Bacteroidetes are reduced. c Significantly over and under-represented bacterial genera from 16S in fecal samples that contained visible fragments of arthropods
In almost all cases, linear mixed effects models including rainfall, monthly fruit biomass, and arthropod consumption substantially outperformed null models. Models with the greatest predictive value (∆AICc < 2) and importance values for individual predictors are presented in Table 1. Rainfall was an important, but not predominant predictor of microbiome beta diversity (NMDS axes 1 and 2); contribution of arthropod consumption had a significant, but weaker, effect (axis 2 only). In contrast, models of Shannon Alpha diversity had a negative association with fruit biomass abundance and positive association with the consumption of arthropods (Importance = 0.88; 0.72). For the two most abundant bacterial genera in the white-faced capuchin gut microbiome, Bifidobacterium and Streptococcus, dietary composition had differing impacts. For Bifidobacterium, fruit abundance was a positive predictor (importance = 0.99), while rainfall had a (importance = 0.88) negative effect. In contrast, Streptococcus had a negative relationship with arthropod consumption (importance = 0.99).
Bacterial genera previously associated with healthful (Bacteroides, Bifidobacterium, Clostridium XIVa, Lactobacillus, and Lactococcus) and harmful (Enterococcus, Escherichia/Shigella, Haemophilus, Helicobacter, Campylobacter, Pseudomonas, and Streptococcus) effects in the human and mouse gut microbiome literature did not show consistent patterns regarding relationships to rainfall or fruit abundance (Table 1). Bifidobacterium, Clostridium XIVa, Lactococcus, and Bacteroides were predicted by fruit biomass, but directionality was not uniform. No model outperformed the null model for Lactobacillus. Additionally, Bacteroides had a positive relationship with arthropod consumption (importance = 0.96), and rainfall was a negative predictor of Bifidobacterium. The abundances of potential bacterial enteric pathogens were associated with a variety of factors: rainfall (Pseudomonas, Helicobacter), arthropod consumption (Enterococcus, Escherichia/Shigella, and Streptococcus), and fruit biomass (Campylobacter, Enterococcus, and Haemophilus).
Functional metagenomics
To evaluate metabolic function, we shotgun sequenced libraries from individuals in each sampling bout. Although one monkey died before the fourth bout, we included it in the analysis. It provided three samples—the bulk of the series. The relative abundance of predicted carbohydrate-active enzymes in the capuchin gut was dominated by carbohydrate-binding modules (CBM, 63%), with lesser contributions from the cellulosome complex (15%), glycoside hydrolase (13%), carbohydrate esterases (4%), glycoslytransferase (4%), and polysaccharide lyase (1%) families (Table S7). We identified 50 CBMs, of which two modules, CBM50 (42% of CBMs; 27% of total CAZymes) and CBM48 (19% of CBMs; 12% of total CAZymes), composed the majority. The pattern of plant CBM relative abundance strongly resembles that of capuchin fruit foraging behavior (Fig. 2a, b, d). During the late dry season (SB1), the amount of both fruit consumption and plant CBMs is highly variable depending on the individual sampled, whereas both are contracted during the remainder of the year (Fig. 2d). In contrast, the relative abundance of chitin CBMs varied widely across sampling bouts (Fig. 2e). We observed a significantly (W = 60, p < 0.05) higher ratio of chitin CBMs to plant CBMs from the samples in which we were able to observe fragments of arthropods (Fig. 2f).
Discussion
Our results lead us to three main conclusions: (1) rainfall and (2) diet are associated with the diversity, composition, and function of the gut microbiome; (3) microbial fluctuations are likely contributing to nutrient uptake and the health of wild primate populations. Furthermore, the gut microbiome of our study population differs substantially from that of other free-ranging primates [15, 22, 23, 52–57]. At SSR, the white-faced capuchin gut microbiome is dominated by Streptococcus and Bifidobacterium. While both are common members of primate gut microbial communities, their consistently high abundances are atypical of what has been observed in other primates (Figure S4), but methodological differences across studies should not be ignored. We caution that our study is observational and limited in its ability to distinguish causality. Furthermore, not all bacteria taxa are necessarily serving functional roles for the host and could be selectively neutral. Future research should include independent metrics of health and strain-specific metagenomic assembly.
Seasonality
Due to an El Niño event, 2014 rainfall was well-below the annual average, falling in the 20th percentile of 39 recorded years from SSR [58]. Nonetheless, seasonal rainfall patterns remained evident, and our hypothesis (1) that rainfall patterns would be associated with the gut microbiome was supported. That samples collected during the wet season (Sampling Bouts 2 and 3) cluster loosely apart from those collected during the dry season (Bouts 1 and 4) on beta diversity ordinations (Fig. 4), indicates that non-temporal factors drive capuchin gut microbiome composition. The broad relationship with seasonality is further supported by rainfall being the strongest predictor of a sample’s position along the first NMDS axis in linear mixed models. Although rainfall is not an important predictor of Shannon Alpha diversity, it is positively associated with Helicobacter and Pseudomonas, two potential pathogens in the human and mouse literature [9].
An association between host social behavior and gut microbial diversity has been reported in several primate species [15, 18–20]; however, we did not observe any distinct effect of social group membership. This is evident in our Beta diversity plots, where points from the different social groups largely overlap in each sampling bout (Fig. 4), and the lack of difference in Shannon Alpha diversity among the three groups (Figure S5). The three social groups observed at SSR have adjacent territories with highly similar food resources, which likely acts to minimize intergroup differences. Future research investigating how social behavior is associated with the gut microbiota of capuchins at Sector Santa Rosa is warranted.
The gut microbiota of white-faced capuchins living in the seasonal forests of SSR have stark compositional differences in comparison to those from a non-seasonal rainforest. Streptococcus and Bifidobacterium are far more abundant in our wild samples than those from other wild white-faced capuchins in a non-seasonal rainforest, which may indicate a strong impact of environment [59]. Furthermore, the rainforest capuchin microbiome is dominated by Firmicutes 41.6%, Proteobacteria 39.2%, and Bacteroidetes 13.2%, with only trace amounts of Actinobacteria [59]. At SSR, we observed a comparable amount of Firmicutes, but substantially less Proteobacteria and Bacteroidetes, and a high amount of Actinobacteria. Interestingly, the log2 fold abundances of ASVs from the latter three phyla are strongly associated with seasonality (particularly rainfall) at SSR (Figs. 4b and 5a, b), which could explain the strikingly high level of Proteobacteria observed in the wet forest [59]. Future sampling in more climatically typical years should be pursued to confirm the consistency of these patterns and assess health impacts. Additionally, microbial profiling of water (and food) sources will help distinguish whether microbes are more prevalent in the environment and being differentially ingested, or fluctuating in abundance through internal mechanisms.
Fruit biomass and carbohydrate digestion
Our hypothesis (2) that habitat-wide fruit biomass would be related to behavior and gut microbiota was also supported. Sampling Bout 1 occurred during the highest period of dietary fruit biomass, consistent with previous seasonal fluctuations [25, 60]. Fruit biomass values aligned with primate behavior—capuchins spent more time foraging for fruit during this period. The dietary importance of fruit in Sampling Bout 1 was also recapitulated by the abundance of CBMs associated with plant carbohydrate degradation, indicating that the capuchin gut microbiome is responsive to ecological and dietary variation. We observed positive associations between available fruit biomass and several bacterial taxa. The Lactococcus result is intuitive, as strains of Lactococcus are established to be involved with sugar digestion [61]. The role of Enterococcus is less clear, although at least one strain of E. faecalis (RKY1) has a commensal role in lactic acid fermentation [62, 63]. Additionally, we found that Bacteroides, although relatively scarce in the SSR capuchin gut, was positively associated with fruit biomass. In the human gut, Bacteroides has a role in polysaccharide digestion of starch glycans, but is more commonly associated with a proteolytic function [64–66]. Given that many reported effects are strain-specific, finer grained taxonomic information in future work may promote further understanding of these interactions.
Strikingly, the gut microbiome of capuchins at SSR exhibits a carbohydrate degrading functional profile similar to that of human hunter-gatherers (enrichment of CBM48 and CBM50), despite resembling westernized humans taxonomically (low alpha diversity, abundant Bifidobacteria, loss/scarcity of Treponema/Succinivibro) [67–69]. The Hadza, whose diet is rich in complex polysaccharides from unrefined plant foods have a gut microbiome enriched in CBM48 and CBM50, which provides functional capacity to degrade α-glucans and peptidoglycan, but Bifidobacterium is notably under-represented [68–70]. One explanation for the abundance of Bifidobacterium in SSR capuchin guts may be that it is engaged in the degradation of sugar-rich (ripe) fruits preferred by capuchins [60]. Fruits rich in sugar and fiber [66, 71] can have a bifidogenic effect, and we observed a clear positive relationship between fruit biomass and Bifidobacterium abundance. Additionally, the dietary spectrum of capuchins is relatively narrow (almost entirely fruit and insects) in comparison to the Hadza, who consume a broader array foods, which might explain the low alpha diversity of the capuchin gut.
Arthropod consumption and chitin-digestion
Additional evidence in support of the hypothesis (2) that diet shapes the microbiome in wild monkeys is seen in the evidence of increased arthropod digestion activity during periods of fruit scarcity. White-faced capuchins consume large amounts of arthropods throughout the year, and during times of fruit scarcity rely on them heavily [30, 33]. Our results, although observational, suggest that the capuchin gut microbiome facilitates arthropod consumption. We observed significant associations between arthropod consumption and bacteria with known chitinolytic or protein-degrading functions, including Enterococcus sp., Serratia marcescens, and Bacteroides [66, 72–74]. From a taxonomic perspective, the capuchin microbiome resembles that of myrmecophageous mammals and insectivorous plants, with Streptococcus, Bifidobacterium, Lactococcus, Lactobacillus, Sarcina, Serratia, Pseudomonas, and Proteobacteria either being commonplace or associated with arthropod consumption (Fig. 5c) [12, 59, 75]. However, we caution that the directionality of these associations is not uniform, likely because it was not possible to identify the physical remains of all arthropods in capuchin feces. Future work including dietary metabarcoding could clarify this relationship.
Our metagenomics results reveal that the gut microbiome likely has diverse and important roles for assisting the digestion of invertebrates. The ratio of chitin degrading to plant degrading CBMs was significantly higher in fecal samples that contained partially digested arthropods. The high proportion of invertebrates in the capuchin CAZyome could explain the abundance of CBM50 in the capuchin CAZyome. CBM50 has been reported to bind to the N-acetylglucosamine (GlcNac) present in chitin [76] and previously shown to function in chitin degradation [77]. Our result—that a dominant module in the capuchin gut microbiota was CBM50—is consistent with the hypothesis that the bacterial microbiome has an important role in digestion of invertebrate exoskeletons, thus augmenting capacities of the host genome for acquiring energy and nutrition from this essential component of their diet [78]. However, given that CBM50 is also involved in polysaccharide digestion and in the breakdown of peptidoglycans, this result warrants further investigation. In vitro assays may help clarify the relative importance of different functions in the future.
Health
Capuchins at SSR have been observed with reduced creatinine concentration in their urine at the start of the wet season, which is associated with reduced muscle mass during this periods of fruit scarcity [30]. Accordingly, we were interested in exploring whether aspects of the gut microbiome composition of white-faced capuchins at SSR exhibit variation consistent with periods of poor health. We acknowledge that comprehensive understandings of interrelationships between gut microbiota and health outcomes are difficult to measure in the wild, and present our results below with caution.
During periods of fruit scarcity and/or high rainfall, we observed increased abundances of several genera associated with human dysbiosis, irritable bowel syndrome, and ill health (Campylobacter, Enterococcus, Helicobacter, Haemophilis, Pseudomonas, and Streptococcus). This supports our hypothesis (3) that bacteria previously associated with healthy gut function in the comparative literature will be more abundant during periods of high fruit availability versus periods of fruit scarcity when animals are more nutritionally stressed [9, 50, 79]. We also observed an unusually high diversity of Proteobacteria during these time periods and reductions in Bacteroidetes abundances relative to Firmicutes, which can be considered a coarse marker of poor digestive health [80–82]. If these bacteria also have deleterious health consequences in capuchins, it is possible that the high abundance of Bifidobacterium (and Clostridium XIVa) in the capuchin gut provides a counterbalancing effect in addition to any role in carbohydrate digestion. The probiotic effects of both genera are well established in multiple species [51, 83, 84] and might help maintain gut homeostasis for capuchins. Alternatively, gut microbial associations with IBS in the white-faced capuchins could be a product of their naturally rapid gut passage [85]. While IBS is an inflammatory state for humans, IBS-associated bacteria might not cause a state of ill health in a primate with naturally inefficient digestion.
In conclusion, seasonality of rainfall and food availability is associated with the composition of the white-faced capuchin monkey gut microbiome. Given the inherent difficulty of collecting replicate samples from multiple free-ranging individuals, most studies of free-ranging mammalian microbiota are taken as single snapshots in time. Our results demonstrate that seasonal fluctuations in local ecosystems can correspond with profound shifts in the composition of a host animal’s gut microbiome and its corresponding function. We contend that future studies of free-ranging mammalian microbiota should account for temporal variation in microbial abundance and diversity.
Electronic supplementary material
Acknowledgements
For assistance and advice, we thank Guatam Dantas, Mrinalini Watsa, Matt Workentine, PJ Perry, Emily Davenport, Urs Kalbitzer, Jeremy Hogan, Xiaoqing Sun, and Emily Walco. Research was supported by The Eppley Foundation for Research, The International Center for Advanced Renewable Energy and Sustainability (I-CARES), Washington University in St. Louis, and the Alberta Children’s Hospital Research Institute (ACHRI). Research protocols were approved by the Animal Studies Committee of Washington University in St. Louis, Approval No. 20140017, and the Animal Care Committee of the University of Calgary, Approval No. AC15-0161. Capuchin feces were collected in Costa Rica under permission from CONEGABIO, Approval No. R-025-2014-OT-CONEGABIO and exported under Área de Conservación Guanacaste permit number DSVS-029-2014-ACG-PI-060-2014. Feces were imported to Washington University in St. Louis under Center for Disease Control (CDC) permit number 2014-03-093, and transferred to the University of Calgary with permission from the Public Health Agency of Canada (PHAC), Permit No. P-15-6481. Raw sequencing reads are available at the NCBI Sequence Read Archive (SRA) under the accession number SRP156892 and the BioProject PRJNA485217.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Joseph D. Orkin, Phone: +1 (403) 220-6516, Email: joseph.orkin@ucalgary.ca
Amanda D. Melin, Phone: +1 (403) 210-7579, Email: amanda.melin@ucalgary.ca
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0256-0) contains supplementary material, which is available to authorized users.
References
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–88. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ochman H, Worobey M, Kuo CH, Ndjango JBN, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 6.Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, et al. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci USA. 2012;109:13034–9. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci USA. 2014;111:16431–5. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutgendorff F, Akkermans LMA, Söderholm JD. The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr Mol Med. 2008;8:282–98. doi: 10.2174/156652408784533779. [DOI] [PubMed] [Google Scholar]
- 9.Maurice CF, Knowles SCL, Ladau J, Pollard KS, Fenton A, Pedersen AB, et al. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015;9:2423–34. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutherland AD, Gearry RB, Frizelle FA. Review of fecal biomarkers in inflammatory bowel disease. Dis Colon Rectum. 2008;51:1283–91. doi: 10.1007/s10350-008-9310-8. [DOI] [PubMed] [Google Scholar]
- 11.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA. 2016;113:10376–81. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol. 2014;23:1301–17. doi: 10.1111/mec.12501. [DOI] [PubMed] [Google Scholar]
- 13.Nelson TM, Rogers TL, Carlini AR, Brown MV. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ Microbiol. 2013;15:1132–45. doi: 10.1111/1462-2920.12022. [DOI] [PubMed] [Google Scholar]
- 14.Becker AAMJ, Hesta M, Hollants J, Janssens GPJ, Huys G. Phylogenetic analysis of faecal microbiota from captive cheetahs reveals underrepresentation of Bacteroidetes and Bifidobacteriaceae. BMC Microbiol. 2014;14:43. doi: 10.1186/1471-2180-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, et al. Social networks predict gut microbiome composition in wild baboons. Elife. 2015;4:e05224. [DOI] [PMC free article] [PubMed]
- 16.Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7:1344–53. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perofsky AC, Lewis RJ, Abondano LA, Di Fiore A, Meyers LA. Hierarchical social networks shape gut microbial composition in wild Verreaux’s sifaka. Proc R Soc B. 2017;284:20172274. doi: 10.1098/rspb.2017.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amato KR, Van Belle S, Di Fiore A, Estrada A, Stumpf R, White B, et al. Patterns in gut microbiota similarity associated with degree of sociality among sex classes of a neotropical primate. Microb Ecol. 2017;74:250–8. doi: 10.1007/s00248-017-0938-6. [DOI] [PubMed] [Google Scholar]
- 19.Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2:e1500997. doi: 10.1126/sciadv.1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc Natl Acad Sci USA. 2017;114:13768–73. doi: 10.1073/pnas.1700122114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orkin JD, Yang Y, Yang C, Yu DW, Jiang X. Cost-effective scat-detection dogs: unleashing a powerful new tool for international mammalian conservation biology. Sci Rep. 2016;6:34758. doi: 10.1038/srep34758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, et al. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra) Microb Ecol. 2015;69:434–43. doi: 10.1007/s00248-014-0554-7. [DOI] [PubMed] [Google Scholar]
- 23.Gomez A, Rothman JM, Petrzelkova K, Yeoman CJ, Vlckova K, Umaña JD, et al. Temporal variation selects for diet-microbe co-metabolic traits in the gut of Gorilla spp. ISME J. 2016;10:514–26. doi: 10.1038/ismej.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amato KR. Co-evolution in context: the importance of studying gut microbiomes in wild animals. Microbiome Sci Med. 2013;1:10–29.
- 25.Melin AD, Young HC, Mosdossy KN, Fedigan LM. Seasonality, extractive foraging and the evolution of primate sensorimotor intelligence. J Hum Evol. 2014;71:77–86. doi: 10.1016/j.jhevol.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Campos FA, Jack KM, Fedigan LM. Climate oscillations and conservation measures regulate white-faced capuchin population growth and demography in a regenerating tropical dry forest in Costa Rica. Biol Conserv. 2015;186:204–13. doi: 10.1016/j.biocon.2015.03.017. [DOI] [Google Scholar]
- 27.Campos FA, Fedigan LM. Behavioral adaptations to heat stress and water scarcity in white-faced capuchins (Cebus capucinus) in Santa Rosa National Park, Costa Rica. Am J Phys Anthropol. 2009;138:101–11. doi: 10.1002/ajpa.20908. [DOI] [PubMed] [Google Scholar]
- 28.Hogan JD, Melin AD, Mosdossy KN, Fedigan LM. Seasonal importance of flowers to Costa Rican capuchins (Cebus capucinus imitator): implications for plant and primate. Am J Phys Anthropol. 2016;161:591–602. doi: 10.1002/ajpa.23059. [DOI] [PubMed] [Google Scholar]
- 29.Mosdossy KN, Melin AD, Fedigan LM. Quantifying seasonal fallback on invertebrates, pith, and bromeliad leaves by white-faced capuchin monkeys (Cebus capucinus) in a tropical dry forest. Am J Phys Anthropol. 2015;158:67–77. doi: 10.1002/ajpa.22767. [DOI] [PubMed] [Google Scholar]
- 30.Bergstrom ML, Emery Thompson M, Melin AD, Fedigan LM. Using urinary parameters to estimate seasonal variation in the physical condition of female white-faced capuchin monkeys (Cebus capucinus imitator) Am J Phys Anthropol. 2017;163:707–15. doi: 10.1002/ajpa.23239. [DOI] [PubMed] [Google Scholar]
- 31.Janzen D. Tropical dry forest: area de Conservación Guanacaste, northwestern Costa Rica. In: Perrow MDA, editor. Handbook of ecological restoration: restoration in practice. Cambridge: Cambridge University Press; 2002. p. 559–83.
- 32.Campos FA, Bergstrom ML, Childers A, Hogan JD, Jack KM, Melin AD, et al. Drivers of home range characteristics across spatiotemporal scales in a neotropical primate, Cebus capucinus. Anim Behav. 2014;91:93–109. doi: 10.1016/j.anbehav.2014.03.007. [DOI] [Google Scholar]
- 33.Melin AD, Hiramatsu C, Parr NA, Matsushita Y, Kawamura S, Fedigan LM. The behavioral ecology of color vision: considering fruit conspicuity, detection distance and dietary importance. Int J Primatol. 2014;35:258–87. doi: 10.1007/s10764-013-9730-8. [DOI] [Google Scholar]
- 34.Fragaszy DM, Visalberghi E, Fedigan LM. The complete capuchin. Cambridge: Cambridge University Press; 2004.
- 35.Melin AD, Fedigan LM, Hiramatsu C, Kawamura S. Polymorphic color vision in white-faced capuchins (Cebus capucinus): Is there foraging niche divergence among phenotypes? Behav Ecol Sociobiol. 2008;62:659–70. doi: 10.1007/s00265-007-0490-3. [DOI] [Google Scholar]
- 36.Rose LM. Sex differences in diet and foraging behavior in white-faced capuchins (Cebus capucinus) Int J Primatol. 1994;15:95–114. doi: 10.1007/BF02735236. [DOI] [Google Scholar]
- 37.Fedigan Linda M., Jack Katharine M. Long-Term Field Studies of Primates. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. Tracking Neotropical Monkeys in Santa Rosa: Lessons from a Regenerating Costa Rican Dry Forest; pp. 165–184. [Google Scholar]
- 38.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Qian PY. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews S. FastQC. A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 41.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 42.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–43. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Hahn AS, Wu S-J. FragGeneScan-Plus for scalable high-throughput short-read open reading frame prediction. In: IEEE conference on computational intelligence in bioinformatics and computational biology (CIBCB);2015. p 1–8.
- 46.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–51. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–11. [PubMed] [Google Scholar]
- 48.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stecher B. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr. 2015;3:MBP-0008-2014. [DOI] [PubMed]
- 51.Wang J, Tang H, Zhang C, Zhao Y, Derrien M, Rocher E, et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015;9:1–15. doi: 10.1038/ismej.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS ONE. 2010;5:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moeller AH, Peeters M, Ndjango JB, Li Y, Hahn BH, Ochman H. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 2013;23:1715–20. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aivelo T, Laakkonen J, Jernvall J. Population- and individual-level dynamics of the intestinal microbiota of a small primate. Appl Environ Microbiol. 2016;82:3537–45. doi: 10.1128/AEM.00559-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hale VL, Tan CL, Niu K, Yang Y, Knight R, Zhang Q, et al. Diet versus phylogeny: a comparison of gut microbiota in captive colobine monkey species. Microb Ecol. 2018;75:515–27. doi: 10.1007/s00248-017-1041-8. [DOI] [PubMed] [Google Scholar]
- 56.Bennett G, Malone M, Sauther ML, Cuozzo FP, White B, Nelson KE, et al. Host age, social group, and habitat type influence the gut microbiota of wild ring-tailed lemurs (Lemur catta) Am J Primatol. 2016;78:883–92. doi: 10.1002/ajp.22555. [DOI] [PubMed] [Google Scholar]
- 57.Amato KR, Martinez-Mota R, Righini N, Raguet-Schofield M, Corcione FP, Marini E, et al. Phylogenetic and ecological factors impact the gut microbiota of two Neotropical primate species. Oecologia. 2016;180:717–33. doi: 10.1007/s00442-015-3507-z. [DOI] [PubMed] [Google Scholar]
- 58.Campos FA. A synthesis of long-term environmental change in Santa Rosa. In: Kalbitzer U, Jack KM, editors. Primate life histories, sex roles, and adaptability - essays in honour of Linda M. Fedigan. New York, NY: Springer; 2018..
- 59.Mallott Elizabeth K., Amato Katherine R., Garber Paul A., Malhi Ripan S. Influence of fruit and invertebrate consumption on the gut microbiota of wild white-faced capuchins (Cebus capucinus ) American Journal of Physical Anthropology. 2018;165(3):576–588. doi: 10.1002/ajpa.23395. [DOI] [PubMed] [Google Scholar]
- 60.Hogan Jeremy, Melin Amanda D. Primate Life Histories, Sex Roles, and Adaptability. Cham: Springer International Publishing; 2018. Intra- and Interannual Variation in the Fruit Diet of Wild Capuchins: Impact of Plant Phenology; pp. 193–212. [Google Scholar]
- 61.Neves A, Pool W, Kok J, Kuipers O, Santos H. Overview on sugar metabolism and its control in—The input from in vivo NMR. FEMS Microbiol Rev. 2005;29:531–54. doi: 10.1016/j.femsre.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Wee YJ, Kim JN, Yun JS, Ryu HW. Utilization of sugar molasses for economical l(+)-lactic acid production by batch fermentation of Enterococcus faecalis. Enzym Microb Technol. 2004;35:568–73. doi: 10.1016/j.enzmictec.2004.08.008. [DOI] [Google Scholar]
- 63.Yun JS, Wee YJ, Ryu HW. Production of optically pure l(+)-lactic acid from various carbohydrates by batch fermentation of Enterococcus faecalis RKY1. Enzym Microb Technol. 2003;33:416–23. doi: 10.1016/S0141-0229(03)00139-X. [DOI] [Google Scholar]
- 64.D’elia JN, Salyers AA. Contribution of a neopullulanase, a pullulanase, and an a-glucosidase to growth of bacteroides thetaiotaomicron on starch. J Bacteriol. 1996;178:7173–9. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–35. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segata N. Gut microbiome: westernization and the disappearance of intestinal diversity. Curr Biol. 2015;25:R611–3. doi: 10.1016/j.cub.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 68.Soverini M, Rampelli S, Turroni S, Schnorr SL, Quercia S, Castagnetti A, et al. Variations in the post-weaning human gut metagenome profile as result of bifidobacterium acquisition in the western microbiome. Front Microbiol. 2016;7:1058. doi: 10.3389/fmicb.2016.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, et al. Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr Biol. 2015;25:1682–93. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 70.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eid N, Enani S, Walton G, Corona G, Costabile A, Gibson G, et al. The impact of date palm fruits and their component polyphenols, on gut microbial ecology, bacterial metabolites and colon cancer cell proliferation. J Nutr Sci. 2014;3:e46. [DOI] [PMC free article] [PubMed]
- 72.Vaaje-Kolstad G, Bøhle LA, Gåseidnes S, Dalhus B, Bjørås M, Mathiesen G, et al. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J Mol Biol. 2012;416:239–54. doi: 10.1016/j.jmb.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 73.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horn SJ, Sørbotten A, Synstad B, Sikorski P, Sørlie M, Vårum KM, et al. Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 2006;273:491–503. doi: 10.1111/j.1742-4658.2005.05079.x. [DOI] [PubMed] [Google Scholar]
- 75.Chan XY, Hong KW, Yin WF, Chan KG. Microbiome and biocatalytic bacteria in monkey cup (Nepenthes pitcher) digestive fluid. Sci Rep. 2016;6:20016. doi: 10.1038/srep20016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akcapinar GB, Kappel L, Sezerman OU, Seidl-Seiboth V. Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr Genet. 2015;61:103–13. doi: 10.1007/s00294-014-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karlsson M. Molecular evolution of trichoderma chitinases. In: Gupta VK, Schmoll M, Herrera‐Estrella A, Upadhyay RS, Druzhinina IS, Tuohy MG, editors. Biotechnology and biology of trichoderma. Oxford: Elsevier; 2014. p 67–78.
- 78.Janiak Mareike C, Chaney Morgan E, Tosi Anthony J. Evolution of Acidic Mammalian Chitinase Genes (CHIA) Is Related to Body Mass and Insectivory in Primates. Molecular Biology and Evolution. 2017;35(3):607–622. doi: 10.1093/molbev/msx312. [DOI] [PubMed] [Google Scholar]
- 79.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76. doi: 10.1186/gb-2014-15-6-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–9. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE. 2011;6:e21490. doi: 10.1371/journal.pone.0021490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Gong J, Wang W, Long Y, Fu X, Fu Y, et al. Are there any different effects of Bifidobacterium, Lactobacillus and Streptococcus on intestinal sensation, barrier function and intestinal immunity in PI-IBS mouse model? PLoS ONE. 2014;9:e90153. doi: 10.1371/journal.pone.0090153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valenta K, Fedigan LM. Effects of gut passage, feces, and seed handling on latency and rate of germination in seeds consumed by capuchins (Cebus capucinus) Am J Phys Anthropol. 2009;138:486–92. doi: 10.1002/ajpa.20982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.