Abstract
Foxp3+CD4+ regulatory T (Treg) cells are essential for preventing fatal autoimmunity and safeguard immune homeostasis in vivo. While expression of the transcription factor Foxp3 and IL-2 signals are both required for the development and function of Treg cells, the commitment to the Treg cell lineage occurs during thymic selection upon T cell receptor (TCR) triggering, and precedes the expression of Foxp3. Whether signals beside TCR contribute to establish Treg cell epigenetic and functional identity is still unknown. Here, using a mouse model with reduced IL-2 signaling, we show that IL-2 regulates the positioning of the pioneer factor SATB1 in CD4+ thymocytes and controls genome wide chromatin accessibility of thymic-derived Treg cells. We also show that Treg cells receiving only low IL-2 signals can suppress endogenous but not WT autoreactive T cell responses in vitro and in vivo. Our findings have broad implications for potential therapeutic strategies to reprogram Treg cells in vivo.
Regulatory T (Treg) cells are developed in the thymus, and are essential for suppressing detrimental autoimmunity. Here the authors show, using mice with dampened interleukin 2 (IL-2) signaling, that IL-2 helps position the pioneer factor SATB1 to control genome-wide chromatin accessibility to facilitate Treg cell lineage commitment in the thymus.
Introduction
Naturally occurring, thymus-derived Foxp3+ Treg cells represent a distinct lineage of CD4+ T cells which major role is to maintain self-tolerance1–4. Foxp3, a forkhead/winged helix X-linked transcription factor (TF), is the major lineage-specifying TF for these cells and is indispensable for their differentiation, long-term maintenance, and suppressive functions5–7. Functional loss of FOXP3 is associated with the rapid onset of fatal T cell-mediated autoimmunity, also known as the IPEX syndrome in humans (immune dysregulation, polyendocrinopathy, enteropathy, X-linked8) and the Scurfy phenotype in mice5–7. Foxp3 is induced in thymocytes undergoing positive selection following T cell receptor (TCR) triggering9,10 and additional signals such as IL-2, which stabilizes Foxp3 and the associated thymic-derived Treg cell program of differentiation11–13. The transcriptional regulation of Foxp3 is complex involving the cooperation with multiple additional TFs to enable and maintain Treg cell functional attributes10,12,14–16. Foxp3 therefore acts as an essential TF, which sustained expression is regulated by three intronic conserved non-coding sequence (CNS) elements (CNS1-3)17. CNS3 for instance, acts as a pioneer element in inducing the expression of Foxp3 while CNS2 is bound by IL-2-activated STAT5, directly enabling the stabilization of Foxp3 expression in Treg cells18,19. The lack of IL-2, its high affinity receptor chain IL-2Rα/CD25 or its transducing chain IL-2Rβ/CD12220, lead to the development of wasting autoimmunity as a result of the loss of stable Foxp3 expression5–7 and subsequent Foxp3+ Treg cells in the periphery21–23.
While the Foxp3 TF is required to maintain peripheral Treg cell identity and functions, it is not sufficient per se to confer the functional attributes of Treg cells12,16,24, also consistent with two-thirds of the Treg cell transcriptional signature that cannot be induced by ectopic expression of Foxp3 in Tconv cells14,25. Studies using Foxp3Gfp knock-in/out reporter mice in which GFP+/+ thymocytes lacked functional Foxp3, showed that Foxp3 acts to amplify and fix pre-established molecular features of Treg cells acquired during thymic selection but prior Foxp3 expression14. Foxp3Gfp/Gfp thymocytes exhibit CpG hypomethylation patterns characteristic of mature peripheral Treg cells24. Foxp3 also overwhelmingly binds to pre-existing genome-wide enhancers in thymocytes committed to become Treg cells during positive selection26. Altogether these findings suggest that TCR-induces epigenetic modifications independently of Foxp3 but likely to involve other transcriptional regulators predefining Treg cell identity. Two recent reports provided mechanistic evidence for this concept27,28 by showing (i) that the epigenetic modifier SATB1 is essential in activating Treg cell-specific super-enhancers associated with Foxp3 and other Treg-cell signature genes in thymic precursor Treg cells27 and (ii) that the methylation enzyme MLL4, which regulates the level of monomethylated H3K4 and chromatin interactions at putative gene enhancers, sets the enhancer landscape for Foxp3 induction via chromatin looping28.
These studies represent important conceptual advances in our understanding of the molecular genetics underlying Treg cell-lineage commitment. However, other signals subsequent or concomitant to TCR triggering that may contribute to setting up the functional identity of Foxp3+ Treg cells are largely unknown. IL-2 is proposed to be essential in this process12,19,21,29, but it is believed to be through the stabilization of the Foxp3 TF. Since TCR signaling leads to CD25 upregulation on thymocytes, it is conceivable that IL-2 contributes to establishing thymic Treg cell identity in vivo.
Herein, we isolate and characterize a mouse model, the Il2ramut/mut mouse that bears a point mutation in the IL-2 receptor high affinity chain CD25 resulting in a selective and quantifiable decrease in response to only high affinity IL-2 signals, to test the hypothesis that IL-2 signals modulate Treg cell epigenetic and transcriptional identity, and subsequent suppressive functions in vivo. Our results suggest that IL-2, possibly through the positioning of the genome organizer SATB1, modulates thymic-derived Treg cell epigenetic identity prior to Foxp3 expression. Treg cells that integrate low IL-2 signals (Il2ramut/mut) only repress endogenous but not WT autoreactive T cells, illustrating further the importance of IL-2 signaling for optimal Treg cell functions. These observations are consistent with the idea that altering Treg cell epigenetic identity, in addition to IL-2 capture and signaling, leads to more rapid autoimmunity, and further raise the possibility that epigenetic reprogramming of Treg cells at the time of their selection in the thymus could improve Treg cell functions in autoimmune patients.
Results
Isolation of a novel IL-2 receptor alpha point mutant mouse
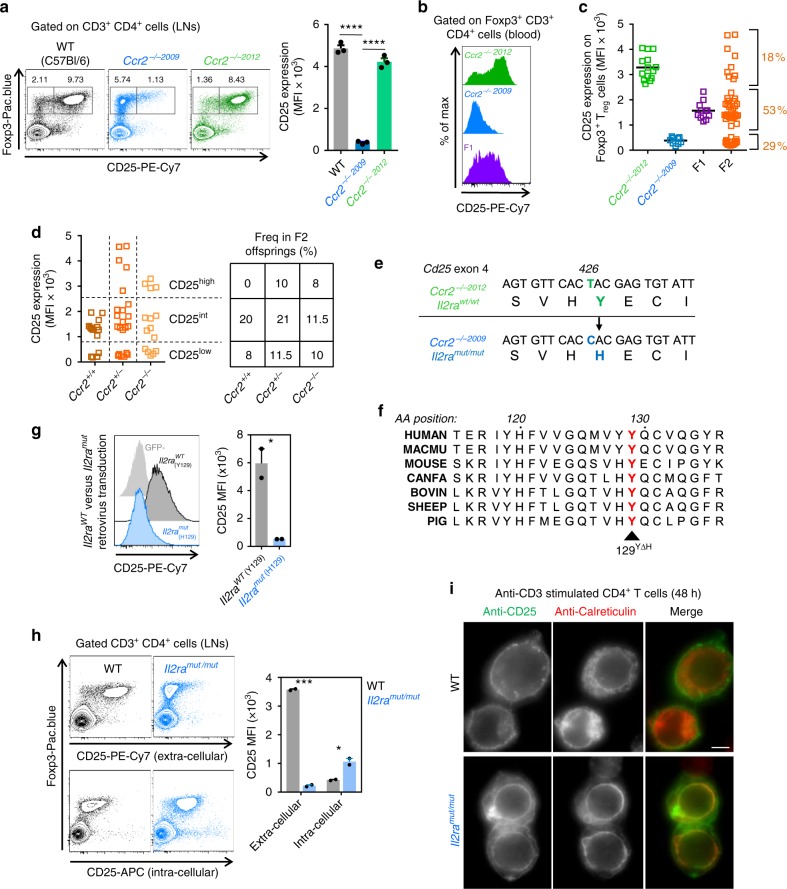
During experiments in Ccr2−/− mice30 obtained from the Jackson Laboratory (JacksLab) in 2009, referred to as Ccr2−/− 2009, we observed that CD4+ Foxp3+ Treg cells in primary and secondary lymphoid organs and blood, displayed low to no cell-surface expression of the IL-2Rα/CD25 compared to Treg cells from wild type (WT) B6 counterparts (WT) (Supplementary Fig. 1a). Treg cell frequency among CD4+ T cells was reduced in spleens and lymph nodes (LNs) while remaining comparable in blood and primary lymphoid organs (Supplementary Fig 1b, c). Since Ccr2−/− mice were neither reported to have different Treg cell frequencies nor distinct expression of CD25 compared to WT mice31,32, we re-acquired Ccr2−/− mice from the JacksLab in 2012, referred to as Ccr2−/− 2012, and compared them to Ccr2−/− 2009 mice (Fig. 1a). As previously documented, and in contrast to Ccr2−/− 2009 mice, Treg cell frequencies and CD25 expression were unaltered in Ccr2−/− 2012 mice. We reasoned that the altered Treg cell phenotype could be accounted for by (i) a difference in the skin and/or gut microbiota between the two Ccr2−/− lines, or (ii) a spontaneous mutation acquired in the Ccr2−/− 2009 colony before 2009. Between 2009 and 2012, Ccr2−/− 2009 mice were backcrossed twice to the WT B6 background and underwent 23 intercrosses (JacksLabs communication). Co-housing of Ccr2−/− 2012 and Ccr2−/− 2009 mice for 3 months neither rescued Treg cell frequencies nor expression of CD25 in Ccr2−/− 2009 mice, likely ruling out a microbiota hypothesis (Supplementary Fig. 1d). However, intercross of Ccr2−/− 2009 and WT B6 mice revealed that Treg cells in the F1 offspring expressed twice as less cell-surface CD25 (MFI) compared to Ccr2−/− 2012 Treg cells, but twice as more compared to Ccr2−/− 2009 Treg cells, suggesting that the Ccr2−/− 2009 trait is haploinsufficient (Fig. 1b, c). Analysis of CD25 expression levels on Treg cells from 51 F2 offspring obtained from intercrossing F1 mice revealed a quasi-Mendelian distribution of the various possible phenotypes, consistent with the Ccr2−/− 2009 trait being controlled by a single gene. Plotting CD25 expression as a function of CCR2 genotypes in the F2 progeny established that CCR2 and CD25 segregated independently of each other with the number of meiosis events close to 50% (Fig. 1d and Supplementary Fig. 1e). This result suggests that the Ccr2−/− 2009 trait depends on a single locus likely located on a distinct chromosome than the 9th that carries Ccr2. Whole exome sequencing of Ccr2−/− 2009 and Ccr2−/− 2012 mouse DNA identified only 5 non-synonymous coding gene variants (Fig. 1e and Supplementary Fig. 1f), with the most relevant coding variant replacing a single thymidine with a cytosine nucleotide on position 426 of exon 4 of the Il2ra gene in the Ccr2−/− 2009 line. This mutation led to (i) the replacement of an evolutionarily highly conserved tyrosine (Y) with a histidine (H) on position 129 of the CD25 protein (Fig. 1f), and (ii) the creation a new BssSI restriction site (Supplementary Fig. 1g) which helped confirm the presence of this mutation in Ccr2−/− 2009 but not Ccr2−/− 2012 mice. Targeted mutagenesis of the Y129 of WT Il2ra to H129 and retroviral transduction of WT and H129 CD25 in 293T cells (Fig. 1g) recapitulated the CD25 cell-surface expression phenotypes reported in Ccr2−/− 2009 vs Ccr2−/− 2012 mice, demonstrating that this point mutation accounted for the mouse phenotype. We next bred mice carrying only the homozygous Y129H mutation from the Ccr2+/+ F2 progeny (Fig. 1c), further referred to as Il2ramut/mut mice.
Fig. 1.
Isolation and characterization of the Il2ramut/mut mouse model. a Lymph node cells isolated from WT, Ccr2−/−2009 and Ccr2−/−2012 (all on the B6 background) mice were stained with mAbs against cell-surface CD3, CD4, CD25 (clone PC61), and intracellular Foxp3. A representative dot plot is shown. Bar graph summarizes CD25 expression levels (MFI) across all mice analyzed. b FACS histograms of cell-surface CD25 levels (MFI) gated on blood Foxp3+ Treg cells (CD3+CD4+) from Ccr2−/−2009, Ccr2−/−2012, Ccr2+/−2009 (F1) mice. c Distribution of CD25 cell-surface expression levels (MFI) in Treg cells from the blood of individual mice of indicated genotypes including Ccr2+/−2009 × Ccr2+/−2009 (F2) mice. d Ccr2 genotypes based on CD25 surface expression levels and experimental frequencies obtained in F2 offsprings. e DNA and corresponding amino-acid sequence alignments of the mutation found in exon 4 of Il2ra gene in Ccr2−/−2009 vs Ccr2−/−2012 mice after whole-exome sequencing. f Alignment of the CD25 amino-acid sequence surrounding tyrosine 129 across multiple species. g FACS histograms of cell-surface expression of CD25 in 293T cells retrovirally transduced with WT or mutagenized Y129H Il2ra cDNA. Bar graph shows CD25 expression (MFI) across two independent transduction experiments. h Dot plots of cell-surface or intracellular expression levels (MFI) of CD25 in WT and mutant Treg cells. Bar graphs average data from one of 2–3 experiments with similar results with >2 mice per group. i Fluorescent microscopy staining of CD25 on anti-CD3 stimulated purified CD4+ T cells isolated from LNs. T cells were fixed and co-stained with anti-CD25 (green) and anti-calreticulin (red) prior to image acquisition. Scale bar is 5 μm. p-values are indicated when relevant with *p < 0.05; **p < 0.01; ***p < 0.001; NS not significant, using two-tailed unpaired Student’s t-test. Error bars are mean ± SEM in all figures
Since the Y129H change impaired cell-surface expression of CD25 on Treg cells, we further hypothesized that CD25 may not be properly folded and/or lack stability. Based on the structure of human CD25, which has ~60% percent homology with mouse CD25, the Y129 likely makes stabilizing hydrogen bonds with a section of CD25 that is in direct contact with IL-2 (Supplementary Fig. 1h). Consistent with this possibility, intracellular staining for CD25 revealed high levels of CD25 inside Treg cells from Il2ramut/mut compared to WT mice, a finding also confirmed in retrovirally transduced 293T cells (Fig. 1h and Supplementary Fig. 1i). CD25 subcellular localization in anti-CD3-stimulated CD4+ T cells from either Il2ramut/mut or WT mice, suggested that mutant CD25 mostly co-localized with calreticulin, an endoplasmic reticulum (ER) resident protein. A strong staining of the nuclear envelope, a subdomain of the ER, was observed in Il2ramut/mut but not WT CD4+ T cells, suggesting that the Y129H mutation prevented proper folding of CD25 and subsequent egress from the ER to the cell surface (Fig. 1i). Thus, we isolated and characterized a mouse mutant line with a point mutation in an evolutionarily highly conserved tyrosine of the high affinity IL-2 receptor alpha chain CD25, likely preventing its optimal folding and subsequent cell-surface expression.
Il2ramut/mut T cells exhibit impaired responses to IL-2
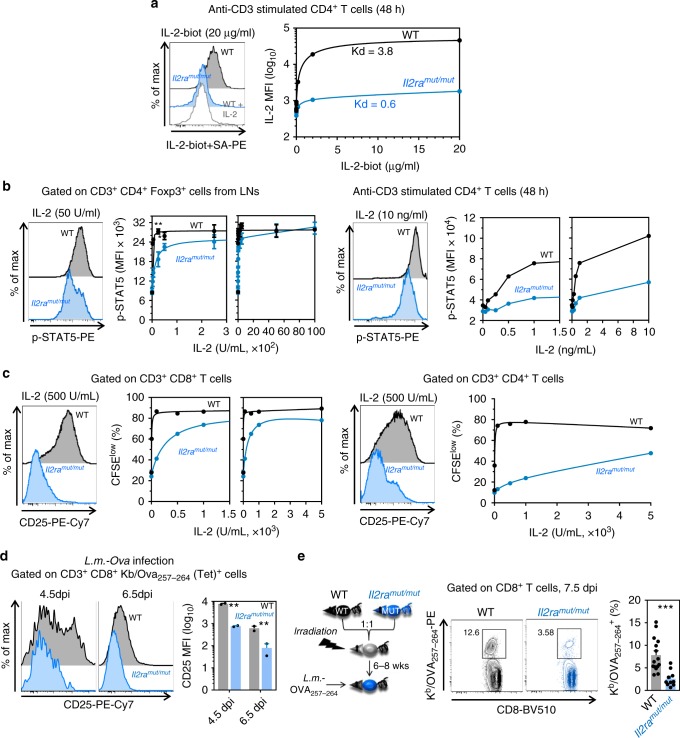
We next assessed whether impaired cell-surface expression of CD25 on T cells translates into decreased IL-2 binding and signaling. Using labeled IL-2 incubated with anti-CD3 stimulated CD4+ T cells, we found higher IL-2 binding to WT compared to either Il2ramut/mut or WT T cells incubated with an excess of non-biotinylated IL-2 (Fig. 2a and Supplementary Fig. 2a). STAT5a phosphorylation, a very early signaling response that follows IL-2 binding to CD25, was also profoundly altered both in Treg cells from thymus and LNs and in conventional CD4+ T (Tconv) cells from Il2ramut/mut mice (Fig. 2b and Supplementary Fig. 2a, b). Consistent with these findings, Il2ramut/mut CD8+ and CD4+ Tconv cells proliferated substantially less compared to WT counterparts as quantified in vitro by CTV dilution of labeled Tconv cells after anti-CD3 and varied amounts of IL-2 (Fig. 2c and Supplementary Fig. 2a, c). To extend findings in vivo, we next monitored CD25 upregulation on Kb/Ova257-264 tetramer+ (Tet+) CD8+ Tconv cells primed in WT or Il2ramut/mut mice infected intravenously (i.v.) with Listeria monocytogenes expressing the model antigen Ovalbumin (Lm-Ova) (Fig. 2d). Expression of CD25 by Ova-specific Tet+ CD8+ Tconv cells was lower in Il2ramut/mut compared to WT mice33, in line with the in vitro data (Fig. 2d). Then, we generated mixed bone-marrow chimeras from lethally irradiated recipient mice reconstituted with WT and Il2ramut/mut donor bone-marrow cells (ratio 1:1) so that we could monitor and compare WT and Il2ramut/mut antigen-specific CD8+ Tconv cell responses in the same host (Fig. 2e and Supplementary Fig. 2d). Reconstituted chimeras were further infected with Lm-Ova or the Herpes Simplex Virus 2 (HSV-2) intravaginally (i.vag.), and both Kb/Ova257-264 and Kb/gB498-505 Tet+ Tconv cells were monitored at the peak of the primary response. In both models, Ova- and gB-specific Il2ramut/mut CD8+ Tconv cells expanded ~3-fold less than WT counterparts. Thus, the CD25 Y129H mutation profoundly alters CD25 expression and functional response to IL-2 in both Tconv and Treg cells.
Fig. 2.
The Y129H mutation in CD25 impairs IL-2 binding, signaling and T cell proliferation. a Recombinant human IL-2 was biotinylated and incubated with anti-CD3 stimulated purified CD4+ T cells from Il2ramut/mut or WT mice prior to staining with streptavidin PE. Unbiotinylated IL-2 was added to WT T cells as a control of binding specificity. Representative FACS histograms of IL-2 or no IL-2 bound to mutant or WT T cells are shown. The graph plots IL-2 binding signals (MFI) with increasing concentration of biotinylated IL-2. IL-2 Kd (50% binding capacity) for Il2ramut/mut or WT T cells are shown. b Purified CD4+ T cells from LNs of Il2ramut/mut or WT mice (n = 3) were incubated in serum-free media (30’) and with IL-2 (20’) before proceeding to intracellular STAT5 phosphorylation staining. FACS histograms of STAT5 phosphorylation levels in Foxp3+ Treg (left) and anti-CD3 activated CD4+ Tconv (right) cells are shown. Graphs plot phosphorylated STAT5 expression with increased IL-2 concentrations. c Purified LN CD4+ or CD8+ T cells from Il2ramut/mut or WT mice were CFSE-labeled and incubated with anti-CD3 and IL-2. The proportion of CFSElow T cells was determined by FACS 4 days later. Graphs plot the % CFSElow T cells as IL-2 increases in 1 of 3 replicate experiments. FACS histograms show cell-surface CD25 expression in Il2ramut/mut or WT T cells 4 days post IL-2 incubation. d Il2ramut/mut or WT mice (n = 2) were immunized i.v. with 104 WT Listeria monocytogenes (Lm) expressing Ovalbumin (Lm-Ova) and spleen cells were stained at indicated days with Ova257-264/Kd tetramers (Tet+) and anti-CD25. Representative FACS histograms of CD25 expression on tet+ CD8+ T cells is shown with a summary bar graph. e WT/Il2ramut/mut mixed bone-marrow (BM) chimeras (ratio 1:1) with discriminative CD45.1/2 congenic markers were immunized i.v. with WT Lm-Ova and 7.5 days later spleen cells stained with Ova257-264/Kd tetramers. Representative FACS dot plots and summary bar graphs of Lm-specific Il2ramut/mut or WT tet+ CD8+ T cells are shown (n = 15 mice). p-values are indicated with *p < 0.05; **p < 0.01; ***p < 0.001; NS not significant, using two-tailed unpaired Student’s t-test. Error bars are mean ± SEM in all figures
IL-2 modulates Treg cell epigenetic landscapes
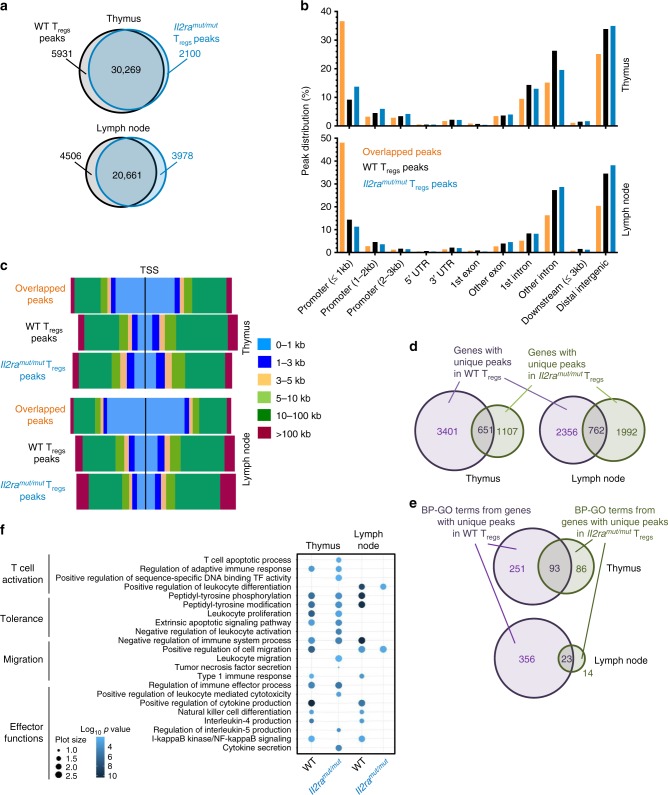
IL-2 signals are postulated to contribute to establishing the Treg cell program of differentiation before Treg cells express Foxp3 in the thymus11–13,34 but only little evidence exist to support this hypothesis. To explore this possibility, we took advantage of the Il2ramut/mut mouse model in which Treg cells receive reduced IL-2 signals, and asked whether their chromatin accessibility differs just after they have committed to the Treg cell lineage (thymus) and in the periphery in the secondary lymphoid organs (LNs). We isolated Foxp3+ Treg cells from thymus and LNs of Il2ramut/mut and WT Foxp3Rfp/Rfp reporter mice and conducted an analysis of the genome-wide open chromatin regions (OCRs) using an assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq35, Fig. 3 and Supplementary Fig. 3). While we found 36,200 and 32,369 OCRs (ATAC-seq peaks) in WT and Il2ramut/mut thymic Treg cells respectively, LN Treg cells exhibited 25,167 and 24,639 OCRs (Fig. 3a). Ninety-five percent of WT LN Treg cell OCRs identified overlapped with those reported in a prior study analyzing LN Treg cell OCRs36 (Supplementary Fig. 3a). As expected, this overlap was lower (75-80%) when we compared WT Treg cell’s OCRs to that reported in CD8+ T cells37. Within the thymic and LN Treg cell OCRs, we respectively identified 8031 and 8484 differentially accessible OCRs in WT (5931 in thymus and 4506 in LN) and Il2ramut/mut (2100 in thymus and 3978 in LN) Treg cells (Fig. 3a). This represents ~41% difference in the epigenetic landscape of WT vs Il2ramut/mut LN Treg cells, which is comparable to what is reported between resting and activated Treg cell OCRs36 (~36%, Supplementary Fig. 3b). These differences are also similar to those measured between effector and memory or exhausted CD8+ T cells (34–41%)37–39 but substantially greater than that reported in effector cells isolated from mice infected with acute vs chronic LCMV (~15%)38. Analysis of the relative localizations of differential OCRs in relation to the mouse ensemble annotated genes revealed a lower proportion of OCRs in the transcription start sites (TSS, ≤1 kb) compared to that of the overlapping peaks both for thymic and LN Treg cells (~9–14% vs 37–48%, Fig. 3b, c and Supplementary Fig. 3c and Data 1). The majority of unique peaks were found in introns and distal intergenic regions (>10 kb), suggesting that IL-2 signals regulate chromatin opening of elements distal to the TSS already at early stages of Treg cell commitment in the thymus. Upon assigning these peaks to the closest gene TSS, 651 (thymus) and 762 (LN) genes with differential OCRs were common to WT and Il2ramut/mut Treg cells while the remaining were within non-overlapping genes (Fig. 3d and Supplementary Data 2). Approximately three times as many genes and OCRs were uniquely present in WT compared to Il2ramut/mut thymic Treg cells (3401 vs 1107 genes) while LN Treg cells had comparable numbers of unique genes and OCRs (2356 vs 1992 genes)(Fig. 3a,d). This result shows that the ratio of differential OCRs between WT and Il2ramut/mut Treg cells from the thymus to the LNs is substantially modulated (from ~3:1 to 1:1), suggesting further functional commitment of Treg cells. Analysis of the biological-process (BP) gene-ontology (GO) pathways of genes containing unique peaks (<20 kb from TSS) in WT thymic and LN Treg cells highlighted a much greater diversity of BP (251 and 356 GO pathways vs only 86 and 14 GO pathways, respectively)(Fig. 3e and Supplementary Data 3). Since the differences in the number of genes with unique peaks between WT and Il2ramut/mut Treg cells is much smaller compared to that of the GO pathways (Fig. 3d, e), it further suggests that the OCRs in Il2ramut/mut Treg cells are more random. While some processes related to T cell activation and tolerance are common to both types of Treg cells in the thymus, others targeting activation, migration and effector mechanisms are only maintained in WT LN Treg cells (Fig. 3f). Conducting the GO pathway analysis on genes containing unique peaks without any limit from TSS revealed the exact same trend (Supplementary Fig. S3d,e and Data 3). Thus, IL-2 signals have a broad impact on genome-wide Treg cell epigenetic landscape, both for the opening and the closing of mostly TSS-distant regulatory regions. IL-2-mediated alterations occur already at early stages of Treg cell commitment to this lineage in the thymus and are sustained throughout LN Treg cells.
Fig. 3.
IL-2 makes substantial modifications in the epigenetic landscape of Treg cells: Treg cells (5 × 104) were sorted by flow cytometry from thymus or LNs of Il2ramut/mut and WT Foxp3Rfp/Rfp reporter mice, lysed and DNA from nucleus extracted for analysis of open chromatic regions by ATAC-seq. a Venn diagram of number of common and unique ATAC-seq peaks (OCRs) in Il2ramut/mut vs WT Treg cells from thymus and LNs. b, c Distribution of common and unique ATAC-seq peaks across gene organization (b) and distance to the transcription start site (TSS, c) in the whole genome. d, e Venn diagrams of number of genes (d) and GO pathways (e) exhibiting unique peaks in Il2ramut/mut vs WT thymic and LN Treg cells on all (d) or within (e) 20 kb from TSS. f Network analysis of biological-process GO term enrichment among genes with unique peaks within 20 kb from TSS in Il2ramut/mut or WT thymic and LN Treg cells. Genes were analyzed for over-represented GO terms using ReviGO. Node color is proportional to the FDR-adjusted p-value of the enrichment. Node size is proportional to gene set size
IL-2-driven epigenetic changes mildly alter gene expression
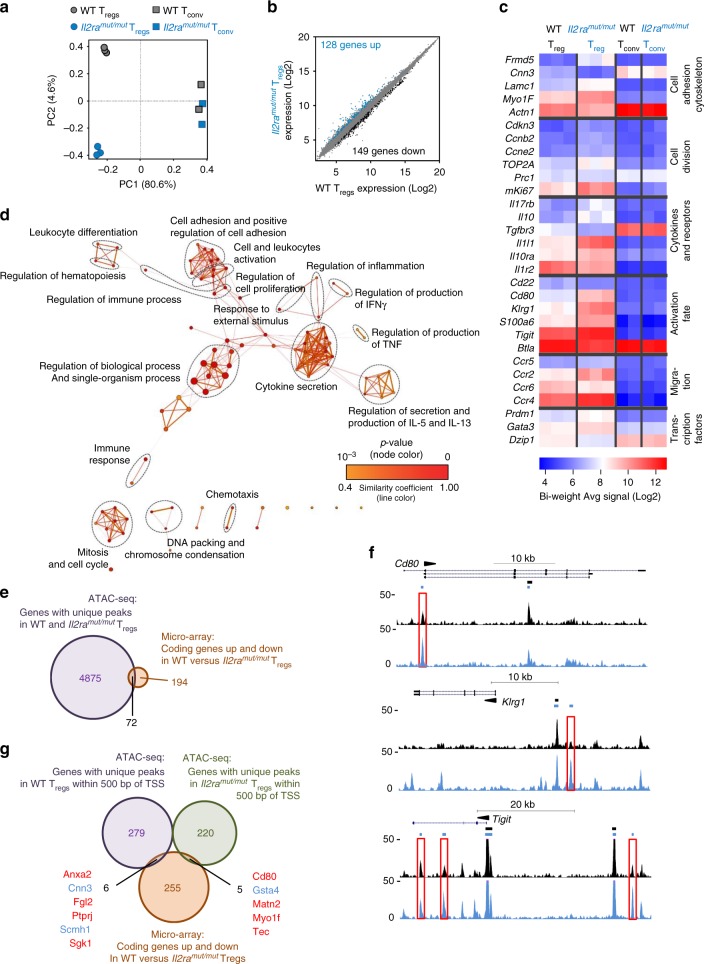
Since IL-2 induces extensive modifications in the chromatin of Treg cells (Fig. 3), we next assessed whether these alterations translated into a distinct program of expression between Il2ramut/mut and WT LN Treg cells. The lack of IL-2 signaling in Treg cells was reported to only alter a relatively focused set of genes implicated in cell growth, metabolism, motility and adhesion, preventing their overall competitive fitness in vivo21,29,40. We purified total RNA from flow-sorted Foxp3+ (RFP+) Treg or naive (CD62LhiCD44lo) Tconv cells isolated from the LNs of WT or Il2ramut/mutFoxp3Rfp/Rfp reporter mice, and analyzed gene expression using microarrays (Fig. 4). While the expression profiles of naive Tconv cells are almost identical between the two groups, that of Il2ramut/mut and WT Treg cells exhibit significantly more differences (Fig. 4a and Supplementary Fig. 4a and Data 4). We note a set of 277 differentially expressed genes among which 128 and 149 are up- or downregulated respectively in Il2ramut/mut compared to WT Treg cells (Fig. 4b and Supplementary Fig. 4b). These genes are involved in cellular adhesion and cytoskeleton rearrangement, cell division and differentiation, cytokines and receptors, cellular activation and chemotaxis (Fig. 4c), consistent with prior reports21,29,40. We checked some of the differentially upregulated genes for which Abs specific for encoded proteins are available, and found a direct correlation with protein expression levels for KLRG1, TIGIT, CCR4 while we did not for CCR5 or BTLA (Supplementary Fig. 4c). Interestingly, increased expression of these markers is detected only in a small subset of CD4+ Treg cells, suggesting that lowering IL-2 signals promotes the onset of a subpopulation of Treg cells with a slightly distinct expression program, rather than altering the whole Treg cell population. Since only a few of the downregulated genes encode for proteins of known functions, we next focused primarily on sets of upregulated genes to run a comparison against known BP-GO databases (Fig. 4d and Supplementary Fig. 4d,e and Data 5). We found statistically over-represented gene sets implicated in cell adhesion, activation, proliferation/cell cycle, cellular differentiation as well as inflammatory processes, transcriptional regulators and chromatin organization/epigenetic gene regulation. While only a limited set of genes are differentially expressed in Foxp3+ Treg cells that receive lower IL-2 signals, the biological functions that are altered seem to directly target their ability to migrate, undergo homeostatic proliferation and express suppressive effector functions. We also noted differences in expression of genes implicated in various steps of chromatin modifications (Supplementary Fig. 4f).
Fig. 4.
The expression of only a small set of genes in Treg cells is altered by low IL-2 signals: a 5 × 104 Treg and Tconv cells were sorted by flow cytometry from LNs of three independent replicate Il2ramut/mut or WT Foxp3Rfp/Rfp reporter mice and total RNA extracted and reverse transcribed to cDNA. Affymetrix mouse expression arrays (Pico 1.0) were then conducted. Principal component analysis (PCA) of expressed genes using the top 10% of genes with the highest variance in analyzed groups with each symbol featuring one mouse. b Scatter plot of individual genes expressed in Il2ramut/mut vs WT Treg cells. Significantly upregulated and downregulated genes, defined as genes with at least 1.5 fold change, p-value ≤ 0.01, are colored blue or black, respectively, and numbers are shown. c Heat map of selected genes grouped under various categories as indicated and for which expression is significantly different between Il2ramut/mut and WT Treg cells. d Network analysis of biological-process gene-ontology (GO) term enrichment among significantly upregulated genes in Il2ramut/mut vs control WT Treg cells. Upregulated genes were analyzed for over-represented GO terms using BiNGO in Cytoscape, and the resulting network was calculated and visualized using EnrichmentMap. Groups of similar GO terms were manually circled. Line thickness and color are proportional to the similarity coefficient between connected nodes. Node color is proportional to the FDR-adjusted p-value of the enrichment. Node size is proportional to gene set size. e Venn diagram comparing differentially expressed genes in Il2ramut/mut vs WT Treg cells to genes with unique peaks called in Il2ramut/mut and WT Treg cells found in ATAC-seq in Fig. 3 based on gene symbols. f ATAC-seq tracks for 3 sample genes with at least 1 unique peak localized in distinct regions of the gene (promotor, intronic, intergenic). g Three-way Venn diagram comparing differentially expressed genes in Il2ramut/mut versus WT Treg cells to genes containing unique peaks close to TSS (<500 bp) in Il2ramut/mut vs WT Treg cells found in ATAC-seq
Most interestingly, although the number (4875) and relative proportion (~40%) of differential OCRs between Il2ramut/mut and WT LN Treg cells is important (Fig. 3), those of differentially expressed genes (266) remain very small (below 1% of genes), suggesting that IL-2 signals play a more prominent role in shaping the epigenetic landscape and identity of Treg cells than their ability to express sets of genes (Fig. 4e and Supplementary Data 6). Among the differentially expressed genes, only 72 exhibited unique OCRs in either WT or Il2ramut/mut LN Treg cells. These OCRs may contribute to the changes in gene expression via chromatin structure modifications while the remaining genes (194) are likely to be controlled at the level of gene transcription with no changes in chromatin structures. For instance, CD80 that is highly expressed in Il2ramut/mut compared to WT LN Treg cells, exhibits greater OCRs heights in Il2ramut/mut vs WT Treg cell promoter regions (Fig. 4f). Differential OCRs are further noted in intergenic regions of KLRG1 or in intronic regions for TIGIT. Interestingly, neither the Foxp3 nor the Helios genes had differential OCRs between Il2ramut/mut and WT Treg cells (Supplementary Fig. 4g and Data 7). Analysis of the potential binding motifs for TFs in the differential OCRs revealed that a different set of TFs such as FOXO and KLF family members, could possibly bind in WT but not Il2ramut/mut Treg cells (Supplementary Fig. 4h). Because chromatin structures in the vicinity of TSS regions play critical roles in the expression status of the gene, we tested if differentially expressed genes contain differential OCRs close to the TSS (Fig. 4g and Supplementary Data 6). Eleven differentially expressed genes contain differential OCRs close to their TSS (±500 bp) while the majority of the remaining genes did not, suggesting that these genes undergo epigenetic modifications mostly occurring on elements distal to the TSS such as gene enhancer regions. In summary, despite establishing major genome wide changes of the Treg cell epigenetic landscape, IL-2 signals on LN Treg cells at steady state only translate in modest modifications of their gene expression profiles.
IL-2 regulates SATB1 positioning prior Treg cell commitment
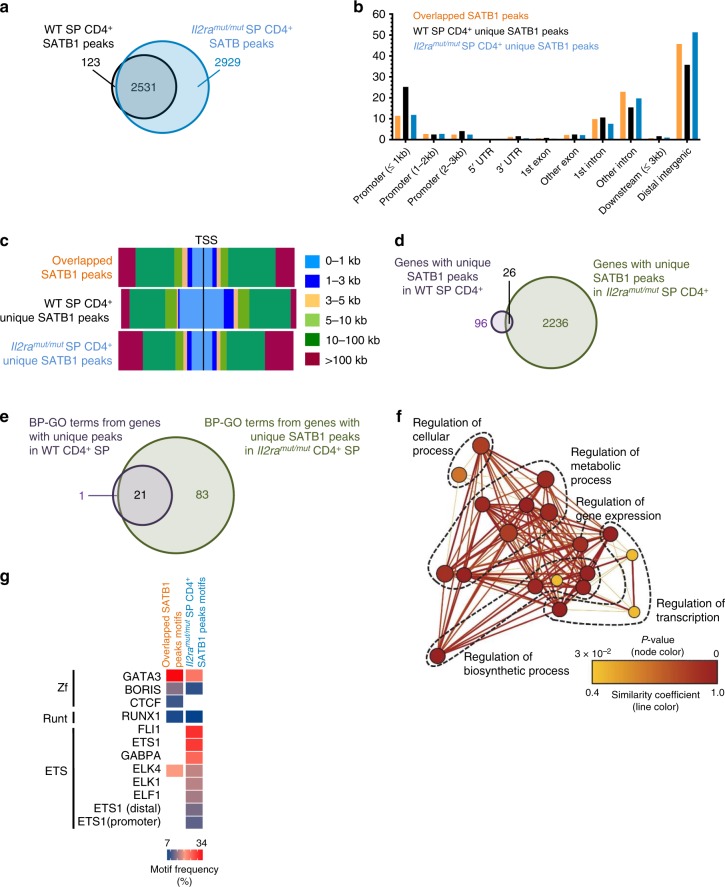
We next investigated how IL-2 signals mediate such large genome-wide changes in the Treg cell epigenome. Given its essential role in Treg cell-lineage specification and functions5–7,14, and its dependence on IL-219,21, one obvious candidate is the Foxp3 TF. If Foxp3 accounted for our observations, we predicted that Foxp3 binding regions would exhibit a very large overlap with the OCRs found in WT Treg cells but less with that of Il2ramut/mut Treg cells (Supplementary Fig. 5a). By aligning published chromatin immunoprecipitation sequencing (ChIP-seq) results of the Foxp3 TF defining the Foxp3 binding sites throughout the genome of LN Treg cells26 with our ATAC-seq data, we found that Foxp3 DNA binding regions largely overlapped with OCRs common between Il2ramut/mut and WT LN Treg cells, suggesting that IL-2-mediated alterations of the Treg cell epigenome are independent of Foxp3. Robust experimental evidence supports the idea that Treg cell epigenetic and functional identity is established in the thymus prior Treg cells express Foxp313,24–26. Notably, the pioneer factor SATB1 was reported to be essential for Treg cell-lineage specification before expression of Foxp3 in the thymus but not after27. Thus, we hypothesized that IL-2 acts through SATB1 before thymocytes commit to the Treg cell fate, at the single positive (SP) CD4+ thymocyte stage11,13,24. We know (Fig. 3) that mature Foxp3+ Il2ramut/mut thymic Treg cells already exhibit differential OCRs compared to WT counterparts, suggesting epigenetic changes may have occurred before this stage. We conducted ChIP-seq experiments of SATB1 on SP CD4+ thymocytes isolated from Il2ramut/mut or WT mice, postulating that SATB1 DNA positioning would differ if indeed this pioneer factor is involved in the regulation of IL-2-mediated Treg cell epigenetic changes (Fig. 5). The vast majority of SATB1-bound regions in WT and Il2ramut/mut SP CD4+ thymocytes overlapped. While only 123 unique DNA regions are bound by SATB1 in WT thymocytes, SATB1 associate to more than 50% additional regions (2929) in Il2ramut/mut thymocytes, showing that ectopic binding of this pioneer factor occurs when IL-2 signals are limiting (Fig. 5a). Interestingly, the 123 unique SATB1-binding regions in WT thymocytes localize closer to promoter areas compared to that of Il2ramut/mut counterparts which are far from TSS (>100 kb) in distal intergenic regions (Fig. 5b, c and Supplementary Fig. 5b). Assigning the 123 SATB1-bound regions to the nearest gene TSS highlighted 96 genes among which 26 are common to Il2ramut/mut Treg cells despite different SATB1-bound areas (Fig. 5d and Supplementary Data 7). Interestingly, 29 of the 96 genes with unique SATB1-binding sites are located within less than 2 kb from the TSS, and a majority encodes for DNA remodeling proteins, cell cycle, signal transduction and metabolism (Supplementary Fig. 5c). These genes exhibit variable levels of expression across thymocyte development41, in particular between the SP CD4+ thymocyte and thymic Treg cell stages, consistent with a possible regulatory role of their expression by promoter-bound SATB1. SATB1 binds ~20 times more genes (2236) in Il2ramut/mut compared to WT thymocytes, illustrating its ectopic binding when IL-2 signals are limiting (Fig. 5d). BP-GO analysis on the common genes between the two conditions revealed that SATB1 targets common processes (~21) involved in DNA remodeling, transcriptional regulation and metabolism (Fig. 5e and Supplementary Fig. 5d and Data 8a,b). However, when IL-2 signaling is impaired, SATB1-associated genes are implicated in 83 additional processes that include immune cell activation, adhesion and differentiation, and may reflect less focused binding. A search for known TF binding motifs within the regions bound by SATB1 highlighted GATA3, RUNX1 and ETS1 among the top ones, which are all associated with the regulation of Foxp3 expression15,17,42,43 (Fig. 5g). These motifs are AT-rich, which represent SATB1 preferential binding motifs44. While no enriched motifs are revealed in the 123 unique regions of WT SP CD4+ thymocytes, all of the prior ones are found in the remaining WT and Il2ramut/mut SATB1-bound regions. The proportion of these TF-binding motifs is enriched in peaks common with WT thymocytes, suggesting a more focused binding of SATB1 when normal IL-2 signals are received. Thus, IL-2 signals have a significant impact on where the pioneer factor SATB1 binds to DNA in SP CD4+ thymocytes, which is likely to contribute to IL-2 modulation of the Foxp3+ Treg cell epigenetic landscape.
Fig. 5.
Ectopic DNA binding of SATB1 in SP CD4+ thymocytes receiving low IL-2 signals: 2 × 106 SP CD4+ thymocytes were sorted in 2 independent duplicate experiments by flow cytometry from the thymus of Il2ramut/mut and WT mice, PFA fixed, lysed, and cross-linked DNA was used for chromatin immunoprecipitation sequencing (ChIP-seq) of the pioneer factor SATB1. a Venn diagram of the number of common and unique SATB1 DNA binding peaks in Il2ramut/mut vs WT SP CD4+ thymocytes. b, c Distribution of common and unique SATB1 DNA binding peaks across gene organization (b) and distance to the transcription start site (TSS, c) in the whole genome. d Venn diagram of the number of genes exhibiting unique SATB1 DNA binding peaks in Il2ramut/mut vs WT SP CD4+ thymocytes. e Venn diagram of biological-process gene-ontology (BP-GO) from genes with unique SATB1 DNA binding peaks in WT vs Il2ramut/mut SP CD4+ thymocytes. f Network analysis of BP-GO term enrichment among genes with unique SATB1 DNA binding peaks in WT SP CD4+ thymocytes. Genes with unique SATB1-binding peaks were analyzed for over-represented GO terms using BiNGO in Cytoskape, and the resulting network was calculated and visualized using EnrichmentMap in Cytoscape. Groups of similar GO terms were manually circled. Line thickness and color are proportional to the similarity coefficient between connected nodes. Node color is proportional to the FDR-adjusted p-value of the enrichment. Node size is proportional to gene set size. g Frequency of known TF binding site motifs enriched in common and unique SATB1 DNA binding peaks in WT and Il2ramut/mut SP CD4+ thymocytes. Motif frequency (%) is shown in heat map
Il2ramut/mut mice lack autoimmunity despite altered chromatin
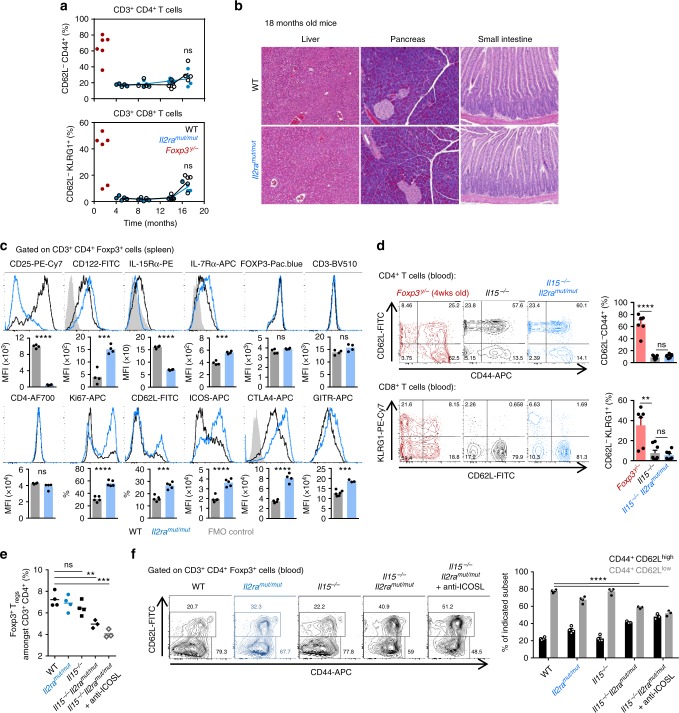
Given the importance of IL-2 in Treg cell stability, immune homeostasis21–23,40 and epigenetic programming (Figs. 3, 5), we postulated that Il2ramut/mut mice may develop symptoms consistent with autoimmunity over time. Monitoring of T cell phenotypes in the peripheral blood of Il2ramut/mut and WT mice over ~1.5 years did not reveal any differences in cell-surface expression of classical activation markers (CD44, CD62L, KLRG1), in contrast to T cells from Foxp3y/− mice analyzed 1.5 months post birth (Fig. 6a and Supplementary Fig. 6a). No T cell infiltrates were detected in the liver, pancreas or small intestine of Il2ramut/mut mice, which all represent peripheral organs often associated with the onset of an autoimmune reaction (Fig. 6b). We next investigated how mutant Treg cells may control autoreactive Tconv cells and maintain immune homeostasis. We first conducted an extensive characterization of Treg cells from Il2ramut/mut compared to WT mice (Fig. 6c and Supplementary Fig. 6b). Il2ramut/mut Treg cells express higher cell-surface levels of IL-7 and IL-15 survival receptors (CD127 and CD122) and lower levels of IL-15Rα, consistent with the idea that Treg cells receiving lower IL-2 signals may rely on the use of compensatory IL-7 and IL-15 cytokines11,19,29. These Treg cells also undergo increased cell proliferation (Ki67+) and upregulated T cell inhibitory receptors (CTLA4, GITR) and ICOS, which are reported to promote efficient Treg cell-mediated suppression45. We did not detect any difference in TCR Vβ usage (Supplementary Fig. 6c), likely ruling out major biases in Il2ramut/mut Treg cell repertoire. Since the common IL-2/IL-15 transducing beta chain CD122 was significantly upregulated on Il2ramut/mut Treg cells compared to WT counterparts (spleen and thymus), we next crossed Il2ramut/mut mice to Il15−/− mice, postulating that IL-15 signals are essential for mutant Treg cells to maintain immune homeostasis in these mice (Fig. 6d) and compensate for impaired IL-2 signals though Il2ramut/mut mice have higher levels of circulating IL-2 (Supplementary Fig. 6d). The proportion of activated CD4+ (CD62LloCD44hi) and CD8+ (CD62loKLRG1+) Tconv cells in Il2ramut/mutIl15−/− and Il15−/− mice was equivalent (Fig. 6d) and similar to that of WT mice (Fig. 6a), but significantly lower than in Foxp3y/− mice. In addition, we did not find any evidence of T cell infiltrates in the liver or the spleen of these mice (Supplementary Fig. 6e). However, the proportion of Foxp3+ Treg cells in Il2ramut/mutIl15−/− mice was reduced compared to WT, Il2ramut/mut or Il15−/− mice (Fig. 6e). Recently, two distinct subsets of Treg cells discriminated on CD44, CD62L and CCR7 expression were proposed to require either IL-2 (CD44loCD62hi) or ICOS (CD44hiCD62lo) signaling for their maintenance46. Since we noted high cell-surface expression of ICOS on Il2ramut/mut Treg cells (Fig. 6c), we reasoned that impaired IL-2 signals might favor the onset of a CD44hiCD62lo Treg cell subset that requires ICOS signaling. Unexpectedly, we only observed lower proportions of the CD44hiCD62lo Treg cell subset in Il2ramut/mut and Il2ramut/mutIl15−/− compared to WT mice, and this proportion was even further decreased by blocking ICOS for 2 weeks (Fig. 6f). The overall proportion of Foxp3+ Treg cells among CD4+ T cells in the blood of ICOS-neutralized Il2ramut/mutIl15−/− mice was also reduced by ~50%, although mice did not develop autoimmunity (Fig. 6e and Supplementary Fig. 6f), suggesting that none of the proposed mechanisms accounts for the absence of autoimmune disease in Il2ramut/mut mice.
Fig. 6.
Il2ramut/mut mice do not develop any signs of autoimmunity: a T cells collected from blood of Il2ramut/mut and WT mice were stained for cell-surface CD3, CD8, CD4 and CD62L, CD44, KLRG1 activation markers at indicated times over an 18 months period. Blood from control Foxp3y/− was analyzed at ~1.5 months post birth. Each symbol represents an individual mouse. b At month 18, liver, pancreas, and small intestine were harvested, fixed/cut, and stained with H/E. c Spleen cells from 8 to 10-weeks-old Il2ramut/mut and WT mice were stained for cell-surface CD3, CD4, intracellular Foxp3 and indicated markers. Overlay of representative FACS histograms after gating on CD3+CD4+Foxp3+ T cells are shown and bar graphs average relative expression levels (MFI, n = 4–5). d Blood from Il2ramut/mutIl15−/−, Il15−/− (6 months old) or Foxp3y/− mice was stained as in a and representative FACS dot plots with bar graphs summarizing average across mice (n = 6–7) are shown. e Proportion of peripheral blood Foxp3+ Treg cells among CD3+CD4+ T cells in indicated mice treated (or not) with anti-ICOSL for 2 weeks. f Blood leukocytes from indicated mice groups were stained for expression of CD3, CD4, intracellular Foxp3, CD62L, CD44. Representative dot plots and summary bar graphs of average CD44/CD62L expression levels across all groups of mice are shown (n = 3–4). p-values are indicated when relevant with *p < 0.05; **p < 0.01; ***p < 0.001; NS not significant, using two-tailed unpaired Student’s t-test
Il2ramut/mut Treg cells cannot repress WT autoimmune T cells
Lack of IL-2 or its receptor chains leads to fatal autoimmunity because Treg cells are unstable and fail to suppress autoreactive Tconv cells11–13. Yet, mice in which IL-2 signaling is genetically altered by targeted mutation in the IL-2Rβ/CD122 transducing chain do not become autoimmune29. Thus, we reasoned that similar to IL-2Rβ mutant Treg cells, Il2ramut/mut Treg cells may repress Il2ramut/mut but not WT autoreactive Tconv cells. As a first step to assess this idea, we conducted a standard in vitro suppression assay using increasing numbers of Il2ramut/mut or control WT Treg cells incubated with CFSE-labeled anti-CD3 activated Tconv cells (Supplementary Fig. 7a). Il2ramut/mut Treg cells inhibit Tconv cell proliferation ~two fold less than WT Treg cells across all ratios, possibly accounting for the lack of autoimmunity in Il2ramut/mut mice. Since Treg cells grown in vitro with anti-CD3/CD28 stimulation and under limited IL-2 availability lose Foxp3 expression and convert to effector T cells18, we next assessed whether Il2ramut/mut Treg cells also lacked stability after TCR triggering in vitro (Supplementary Fig. 7b). While ~60% of WT Treg cells lost Foxp3 expression, ~85% of Il2ramut/mut converted to Foxp3neg T cells, consistent with impaired lineage stability when IL-2 signals are limiting. Thus, even if they can prevent autoimmunity in Il2ramut/mut mice, Il2ramut/mut Treg cells may not be able to suppress WT autoreactive Tconv cells to the extent to which WT Treg cells can.
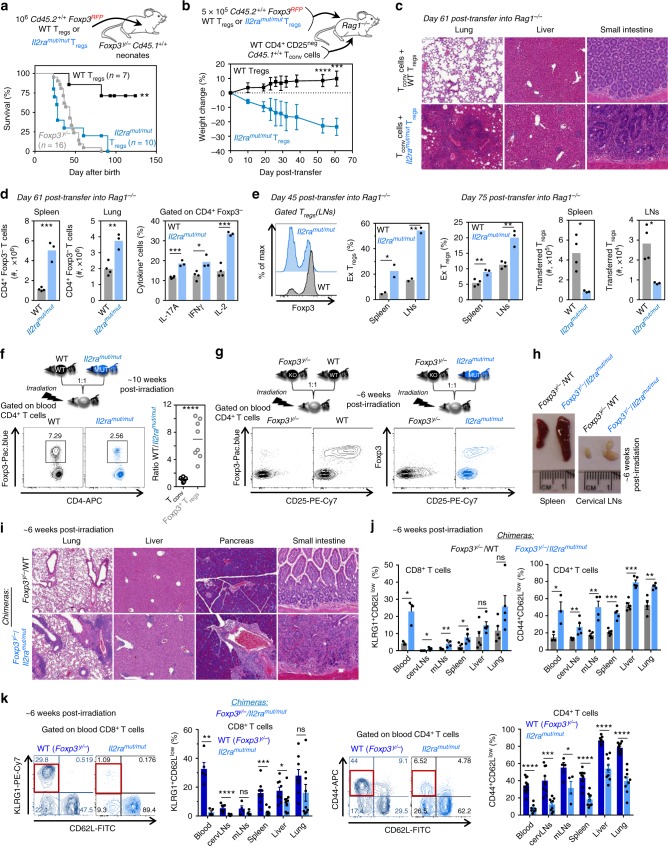
To further evaluate their suppressive capacity in vivo, we used two distinct but complementary models of acute autoimmunity development, and a model of reconstitution at homeostasis. We first conducted rescue experiments of Foxp3y/− mice5 which develop fatal autoimmunity within ~3–4 weeks post birth, by transferring Treg cells sorted from the secondary lymphoid organs (SLOs) of WT or Il2ramut/mut Foxp3Rfp/Rfp reporter mice to recipient neonates (day 1–3) (Fig. 7a). As expected, Foxp3y/− mice receiving WT Treg cells survived, while those transferred with Il2ramut/mut Treg cells did not control the development of wasting disease and ultimately succumbed, as did untransferred counterparts. Second, we adoptively transferred CD4+CD25neg Tconv cells to Rag1−/− mice, which also results in a rapid wasting disease unless functional Treg cells are co-transferred (Fig. 7b). As predicted from the first model, Il2ramut/mut Treg cells fail to suppress autoreactive Tconv cell-mediated autoimmunity with a rapid loss of weight and massive infiltrates of T cells in multiple organs (lung, liver, small intestine) in contrast to control groups transferred with WT Treg cells or only CD4+ Tconv cells (Fig. 7c and Supplementary Fig. 7c). Consequently, the absolute numbers of CD4+ Tconv cells were two (lung) to five (spleen) fold increased in autoimmune mice with a higher proportion secreting IL-2 (factor of 2), IL-17A, and IFNγ (Fig. 7d). Fate mapping of Il2ramut/mut or WT Treg cells transferred to Rag1−/− (Fig. 7e) or Foxp3y/− (Supplementary Fig. 7d) mice further showed that Foxp3+ Il2ramut/mut Treg cells become ex-Treg cells and are lost compared to WT counterparts, both in proportion and absolute numbers. Thus, a threshold of IL-2 signals to Treg cells is needed to program their functional attributes, for their maintenance, and to prevent wasting autoimmunity. Consistent with this conclusion and the lack of competitive fitness21 of Il2ramut/mut Treg cells, a significantly lower proportion of Il2ramut/mut compared to WT Treg cells was measured in WT B6 chimeric mice reconstituted with an equal ratio of Il2ramut/mut and WT bone-marrow donor cells while the proportion of CD4+ Tconv cells was maintained (Fig. 7f).
Fig. 7.
Treg cells receiving limited IL-2 signals cannot suppress autoreactive WT Tconv cells: a Foxp3y/− Cd45.1+/+ neonate mice were transferred i.p. with 106 Il2ramut/mut or WT flow-purified Foxp3Rfp/Rfp CD3+CD4+ Treg cells and survival was monitored over 120 days. Graphs average results over multiple mice in each group and 7-16 replicate experiments. b, c Rag1−/− mice received 5 × 105 flow-purified Foxp3Rfp/Rfp CD3+CD4+ Treg cells together with 1.5 × 106 WT CD3+CD4+CD25neg Tconv cells i.v. Weight loss was monitored up to 61 days post transfer and immune cell infiltrates on H/E stained sections is shown at day 61 (n = 4). d Numbers of transferred Foxp3Rfp/Rfp CD3+CD4+ Treg cells in the spleen and lung of Rag1−/− mice (left bar graphs) and proportion of cytokine-secreting CD3+CD4+ Tconv cells 4 h post PMA/ionomycin stimulation. e Foxp3 expression in Foxp3Rfp/Rfp CD3+CD4+ Treg cells 45 and 75 days post transfer, proportion and absolute numbers of ex-Treg cells in LNs and spleens. f, g Schematic of mixed bone-marrow chimera generation in lethally irradiated WT B6 recipient mice with discriminative congenic markers. f, g Representative FACS dot plots of Foxp3 and CD4 expression in blood T cells of f WT/Il2ramut/mut and g Foxp3y/−/WT and Foxp3y/−/Il2ramut/mut chimeras 10 and 6 weeks post reconstitution, respectively. Graph in f shows the ratio of WT vs Il2ramut/mut Tconv and Treg cells for each individual chimera. Data are across several chimeras (5–9 mice/group) in 3–4 replicate experiments. h, i Spleen, LN pictures, and H/E stained sections of lung, liver, pancreas, and small intestine from Foxp3y/−/WT and Foxp3y/−/Il2ramut/mut chimeras. j Cells from indicated organs from the two groups of chimeras were stained for cell-surface expression of CD3, CD4, CD8, CD62L, KLRG1, and CD44. Bar graphs show the proportion of activated CD4+ (CD44+CD62Llo) or CD8+ (KLRG1+CD62Llo) T cells (n = 5–9). k Proportion of activated CD3+CD4+ or CD8+ Tconv cells from Il2ramut/mut or WT BM in Foxp3y/−/Il2ramut/mut chimeras. Bar graphs summarize average proportion across mice (n = 5–9). p-values are indicated when relevant with *p < 0.05; **p < 0.01; ***p < 0.001; NS not significant, using two-tailed unpaired Student’s t-test. Symbols on bar graphs feature individual mice
As a last step to assess Il2ramut/mut Treg cells functional attributes, we created another chimeric mouse model at homeostasis in which Treg cells lacked full IL-2 responsiveness while Tconv cells responded normally. We reconstituted lethally irradiated WT mice with bone marrow cells from Foxp3y/− and Il2ramut/mut or control WT mice (Fig. 7g). In such Foxp3y/−/Il2ramut/mut chimeras, only Treg cells lacked CD25 expression and mice exhibited enlarged spleens and LNs compared to the Foxp3y/−/WT control chimeras (Fig. 7h). Moreover, massive infiltrates of T cells were noted in lung, liver, pancreas and small intestine of Foxp3y/−/Il2ramut/mut mice, with gross architectural tissue alterations compared to the control group (Fig. 7i). In line with these findings, both CD4+ and CD8+ T cells in Foxp3y/−/Il2ramut/mut but not Foxp3y/−/WT chimeras underwent robust activation (CD62Llow, KLRG1+, CD44hi, CD69hi) across multiple tissues (Fig. 7j and Supplementary Fig. 7e, f). Only WT but not Il2ramut/mut Tconv cells in Foxp3y/−/Il2ramut/mut chimeras were activated (Fig. 7k and Supplementary Fig. 7g), suggesting that equal responsiveness to IL-2 by Tconv cells and Treg cells represent a major parameter safeguarding immune homeostasis. In summary, Treg cells that can only integrate lower IL-2 signals than their Tconv cell counterpart, acquire a suboptimal epigenetic program, are less stable (Foxp3) and are impaired in their ability to deprive autoreactive Tconv cells of IL-2.
Discussion
While the Foxp3 TF is required to maintain peripheral Treg cell identity and functions, it is not sufficient to secure their stable epigenetic identity and program their functional features12,16,24. But how exactly are Treg cell chromatin accessibility characteristics determined before Foxp3 is expressed is still hotly debated. TCR signaling represents the major cue driving thymic selection, and this plays a determining role in thymocytes commitment to the Treg cell lineage47, however this is likely not sufficient12,14,25. Signals such as IL-2 and TGFβ cytokines, are suggested to contribute to the epigenetic modeling of Treg cells2,3, yet current evidence for IL-2 mostly pointed to its role as a major stabilizer of Foxp3 expression. Here, our data support a role for IL-2 in defining the epigenetic identity of Foxp3+ Treg cells, which appears largely independent of Foxp3. In fact, genome-wide chromatin accessibility changes when IL-2 signals are impaired are much wider than the regions where Foxp3 binds. Our whole genome analysis of OCRs in Il2ramut/mut vs WT Treg cells from the LNs reveals that the epigenetic landscapes are widely distinct (~41%). Consistent with this result, constitutive STAT5 expression, which forces IL-2 signaling, is shown to divert thymocyte selection to the Treg cell lineage11. Using data from a prior report on Treg cell epigenetic signature under various conditions of activation36, we analyzed that ~36% of OCRs differed in activated vs resting Foxp3+ Treg cells, a proportion comparable to the extent of IL-2-mediated remodeling observed in WT vs IL2ramut/mut LN Treg cells. These differences are equivalent to what is documented between effector and memory or exhausted CD8+ T cells38, which are undergoing extensive chromatin remodeling upon differentiation37–39, overall supporting the conclusion that IL-2 signals significantly shape the epigenetic landscape of Treg cells in vivo.
Because IL-2 signals mediate genome-wide chromatin remodeling beyond Foxp3, and already on mature Foxp3+ Treg cells in the thymus, we postulated that this may occur in the thymus at the SP CD4+ thymocyte stage preceding Foxp3 expression in committed Treg cells. The genome organizer SATB1, which is highly expressed during most stages of T cell thymic selection, was recently involved in setting-up thymic-derived Treg cell functional features and ability to prevent autoimmunity before Foxp3 is turned on27. We further reveal that SATB1 positioning in the genome of SP CD4+ thymocytes impaired in IL-2 signaling, is much broader than in WT thymocytes, likely reflecting ectopic binding of this pioneer factor when IL-2 signals are limiting. Our finding that more than 20 times as many GO pathways are associated with the genes exhibiting unique OCRs in WT LN Treg cells compared to IL2ramut/mut counterparts supports the idea that such ectopic binding of SATB1 lead to unfocused Treg cell chromatin modeling. In WT thymocytes, SATB1 is more selectively bound to the TSS of genes encoding for TFs and nucleic acid binding proteins, which may in turn control the correct Treg cell epigenetic landscape. Several TFs such as STAT5 and PI3/MAP kinase-dependent TFs, are activated downstream of IL-2 signaling. From our analysis of the SATB1 DNA binding regions and TFs known to bind to these motifs, we noted GATA3, RUNX1 and ELF1 that are all reported to be present prior to Foxp3 expression to enable its future recruitment and optimal function15,17,26,43. SATB1 is also described among the “quintet” TFs—together with Foxp3—that are essential for in vitro re-programing of Tconv cells to Treg cells16. While still speculative, it is possible that IL-2 signals, in addition to TCR triggering, are essential to promote the expression/activation of the previous TFs and of SATB1 and/or other pioneer factors, to set-up the appropriate epigenetic landscape on which Foxp3 and these partner TFs act to achieve the final Treg cell identity. Binding to the Foxp3 CNS3 and CNS2 elements for instance is essential to initiate and then stabilize expression of Foxp317,18,26,48. The NF-kB subunit c-Rel binds CNS3, and STAT5 and GATA349 stabilize CNS2. While we did not find evidence for SATB1-binding on c-Rel or STAT5 motifs, SATB1 may associate with GATA3 to help focus its binding to DNA area relevant to the acquisition of the Treg cell signature prior Foxp3 is expressed. Such mechanism would prevent ectopic binding of SATB1 throughout the genome when IL-2 signals are limiting.
The consequence of lower IL-2 signaling in thymic-derived peripheral Treg cells is a different epigenetic landscape, which we suggest is being set in the thymus through SATB1 together with other IL-2-dependent TFs. In the periphery, however, SATB1 is not required to sustain functional thymic-derived Treg cells27. Some TF binding motifs found in SATB1-binding regions like RUNX1, which is likely to interact with SATB1 in SP CD4+ thymocytes, are also among the motifs found in the differential OCRs of WT compared to Il2ramut/mut Treg cells in the periphery. RUNX1 binds to the IL-2 promoter and promotes its transcription, but is repressed by Foxp3 through physical interactions42, preventing the expression of an effector T cell program by Treg cells. Other TF binding motifs only found in WT peripheral Treg cells, such as FOXO1 and FOXO3, also bind to the Foxp3 promoter and conserved intronic enhancer regions, allowing for Foxp3 expression and Treg cell-lineage specification50. IL-2-mediated induction of all these TFs at the pre-Treg cell stage, via SATB1 or not, may help maintain the Treg cell program in the periphery. Along these lines, we noted multiple differentially expressed TF-encoding genes (Prdm1, Gata3, Dzip1, Irf4), chromatin remodeling- and histone-encoding genes between WT and Il2ramut/mut LN Treg cells.
Given that our data support a model in which IL-2 alters Treg cell epigenetic identity early during thymic selection when thymocytes commit to this lineage, it is unlikely that this mechanism will be at work for induced Treg cells in peripheral tissues51. Our analysis in WT vs Il2ramut/mut Treg cells failed to reveal any differences in OCRs of genes encoding Foxp3 or HELIOS that were reported to exhibit differential methylation profiles in induced/peripheral Treg cells52.
We report a role for IL-2 on Foxp3+ Treg cells that is largely independent of Foxp3-mediated stabilization of Treg cells and of their ability to capture IL-2 from activated Tconv cells. The Y129H mutation in CD25 prevents optimal folding and/or egress of CD25 to the surface of T cells, translating into decreased binding of IL-2 and subsequent signaling and proliferative response. Two closely comparable models were previously reported: in the first, mutations inside several of key signal transducing cytoplasmic tyrosines of the IL-2Rβ chain disrupted both IL-2 or IL-15 signaling29. The second model overexpressed a constitutively active form of the b subunit of the STAT5 TF51 on either IL-2Rβ (Cd122−/−)19 or IL-2Rγ (γc−/−) knockout backgrounds11. None of these models could differentiate between IL-2 and IL-15, or other γ-chain-dependent cytokine signaling, confounding interpretation on the roles of high affinity IL-2 signaling. Nevertheless, a rather intriguing finding in both of these settings, as in our model, is that receptor deficient mice did not develop autoimmunity, prompting the idea that Treg cells were set to a low IL-2 receptor signaling threshold sufficient for Foxp3 induction and maintenance. Our current results, however, favor a different interpretation in which, when comparable IL-2 signals are received by Treg and Tconv cells, the development of autoimmunity can be prevented. Yet, if Treg cells cannot receive as much IL-2 signals as Tconv cells, mice will develop fatal wasting disease. A third model recently developed by Rudensky and colleagues, enabled to discriminate the contribution of IL-2 capture and Treg cell stability in preventing fatal autoimmunity40. As Treg cells cannot secrete IL-26 but constitutively express the high affinity IL-2Rα/CD25 chain, a large conundrum in the field is their reliance on cell-extrinsic IL-2 from activated -potentially autoreactive- Tconv cells, thus preventing excessive Tconv cell proliferation and activation by physically capturing the IL-2 they secrete. This study brought strong evidence that IL-2 capture does represent a key mechanism of suppression used by Treg cells. While the Il2ramut/mut mouse does not allow to assess the role of IL-2 capture by Treg cells in maintaining immune homeostasis, the progression of disease and survival in Foxp3y/− mice rescued with Il2ramut/mut Treg cells, the Rag−/− transfer experiments and the Foxp3y/−/Il2ramut/mut chimeras, are close to that observed in Foxp3y/− mice. Length of survival is, however, distinct to that of the Rudensky model, in which mouse survival is approximately twice increased. In this latter model too, CD8+ but not CD4+ T cells, exhibit robust activation whereas in our Foxp3y/−/Il2ramut/mut chimeras, both subsets of Tconv cells are highly activated. Altogether, these observations support the idea that altering Treg cell epigenetic identity, IL-2 capture and signaling leads to more rapid autoimmunity.
IL-2 therapy has been used for over two decades in patients, initially with the goal to boost anti-tumoral and HIV-specific immunity52,53. High doses of IL-2 were mostly used with important side effects and mitigated efficiencies. Yet the use of lower doses of IL-2, with the underlying rationale that Treg cells would be more efficiently targeted than effector T cells, showed a preferential expansion of Treg cells both in preclinical models and in type 1 diabetes patients, which was associated with better prognostic markers54,55. While the mechanism underlying these promising outcomes in patients may be accounted for by Treg cell expansion, our results further raise the possibility that qualitative changes at the levels of chromatin accessibility and epigenetic reprogramming of Treg cells during thymic selection, may contribute to ameliorate Treg cell functions in autoimmune patients.
Methods
Ethics statement
This study was carried out in strict accordance with the recommendations by the animal use committees at the Albert Einstein College of Medicine. All efforts were made to minimize suffering and provide humane treatment to the animals included in the study.
Mice
All mice were bred in our SPF animal facility at the Albert Einstein College of Medicine. We used wild-type (WT) C57BL/6J (B6) 6-8 weeks old mice, congenic CD45.1+/+ (#2014), Ccr2−/−2009 (#4999), Ccr2−/−2012 (#4999), Foxp3+/− (#19933), Foxp3Rfp/Rfp (#8374), Rag1−/− (#2216) from the Jackson labs all on the B6 genetic background. Ccr2+/+2009 Il2ramut/mut (called Il2ramut/mut) were generated by intercrossing to the B6. Il15−/− (#4269) mice were purchased from Taconic.
Microbial pathogens and mice infections
We used wild type Listeria monocytogenes expressing the Ovalbumin (Ova) model antigen (Lm-Ova) on the 10403s genetic background56. For infections, bacteria were grown to a logarithmic phase in broth heart infusion medium, diluted in PBS to infecting concentration (104) and injected i.v. For Herpes Simplex Virus 2 (HSV-2) infection, female mice treated with 2 mg medroxyprogesterone acetate subcutaneously (s.c.) were immunized or not 5 days later intravaginally with 2 × 105 plaque forming units (PFU) of 186ΔKpn HSV-2 (TK− HSV-2) and organs (spleen and draining LNs) were harvested 7 days later.
Cell suspensions for flow cytometry and adoptive transfers
Spleens, lymph nodes (inguinal and cervical), or thymuses were dissociated on nylon mesh while lungs, liver, and pancreas were cut. All organs were incubated in HBSS medium with 4000 U/mL collagenase I and 0.1 mg/mL DNase I, and red blood cells (RBC) lysed with NH4Cl buffer (0.83% vol/vol). Blood was harvested into heparin tubes and RBC lysed as before. Bone marrow cells were obtained from flushing femur with complete RPMI with 10% FCS.
Cell-staining for FACS analysis
Cell suspensions were incubated with 2.4G2 Fc Block and stained with fluorescently tagged antibodies (Abs) purchased from eBioscience, BD Biosciences, Tonbo Bioscience, or BioLegend (Supplementary Table1) in PBS 1% FCS, 2 mM EDTA, 0.02% sodium azide. Biotinylated monomers (1 mg/mL) obtained from the NIH tetramer Core Facility, were conjugated with PE-labeled Streptavidin (1 mg/mL) as follow: 6.4 μL of PE-Streptavidin were added to 10 μL of monomers every 15 min four times on ice. Newly generated tetramers (1/400-1/500 dilution) were then used to stain spleen/LN cells for 1 h at 4 °C. For transcription factor (TF) intracellular staining (Foxp3, Blimp-1), cells were fixed in eBioscience Fixation/Permeabilization buffer prior to TF Ab staining in eBioscience Permeabilization buffer for 30 min. For intracellular cytokine staining (ICS), cells were incubated 4 h at 37 °C/5%CO2 in RPMI1640 10%FCS, Golgi Plug/Golgi Stop (BD), fixed in IC fixation buffer (eBioscience), and permeabilized prior to 30 min staining with Abs against indicated intracellular markers. For intracellular phosphorylated STAT5 staining, cells were starved in RPMI w/o FCS for 30 min before stimulation with variable concentration of recombinant human IL-2 (Gemini) for 20 min. Cells were then fixed and permeabilized with 4% PFA followed by 90% methanol, and stained with anti-pY-STAT5 antibody (BD Biosciences). Data acquisition was done using a BD LSR II or a FACS Aria III flow cytometer. All flow cytometry data were analyzed using FlowJo v9 software (TreeStar). Cell sorting of Foxp3+ and Foxp3− cells was performed based on RFP expression in Foxp3Rfp/Rfp reporter mice using a BD FACS Aria III cell sorter. The FACS gating strategies for the experiments shown and T cell sorting are provided in Supplementary Fig. 8.
Transfection of wild-type and Y129H Il2ra in 293T cells
The Il2ra cDNA was amplified by PCR and cloned into pMSCV-IRES-GFP (pMIG, kind gift from Guy Sauvageau) to generate pMIG-Il2ra. PCR was performed on pMIG-Il2ra to introduce a point mutation on nucleotide 426 (thymidine to a cytosine), which results in a tyrosine-histidine conversion at amino-acid position 129. The Il2ramut cDNA was then cloned into pMIG to generate pMIG-Il2ramut. The presence of the introduced point mutation was confirmed by sequencing.
293T cells were plated 2 days before transfection on 100 mm adherent petri dish at 106 cells per plates. Cells at 70% confluence were transfected with 30 µg of the plasmid pMIG, pMIG-Il2ra or pMIG-Il2ramut using 45 µl Lipofectamine 2000 in 4.5 mL OptiMEM (ThermoFisher) overnight in antibiotic-free medium. Medium was replaced the day after transfection and 3 days later, cells were trypsinized and analyzed for CD25 extra- and intracellular expression by flow cytometry as described before. The 293T cells were a kind gift from Heather Melichar (University of Montreal, Canada) and were tested mycoplasma negative.
In vitro Tconv and Treg cell assays
For all assays, naive CD4+ or CD8+ Tconv cells were negatively selected from spleen and LNs using either anti-CD8β (H35) or anti-CD4 (GK1.5), anti-CD11b (M1/70), anti-MHC-II (M5/114), anti-TER119, anti-B220 (RA3-6B2) and anti-CD25 (PC61), all at 5 μg/mL for 30 min at 4 °C. Cells were then washed and incubated with anti-rat magnetic beads at 1 bead/target cell for 30 min at 4 °C (Dynabeads sheep anti-rat IgG, Invitrogen). For suppression assays, purified naive WT Cd45.1+/+ CD4+ Tconv cells were stained with 1–5 μM of CellTrace Violet (CTV, Invitrogen) according to the manufacturer’s protocol. WT and Il2ramut/mut Foxp3Rfp/Rfp Treg cells were sorted by flow cytometry (Aria III). 5 × 104 CTV-labeled naive CD4+ Tconv cells were cultured with increasing numbers of Treg cells in the presence of 105 irradiated, T cell-depleted WT B6 splenocytes and 1 μg/mL anti-CD3ε (clone 145-2C11, BD) in a 96 round-bottom plate for 72 h. Cell proliferation of Tconv cells (live CFSE−CD4+Foxp3−) was determined by flow cytometry based on CTV fluorescence intensity dilution of Tconv cells. For proliferation assays, CTV-labeled purified WT and Il2ramut/mut naive CD4+ or CD8+ Tconv cells were incubated for 24 h on anti-CD3ε (10 μg/mL) pre-coated wells before co-culture for 96 h with varied concentrations (2.5-104 U/mL) of human recombinant IL-2 (Gemini Bio-product). Cell proliferation was determined by flow cytometry and CTV fluorescence dilution of Tconv cells. For short-term activation assays for immunofluorescence, purified WT and Il2ramut/mut naive CD4+ T cells were incubated for 48 h with anti-CD3ε (10 μg/mL) pre-coated wells to induce CD25 upregulation. At 48 h, cells were transferred in polylysine pre-coated chambers, left to adhere for 1 h at 37 °C, fixed with 1% PFA and permeabilized with 0.1% Triton X-100 prior to OVN staining in PBS 0.5% BSA, 0.05% Triton X-100 containing polyclonal goat anti-mouse CD25 (RD systems) and rabbit anti-calreticulin Abs (1/300 dilution, ThermoFisher, PA3-900). Staining was revealed by staining for 2 h with secondary anti-goat-Alexa 488 and anti-rabbit-Alexa 546 Abs (Invitrogen). Cells were then covered with Fluoromount-G (SouthernBiotech) and imaged using a Zeiss Axiovert microscope (Carl Zeiss Microimaging Inc., Thornwood, NY) with a ×63 NA 1.4 objective and a Retiga 2000 camera. Green channel images using a 450–490 excitation/500–550 emission bandpass filter and red staining was imaged with a 565/30 excitation-620/60 emission bandpass filter. Images were processed using Adobe Photoshop CS 4 (Adobe Systems, Inc. San Jose, CA).
In vivo Treg cell functional assays
For mixed bone-marrow chimera mice, Rag1−/−Cd45.2+/+ mice were lethally irradiated with 1200 rads before immediate reconstitution with 5 × 106 T cell-depleted (as for the in vitro assays) bone marrow from Foxp3−/yCd45.1+/+ and Il2ramut/mutCd45.1+/− or congenic Cd45.1+/+ and Il2ramut/mutCd45.1+/− at a 1:1 ratio. Mice were placed under antibiotics for 2 weeks and reconstitution ratios were checked by FACS 4–6 weeks later. In some experiments, C57BL/6 Cd45.2+/+ were used as recipient. In such case, mice received 150 μg of anti-CD8β and anti-CD4 i.v. with for two consecutive days to deplete T cells prior irradiation.
For Rag1−/− T cell transfers, LN- and spleen-purified naive CD4+ Tconv cells (by negative selection as above) isolated from Cd45.1+/+ male mice were transferred to Rag1−/− recipients alone or mixed at a 5:1 ratio with flow-sorted sorted (Aria III) RFP+ Treg cells from spleen and LNs of WT or Il2ramut/mut Foxp3Rfp/Rfp reporter mice (2.5 × 106 cells per recipient). In some experiments (when indicated), naive CD4+ T cells were labeled with 10 nM of CTV prior adoptive transfer. Recipient mice were monitored for body weight changes and transferred lymphocyte subsets were monitored by flow cytometry 45 and 75 days later.
For Foxp3−/y mice rescue experiments, 106 flow-sorted Treg cells from WT or Il2ramut/mut Foxp3Rfp/Rfp reporter mice were injected i.p. into 1–2-day-old pups (Foxp3y/− males, genotyped after birth) from females Foxp3+/− mice bred with Cd45.1+/+ congenic males. Male pups were assessed for the development of lymphoproliferative syndrome in blood and LNs at ~30 days of age.
For the antibody neutralization treatment, mice were given 150 µg of anti-ICOSL (clone HK5.3, BioXcell) by i.v. injection on days 0, 3, 6, 9, and 12 and sacrificed on day 14.
Histological analysis
Tissue samples were fixed in 10% neutral buffered formalin and processed for hematoxylin and eosin staining. For each organ collected and for each genotype, two sections were cuts at 100 μm apart and all slides were scanned using a P250 High Capacity Slide Scanner (Perkin Elmer).
Microarrays
Overall, 50,000 RFP+ Treg or naive (CD62LhiCD44lo) RFPnegCD25negCD4+ Tconv cells from LNs of WT or Il2ramut/mut Foxp3Rfp/Rfp mice were flow-sorted based on the RFP signal and indicated markers after enrichment for CD4+ T cells (using negative selection, as described above). Pelleted cells were stored in 700 μL of TRIzol (Life Technologies) at −80 °C until RNA extraction. Total RNA was extracted using the RNAeasy Micro kit with RNase-Free DNase Set (Qiagen) according to the manufacturer protocol. The quality score and quantity of purified RNA was assessed with a Bioanalyzer RNA 6000 Pico Chip (Agilent). Total RNA was then converted to cDNA, amplified and hybridized to Affymetrix Mouse Transcriptome Array 1.0 Pico. Raw CEL files were preprocessed and normalized using Affymetrix Expression Console (version 1.4.1.46) and resulting data were analyzed with the Affymetrix Transcriptome Analysis Console (version 3.1.0.5). We calculated fold-differences between experimental groups and tested significance using one-way ANOVA (unpaired). Significantly up and downregulated genes were defined with at least a 1.5-fold expression difference and a p-value ≤ 0.01. Over-representation of biological-process (BP) gene-ontology (GO) terms was calculated using BiNGO (version 3.0.3) in Cytoscape (version v3.4.0), employing the hypergeometric test and applying a significance cutoff of FDR-adjusted p-value ≤ 0.05. The GO ontology and annotation files used were downloaded on May. 25, 2017. The output of BiNGO was imported into EnrichmentMap (version 2.1.0) in Cytoscape to cluster redundant GO terms and visualize the results. An EnrichmentMap was generated using a Jaccard similarity coefficient cutoff of 0.4, a p-value cutoff of 0.001, an FDR-adjusted cutoff of 0.005, and excluding gene sets with fewer than 5 genes. The network was visualized using a perfuse force-directed layout with default settings and 500 iterations. Groups of similar GO terms were manually circled. Finally, for analysis of gene expression in thymocyte fractions shown in Supplementary Fig. 5c, raw data from the NCBI database (GEO GSE15907) were analyzed41.
Epigenetic profiling
For ATAC-seq experiments, we performed the analysis on two (LNs) or four (thymus) biological replicates per group as previously described35. Briefly, nuclei were isolated from 50,000 CD4+ T cell-enriched (by negative selection) flow-sorted RFP+ Treg cells from WT or Il2ramut/mut Foxp3Rfp/Rfp mice (LNs or thymus) using a solution of 10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630. Immediately following nuclei isolation, the transposition reaction was conducted using Tn5 transposase and TD buffer (Illumina) for 45 min at 37 °C. Transposed DNA fragments were purified using Qiagen Mini-Elute Kit and PCR amplified using NEB Next High Fidelity 2× PCR master mix (New England Labs) with dual indexes primers (Illumina Nextera).
For SATB1 ChIP-seq experiments, we performed the ChIP-seq following the Mayers Lab ChIP-seq Protocol v011014 which is one of the suggested protocols by ENCODE project with slight modifications (https://www.encodeproject.org/documents/6ecd8240-a351-479b-9de6-f09ca3702ac3/@@download/attachment/ChIP-seq_Protocol_v011014.pdf). Single positive (SP) CD4+ thymocytes from thymus of WT and Il2ramut/mut mice on two biological replicates were pre-enriched by negative selection using anti-CD8β (H35), anti-MHC-II (M5/114), anti-CD11b (M1/70) and anti-TER119. After staining the remaining cells with fluorescent labeled antibodies against CD4 (GK1.5) and CD8α (53-6.7), we flow-sorted 2 × 106 CD4+ CD8αneg SP thymocytes. Sorted cells were next cross-linked in 1% (wt/vol) formaldehyde solution for 30 min. Cross-linked DNA was lysed in Farnham lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl and 0.5% NP-40), fragmented in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS in 1×PBS) using Bioruptor (Diagenode), and incubated overnight at 4 °C with 100 µL of DynaBeads M-280 Sheep anti-Rabbit IgG magnet beads (Invitrogen) preincubated with 20 µg of anti-SATB1 antibody (Abcam, ab70004). Beads were washed five times with LiCl wash buffer (100 mM Tris pH 7.5, 500 mM LiCl, 1% NP-40 and 1% sodium deoxycholate) and one time with 10 mM Tris/0.1 M EDTA. SATB1-bound DNA was then eluted from the beads and reverse cross-linked by incubating the beads pellet in 200 µl of IP elution buffer (1% SDS and 0.1 M NaHCO3) at 65 °C overnight and further purified with MinElute PCR Purification Kit (QIAGEN). The library was prepared using Accel-NGS 2 S Plus DNA library kit according to the manufacturer’s instructions.
Sequencing libraries processing
The size distribution and molarity of the sequencing library were determined by using an Bioanalyzer analysis (High Sensitivity DNA chip, Agilent). Sequencing was performed using a HiSeq 2500 system (Illumina). Obtained sequences were mapped to the mouse mm10 reference genome using BWA mem57,58. After eliminating duplicated reads and the reads aligned to mitochondrial DNA, we kept only concordantly mapped pairs for further analysis. Peak calling was performed on shifted reads using MACS v2.1 with narrow peaks option to identify areas of sequence tag enrichment following the original report35. For ATAC-seq, read1 reads were shifted using bedtools2, as previously performed in Buenestro et al.35, before calling peaks for each replicate. We performed Irreproducible Discovery Rate analysis for finding reproducible peaks among the biological replicates (0.05 cutoff)59. Analysis was performed in R (the R project) using Bioconductor packages. The genomic locations, overlapping or nearest genes and finding overlapping peaks between groups were annotated with ChIPpeakAnno60 and ChIPseeker61 using TxDb.Mmusculus.UCSC.mm10.knownGene annotation (Bioconductor). The motif analysis was performed on the genomic sequences of peaks using MEME-ChIP 63 and HOMER62. Over-representation of biological-process BP-GO terms was calculated using BiNGO as described above. For the ATAC-seq data analysis, the output of BiNGO was further filtered by selecting GO with an FDR-adjusted cutoff < 0.001 and excluding gene sets with fewer than five genes. The list of BP-GO was then imported into REViGO using a similarity coefficient of 0.7 and the SimRel columns to generate semantic similarities scores. The scored terms were visualized in semantic similarity-based scatterplots. For the SATB1 ChIP-seq data analysis, the output of BiNGO was imported into EnrichmentMap (version 2.1.0) in Cytoscape to cluster redundant GO terms and visualize the results as described above.
Statistics
Statistical significance was calculated using an unpaired Student t test with GraphPad Prism software and two-tailed P values are given as: (*) p < 0.1; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001 and (ns) p > 0.1. All p values of 0.05 or less were considered significant and are referred to as such in the text. Error bars are mean ± SEM in all figures.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
H2-Kb/OVA257-264 and H2-Kb/gB498-505 tetramers were obtained from the NIH Tetramer Facility. We are grateful to M. Levy (Einstein) for privileged access to the FACS Aria III and thank the AECOM FACS and genomic core facilities. We thank the Lauvau laboratory members and S. Soudja for critical comments, and JF Daudelin for characterizing the transient CD25 transfectants. This work was funded by the National Institute of Health Grants (NIH) AI103338, Hirschl Caulier Award to G.L.; the Canadian Institutes of Health Research Grants PJT-149023 and PJT-152988 to N.L. L.C. received fellowships from ARC, Fondation Bettencourt-Schuller, and the American Association of Immunology (AAI). T.M.W. and S.S.C. were supported by NIH training Grant T32A170117. L.O. received a studentship from the Fonds de recherche du Québec-Santé. Core resources for FACS were supported by the Einstein Cancer Center (NCI cancer center support grant 2P30CA013330).
Author contributions
L.C. designed, performed, and interpreted most experiments, designed and assembled figures, and contributed to editing of the manuscript. M.S. conducted and guided ChIP-seq experiments with L.C., performed analysis of all ATAC-seq, ChIP-seq, and microarray data, contributed to data production and interpretation for the corresponding figures and manuscript editing. S.S.C. contributed to sequencing sample processing, manuscript, and figure editing and reorganization. T.M.W. and E.L.S. conducted the fluorescent microscopy experiments and image acquisition. L.O. and N.L. did the Y129H mutagenesis of WT Il2ra and transient transfections and analysis. N.L. contributed to manuscript editing and discussions. G.L. designed and interpreted all experiments and data with L.C. and all authors, contributed to figure design and editing, and wrote the paper.
Data availability
The accession number for microarrays, ATAC-seq, and ChIP-seq data reported in this paper underlying Figs. 3–5 and Supplementary Figs 3–5 is GEO: GSE103217. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files. A reporting summary Article is available as a Supplementary Information file. The source data underlying Figs. 1a, c, d, g, h, 2a–e, 6a, c–f and 7a, b, d, e, j, k and Supplementary Figs 1b, d, 2b, d, 4c, 6b-d, f and 7a, b, f, g are provided as a Source Data file.
Competing interests
The authors declare no competing interests.
Footnotes
Journal peer review information: Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. [Peer reviewer reports are available.]
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-07806-6.
References
- 1.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.van der Veeken J, Arvey A, Rudensky A. Transcriptional control of regulatory T-cell differentiation. Cold Spring Harb. Symp. Quant. Biol. 2013;78:215–222. doi: 10.1101/sqb.2013.78.020289. [DOI] [PubMed] [Google Scholar]
- 3.Benoist C, Mathis D. Treg cells, life history, and diversity. Cold Spring Harb. Perspect. Biol. 2012;4:a007021. doi: 10.1101/cshperspect.a007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+ CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 6.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 7.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 8.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 9.Sekiya T, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 10.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burchill MA, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 15.Rudra D, et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu W, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat. Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 20.Malek TR. The biology of interleukin-2. Annu. Rev. Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 21.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 22.D’Cruz LM, Klein L. Development and function of agonist-induced CD25+ Foxp3+ regulatory T cells in the absence of interleukin 2 signaling. Nat. Immunol. 2005;6:1152–1159. doi: 10.1038/ni1264. [DOI] [PubMed] [Google Scholar]
- 23.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 26.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017;18:173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Placek K, et al. MLL4 prepares the enhancer landscape for Foxp3 induction via chromatin looping. Nat. Immunol. 2017;18:1035–1045. doi: 10.1038/ni.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu A, Zhu L, Altman NH, Malek TR. A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells. Immunity. 2009;30:204–217. doi: 10.1016/j.immuni.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boring L, Gosling J, Cleary M, Charo IF. Decreased lesion formation in CCR2-/- mice reveals a role for chemokines in the initiation of atherosclerosis. Nature. 1998;394:894–897. doi: 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- 31.Bruhl H, et al. Dual role of CCR2 during initiation and progression of collagen-induced arthritis: evidence for regulatory activity of CCR2+ T cells. J. Immunol. 2004;172:890–898. doi: 10.4049/jimmunol.172.2.890. [DOI] [PubMed] [Google Scholar]
- 32.Zhang N, et al. Regulatory T cells sequentially migrate from inflamed tissues to draining lymph nodes to suppress the alloimmune response. Immunity. 2009;30:458–469. doi: 10.1016/j.immuni.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obar JJ, et al. CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8 + T cells in primary and secondary responses. Proc. Natl Acad. Sci. USA. 2010;107:193–198. doi: 10.1073/pnas.0909945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng G, Yu A, Dee MJ, Malek TR. IL-2R signaling is essential for functional maturation of regulatory T cells during thymic development. J. Immunol. 2013;190:1567–1575. doi: 10.4049/jimmunol.1201218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Veeken J, et al. Memory of inflammation in regulatory T cells. Cell. 2016;166:977–990. doi: 10.1016/j.cell.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott-Browne JP, et al. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity. 2016;45:1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chinen T, et al. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 2016;17:1322–1333. doi: 10.1038/ni.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mingueneau M, et al. The transcriptional landscape of alphabeta T cell differentiation. Nat. Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 43.Rudra D, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 45.Herman AE, Freeman GJ, Mathis D, Benoist C. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 2004;199:1479–1489. doi: 10.1084/jem.20040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smigiel KS, et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity. 2012;37:475–486. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuster M, et al. IkappaB(NS) protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity. 2012;37:998–1008. doi: 10.1016/j.immuni.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 49.Wohlfert EA, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouyang W, et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 51.Burchill MA, et al. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J. Immunol. 2003;171:5853–5864. doi: 10.4049/jimmunol.171.11.5853. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg SA, et al. Observations on the systemic administration of autologous lymphokine-activated killer cells and recombinant interleukin-2 to patients with metastatic cancer. N. Engl. J. Med. 1985;313:1485–1492. doi: 10.1056/NEJM198512053132327. [DOI] [PubMed] [Google Scholar]
- 53.Group IES, et al. Interleukin-2 therapy in patients with HIV infection. N. Engl. J. Med. 2009;361:1548–1559. doi: 10.1056/NEJMoa0903175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dwyer CJ, Ward NC, Pugliese A, Malek TR. Promoting immune regulation in type 1 Diabetes using low-dose interleukin-2. Curr. Diab. Rep. 2016;16:46. doi: 10.1007/s11892-016-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenzwajg M, et al. Low-dose interleukin-2 fosters a dose-dependent regulatory T cell tuned milieu in T1D patients. J. Autoimmun. 2015;58:48–58. doi: 10.1016/j.jaut.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pope C, et al. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J. Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 57.Heng, L. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM (Oxford University Press, Oxford, 2013).
- 58.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 62.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The accession number for microarrays, ATAC-seq, and ChIP-seq data reported in this paper underlying Figs. 3–5 and Supplementary Figs 3–5 is GEO: GSE103217. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files. A reporting summary Article is available as a Supplementary Information file. The source data underlying Figs. 1a, c, d, g, h, 2a–e, 6a, c–f and 7a, b, d, e, j, k and Supplementary Figs 1b, d, 2b, d, 4c, 6b-d, f and 7a, b, f, g are provided as a Source Data file.