Abstract
For long-duration space missions, it is critical to maintain health-associated homeostasis between astronauts and their microbiome. To achieve this goal it is important to more fully understand the host–symbiont relationship under the physiological stress conditions of spaceflight. To address this issue we examined the impact of a spaceflight analog, low-shear-modeled microgravity (LSMMG), on the transcriptome of the mutualistic bacterium Vibrio fischeri. Cultures of V. fischeri and a mutant defective in the global regulator Hfq (∆hfq) were exposed to either LSMMG or gravity conditions for 12 h (exponential growth) and 24 h (stationary phase growth). Comparative transcriptomic analysis revealed few to no significant differentially expressed genes between gravity and the LSMMG conditions in the wild type or mutant V. fischeri at exponential or stationary phase. There was, however, a pronounced change in transcriptomic profiles during the transition between exponential and stationary phase growth in both V. fischeri cultures including an overall decrease in gene expression associated with translational activity and an increase in stress response. There were also several upregulated stress genes specific to the LSMMG condition during the transition to stationary phase growth. The ∆hfq mutants exhibited a distinctive transcriptome profile with a significant increase in transcripts associated with flagellar synthesis and transcriptional regulators under LSMMG conditions compared to gravity controls. These results indicate the loss of Hfq significantly influences gene expression under LSMMG conditions in a bacterial symbiont. Together, these results improve our understanding of the mechanisms by which microgravity alters the physiology of beneficial host-associated microbes.
Introduction
All animals form beneficial relationships with microbes.1 The normal microbiota of animals is extremely important for maintaining almost every aspect of animal fitness including host development, behavior, and immune system homeostasis.2,3 Understanding how these beneficial microbes respond to their continually changing environments represents an important area in animal microbiome research. One particular environment that presents numerous physiological challenges to animals and their microbiomes is spaceflight.4–8 During spaceflight, the reduction in gravity, or microgravity, can have widespread health impacts to the host including bone loss, alterations to the genome, neurovestibular, and immune systems.9–13 In particular, animal immune systems are highly dysregulated and host–microbe interactions have now been shown to play a significant role in maintaining healthy immune function during spaceflight.14
In addition to physiological changes in human and animal hosts, microbes are also impacted by microgravity. Some microbes exhibit altered growth rates and cell densities grown under both natural and analog microgravity conditions.15–20 Although this is not a universal response as several taxa, including pathogenic Streptococcus mutants and Salmonella enterica Serovar Typhimurium, exhibit no changes to growth rates under modeled microgravity conditions.21,22 For many taxa, however, there is an increased growth rate under both natural and simulated microgravity conditions,23,24 which can be highly dependent on the growth media used.19 Although the precise mechanisms underlying the increased growth rate in certain taxa have not been fully elucidated, research has indicated that in some bacteria the lag phase of growth is shortened and the exponential growth phase is lengthened.15
Microbes also respond to changes in the mechanical and physical forces (e.g., low-shear) associated with microgravity by modifying their gene expression,19,22,23,25–43 secondary metabolism,44,45 biofilm formation,26,28,34,40,43,46,47 and pathogenesis.48,49 Many pathogenic microbes under microgravity conditions exhibit altered virulence,21,39,47,50,51 resistance to environmental stress and antibiotics,27,29,34,35,37,40,42,48,52 as well as increased survival in host macrophages.21,39,42,49 Previous studies have shown that these changes in virulence are environment-dependent and in some cases can be attenuated through media supplementation, such as inorganic phosphate.51 These same studies have also determined there are extensive changes in microbial gene expression both at the transcriptional and translational levels.
One key finding is that microgravity alters the expression of the global regulator Hfq, an RNA-binding protein that stabilizes an interaction between small RNAs (sRNAs) and their target message RNAs (mRNAs) to influence gene expression53 and has been found in about half of all known bacterial genomes.54 This protein has been implicated as an important mechanism involved in bacterial stress response, and therefore, may be especially important in microgravity conditions.23,47,55 Several studies have shown that the hfq gene is down-regulated in bacteria under natural and modeled microgravity conditions, including beneficial microbes.23,47
Although significant progress has been made in understanding microbial responses to microgravity, most of these studies have focused on pathogenic strains of microbes.27,34,40,47,49 The effects of microgravity and low shear fluid dynamics on mutualistic bacteria are relatively unknown. Two recent studies on gut-associated Lactobacillus acidophilus revealed relatively few transcriptional and physiological differences when cultures were grown under low-shear-modeled microgravity (LSMMG) conditions.25,56 For example, no transcriptomic or growth changes were observed when the cultivars were grown under anaerobic conditions,56 however, some increased acid stress resistance and antimicrobial activity was observed when grown under aerobic conditions,25 suggesting more investigations in how mutualistic bacteria respond to the stress of microgravity are needed.
In this study, we investigate the impact of LSMMG on the beneficial symbiont, Vibrio fischeri, which forms a simplified binary relationship with the bobtail squid Euprymna scolopes. V. fischeri colonizes the epithelial-lined crypt spaces of a specialized light organ in the squid and induces a series of rapid immunological and developmental changes in the host tissues.57–60 This type of colonization of host epithelial tissues represents the most common form of symbioses in animals.61 Previous research on the effects of modeled microgravity on the squid-vibrio system has identified several microgravity-induced phenotypes in the host tissue,14,23,62,63 however, the effects of LSMMG on the V. fischeri transcriptome has not been explored.
To address this issue, we examined the transcriptional response of V. fischeri cultures to LSMMG at both exponential (12 h) and stationary (24 h) growth phases. Additionally, the transcriptome of a V. fischeri mutant defective in hfq was also compared to determine the role of this transcriptional regulator in V. fischeri physiology under LSMMG conditions. Previous work has shown that the hfq gene is down regulated in V. fischeri during LSMMG and squid infected with ∆hfq mutants exhibited several altered developmental phenotypes.23 Together, this work helps elucidate the impact of microgravity and the importance of Hfq in a beneficial microbe. By understanding the effects that spaceflight has on beneficial microbes critical insight can be inferred into maintaining healthy astronaut microbiomes and decrease the potential health risks associated with the exploration of space.
Results
Overview of transcriptome analysis of V. fischeri cultivars under gravity and LSMMG conditions
RNA-seq was used to evaluate the transcriptional changes of wild type V. fischeri ES114 (WT) and a ∆hfq deletion mutant (KV7142) at two key time points during bacterial growth. Strains were grown aerobically in a rotary culture system using high aspect ratio vessels (HARVs) in both gravity and LSMMG positions and their transcriptomes were examined during exponential (12 h) and stationary (24 h) growth phases. Growth curves for all strains, including ∆hfq complementation mutants (KV148, KV149) are shown in Supplemental Fig. S1 and correlate with previously published studies.23,62 There was a statistically higher number of colony forming units per ml at 12 h in LSMMG conditions, but the growth curves suggest both the LSMMG-treated and gravity-treated cultures were in log phase growth. Three libraries were generated for each treatment (note: only two libraries were created for the ∆hfq gravity controls). An average of 11.19 million high-quality reads that consistently mapped (>95%) to V. fischeri ES114 genome were obtained for each treatment (Table 1). This level of sequencing depth in RNA-seq analyses has been shown to be effective in detecting the majority of significant changes to gene expression profiles in bacteria.27,64
Table 1.
Overview of recovered transcriptome sequencing results from V. fischeri wild type (WT) and ∆hfq mutant exposed to low-shear-modeled microgravity (LSMMG) and gravity conditions
| Time point (h) | Treatment | Strain | Total readsa per treatment (million)a | Average reads per library (million) | Mapped reads (% mapped) |
|---|---|---|---|---|---|
| 12 | Gravity | WT | 11.18 | 3.73 | 96.31 |
| 12 | Gravity | ∆hfq | 7.20 | 3.60 | 96.20 |
| 12 | LSMMG | WT | 11.73 | 3.91 | 96.17 |
| 12 | LSMMG | ∆hfq | 11.25 | 3.75 | 96.08 |
| 24 | Gravity | WT | 13.03 | 4.34 | 95.86 |
| 24 | Gravity | ∆hfq | 8.40 | 4.20 | 96.49 |
| 24 | LSMMG | WT | 12.66 | 4.22 | 96.57 |
| 24 | LSMMG | ∆hfq | 14.04 | 4.68 | 97.10 |
aHigh-quality reads were filtered using Trimmomatic default parameters
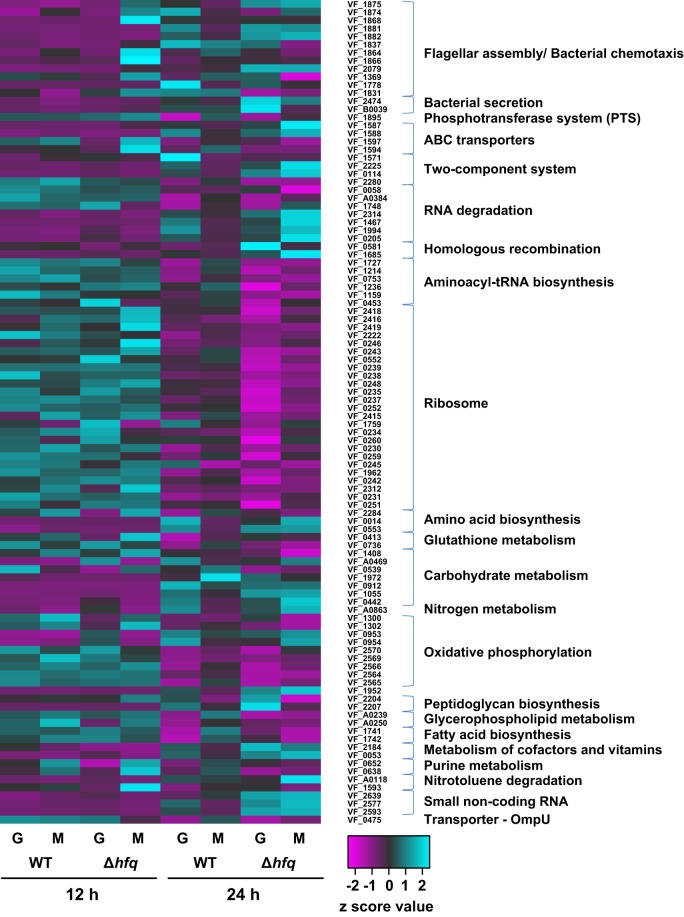
For control purposes, we first examined the transition between exponential and stationary phase in both WT and ∆hfq to ensure key metabolic transitions were being captured with the RNA-seq analyses in the HARV environment (Figs. 1 and 2). During growth under both LSMMG and gravity conditions the V. fischeri strains exhibited several typical responses of bacterial populations during stationary phase, including an overall down-regulation of genes associated with the translational apparatus, such as ribosomal proteins (e.g. rpsB, rpsG, rpsL, rpsM, rplM), tRNA synthases (e.g., tyrS, leuS, lysS), and translation factors (e.g., tufAB, infC, miaA) (Fig. 2; Supplemental Tables S1 and S2). In each treatment during stationary phase there was also an increase in the expression of several genes typically associated with stress responses, such as oxidative (e.g. VF_A0005, VF_A0335) and heat shock chaperones (e.g. dnaK1, dnaK2, htpG, hslO, hslV, ibpA, VF_1466) (Supplemental Tables S1–S3). These results are consistent with numerous studies indicating that in stationary phase bacteria become resistant to a wide range of environmental stresses65,66 and down-regulate their translational apparatus during nutrient limiting conditions.67 Together, the results indicate that the RNA-seq libraries were capturing the major transcriptional changes in V. fischeri during the different treatments.
Fig. 1.

Overview of the differentially expressed genes associated to each of the eight transcriptomic comparisons in Vibrio fischeri. a Matrix of the significant differentially expressed genes (DEGs) in WT and ∆hfq mutants under LSMMG (M) and gravity (G) conditions with the colors reflecting the relative abundance. b Venn diagrams indicating the intersections of the significant DEGs shared between the different transcriptomic comparisons in this study at 12 and 24 h
Fig. 2.
Heat map depicting the clustering patterns of the eight treatments by KEGG pathways associated with the proposed function of the V. fischeri genes at 12 and 24 h. Colors represent the differential abundance of individual genes listed by V. fischeri identification number (VF-ID) for both WT and ∆hfq mutants under low-shear modeled microgravity (M) and gravity (G) conditions
LSMMG-specific changes in V. fischeri transcriptome
Pairwise comparisons between the WT libraries revealed no significant differentially expressed genes (DEGs; adjusted p-value < 0.05) between the LSMMG and gravity treatments for WT at each of the time points tested, suggesting that the modeled microgravity environment had an overall minimal impact on the transcriptome of WT V. fischeri (Fig. 1). However, a comparison between the time points revealed five LSMMG-specific upregulated DEGs in both the WT and ∆hfq cultivars at 12 h when compared to 24 h (Fig. 1b; Supplemental Table S3). Two of these DEGs were associated with stress responses, including open-reading frame (ORF) VF_2561, whose gene product was annotated as a cold shock protein, and yceD, which encodes for a hypothetical protein that has been implicated in oxidative stress resistance in Bacillus subtilis.68
At 24 h there was an up-regulation of seven LSMMG-specific genes in both V. fischeri WT and ∆hfq strains when compared to 12 h libraries (Fig. 1b; Supplemental Table S3), several of which are known to be critical for stress resistance and microbial pathogenesis. For example, there was an up-regulation of yghU, which encodes for glutathione S-transferase and is essential for the detoxification of reactive oxygen species (ROS) in a wide range of taxa69 including several symbiotic taxa.70,71 There was also an increase in expression of blc, which encodes for the outer membrane lipoprotein lipocalin that is upregulated under high osmotic stress conditions in Escherichia coli and thought to play a role in antimicrobial resistance in several other bacteria.72 Additionally, there was an increase in expression of zwf, which encodes for glucose 6-phosphate dehydrogenase (G6PD), and has been shown to be required for virulence in Salmonella Typhimurium and protects against reactive oxygen and nitrogen species in both S. Typhimurium and E. coli.73,74 There was also up-regulation of katA, which encodes for the only periplasmic catalase present in the V. fischeri genome and is induced under oxidative stress conditions, as well as required for symbiosis competence in V. fischeri.75
Differential gene expression changes in ∆hfq mutant under both gravity and LSMMG conditions during exponential phase
In gravity conditions, there were few significant DEGs upregulated in the ∆hfq mutant compared to LSMMG at 12 h (Figs. 1 and 3a; Supplemental Table S1). One DEGs upregulated in gravity conditions was gnd, which encodes for 6-phosphogluconate dehydrogenase (6PGD), a key enzyme in the pentose phosphate pathway. The 6PGD enzyme produces NADPH, which provides the reducing power to several antioxidant proteins.76 Additionally, in the ∆hfq mutant, there was an increase in the expression of katA in gravity compared to LSMMG. The RNA-seq trends for katA were independently confirmed with qRT-PCR, although different transcript abundances were observed between the two methodologies for katA likely due to the differences in resolution between the approaches (Fig. 3b). At 12 h there were also three ORFs with unknown function upregulated in the ∆hfq gravity conditions (VF_2662, VF_A0979, and VF_A1190) (Supplemental Table S1).
Fig. 3.
Differential gene expression between LSMMG and gravity (G) in a ∆hfq mutant at 12 h. a Volcano plot visualizing the global transcriptional changes within the mutant cells. All normalized transcripts were plotted and each point reflects one gene with those in red indicating significance (adjusted p-value < 0.05). Full details of the significant differentially expressed genes are listed in Supplemental Table S1. b Quantitative real-time PCR of selected genes compared under LSMMG and gravity conditions in WT and ∆hfq mutants. Genes selected were significant in the RNA-seq results and included genes encoding a flagellin structural protein (flaA), flagellar hook-associated protein (flgK), and catalase (katA). Asterisk indicates significance between comparisons and error bars reflect the standard error of the mean
Under LSMMG conditions, however, the ∆hfq mutant exhibited a pronounced change to its transcriptome compared to gravity controls at 12 h (Figs. 1a and 3a; Supplemental Tables S1 and S2). During exponential phase in the ∆hfq mutant there was an accumulation of transcripts that encode for several components of the tricarboxylic acid (TCA) cycle including succinate dehydrogenase (sdhAB), aconitate hydratase (acnB), succinyl-CoA synthetase (sucCD), fumarate hydratase (fumB), and fumarate reductase (frdA) (Fig. 3a; Supplemental Table S1), all of which have been shown to be repressed by the sRNA RyhB in other taxa.77,78 Hfq is required for the stability and pairing of the sRNA RhyB to mRNA.78 RhyB has been identified in the V. fischeri genome (VF_2578), however, it was not significantly differentially expressed in this study. Additionally, there was an enrichment of transcripts associated with fatty acid synthesis (e.g. fabDFH), which in Salmonella Typhimurium is dependent on the sRNA SmpP,79 as well as oligopeptide transport (e.g., oppADF), which is regulated by the Hfq-dependent small RNA GcvB in a number of taxa including several vibrios.80 Homologs to SmpP and GcvB have not yet been reported in V. fischeri.
In LSMMG conditions there was also an increase in transcripts associated with flagella synthesis in exponential phase including genes that encode for both structural (e.g. flaACE), basal body rod (e.g. flgD), and hook-associated (e.g. flgEK) proteins81 (Fig. 3a; Supplementary Tables S1 and S2). The differential expression of flaA and flgK were confirmed with qRT-PCR in the ∆hfq (Fig. 3b). Hfq has been associated with flagellar synthesis in a wide range of taxa, including both pathogenic (e.g., Salmonella) and mutualistic bacteria (e.g., Sinorhizobium meliloti), however, in most cases mutants defective in hfq exhibit a repression of flagellar synthesis genes and in some cases are non-motile.82,83
Differential gene expression changes in ∆hfq mutant under both gravity and LSMMG conditions during stationary phase
The ∆hfq mutants exhibited extensive transcriptional changes during stationary phase under both gravity and LSMMG conditions (Figs. 1, 2, and 4; Supplemental Tables S1 and S2). One pronounced characteristic of the ∆hfq transcriptomes was the up-regulation of numerous transcriptional regulators during stationary phase (Fig. 4; Supplemental Tables S1 and S2). In both gravity and LSMMG conditions there was increased expression of agaR, which encodes for a putative transcriptional repressor of N-acetyl galactosamine (GalNAc) transport and metabolism in a wide range of bacterial taxa;84 iscR, a transcriptional repressor of genes associated Fe–S cluster assembly proteins;85 and yqhC, whose gene product regulates aldehyde reductase.86
Fig. 4.
Differential gene expression between WT and ∆hfq mutant under LSMMG at 24 h. a Volcano plot visualizing the global transcriptional changes within the V. fischeri cells. All normalized transcripts were plotted and each point reflects one gene with those in red indicating significance (adjusted p-value < 0.05). Full details of the significant differentially expressed genes are listed in Supplemental Table S1. b Quantitative real-time PCR of selected genes compared under low-shear modeled microgravity (LSMMG) and gravity conditions in wild type (WT) and ∆hfq mutants. Genes selected included dnaK, which encodes for a stress-associated chaperone protein and lpxD, which assists in the biosynthesis of lipid A. Asterisk indicates significance between comparisons and error bars reflect the standard error of the mean
In gravity conditions, there was differential expression of VF_1401, which encodes for a Fis family transcriptional regulator, and cysB, which belongs to the LysR family of regulators and is a global transcriptional activator of cysteine biosynthesis and sulfur metabolism.87 CysB is also the only known negative regulator of HslJ, a heatshock/heat-inducible outermembrane lipoprotein.88 The hslJ gene is upregulated in the ∆hfq mutant under both gravity and LSMMG in stationary phase (Fig. 4a; Supplemental Tables S1 and S2). Additionally, in gravity conditions, the ∆hfq mutant had increased expression of genes associated with the Type II section pathway (e.g. gspD, mshQ2) and several transport proteins (e.g. ybhG, argT, hisP) (Supplemental Tables S1 and S2).
Under LSMMG conditions, the genes of several different transcriptional regulators were upregulated during stationary phase. For example, VF_2037, which shares similarity to Cro/Ci family transcriptional regulators, was upregulated but has unknown function in V. fischeri. Additionally, nrdR was also upregulated and its gene product represses the ribonucleotide reductase production (i.e., nrdHIEF), bacterial chemotaxis, and more recently has been shown to inhibit cell adhesion to epithelial cells in E. coli.89,90 In addition to transcriptional regulators, there was also a differential expression of genes associated with modifications to the outer membrane in ∆hfq under LSMMG (Fig. 4a; Supplemental Tables S1 and S2). For example, there was increased expression of the gene slp, which encodes for an outer membrane lipoprotein associated with stress responses during stationary phase and is typically repressed by the Hfq-dependent sRNA GcvB.91 There was also an increased expression of skp, a periplasmic chaperone protein that is associated with the RpoE regulon and is involved in the folding of intermediates of outer membrane proteins.92 Interestingly, rpoE transcription was down-regulated at 24 h compared to 12 h in the ∆hfq mutants irrespective of the gravity or LSMMG treatment (Supplemental Table S1).
Discussion
To prepare for long-duration space travel it is essential to have a comprehensive understanding of the impact that spaceflight has on the physiology of host-associated microbiomes to promote and maintain astronaut health. There has been an extensive focus on the effects of spaceflight and simulated microgravity environments on bacterial pathogens,19,21,22,27,32,34,42,43,47,52 however, only a few studies have begun to examine the impact on beneficial microbes that promote the health of the host organism.14,23,25,56,62 In this study, we expand on this recent work and examine the effects of an LSMMG environment on the transcriptome of the beneficial symbiont V. fischeri, which forms a mutualistic association with the bobtail squid E. scolopes, and is critical for the host’s normal development. The results of this study suggest that there were few transcriptional changes in the WT V. fischeri under LSMMG and that most changes in the bacterium were attributed to the growth phase transition between exponential and stationary phase. Additionally, RNA-seq analyses revealed that mutants defective in the global regulator Hfq exhibited a pronounced change in transcriptional profiles under LSMMG, providing new insight into the role this regulator plays in the symbiotic V. fischeri under analog microgravity conditions.
Previous studies have shown that V. fischeri exhibits an altered growth response in simulated microgravity conditions, with cultures reaching higher cell densities compared to gravity controls.23 This altered growth response under LSMMG has been observed in many, but not all, taxa18,22,32,42 and is thought to reflect the selected growth medium. The nutritional micro-environment of the cells in LSMMG has been shown to significantly impact microbial physiology.18,19 For example, under low phosphate conditions some microbes, such as Salmonella Typhimurium, exhibit increased virulence.51 Despite the change in growth phenotype in V. fischeri under LSMMG conditions, no significant DEGs were observed when the transcriptomes of 12 h LSMMG-treated WT cells were compared to 12 h gravity controls, even under the low phosphate conditions of SWT media (Fig. 1). Similar results were observed when 24 h LSMMG-treated libraries were compared to 24 h gravity-treated libraries, suggesting that modeled microgravity itself does not significantly alter transcription within V. fischeri compared to gravity controls. These results are comparable to several recent studies on the effects of LSMMG on the probiotic strain L. acidophilus.25,56 Under the anaerobic conditions, transcriptomes of the L. acidophilus cultivar showed no DEGs in LSMMG when compared to gravity controls at stationary phase.56 As both L. acidophilus and V. fischeri typically form associations with host epithelium and are regularly exposed to low shear conditions in their natural environments, the modeled microgravity environment does not likely impose a significant stress for these taxa. Additionally, recent studies have shown that under LSMMG conditions V. fischeri exhibits no delay in colonizing host tissues62 and that during spaceflight V. fischeri reached the same colonization densities as under gravity controls.14 Together, these results suggest that microgravity conditions do not negatively impact V. fischeri.
Although the overall transcriptional response in V. fischeri was typical of the normal transition to stationary phase, there were several stress-associated genes differentially expressed under LSMMG conditions in both the WT and ∆hfq mutant. Of the DEGs differentially upregulated in LSMMG during the transition to stationary phase, three of the observed genes (i.e., yceD, yghU, and katA) are associated with stress responses and have been observed in E. coli K12 under modeled microgravity conditions.38 In E. coli these genes are associated with both oxidative and osmotic stress responses and may suggest that under LSMMG a small microenvironment of increased stress may occur around the V. fischeri cells. The formation of nutrient-depleted microenvironments has long been postulated under LSMMG conditions, which may simulate the genomic and physiological responses of cells as they transition to stationary phase.15,38 During exponential phase, V. fischeri cells are flagellated, as opposed to stationary phase when production of flagella is decreased and cells are non-motile (Edward Ruby, personal communication). The flagella during exponential phase may be disrupting the low shear environment thereby minimizing the effects of LSMMG on the cells and resulting in very few stress-associated genes being differentially regulated under the LSMMG environments at 12 h.
At 24 h, there was also expression of several other stress-associated genes (e.g. blc, zwf, katA), however, only katA has been previously described in V. fischeri. The katA gene encodes for a periplasmic catalase that is essential for the normal colonization of the host squid and is typically induced as cells approach stationary phase.75 The higher expression of katA during LSMMG in stationary phase suggests the cells are experiencing a more pronounced oxidative stress environment compared to the gravity controls. Similar results have been observed in several Salmonella spp., where bacteria grown under LSMMG conditions exhibited a higher resistance to hydrogen peroxide and increased catalase activity.52 Interestingly, although there was up-regulation of blc, which encodes for an outer membrane lipoprotein in E. coli expressed during osmotic stress,72 there were no observed significant DEGs associated with lipopolysaccharide biosynthetic genes or other cell membrane modifications, which have been reported to be differentially regulated under LSMMG.18,22,47 Together, the results reinforce the interpretation that the low shear environment of modeled microgravity does not significantly alter the transcriptional response V. fischeri cultivars, but that the few genes that are differentially expressed are primarily associated with environmental stress responses.
Although the transcriptome of WT cells did not display extensive changes in response to LSMMG, mutants defective in the global regulator Hfq exhibited a pronounced transcriptional response to LSMMG conditions (Fig. 1; Supplemental Table S1). The RNA-binding protein Hfq has been identified as an important transcriptional regulator in several pathogenic taxa in response to both spaceflight and microgravity analog environments.33,34,47 Additionally, the gene encoding Hfq has been shown to be down-regulated in several taxa, including V. fischeri, under LSMMG conditions.23,47 One of the major functions of Hfq is to bind to sRNAs, which then together subsequently target various mRNAs, thereby regulating or modulating the stability of the mRNAs.93 The sRNAs can also serve as activators or repressors of mRNA translation.94 During exponential phase, there was a significant accumulation of transcripts associated with the TCA cycle in the ∆hfq mutant. Up-regulation of genes associated with the TCA cycle has been observed under modeled microgravity27,35,95 and many TCA cycle genes are typically repressed by the Hfq-dependent sRNA RhyB,77 which recruits RNase E and facilitates mRNA degradation.78 In the ∆hfq mutant, the increase in recovered TCA cycle transcripts under LSMMG conditions likely reflects an inhibition of mRNA degradation rather than an up-regulation of these genes during LSMMG.
The ∆hfq mutants also exhibited an increase of transcripts associated with flagellar assembly under LSMMG conditions compared to WT cells during exponential phase growth (Fig. 3a; Supplemental Tables S1 and S2). The expression of flagellar assembly genes under microgravity conditions appears to be highly variable based on the taxa and whether the cells were exposed to actual spaceflight or analog conditions.19,51,95,96 The regulation of flagellar synthesis is complex and is not fully delineated in V. fischeri,97 although in many taxa it occurs both at the transcriptional and translational levels.98 In E. coli there are numerous Hfq-dependent sRNAs involved in the positive (e.g. McaS) and negative (e.g. ArcZ, OmrA, OmrB, SdsR, GadY, and OxyS) regulation of flagella synthesis, however, none of these Hfq-dependent sRNAs have been reported in the V. fischeri genome. The up-regulation of flagellar synthesis transcripts during exponential phase in LSMMG may suggest the cells are attempting to move out of potential zones of nutritional depletion. Alternatively, as there is a lack of differentially expressed flagellar synthesis transcripts during stationary phase when nutritional depletion is more severe, the results may simply suggest a lack of negative repression of the flagellar synthesis in the ∆hfq mutants during cell growth. A more detailed analysis of transcriptional and translational regulation of flagella synthesis in V. fischeri is needed.
In stationary phase growth, the ∆hfq mutants exhibited a pronounced increase in the expression of transcriptional regulators under both gravity-specific (e.g. Fis-family regulator, VF_1401; cysB), and LSMMG-specific (Cro/Ci family regulator VF_2037; nrdR) conditions. To our knowledge, none of these regulators have been reported to be differentially expressed during spaceflight or modeled microgravity conditions. For example, nrdR was first shown to positively regulate synthesis of ribonucleotide reductases in response to DNA damage and oxidative stress in Streptococcus pyrogens99 and more recently has been shown in E. coli to be involved in responding to iron starvation100 and the host immune system.101 As the stress responses of several taxa are altered under spaceflight and analog conditions,95 the role of these transcriptional regulators under microgravity-like stress conditions needs to be investigated further.
The ∆hfq mutant also exhibited significant DEGs associated with outer membrane proteins that are differentially expressed during stress conditions in a wide range of taxa. For example, slp is a carbon-starvation-induced gene that has been shown to be upregulated in V. cholerae ∆hfq mutants102 and is released in outer membrane vesicles.103 In E. coli the Slp lipoprotein is essential for acid and metabolic stress and is negatively regulated by the Hfq-dependent GvcB104. Although GvcB has not been reported in V. fischeri it has been reported in several environmental vibrios.80 These differential transcriptional responses in genes encoding for outer membrane proteins in the ∆hfq mutants may indicate that the outer membrane of the mutants may have a different composition compared to the WT cells under both gravity and LSMMG conditions. Recent studies have shown that V. fischeri-derived outer membrane vesicles can induce full developmental remodeling of the host light organ tissues,105 however, mutants defective in Hfq induce an altered phenotype including a decrease in the number of dying apoptotic cells in the host tissues under both LSMMG and gravity conditions.23 The mechanism for these decreased levels of apoptotic cells in ∆hfq mutants is not clear but may be the product of a remodeled outer membrane.
Although the full range of environmental factors that impact a host’s microbiome in the space environment has yet to be fully understood, this study provides insight into the role that microgravity may have on those beneficial microbes that typically associate with animal tissues. The results suggest that under normal growth conditions modeled microgravity does not negatively impact the transcriptional activities of V. fischeri indicating that the beneficial, mutualistic lifestyle of the bacterium is maintained under analog microgravity conditions. The results also deepen our understanding of the mechanisms by which organisms are adapting to changes in their nutritional environment and how the global regulator Hfq impacts the regulatory processes of V. fischeri in both LSMMG and gravity conditions. These results indicate that Hfq serves as an important mechanism by which V. fischeri regulates responses to external stimuli. As many of the mechanisms by which pathogenic and beneficial microbes sense and respond to their ever-changing environment are shared, it will be critical to continue to explore the processes by which microbes form complex communities and interact with their hosts during spaceflight to help mitigate any potential health threats during long-term missions.
Methods
Bacterial strains and growth conditions
The wild type strain V. fischeri ES114 (WT), which was isolated from an adult host squid E. scolopes106 was used as the parent strain for the deletion ∆hfq mutant and complementation (KV7142, ∆hfq; KV148, ∆hfq attTn7::ermR; KV149, ∆hfq attTn7::hfq; courtesy of K. Visick, Loyola University Chicago). The strains were grown aerobically overnight in seawater tryptone (SWT) agar at 28 °C, in which trace elements are at low concentration (e.g. phosphate (0.1 ppm)).23 High aspect ratio rotating vessels (HARVs; Synthecon, Houston, TX, USA) were used to model the microgravity environment as previously described.49,62 Briefly, each HARV was filled with 50 ml of SWT broth inoculated with V. fischeri culture at a concentration of 1 × 105 cells per ml of SWT. The HARVs were either rotated around a horizontal axis to simulate microgravity (LSMMG) or a vertical axis to serve as a normal gravity (1 × g) control. The cultures were incubated in the HARVs at 12 and 24 h in the vertical LSMMG and horizontal gravity control positions at 23 °C to replicate temperatures cells would experience in the natural environment. The HARVs were rotated at a constant velocity of 13 rpm, which prevented V. fischeri cells from forming sedimentary aggregates and to match rotation speed used in comparable squid-vibrio experiments.62 Experiments were conducted in triplicate for each condition, strain, and time. Growth curves of all strains used in this experiment are visualized in Supplemental Fig. S1 and corresponded to previously published results.23,62 At the end of each HARV experiment V. fischeri were flash frozen in liquid nitrogen to halt gene expression and stored at −80 °C until RNA extraction.
RNA extraction, cDNA synthesis, and sequencing
Each replicate V. fischeri WT and ∆hfq culture was thawed on ice and pelleted for RNA extraction. Total RNA was extracted in triplicate for each treatment using PowerSoil® Total RNA Isolation Kit (Qiagen, Germantown, MD) according to manufacturer’s protocol and was treated with TURBO DNase (Thermo Fisher Scientific, Waltham, MA) to remove potential contaminating DNA. The RiboMinus rRNA removal kit (Thermo Fisher Scientific, Waltham, MA) was used to deplete large rRNAs and samples were processed with the Zymo RNA Clean & Concentrator kit (Zymo Research, Irvine, CA). The remaining mRNA was pooled between replicates, the concentration was determined by Qubit 2.0 (Thermo Fisher Scientific, Waltham, MA) and quality was evaluated with a 2100 Bioanalyzer generating RIN factor > 9 (Agilent Technologies, Santa Clara, CA). High-quality RNA was converted to cDNA using a modified SuperScript Double Stranded cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA). A total of three replicate cDNA libraries were generated for each treatment (note: only two libraries were generated for ∆hfq gravity controls) and sequenced using the Illumina NextSeq500 platform (2 × 150 bp paired-end reads; Illumina, San Diego, CA).
Bioinformatic analysis
Sequences were quality trimmed and filtered with Trimmomatic v0.32 using default parameters.107 The quality of the output files was then analyzed using FastQC v0.10.1.108 Reads were then aligned to the V. fischeri ES114 reference genome (GenBank ID: ASM11800v1) using Bowtie 2 v2.2.8.109 Gene counts were obtained using HTSeq-count v 0.6.1.110 Genes with no expression across all conditions were removed. Differential expression analysis was conducted in R using the package DESeq2.111 Genes were considered significantly differentially expressed at adjusted p-value (padj) < 0.05. UpSetR was used to visualize the intersection of DEGs.112 The most recent KEGG database was accessed through the R package KEGGREST v1.16.1 to determine KEGG functional pathways and higher KEGG level classification.113 The top 103 DEGs among time treatment comparisons with one defined KEGG pathway were visualized in a heatmap. Expression values for heatmap were normalized with trimmed mean of M-values (TMM) using the NOISeq package114 and scaled by the sum of each row (z-score) using heatmap.2 in the ggplots package in R.115 Genes associated with multiple pathways at KEGG level 2, or had no specific KEGG pathway association, were not displayed in the heatmap.
Real-time quantitative PCR (qRT-PCR)
Several significantly DEGs were chosen for targeted qRT-PCR confirmation. Primers are listed in Supplemental Table S4. The qRT-PCR reactions were prepared using the iTaq Universal SYBR Green One-Step Kit (Biorad, Hercules, CA) with 10 ng of RNA per reaction. Amplification and quantification were completed using a Biorad SCX9600 Real Time System (Biorad, Hercules, CA). The amplification conditions were as follows: an initial incubation at 50 °C for 10 min then 1 min at 95 °C followed by 39 cycles of 95 °C for 10 s and 60 °C for 15 s. Each comparison was run in triplicate and three technical replicates were run for each biological replicate. The relative expression of each gene was analyzed using the comparative Cq method (ΔΔCq) on the Biorad system. The gene rpoD was chosen as the housekeeping reference gene for normalization of transcript abundances as previously described.116
Electronic supplementary material
Acknowledgements
The authors thank Dr. Artemis Louyakis for technical support in data analysis and Hadrien Gourlé for his assistance with KEGG pathway analysis. This work was supported in part by the NASA Space Biology program (NNX13AM44G) and Florida Space Institute Space Research Initiative awarded to J.S.F. A.A.D. was supported in part by a Dissertation Improvement Fellowship from the NASA Florida Space Grant Consortium. S.D.Z. was supported by a National Science Foundation STEP award (DUE 1161177). M.B. and M.M.V. were supported by a NASA Florida Space Grant Consortium Scholarship award.
Author contributions
A.A.D., A.C., M.B., M.M.V., S.D.Z., and J.S.F. performed the experiments and analyzed the data. A.A.D. and J.S.F. wrote the manuscript.
Data availability
All reads have been deposited in the NASA GeneLab database as well as NCBI under Bioproject PRJNA357702.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies the paper on the npj Microgravity website (10.1038/s41526-018-0060-1).
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35:507–517. doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voorhies AA, Lorenzi HA. The challenge of maintaining a healthy microbiome during long-duration space missions. Front. Astron. Space Sci. 2016;3:23. doi: 10.3389/fspas.2016.00023. [DOI] [Google Scholar]
- 5.Mardanov AV, et al. Metagenomic analysis of the dynamic changes in the gut microbiome of the participants of the MARS-500 experiment, simulating long term space flight. Acta Nat. 2013;5:116–125. [PMC free article] [PubMed] [Google Scholar]
- 6.Guéguinou N, et al. Could spaceflight-associated immune system weakening preclude the expansion of human presence beyond Earth’s orbit? J. Leukoc. Biol. 2009;86:1027–1038. doi: 10.1189/jlb.0309167. [DOI] [PubMed] [Google Scholar]
- 7.Barrila J, et al. Spaceflight modulates gene expression in the whole blood of astronauts. NPJ Microgravity. 2016;2:16039. doi: 10.1038/npjmgrav.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terada M, et al. Effects of a closed space environment on gene expression in hair follicles of astronauts in the International Space Station. PLoS One. 2016;11:e0150801. doi: 10.1371/journal.pone.0150801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervantes JL, Hong BY. Dysbiosis and immune dysregulation in outer space. Int. Rev. Immunol. 2016;35:67–82. doi: 10.3109/08830185.2015.1027821. [DOI] [PubMed] [Google Scholar]
- 10.Crucian B, et al. Alterations in adaptive immunity persist during long-duration spaceflight. NPJ Microgravity. 2015;1:15013. doi: 10.1038/npjmgrav.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams D, Kuipers A, Mukai C, Thirsk R. Acclimation during space flight: effects on human physiology. CMAJ. 2009;180:1317–1323. doi: 10.1503/cmaj.090628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selch F, et al. Genomic response of the nematode Caenorhabditis elegans to spaceflight. Adv. Space Res. 2008;41:807–815. doi: 10.1016/j.asr.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borchers AT, Keen CL, Gershwin ME. Microgravity and immune responsiveness: implications for space travel. Nutrition. 2002;18:889–898. doi: 10.1016/S0899-9007(02)00913-9. [DOI] [PubMed] [Google Scholar]
- 14.Casaburi G, Goncharenko-Foster I, Duscher AA, Foster JS. Transcriptomic changes in an animal-bacterial symbiosis under modeled microgravity conditions. Sci. Rep. 2017;7:46318. doi: 10.1038/srep46318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaus D, Simske S, Todd P, Stodieck L. Investigation of space flight effects on Escherichia coli and a proposed model of underlying physical mechanisms. Microbiology. 1997;143:449–455. doi: 10.1099/00221287-143-2-449. [DOI] [PubMed] [Google Scholar]
- 16.Ciferri O, Tiboni O, Di Pasquale G, Orlandoni AM, Marchesi ML. Effects of microgravity on genetic recombination in Escherichia coli. Naturwissenschaften. 1986;73:418–421. doi: 10.1007/BF00367284. [DOI] [PubMed] [Google Scholar]
- 17.Taylor GR. Space Microbiology. Annu. Rev. Microbiol. 1974;28:121–137. doi: 10.1146/annurev.mi.28.100174.001005. [DOI] [PubMed] [Google Scholar]
- 18.Arunasri K, et al. Effect of simulated microgravity on E. coli K12 MG1655 growth and gene expression. PLoS One. 2013;8:e57860. doi: 10.1371/journal.pone.0057860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tucker DL, et al. Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol. 2007;7:15. doi: 10.1186/1471-2180-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fajardo-Cavazos P, Nicholson WL. Cultivation of Staphylococcus epidermidis in the human spaceflight environment leads to alterations in the frequency and spectrum of spontaneous rifampicin-resistance mutations in the rpoB gene. Front. Microbiol. 2016;7:999. doi: 10.3389/fmicb.2016.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nickerson CA, et al. Microgravity as a novel environmental signal affecting Salmonella enterica Serovar Typhimurium virulence. Infect. Immun. 2000;68:3147–3152. doi: 10.1128/IAI.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orsini SS, Lewis AM, Rice KC. Investigation of simulated microgravity effects on Streptococcus mutans physiology and global gene expression. NPJ Microgravity. 2017;3:4. doi: 10.1038/s41526-016-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant KA, Khodadad CL, Foster JS. Role of Hfq in an animal-microbe symbiosis under simulated microgravity conditions. Int. J. Astrobiol. 2014;13:53–61. doi: 10.1017/S1473550413000359. [DOI] [Google Scholar]
- 24.Kacena MA, et al. Bacterial growth in space flight: logistic growth curve parameters for Eschechia coli and Bacillus subtilus. Appl. Microbiol. Biotech. 1999;51:229–234. doi: 10.1007/s002530051386. [DOI] [PubMed] [Google Scholar]
- 25.Shao D, et al. Simulated microgravity affects some biological characteristics of Lactobacillus acidophilus. Appl. Microbiol. Biotech. 2017;101:3439–3449. doi: 10.1007/s00253-016-8059-6. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, et al. Increased biofilm formation ability in Klebsiella pneumoniae after short-term exposure to a simulated microgravity environment. Microbiologyopen. 2016;5:793–801. doi: 10.1002/mbo3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abshire CF, et al. Exposure of Mycobacterium marinum to low-shear modeled microgravity: effect on growth, the transcriptome and survival under stress. NPJ Microgravity. 2016;2:16038. doi: 10.1038/npjmgrav.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dingemans J, et al. Effect of shear stress on Pseudomonas aeruginosa isolated from the cystic fibrosis lung. mBio. 2016;7:e00813–e00816. doi: 10.1128/mBio.00813-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soni A, et al. Conservation of the low-shear modeled microgravity response in Enterobacteriaceae and analysis of the trp genes in this response. Open Microbiol. J. 2014;8:51–58. doi: 10.2174/1874285801408010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mastroleo F, et al. Modelled microgravity cultivation modulates N-acylhomoserine lactone production in Rhodospirillum rubrum S1H independently of cell density. Microbiology. 2013;159:2456–2466. doi: 10.1099/mic.0.066415-0. [DOI] [PubMed] [Google Scholar]
- 31.A. C, et al. Spaceflight enhances cell aggregation and random budding in Candida albicans. PLoS One. 2013;8:e80677. doi: 10.1371/journal.pone.0080677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vukanti R, Model MA, Leff LG. Effect of modeled reduced gravity conditions on bacterial morphology and physiology. BMC. Microbiol. 2012;12:4. doi: 10.1186/1471-2180-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabbé A, et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl. Environ. Microbiol. 2011;77:1221–1230. doi: 10.1128/AEM.01582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro SL, Nelman-Gonzalez M, Nickerson CA, Ott CM. Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl. Environ. Microbiol. 2011;77:6368–6378. doi: 10.1128/AEM.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crabbé A, et al. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 2010;12:1545–1564. doi: 10.1111/j.1462-2920.2010.02184.x. [DOI] [PubMed] [Google Scholar]
- 36.Rosado H, Doyle M, Hinds J, Taylor PW. Low-shear modelled microgravity alters expression of virulence determinants of Staphylococcus aureus. Acta Astronaut. 2010;66:408–413. doi: 10.1016/j.actaastro.2009.06.007. [DOI] [Google Scholar]
- 37.Altenburg SD, Nielsen-Preiss SM, Hyman LE. Increased filamentous growth of Candida albicans in simulated microgravity. Genom. Proteom. Bioinform. 2008;6:42–50. doi: 10.1016/S1672-0229(08)60019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vukanti R, Mintz EM, Leff LG. Changes in gene expression of E. coli under conditions of modeled reduced gravity. Microgravity Sci. Technol. 2008;20:41. doi: 10.1007/s12217-008-9012-9. [DOI] [Google Scholar]
- 39.Chopra V, et al. Alterations in the virulence potential of enteric pathogens and bacterial–host cell interactions under simulated microgravity conditions. J. Toxicol. Environ. Health A. 2006;69:1345–1370. doi: 10.1080/15287390500361792. [DOI] [PubMed] [Google Scholar]
- 40.Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl. Environ. Microbiol. 2006;72:7701–7710. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purevdorj-Gage B, Sheehan KB, Hyman LE. Effects of low-shear modeled microgravity on cell function, gene expression, and phenotype in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006;72:4569–4575. doi: 10.1128/AEM.03050-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson JW, et al. Low-Shear modeled microgravity alters the Salmonella enterica serovar typhimurium stress response in an RpoS-independent manner. Appl. Environ. Microbiol. 2002;68:5408–5416. doi: 10.1128/AEM.68.11.5408-5416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim W, et al. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS One. 2013;8:e62437. doi: 10.1371/journal.pone.0062437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demain AL, Fang A. Secondary metabolism in simulated microgravity. Chem. Rec. 2001;1:333–346. doi: 10.1002/tcr.1018. [DOI] [PubMed] [Google Scholar]
- 45.Fang A, Pierson DL, Koenig DW, Mishra SK, Demain AL. Effect of simulated microgravity and shear stress on microcin B17 production by Escherichia coli and on its excretion into the medium. Appl. Environ. Microbiol. 1997;63:4090–4092. doi: 10.1128/aem.63.10.4090-4092.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crabbé A, et al. Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ. Microbiol. 2008;10:2098–2110. doi: 10.1111/j.1462-2920.2008.01631.x. [DOI] [PubMed] [Google Scholar]
- 47.Wilson JW, et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl Acad. Sci. USA. 2007;104:16299–16304. doi: 10.1073/pnas.0707155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor PW. Impact of space flight on bacterial virulence and antibiotic susceptibility. Infect. Drug Resist. 2015;8:249–262. doi: 10.2147/IDR.S67275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbial responses to microgravity and other low-shear environments. Microbiol. Mol. Biol. Rev. 2004;68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Barrila J, Roland KL, Ott CM, Nickerson CA. Physiological fluid shear alters the virulence potential of invasive multidrug-resistant non-typhoidal Salmonella Typhimurium D23580. NPJ Microgravity. 2016;2:16021. doi: 10.1038/npjmgrav.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson JW, et al. Media ion composition controls regulatory and virulence response of Salmonella in spaceflight. PLoS One. 2008;3:e3923. doi: 10.1371/journal.pone.0003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pacello F, Rotilio G, Battistoni A. Low-shear modeled microgravity enhances Salmonella enterica resistance to hydrogen peroxide through a mechanism involving KatG and KatN. Open Microbiol. J. 2012;6:53–64. doi: 10.2174/1874285801206010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 55.Mukhopadhyay S, et al. A systems biology pipeline identifies new immune and disease related molecular signatures and networks in human cells during microgravity exposure. Sci. Rep. 2016;6:25975. doi: 10.1038/srep25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro-Wallace SL, Stahl SE, Voorhies AA, Lorenzi HA, Douglas GL. Response of Lactobacillus acidophilus ATCC 4356 to low-shear modeled microgravity. Acta Astronaut. 2017;139:463–468. doi: 10.1016/j.actaastro.2017.07.033. [DOI] [Google Scholar]
- 57.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 58.Foster JS, McFall-Ngai MJ. Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev. Genes Evol. 1998;208:295–303. doi: 10.1007/s004270050185. [DOI] [PubMed] [Google Scholar]
- 59.Montgomery MK, McFall-Ngai M. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- 60.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 61.McFall-Ngai M. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foster JS, Khodadad CL, Ahrendt SR, Parrish ML. Impact of simulated microgravity on the normal developmental time line of an animal-bacterial symbiosis. Sci. Rep. 2013;3:1340. doi: 10.1038/srep01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foster JS, Kerney KR, Parrish ML, Khodadad CLM, Ahrendt SR. Potential of the Euprymna/Vibrio symbiosis as a model to assess the impact of microgravity on bacteria-induced animal development. Grav. Space Biol. 2011;25:44–47. [Google Scholar]
- 64.Haas BJ, Chin M, Nusbaum C, Birren BW, Livny J. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genom. 2012;13:734. doi: 10.1186/1471-2164-13-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazazzera BA. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 2000;3:177–182. doi: 10.1016/S1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 66.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 67.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 68.Hoper D, Volker U, Hecker M. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 2005;187:2810–2826. doi: 10.1128/JB.187.8.2810-2826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanai T, Takahashi K, Inoue H. Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J. Biochem. 2006;140:703–711. doi: 10.1093/jb/mvj199. [DOI] [PubMed] [Google Scholar]
- 70.Pontes MH, et al. Quorum sensing primes the oxidative stress response in the insect endosymbiont, Sodalis glossinidius. PLoS One. 2008;3:e3541. doi: 10.1371/journal.pone.0003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G+C content of an endocytobiotic DNA. J. Mol. Evol. 1998;47:52–61. doi: 10.1007/PL00006362. [DOI] [PubMed] [Google Scholar]
- 72.Bishop RE. The bacterial lipocalins. Biochim. Biophys. Acta. 2000;1482:73–83. doi: 10.1016/S0167-4838(00)00138-2. [DOI] [PubMed] [Google Scholar]
- 73.Sandoval JM, Arenas FA, Vasquez CC. Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS One. 2011;6:e25573. doi: 10.1371/journal.pone.0025573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lundberg BE, Wolf RE, Jr., Dinauer MC, Xu Y, Fang FC. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 1999;67:436–438. doi: 10.1128/iai.67.1.436-438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Visick KL, Ruby EG. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J. Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ezraty B, Gennaris A, Barras F, Collet JF. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017;15:385–396. doi: 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 77.Desnoyers G, Masse E. Activity of small RNAs on the stability of targeted mRNAs in vivo. Methods Mol. Biol. 2012;905:245–255. doi: 10.1007/978-1-61779-949-5_16. [DOI] [PubMed] [Google Scholar]
- 78.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ansong C, et al. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silveira AC, et al. Identification of non-coding RNAs in environmental vibrios. Microbiology. 2010;156:2452–2458. doi: 10.1099/mic.0.039149-0. [DOI] [PubMed] [Google Scholar]
- 81.Millikan DS, Ruby EG. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torres-Quesada O, et al. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol. 2014;11:563–579. doi: 10.4161/rna.28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sittka A, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leyn SA, Gao F, Yang C, Rodionov DA. N-acetylgalactosamine utilization pathway and regulon in proteobacteria: genomic reconstruction and experimental characterization in Shewanella. J. Biol. Chem. 2012;287:28047–28056. doi: 10.1074/jbc.M112.382333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwartz CJ, et al. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl Acad. Sci. USA. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee C, et al. Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J. Bacteriol. 2010;192:4205–4214. doi: 10.1128/JB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 88.Jovanovic M, Lilic M, Savic DJ, Jovanovic G. The LysR-type transcriptional regulator CysB controls the repression of hslJ transcription in Escherichia coli. Microbiology. 2003;149:3449–3459. doi: 10.1099/mic.0.26609-0. [DOI] [PubMed] [Google Scholar]
- 89.Naveen V, Hsiao CD. NrdR transcription regulation: global proteome analysis and its role in Escherichia coli viability and virulence. PLoS One. 2016;11:e0157165. doi: 10.1371/journal.pone.0157165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Torrents E, et al. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 2007;189:5012–5021. doi: 10.1128/JB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stauffer, L. T. & Stauffer, G. V. Multiple roles for the sRNA GcvB in the regulation of Slp levels in Escherichia coli. ISRN Bacteriology2013, (2013).
- 92.Schafer U, Beck K, Muller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 93.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roy R, Shilpa PP, Bagh S. A systems biology analysis unfolds the molecular pathways and networks of two Proteobacteria in spaceflight and simulated microgravity conditions. Astrobiology. 2016;16:677–689. doi: 10.1089/ast.2015.1420. [DOI] [PubMed] [Google Scholar]
- 96.Benoit M, Klaus DM. Microgravity, bacteria, and the influence of motility. Adv. Space Res. 2007;39:1225–1232. doi: 10.1016/j.asr.2006.10.009. [DOI] [Google Scholar]
- 97.Norsworthy AN, Visick KL. Gimme shelter: how Vibrio fischeri successfully navigates an animal’s multiple environments. Front. Microbiol. 2013;4:356. doi: 10.3389/fmicb.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Borovok I, et al. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 2004;54:1022–1035. doi: 10.1111/j.1365-2958.2004.04325.x. [DOI] [PubMed] [Google Scholar]
- 100.Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y, Wang L, Xie J, Zheng H. Differential protein expression profiling of Arabidopsis thaliana callus under microgravity on board the Chinese SZ-8 spacecraft. Planta. 2015;241:475–488. doi: 10.1007/s00425-014-2196-x. [DOI] [PubMed] [Google Scholar]
- 102.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 103.Lee EY, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 104.Sharma CM, et al. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- 105.Aschtgen MS, et al. Rotation of Vibrio fischeri flagella produces outer membrane vesicles that induce host development. J. Bacteriol. 2016;198:2156–2165. doi: 10.1128/JB.00101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andrews, S. FastQC a quality-control tool for high-throughput sequence data (2014).
- 109.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Prot. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 111.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tenenbaum, D. KEGGREST: Client-side REST Access to KEGG. R package version 1.16.1 (2017).
- 114.Tarazona S, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Warnes, M. G. R., Bolker, B., Bonebakker, L. & Gentleman, R. Package gplots. Various R Programming Tools for Plotting Data (2016).
- 116.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol. Microbiol. 2010;77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reads have been deposited in the NASA GeneLab database as well as NCBI under Bioproject PRJNA357702.