Abstract
N-Acyl homoserine lactone (AHL)-mediated Quorum sensing (QS) is one of the most studied social behavior among Proteobacteria. However, despite the current knowledge on QS-associated phenotypes such as bioluminescence, biofilm formation, or pathogenesis, the characterization of environmental factors driving QS in realistic ecological settings remains scarce. We investigated the dynamics of AHL and AHL-producing Vibrio among 840 isolates collected fortnightly from the Salses-Leucate Mediterranean lagoon in spring and summer 2015 and 2016. Vibrio isolates were characterized by gyrB gene sequencing, Enterobacterial repetitive intergenic consensus polymerase chain reaction, and genome sequencing, and AHL production was investigated by a biosensors-based UHPLC–HRMS/MS approach. Our results revealed, for the first time, a succession of V. mediterranei isolates with different AHL production phenotypes over time and this dynamics was observed in a single genotype (average genomic nucleotide identity >99.9). A multivariate DistLM analysis revealed that 83.4% of the temporal variation of V. mediterranei QS phenotypes was explained by environmental variables. Overall, our results suggest that isolates of a single genotype are able to change their QS phenotypes in response to environmental conditions, highlighting the phenotypic plasticity of bacterial communication in the environment.
Subject terms: Microbial ecology, Water microbiology
Introduction
Bacterial communities inhabiting marine coastal areas are subjected to rapid and somewhat unpredictable changes in their environment [1]. In order to cope with these changes, bacteria implement flexible strategies supporting a certain degree of phenotypic plasticity [2–4]. Vibrio are well known for their capacity of rapid physiological adaptation in response to changing environmental conditions, making them highly dynamic over short-term and seasonal scales [5]. These opportunistic bacteria are sensitive to unfavorable conditions by switching from an active free-living state into a “dormant” viable but not culturable (VBNC) phenotype, or by colonizing viscous surfaces such as biofilms [6–9]. When favorable conditions reappear, their high reactivity allows them to colonize new substrates and to be part of the microbial community associated with zooplankton, phytoplankton, and marine vertebrates and invertebrates [10–13].
In the last decades, our vision of bacterial communities has significantly changed, and microbial cells are no longer considered to behave individually, but rather to act socially [14–16]. It is now clear that micro-organisms perform social behaviors, synchronizing the expression of functional genes at high cell density by sensing their surrounding environment. This mechanism is generally known as Quorum sensing (QS) [17], and type 1 auto-inducers (AI-1) also called N-acyl-homoserine lactones (AHL) are important signal molecules for the communication between closely related members of the Proteobacteria. Among Vibrio species, most studies highlighted the processes controlled by QS, i.e., QS-associated phenotypes, such as bioluminescence and symbiosis [18, 19], biofilm formation [20], toxin production, or expression of virulence factors [21–23]. In vitro studies have demonstrated the involvement of QS in the “resuscitation” of VBNC Vibrio cells [24, 25]. However, Platt and Fuqua [26] presented the QS process themselves (i.e., the production of signal molecules), as impacted by multiple aspects of natural environments. In support of this idea, research on the AHL biosynthetic pathways has suggested that any environmental factor likely to affect the primary metabolism of bacteria could modify the “language” spoken by the bacteria [27, 28]. Finally, several studies have reported heterogeneous AHL production phenotypes (APP) among different isolates of a single Vibrio species, i.e., the production of different sets of AHLs according to their origin [29–32].
Despite all this previous knowledge, the question of whether APP form coherent populations in the environment, show temporal dynamics, or respond to different environmental conditions, remains largely underexplored. Therefore, in the present study, we investigated over a 2-year period the temporal dynamics of AHL-producing Vibrio isolates from a French Mediterranean lagoon and evaluated the possible links between phenotypic plasticity of AHL production and environmental conditions.
Materials and methods
Sample collection
A 2-year study was carried out during the summer in 2015 and from spring to summer (April to July) in 2016 at one sampling site in the south basin of the Salses-Leucate lagoon, on the Mediterranean coast of France (42° 50'54.6" N, 3°00'08.7" E; Figure S1). Surface water was collected in sterile bottles and prefiltered through a 50-µm mesh. Plankton was collected using a 50-µm plankton net (30 cm diameter, 1 m length, 500 mL cod-end) by carrying out a 5-min horizontal tow at two knots. All samples were processed within 3 h. For each sampling date, triplicate plankton aliquots of 15 mL were fixed with 4% neutral Lugol’s solution (0.68% potassium iodide and 0.34% iodine in distilled water) and stored in the dark at 4 °C. Taxonomic identification and enumeration were performed within 2 months. Surface temperature (°C), salinity (psu), dissolved oxygen (DO, % saturation), conductivity (mS/cm), and pH were recorded at each sampling date using an HQ40d multiparametric profiler (Hach). Physicochemical parameters, as well as pigments were analyzed using standard methods (the methodological details can be found in Table 1).
Table 1.
Environmental parameters measurements
| Parameter | Methods/instrumentation | Reference | Unit | Ranges | |
|---|---|---|---|---|---|
| − | + | ||||
| Temperature | Hach HQ40d multi | [72, 73] | °C | 14.2 | 26.4 |
| Salinity | ‰ | 32.8 | 40.1 | ||
| Conductivity | mS/cm | 39.9 | 55.7 | ||
| DO concentration | mg/L | 7.82 | 10.09 | ||
| DO saturation | % | 91.7 | 116 | ||
| pH | * | 7.32 | 8.5 | ||
| Nitrate | Hach DR/890 Colorimeter | [73, 74] | mg/L | 1.89 | 9.26 |
| Nitrite | mg/L | 0.002 | 0.043 | ||
| Ammonium | mg/L | <0.03 | |||
| Phosphate | mg/L | 0.05 | 0.15 | ||
| DOC | VCSN/TNM-1 Shimadzu TOC/TN analyzer | [73, 75] | mg/L | 2.5 | 6.69 |
| TDN | mg/L | 0.15 | 0.59 | ||
| POC | Perkin Elmer C,H,N 2400 | [76] | mg/L | 217.635 | 453.27 |
| PON | mg/L | 36.99 | 92.40 | ||
| Chlorophyll a | Lorenzen | [77, 78] | µg/L | 0.08 | 1.23 |
| Phaeophytin a | µg/L | 0.021 | 1.34 | ||
| Cryptophyta | FACSCantoII | [34] | cells/L | 6.74 × 102 | 1.65 × 106 |
| Nanoeukaryote | cells/L | 6.13 × 105 | 1.01 × 107 | ||
| Picoeukaryote | cells/L | 1.84 × 105 | 2.55 × 107 | ||
| Cyanobacteria | cells/L | 3.86 × 105 | 2.88 × 108 | ||
| Bacteria | cells/L | 3.04 × 109 | 2.14 × 1010 | ||
DO dissolved oxygen; DOC dissolved organic carbon; TDN total dissolved nitrogen; POC particulate organic carbon; PON particulate organic nitrogen
Large phytoplankton and picoplankton counts
Phytoplankton counts were carried out with an Olympus IMT inverted microscope in 5, 10, 25, or 50-ml plate chambers, using the Utermöhl method updated by Karlson et al. [33]. Replicates of different volumes were counted according to the size and abundance of the cells. Small-sized species were mainly counted in diagonal transects at 300× magnification, and for the large or less abundant species half or whole chambers were scanned at 100× and 40× magnifications. A minimum of 100 total cells were counted for each enumeration. Different settling times were applied according to chamber volume in a temperature-controlled room [33]. For picoplankton enumeration, seawater samples were collected (1.5 ml in triplicate, fixed on board with 1% glutaraldehyde and then quickly frozen at −80 °C). The picoplankton abundances, including cyanobacteria (mainly represented by Synechococcus), pico- and nano-eukaryotes, and cryptophytes were measured with a FACSCanto II (Becton Dickinson) flow cytometer. Analyses were performed within 1 month of sampling. Heterotrophic bacterial cells were stained with SYBR-Green I (Molecular Probes) before flow cytometry analysis. Reference beads (Fluoresbrite YG Microspheres, calibration grade 1.00 and 10.00 µm, PolySciences, Inc.) were added to each sample before acquiring the data. Each planktonic group was analyzed according to Marie et al. [34] with the Cell Quest Pro software (Becton Dickinson), in logarithmic mode, to separate the populations based on their scattering and fluorescence signals.
Vibrio isolation and characterization
For the enumeration of Vibrio, water samples (0.1, 1, and 10 mL) were filtered onto 0.45-µm pore-size nitrocellulose filters (Millipore, 47 mm), plated onto the selective medium thiosulfate–citrate–bile–sucrose (TCBS) [35] and incubated for 24 h at 20 °C. Vibrio strains were isolated from 2 fractions: less than 50 µm, assumed to mostly represent Vibrio in the water column and greater than 50 µm, corresponding to large phytoplankton and zooplankton-associated Vibrio. The <50-µm fraction consisted of 20 L of 50 µm prefiltered water further concentrated using a hollow fiber filter HF80S (Hemoflow, Fresenius Medical Care). The concentrate was backwashed with 500 mL of wash solution (0.01% sodium hexametaphosphate and 0.5% Tween 80; Sigma-Aldrich). The >50-µm fraction consisted of a plankton net tow concentrate that was split into five subsamples of 15 mL and homogenized by gentle sonication for 5 min in order to unbind the attached bacteria. For both fractions, a tenfold serial dilution was made in artificial seawater and 100 µL of each was plated in triplicate on TCBS agar. Thirty colonies per fraction and per sampling date were randomly picked using a gridded petri dish.
Isolate identification
A total of 840 isolates were identified based on gyrB gene sequences. PCR was performed using cells from a single colony and amplification reactions consisted of 12.5 µL of Kapa2G Master Mix (KAPA2G Fast Hotstart ReadyMix PCR kit, KapaBiosystems), 6.25 µL of 10 mM MgCl2, and 1.25 µL of each universal gyrB primer UPIE (5′ GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTYRA 3′) and UP2AR (3′ AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGYCAT 5′) [36]. The PCR conditions were slightly optimized for the KAPA2G Fast DNA Polymerase and were as follows: a 5-min initial denaturation step at 95 °C, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s, and a final elongation step of 2 min at 72 °C. PCR products were sent to Macrogen Europe for sequencing using the dideoxy-termination (Sanger) reaction with the primer gyrB UP1S (5′ GAAGTCATCATGACC-GTTCTGCA 3′) [37] to obtain sequences around 800 bp in length.
Detection of QS-active isolates
The methodological approach for the screening of AHL producers and the identification of AHL compounds by ultra high performance liquid chromatography (UHPLC) coupled with high-resolution tandem mass spectrometry UHPLC–HRMS/MS were strictly identical to those previously described in Girard et al. [38]. Briefly, two biosensors were used, Pseudomonas putida F117 (pKR-C12) for long acyl-side chain AHLs (C8–C18) and Escherichia coli MT102 (pJBA-132) for short acyl-side chain AHLs (C6–C10) [38]. Isolates were tested, in triplicate, on each biosensor immediately after isolation from the environmental samples and before mass spectrometry analysis, several months after isolation and preservation at −80 °C.
Genetic and genomic characterization of V. mediterranei isolates
The genetic diversity of V. mediterranei isolates was evaluated by Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) [39]. Briefly, genomic DNA from 253 isolates was obtained by cellular lysis (three rapid freeze/thaw cycles with liquid nitrogen/70 °C incubation), and DNA concentrations were estimated spectrophotometrically in order to dilute each lysate to 100 ng/µL. The ERIC-PCR reactions were performed using previously described protocols employing the ERIC1 and ERIC2 primers [40, 41]. PCR amplifications were performed with the GoTaq G2 Flexi polymerase (Promega), and fingerprinting profiles were visualized using a Typhoon FLA 9000 imager (GE Life Sciences). Gel image analysis was performed using the GelJ software [42]. For whole-genome sequencing, genomic DNA of two V. mediterranei isolates (17LN 0615E and 21LS 0615E) was obtained with a classical CTAB-β-mercaptoethanol extraction protocol and sent for DNA sequencing (Illumina MiSeq) by Mr. DNA (Shallowater, TX, USA), as described by Doberva et al. [43]. The genomes of the isolates 17LN 0615E and 21LS 0615E were assigned the following NCBI accession numbers NZ_NWTN00000000 and NZ_NWTO00000000, respectively. Genomes were automatically annotated using the RAST server [44].
Multivariate analyses
A preliminary PERMANOVA analysis [45] was used to test for significant differences among Vibrio assemblages and V. mediterranei APP between the two-size fractions (<50 µm and >50 µm). As the results indicated that there were no significant differences between the two-size fractions, the remainder of the analysis was then performed by pooling the counts from both fractions. The relationships between the G1 V. mediterranei assemblage (i.e., the abundance of each AHL production phenotypes over time, Square-root transformed and Bray–Curtis distance matrix) and all measured environmental variables (Normalization and Euclidean distance matrix) were investigated using a distance-based linear model [46]. In order to visualize if the temporal variations of G1 phenotypes co-occurred with specific phytoplankton taxa, we conducted a second step of analysis consisting of a principal component ordination (PCO) of G1 phenotypes using phytoplankton counts (square-root transformed and Bray–Curtis distance matrix) as predictor variables (non-parametric Spearman's rank correlation). All the multivariate analyses were performed using PRIMER v.7 and its add-on package PERMANOVA+ [45–47].
Results
Phenotypic and genetic temporal variation of V. mediterranei isolates
The partitioning of Vibrio isolates, between the water column (<50 µm) and plankton (>50 µm), was monitored during the spring and summer 2015/2016, and a total of 840 isolates were identified as Vibrio spp. We did not observe differences in Vibrio assemblages between the fractions over time (data not shown, PERMANOVA, p value = 0.904; the temporal variation of the total Vibrio spp. can be found in Figure S2). Isolates identification, based on gyrB gene sequences, from both size fractions, showed a seasonal dynamics of V. mediterranei isolates (Fig. 1) in particular for 2016, starting from 0 to 28% of total Vibrio isolates during spring and increasing gradually to up to 97% (Summer 2016). Interestingly, the phenotyping of these isolates, under identical culture and screening conditions, revealed a temporal variation of APPs in time. APPs varied, for example, from strains producing only long-chain AHLs (F117+/MT102−) to those producing long- and short-chain AHLs (F117+/MT102+) in 2015 or from a predominance of isolates exhibiting no production (F117−/MT102−) to an exclusive production of long-chain AHLs (F117+/MT102−) in 2016 (Fig. 1b).
Fig. 1.
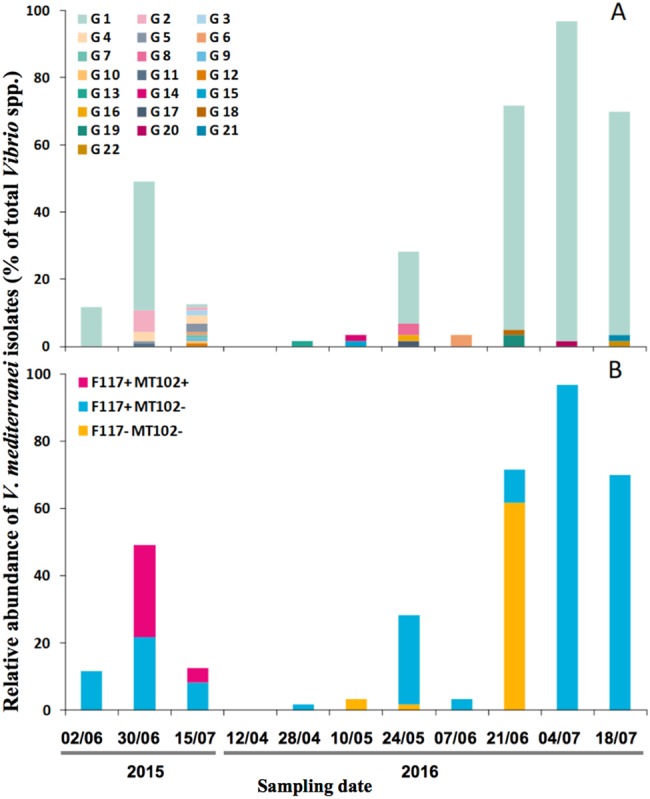
Temporal dynamics of V. mediterranei isolates relative to all Vibrio spp. isolates. a G1–G22 are genotypes based on ERIC-PCR. b APP: AHL production phenotypes, where F117+ and F117− are isolates detectable or not by the biosensor Pseudomonas putida (pKR-C12) and MT102+ and MT102− are detectable or not by the biosensor Escherichia coli (pJBA-132)
In order to determine if these phenotypic changes were due to a shift between different genotypes of V. mediterranei, we used ERIC-PCR [39] to genotype 253 V. mediterranei strains. A total of 22 genotypes were identified, G1–G22 (i.e., Figure 1a and Supplementary Figure 3) and remarkably, 83% of the V. mediterranei isolates, over the 2 years period, belonged to a single genotype, genotype 1 (G1). To further examine the genetic basis of phenotypic variations in V. mediterranei G1 isolates, we obtained a draft whole-genome sequence for two isolates with different phenotypes (17LN 0615E with F117+/MT102− phenotype and 21LS 0615E with F117+/MT102+ phenotype). The comparison between these genomes yielded an average nucleotide identity (ANI) value of 99.96%. Since, as previously proposed, two strains can be considered identical if they have a ANI value > 99.90% [48, 49], we consider that we observed a phenotypic plasticity of AHL production within a single strain of V. mediterranei. To summarize, we observed in 2016, the predominance of a single strain of V. mediterranei, up to 97% of the total cultivable Vibrio spp. at some time point, and showing different APPs over time.
AHL characterization of G1 V. mediterranei APP
To confirm that the observed APPs indeed corresponded to different AHL patterns, we analyzed the produced AHLs among two representative isolates of each phenotype (F117+/MT102−) and (F117+/MT102+) by UHPLC–HRMS/MS. AHL characterization was carried out according to the procedure described in Girard et al. [38]. Briefly, “anticipated” AHLs were identified by comparison with their corresponding standards (Table S8), while “unanticipated” AHLs without commercially available standards were identified based on their fragmentation patterns, predicted retention time, and molecular weight (i.e., Tables S6 and S7). A total of 13 AHLs were detected and 8 were common to the 4 isolates, namely OH-C10-HSL, oxo-C10-HSL, OH-C11-HSL, OH-C12-HSL, oxo-C12-HSL, C12-HSL, C13-HSL, and oxo-C13-HSL (Fig. 2 and Figure S4). Three short-chain AHLs, C6-HSL, oxo-C6-HSL, and oxo-C8-HSL were putatively identified as being responsible for the F117+/MT102+ phenotype. Interestingly, we observed that two long-chain AHLs, oxo-C11-HSL and C14-HSL, were produced by the (F117+/MT102−) isolates, but were not detected in the (F117+/MT102+) isolates. Summarizing, the AHL analysis confirmed that the different APPs detected by the biosensors are producing different AHLs patterns, with (F117+/MT102−) producing long acyl-side chains and (F117+/MT102+) producing long plus short acyl-side chains AHLs.
Fig. 2.
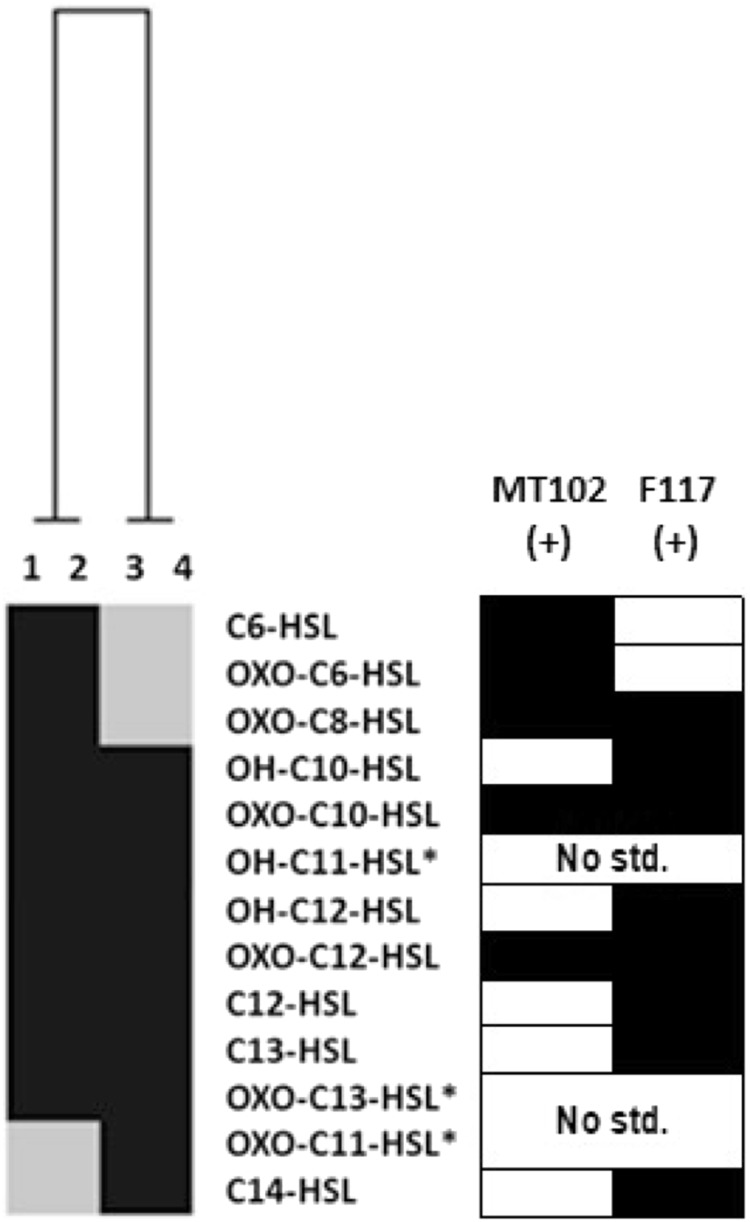
Heatmap of AHL diversity in V. mediterranei G1 isolates by UHPLC–HRMS/MS (Ward’s classification based on Jaccard index; [71]). 1 17 LN0615E, and 2 2 LS0615E corresponding to the F117+/MT102+ phenotype; 3 21 LS0615E and 4 9 LS0615E corresponding to the F117+/MT102− phenotype. (*) Unanticipated AHLs corresponding to any analyzed AHL standards. Gray squares represent the absence of AHL
Genetic basis of AHL production
One explanation for the different APPs with highly similar genomes could be that these differences were carried by different AHL synthase genes. Annotation and exploration of the two V. mediterranei G1 genomes for these genes revealed a single LuxI/LuxR QS system. However, as it has been shown that some Vibrio species can harbor two AHL production pathways (LuxI/R and LuxM/N), we also searched for other putative N-acetyltransferases in our two genomes. The comparison between the two strains (17 LN0615E and 21 LS0615E) revealed a 100% protein identity not only for the AHL synthase LuxI (accession numbers PRQ68803.1 and PCD89229.1, respectively), but also for all the 15 putative N-acetyltransferases (Table S9). Altogether these results strongly suggest that the differences observed in terms of AHL production patterns are not explained by different putative AHL synthases.
Factors affecting the dynamics of G1-AHL production phenotypes
To investigate the relationships between the dynamic of G1 APPs and the environmental variables, we conducted a distance-based linear modeling approach (DistLM). A total of seven variables explained 83.4% of the temporal variation of G1 APPs, and among those, phosphate concentration was the variable with the highest explanatory value (37.4 %) and ammonium concentration was the lowest at 4.5% (Fig. 3 and sequential tests, Table 2). Axis 1 (db RDA1) represented essentially a gradient in the abundance of G1, varying from high abundances at the left end to low abundances or no counts at the right end (July 2015 and April/May 2016). Samples with high counts of V. mediterranei G1 (except 2016/06/21) were associated with higher conductivity (an integrated measure of higher temperature and salinity), total dissolved nitrogen, and low phaeophytin concentrations, while an inverted trend was observed for samples with low counts or where V. mediterranei G1 was not retrieved. Axis 2 (db RDA2) reflected the distribution of different APPs. A peak of phosphates and nitrites were associated with an increase of non-AHL producers in the higher end of the axis. As the (F117+/MT102−) phenotype was dominant at dates with high counts of G1 isolates, the same parameters are linked to counts of the (F117+/MT102−) phenotype at the lower end of db RDA2 axis.
Fig. 3.
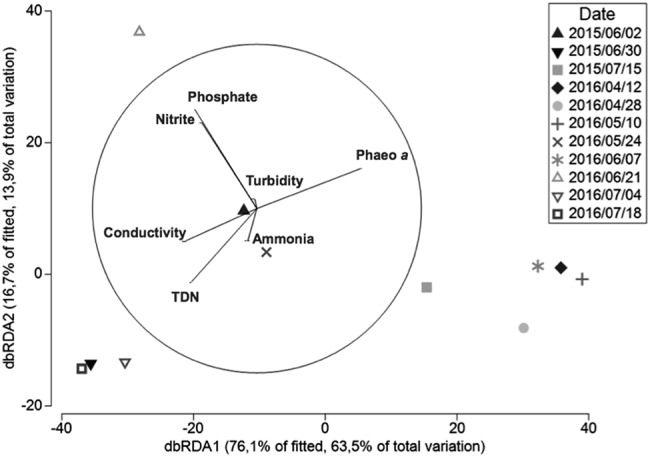
The dbRDA ordination representing the results of the DistLM analysis in 2D. DistLM is used to examine the relationship between the distribution and temporal dynamics of V. mediterranei G1 APP and all-measured environmental variables. Vectors represent predictor variables used for the construction of the constrained ordination (the dbRDA diagram), and the length of the vectors represents the explanatory percentage of each variable. The analysis is detailed in Table 2
Table 2.
Test statistics for distance-based linear model (DISTLM) analyses marginal and sequential tests based on ‘Forward’ procedure and AIC criteria of V. mediterranei G1 phenotypes abundance at the 11 sampling dates
| Variable | AIC | SS (trace) | Pseudo-F | P | Prop. | Cumul. | Res.df |
|---|---|---|---|---|---|---|---|
| Marginal tests | |||||||
| Phosphate | – | 11291.0 | 11.953 | 0.002 | 0.3741 | – | – |
| Conductivity | – | 9146.6 | 8.696 | 0.002 | 0.3030 | – | – |
| TDN | – | 2788.2 | 2.035 | 0.137 | 0.0924 | – | – |
| Nitrite | – | 4424.8 | 3.435 | 0.039 | 0.1466 | – | – |
| Phaeo a | – | 8055.3 | 7.280 | 0.006 | 0.2669 | – | – |
| Turbidity | – | 1490.9 | 1.039 | 0.322 | 0.0494 | – | – |
| Ammonia | – | 2821.9 | 2.062 | 0.138 | 0.0935 | – | – |
| Sequential tests | |||||||
| +Phosphate | 152.62 | 11291.0 | 11.953 | 0.001 | 0.3741 | 0.3741 | 20 |
| +Conductivity | 148.16 | 4810.1 | 6.490 | 0.008 | 0.1594 | 0.5335 | 19 |
| +TDN | 145.80 | 2530.8 | 3.944 | 0.014 | 0.0838 | 0.6173 | 18 |
| +Nitrite | 143.97 | 1846.0 | 3.234 | 0.057 | 0.0612 | 0.6785 | 17 |
| +Phaeo a | 141.94 | 1624.1 | 3.216 | 0.052 | 0.0538 | 0.7323 | 16 |
| +Turbidity | 138.66 | 1723.3 | 4.066 | 0.031 | 0.0571 | 0.7894 | 15 |
| +Ammonia | 135.41 | 1350.8 | 3.777 | 0.032 | 0.0448 | 0.8341 | 14 |
Marginal tests show how much variation each variable explains when considered alone, ignoring other variables. Sequential tests explain the cumulative variation attributed to each variable fitted to the model in the order specified, taking previous variables into account
TDN total dissolved nitrogen
A PCO was performed in order to visualize which phytoplankton species co-occurred with different APP (Fig. 4). As observed in db RDA1, Axis 1 (PCO1) reflected the gradient in G1 abundances among the sampling dates with the lowest counts of G1 in the right end (July 2015 and April/May 2016) and where picoeukaryotes, Cryptophyta, unidentified nanoeukaryotes, Dinophysis sp., and Prorocentrum micans were present in their highest abundances. In the left end of PCO axis 1 were samples with the highest G1 counts (June 2015 and June/July 2016) associated to the highest counts of Grammatophora sp. and Scrippsiella sp., and bacteria abundances. Axis 2 (PCO2) mostly separates dates based on the presence of V. mediterranei G1 AHL producers and nonproducers. While higher abundances of Gonyaulax sp. and Amylax sp. seemed to be associated to sampling dates with the highest counts of non-AHL producers (2016/06/21), Cyanobacteria and Pyrophacus sp. abundances were linked to the samples dominated by AHL producers.
Fig. 4.
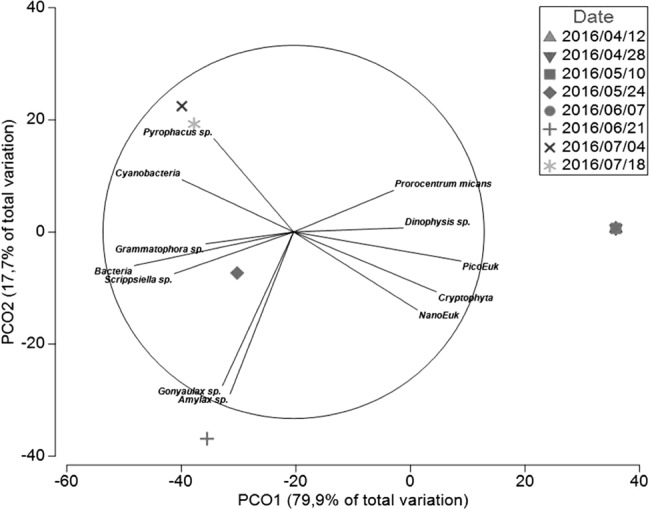
Principal co-ordinate plot (PCO) of V. mediterranei G1 phenotypes for the first and second principal co-ordinates using the Bray–Curtis similarity matrix. Phytoplankton and bacterial counts were used as predictor variables, are shown here only the variable with Pearson correlation > 0.45. The analysis is detailed in Supplementary Table 4 and the contribution of each variable in the construction of PCO axis (Pearson correlation) is detailed in Supplementary Table 5
Discussion
It is well known that the ability of Vibrio, as well of other Gram-negative bacteria to survive by making biofilms, infecting hosts, modifying their metabolism, or to enter into a VBNC state is modulated by QS [14, 24, 50, 51]. In addition to the fact that QS allows the bacteria to sense their number and co-ordinate their actions, the variability of auto-inducers production might offer an advantageous response to adapt their actions according to environmental fluctuations.
In order to survive in a changing environment, bacteria have evolved to set up various adaptive responses. Phenotypic plasticity is one of them and it reflects the direct influence of the environment on the development of individual phenotypes. In other words, bacteria can adapt to environmental selective pressures to maintain their own fitness. Phenotypic diversity of QS systems has already been described for different strains of a single Vibrio species [29–31, 52], and temporal changes in AHL production during the development of natural biofilms including Vibrio species have been shown [53]. However, the results of these earlier studies did not show clear evidence of QS phenotypic plasticity. Our study shows a clear short-term temporal variation in APPs among environmental isolates of a single Vibrio species (V. mediterranei) that were remarkably grouped into a single genotype (G1) based on gyrB sequences, ERIC-PCR, and genomic analysis. This change of AHL production patterns was temporally coherent and in some cases reached the near totality of Vibrio isolates in a sample. As the ANI value among the two sequenced genomes was very high (99.96%) but not identical, it is always possible that the residual 0.04% could be responsible for the differences of APPs by some unknown mechanism. Nonetheless, the luxI sequences, coding for the AHL synthase, as well as other putative N-acetyltransferases sequences were identical.
While it appears that we have different APPs belonging to a single genotype, the temporal sampling of Vibrio isolates in parallel to physicochemical parameters measurements allowed us to evaluate the variation in the environment, leading to the phenotypic plasticity of AHL production. However, as sampling was originally designed to evaluate the diversity of total Vibrio spp. (and not just that of V. mediterranei G1), the total number of studied strains are limited, and thus in the following discussion we only point to major trends and propose ideas to be tested by more targeted later studies.
Based on our DistLM analysis, the phosphate concentration appeared as the most important explanatory variable correlated with high abundances of non-AHL producers. This was particularly striking for the 21 June 2016, where the near totality of the strains belonged to the (F117−/MT102−) phenotype which coincided with a peak of phosphates likely associated with sediment resuspension. In order to explain this potential link between non-AHL producers and high phosphate concentration, we propose a possible implication of the polyphosphate kinase activity. The whole-genome analysis of the G1 isolates revealed multiple copies of the ppk gene, and several studies have demonstrated that polyphosphate kinase (ppk) enhances the ability of bacteria to survive under environmental stresses. Since this enzyme has a negative impact on AHLs production in P. fluorescens and is repressed at high phosphate concentrations [54–56], it is tempting to link the lack of AHL production to a repression of ppk by phosphate. As the peak of non-AHL producers, the 2016/06/21, occurred at the highest phosphate concentration in the 2-year sampling period, we hypothesize that a low expression of the ppk gene could have downregulated the expression of the AHL synthase gene [55].
The Salses-Leucate lagoon was also characterized by high cyanobacteria concentrations, mainly attributed to Synechococcus species, ranging between 3.86 × 105 to 2.88 × 108 cells/L, making them an important part of the microbial community. Over the study, the cyanobacteria counts were highly correlated to the (FF17+/MT102+) phenotype abundances (Pearson correlation 77.3%; p value = 0.002; June/July 2015), but also to the peak of the (F117+/MT102−) phenotype (July 2016, Fig. 4). In the past, Vibrio species have been previously associated to cyanobacterial blooms [57], especially by benefiting from their derived organic matter [58]. Mutualistic interactions were described between Synechococcus species and heterotrophic bacteria [59], and coculture experiments between Vibrio species and Synechococcus revealed a deleterious effect on the cyanobacteria physiology especially on the iron, phosphate, and nitrogen pathways [60]. In the other hand, since Synechococcus species harbor “orphan” luxR genes, possibly encoding for the receptor LuxR involved in AHL signal reception, and also shows the quorum quenching activity against Vibrio species, complex QS-based relationships might occur between these two genera [61–65]. Thereby, the temporal dynamics of different APPs can be related to changes in the cyanobacterial assemblages, switching between commensal/consumers, probably benefiting from the cyanobacterial dissolved organic matter and behaving as competitors for inorganic nutrients, such as phosphate, nitrogen, and iron. Understanding the in situ interactions between APPs of V. mediterranei and cyanobacterial assemblages remains an interesting subject for future studies.
Overall, our results indicate a phenotypic plasticity of AHL production among isolates of a single genotype and converge toward the fact that our strains are harboring a single AHL synthase gene (luxI). The underlying physiological mechanisms leading to this phenotypic plasticity remains to be described. However, it is well known that AHL synthases are catalyzing the reaction between S-adenosylmethionine and acyl-ACPs to produce AHLs [66] and that the available acyl-ACP pools in bacteria may be susceptible to metabolic changes [27]. Considering that it has been recognized that AHL production can be altered though the modulation of the fatty acid metabolic pathway, and that the variation of APPs was explained at 83.4% by the environmental variables, it seems likely that fluctuations in the environment can modify the metabolism of bacteria and thus the nature of the produced AHLs [27, 28]. However, the fact that (1) the near totality of the isolates were non-AHL producers at one time point and AHL producers in the subsequent time point, and (2) that these phenotypes were observed after two rounds of isolation on rich medium raises the intriguing possibility that epigenetic regulation might also be at play. It is well-known that the regulation of phenotypic variation is not always linked to changes in DNA sequence and can be epigenetic in nature [67]. Interestingly, Kurz et al. [68] have shown that epigenetic mechanisms can be involved in the regulation of AHL-based QS system. We hypothesize that V. mediterranei strains could keep a memory of the social traits expressed in the environment. However, as this is the first report on QS mechanisms of V. mediterranei, a consequent work on the description of the QS pathways and QS-associated phenotypes still need to be achieved to confirm or deny this possible epigenetic control. Regardless of the mechanisms leading to the observed phenotypic variation, phenotypic plasticity should confer an increased fitness, as heterogeneous populations should be better to adapt to rapid changes in the environment [69]. The fact that the studied phenotypically plastic V. mediterranei populations thrive in a shallow lagoon subjected to relatively rapid variations in physical and biogeochemical conditions supports this idea. Understanding the possible mechanisms of regulation of AHL-based QS in V. mediterranei remains a fascinating avenue for further studies.
Finally, our work highlights the phenotypic plasticity of AHL production among environmental Vibrio isolates. These results were obtained on strains after isolation and culture on rich media, which underlines the fact that Vibrio isolates could retain a memory of in situ AHL production status. Considering the fact that the quantification of AHL synthases expression by RTqPCR is still difficult due to a very low sequence homology among vibrios [70], the study of AHL-meditated QS by a biosensor-based approach seems an interesting alternative to estimate whether or not AHL synthesis occurred in the environment.
Conclusion
QS is a well-studied social trait among Vibrio species, however, despite the current knowledge obtained by in vitro studies on few species, there is a lack of studies regarding QS in complex ecological communities. Our study provides a first investigation on the seasonal dynamics of the AHL-producing Vibrio in a changing coastal environment and open new perspectives for outgoing studies regarding QS mechanisms in natural habitats and a possible epigenetic regulation of QS phenotypic plasticity.
Electronic supplementary material
Acknowledgements
We would like to thank Professor Marti Anderson for her support concerning the PRIMER 7 statistical analyses. We are thankful to Christophe Canal for the sampling campaigns at Leucate lagoon and for the physicochemical analysis performed in water samples. We are also grateful to Jocelyne Caparros for inorganic nutrient analysis, Nicole Batailler for her technical assistance in some steps of the experimental work, and the Bio2Mar platform for providing access to the analytical chemistry and molecular biology equipment. Finally, we would like to thank Laurence Fonbonne from the “Syndicat RIVAGE” for her help and for giving us the access to the lagoon and Nyree West for the careful reading of the final manuscript. This work was supported by a Ph.D. grant from doctoral school ED227 Sciences de la Nature et de l’Homme: évolution et écologie associating Sorbonne Université and Museum National d’Histoire Naturelle (MNHN), France.
Funding
This work was supported by a Ph.D. grant from the UPMC Doctoral School 227, Natural and Human Sciences: Ecology and Evolution of the Sorbonne University and the Natural History Museum of Paris, France.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0260-4) contains supplementary material, which is available to authorized users.
References
- 1.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal AA. Phenotypic plasticity in the interactions and evolution of species. Science. 2001;294:321–6. doi: 10.1126/science.1060701. [DOI] [PubMed] [Google Scholar]
- 3.Chevin LM, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: Towards a predictive theory. PLoS Biol. 2010;8:e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West-Eberhard MJ. Developmental plasticity and evolution. Oxford; New York, NY: Oxford University Press; 2003. [Google Scholar]
- 5.Takemura, AF, Chien, DM, Polz, MF. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 2014;11:5–38. [DOI] [PMC free article] [PubMed]
- 6.Chimetto Tonon LA, Silva BS, de O, Moreira APB, Valle C, Alves N, et al. Diversity and ecological structure of vibrios in benthic and pelagic habitats along a latitudinal gradient in the Southwest Atlantic Ocean. PeerJ. 2015;3:e741. doi: 10.7717/peerj.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, L, Mendis, N, Trigui, H, Oliver, JD, Faucher, SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014;5:258. [DOI] [PMC free article] [PubMed]
- 8.Vezzulli L, Pezzati E, Stauder M, Stagnaro L, Venier P, Pruzzo C. Aquatic ecology of the oyster pathogens Vibrio splendidus and Vibrio aestuarianus. Environ Microbiol. 2015;17:1065–80. doi: 10.1111/1462-2920.12484. [DOI] [PubMed] [Google Scholar]
- 9.Whitesides MD, Oliver JD. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl Environ Microbiol. 1997;63:1002–5. doi: 10.1128/aem.63.3.1002-1005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Haim Y. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol. 2003;53:309–15. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- 11.Heidelberg JF, Heidelberg KB, Colwell RR. Bacteria of the γ-subclass proteobacteria associated with zooplankton in Chesapeake bay. Appl Environ Microbiol. 2002;68:5498–507. doi: 10.1128/AEM.68.11.5498-5507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soto W, Gutierrez J, Remmenga MD, Nishiguchi MK. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microb Ecol. 2009;57:140–50. doi: 10.1007/s00248-008-9412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendling CC, Wegner KM. Adaptation to enemy shifts: rapid resistance evolution to local Vibrio spp. in invasive Pacific oysters. Proc R Soc B Biol Sci. 2015;282:20142244. doi: 10.1098/rspb.2014.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goo E, An JH, Kang Y, Hwang I. Control of bacterial metabolism by Quorum sensing. Trends Microbiol. 2015;23:567–76. doi: 10.1016/j.tim.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 15.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu Rev Ecol Evol Syst. 2007;38:53–77. doi: 10.1146/annurev.ecolsys.38.091206.095740. [DOI] [Google Scholar]
- 16.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 17.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–86. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 19.Schwartzman Julia A., Ruby Edward G. A conserved chemical dialog of mutualism: lessons from squid and vibrio. Microbes and Infection. 2016;18(1):1–10. doi: 10.1016/j.micinf.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholera. Mol Microbiol. 2003;50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 21.Ha C, Kim SK, Lee MN, Lee JH. Quorum sensing-dependent metalloprotease VvpE is important in the virulence of Vibrio vulnificus to invertebrates. Microb Pathog. 2014;71–72:8–14. doi: 10.1016/j.micpath.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Natrah FMI, Ruwandeepika HAD, Pawar S, Karunasagar I, Sorgeloos P, Bossier P, et al. Regulation of virulence factors by Quorum sensing in Vibrio harveyi. Vet Microbiol. 2011;154:124–9. doi: 10.1016/j.vetmic.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci. 2002;99:3129–34. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayrapetyan M, Williams TC, Oliver JD. Interspecific Quorum sensing mediates the resuscitation of viable but nonculturable vibrios. Appl Environ Microbiol. 2014;80:2478–83. doi: 10.1128/AEM.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bari SMN, Roky MK, Mohiuddin M, Kamruzzaman M, Mekalanos JJ, Faruque SM. Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc Natl Acad Sci. 2013;110:9926–31. doi: 10.1073/pnas.1307697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt TG, Fuqua C. What’s in a name? The semantics of Quorum sensing. Trends Microbiol. 2010;18:383–7. doi: 10.1016/j.tim.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould TA, Herman J, Krank J, Murphy RC, Churchill MEA. Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J Bacteriol. 2006;188:773–83. doi: 10.1128/JB.188.2.773-783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang TT, Sullivan SA, Cusick JK, Schweizer HP. Beta-ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiol Read Engl. 2002;148:3849–56. doi: 10.1099/00221287-148-12-3849. [DOI] [PubMed] [Google Scholar]
- 29.Buchholtz C, Nielsen KF, Milton DL, Larsen JL, Gram L. Profiling of acylated homoserine lactones of Vibrio anguillarum in vitro and in vivo: influence of growth conditions and serotype. Syst Appl Microbiol. 2006;29:433–45. doi: 10.1016/j.syapm.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 30.García-Aljaro C, Vargas-Cespedes GJ, Blanch AR. Detection of acylated homoserine lactones produced by Vibrio spp. and related species isolated from water and aquatic organisms. J Appl Microbiol. 2012;112:383–9. doi: 10.1111/j.1365-2672.2011.05199.x. [DOI] [PubMed] [Google Scholar]
- 31.Purohit AA, Johansen JA, Hansen H, Leiros HKS, Kashulin A, Karlsen C, et al. Presence of acyl-homoserine lactones in 57 members of the Vibrionaceae family. J Appl Microbiol. 2013;115:835–47. doi: 10.1111/jam.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen B, Nielsen K, Machado H, Melchiorsen J, Gram L, Sonnenschein E. Global and phylogenetic distribution of Quorum sensing signals, acyl homoserine lactones, in the family of Vibrionaceae. Mar Drugs. 2014;12:5527–46. doi: 10.3390/md12115527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlson, B, Cusack, C, Bresnan, E. Microscopic and molecular methods for quantitative phytoplankton analysis. IOC Manuals and Guides. 2010;55.
- 34.Marie D, Rigaut-Jalabert F, Vaulot D. An improved protocol for flow cytometry analysis of phytoplankton cultures and natural samples: an improved protocol for flow cytometry analysis. Cytometry A. 2014;85:962–8. doi: 10.1002/cyto.a.22517. [DOI] [PubMed] [Google Scholar]
- 35.Pfeffer C, Oliver JD. A comparison of thiosulphate-citrate-bile salts-sucrose (TCBS) agar and thiosulphate-chloride-iodide (TCI) agar for the isolation of Vibrio species from estuarine environments. Lett Appl Microbiol. 2003;36:150–1. doi: 10.1046/j.1472-765X.2003.01280.x. [DOI] [PubMed] [Google Scholar]
- 36.Le Roux F, Gay M, Lambert C, Nicolas JL, Gouy M, Berthe FCJ. Phylogenetic study and identification of Vibrio splendidus-related strains based on gyrB gene sequences. Dis Aquat Organ. 2004;58:143. doi: 10.3354/dao058143. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61:1104–9. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girard L, Blanchet E, Intertaglia L, Baudart J, Stien D, Suzuki M, et al. Characterization of N-acyl homoserine lactones in Vibrio tasmaniensis LGP32 by a biosensor-based UHPLC-HRMS/MS method. Sensors. 2017;17:906. doi: 10.3390/s17040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera IG, Chowdhury MA, Huq A, Jacobs D, Martins MT, Colwell RR. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl Environ Microbiol. 1995;61:2898–904. doi: 10.1128/aem.61.8.2898-2904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hulton CS, Higgins CF, Sharp PM. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium, and other enterobacteria. Mol Microbiol. 1991;5:825–34. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 41.Khan AA, McCarthy S, Wang RF, Cerniglia CE. Characterization of United States outbreak isolates of Vibrio parahaemolyticus using enterobacterial repetitive intergenic consensus (ERIC) PCR and development of a rapid PCR method for detection of O3: K6 isolates. FEMS Microbiol Lett. 2002;206:209–14. doi: 10.1111/j.1574-6968.2002.tb11011.x. [DOI] [PubMed] [Google Scholar]
- 42.Heras, J, Domínguez, C, Mata, E, Pascual, V, Lozano, C, Torres, C et al. GelJ—a tool for analyzing DNA fingerprint gel images. BMC Bioinform 2012;16:270. [DOI] [PMC free article] [PubMed]
- 43.Doberva M, Sanchez-Ferandin S, Ferandin Y, Intertaglia L, Joux F, Lebaron P, et al. Genome sequence of Maribius sp. strain MOLA 401, a marine Roseobacter with a Quorum-sensing cell-dependent physiology. Genome Announc. 2014;2:e00997-14. doi: 10.1128/genomeA.00997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson MJ. Permutation tests for univariate or multivariate analysis of variance and regression. Can J Fish Aquat Sci. 2001;58:626–39. doi: 10.1139/f01-004. [DOI] [Google Scholar]
- 46.McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2. [DOI] [Google Scholar]
- 47.Anderson MJ. A new method for non-parametric multivariate analysis of variance: non-parametric MANOVA for ecology. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 48.Olm, MR, Brown, CT, Brooks, B, Firek, B, Baker, R, Burstein, D. et al. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 2017;gr.213256.116. [DOI] [PMC free article] [PubMed]
- 49.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative sequencing program group. Henderson DK, Palmore TN, Segre JA. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bondí R, Messina M, De Fino I, Bragonzi A, Rampioni G, Leoni L. Affecting Pseudomonas aeruginosa phenotypic plasticity by Quorum sensing dysregulation hampers pathogenicity in murine chronic lung infection. PLoS ONE. 2014;9:e112105. doi: 10.1371/journal.pone.0112105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, et al. The mechanical world of bacteria. Cell. 2015;161:988–97. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joelsson A, Liu Z, Zhu J. Genetic and phenotypic diversity of Quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect Immun. 2006;74:1141–7. doi: 10.1128/IAI.74.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang YL, Ki JS, Lee OO, Qian PY. Evidence for the dynamics of acyl homoserine lactone and AHL-producing bacteria during subtidal biofilm formation. ISME J. 2009;3:296–304. doi: 10.1038/ismej.2008.105. [DOI] [PubMed] [Google Scholar]
- 54.Jahid IK, Silva AJ, Benitez JA. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl Environ Microbiol. 2006;72:7043–9. doi: 10.1128/AEM.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rashid M. H., Rumbaugh K., Passador L., Davies D. G., Hamood A. N., Iglewski B. H., Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2000;97(17):9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silby MW, Nicoll JS, Levy SB. Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile. Soil Appl Environ Microbiol. 2009;75:3872–81. doi: 10.1128/AEM.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berg KA, Lyra C, Sivonen K, Paulin L, Suomalainen S, Tuomi P, et al. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 2009;3:314–25. doi: 10.1038/ismej.2008.110. [DOI] [PubMed] [Google Scholar]
- 58.Eiler A, Gonzalez-Rey C, Allen S, Bertilsson S. Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water: cyanobacterial DOM, temperature and Vibrio growth. FEMS Microbiol Ecol. 2007;60:411–8. doi: 10.1111/j.1574-6941.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi S, Itoh K, Suyama K. Growth of the cyanobacterium Synechococcus leopoliensis CCAP1405/1 on agar media in the presence of heterotrophic bacteria. Microbes Environ. 2011;26:120–7. doi: 10.1264/jsme2.ME10193. [DOI] [PubMed] [Google Scholar]
- 60.Tai V, Paulsen IT, Phillippy K, Johnson DA, Palenik B. Whole-genome microarray analyses of Synechococcus—Vibrio interactions. Environ Microbiol. 2009;11:2698–709. doi: 10.1111/j.1462-2920.2009.01997.x. [DOI] [PubMed] [Google Scholar]
- 61.Honda T, Liang Y, Arai D, Ito Y, Yoshino T, Tanaka T. Draft genome sequence of marine cyanobacterium Synechococcus sp. strain NKBG042902, which harbors a homogeneous plasmid available for metabolic engineering. Genome Announc. 2014;2:e00704-14. doi: 10.1128/genomeA.00704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marsan D, Wommack KE, Ravel J, Chen F. Draft genome sequence of Synechococcus sp. strain CB0101, isolated from the chesapeake bay estuary. Genome Announc. 2014;2:e01111-13. doi: 10.1128/genomeA.01111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santhakumari S, Kannappan A, Pandian SK, Thajuddin N, Rajendran RB, Ravi AV. Inhibitory effect of marine cyanobacterial extract on biofilm formation and virulence factor production of bacterial pathogens causing vibriosis in aquaculture. J Appl Phycol. 2016;28:313–24. doi: 10.1007/s10811-015-0554-0. [DOI] [Google Scholar]
- 64.Shimura Y, Hirose Y, Misawa N, Wakazuki S, Fujisawa T, Nakamura Y, et al. Complete genome sequence of a coastal cyanobacterium, Synechococcus sp. strain NIES-970. Genome Announc. 2017;5:e00139–17. doi: 10.1128/genomeA.00139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshino T, Honda T, Tanaka M, Tanaka T. Draft genome sequence of marine cyanobacterium Synechococcus sp. Strain NKBG15041c. Genome Announc. 2013;1:e00954-13–e00954-13. doi: 10.1128/genomeA.00954-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–58. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 67.Smits WK, Kuipers OP, Veening JW. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol. 2006;4:259–71. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 68.Kurz V, Nelson EM, Perry N, Timp W, Timp G. Epigenetic memory emerging from integrated transcription bursts. Biophys J. 2013;105:1526–32. doi: 10.1016/j.bpj.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feinberg A. P., Irizarry R. A. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proceedings of the National Academy of Sciences. 2009;107(suppl_1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tait K, Hutchison Z, Thompson FL, Munn CB. Quorum sensing signal production and inhibition by coral-associated vibrios. Environ Microbiol Rep. 2010;2:145–50. doi: 10.1111/j.1758-2229.2009.00122.x. [DOI] [PubMed] [Google Scholar]
- 71.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 72.Al-Bairuty GA, Shaw BJ, Handy RD, Henry TB. Histopathological effects of waterborne copper nanoparticles and copper sulphate on the organs of rainbow trout (Oncorhynchus mykiss) Aquat Toxicol. 2013;126:104–15. doi: 10.1016/j.aquatox.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 73.Vouvé F, Buscail R, Aubert D, Labadie P, Chevreuil M, Canal C, et al. Bages-Sigean and Canet-St Nazaire lagoons (France): physico-chemical characteristics and contaminant concentrations (Cu, Cd, PCBs, and PBDEs) as environmental quality of water and sediment. Environ Sci Pollut Res. 2014;21:3005–20. doi: 10.1007/s11356-013-2229-1. [DOI] [PubMed] [Google Scholar]
- 74.Aryal DR, Geissen V, Ponce-Mendoza A, Ramos-Reyes RR, Becker M. Water quality under intensive banana production and extensive pastureland in tropical Mexico. J Plant Nutr Soil Sci. 2012;175:553–9. doi: 10.1002/jpln.201100117. [DOI] [Google Scholar]
- 75.Romero E, Peters F, Guadayol Ograve. The interplay between short-term, mild physicochemical forcing and plankton dynamics in a coastal area. Limnol Oceanogr. 2013;58:903–20. doi: 10.4319/lo.2013.58.3.0903. [DOI] [Google Scholar]
- 76.Rembauville M, Blain S, Caparros J, Salter I. Particulate matter stoichiometry driven by microplankton community structure in summer in the Indian sector of the Southern Ocean: particulate matter stoichiometry. Limnol Oceanogr. 2016;61:1301–21. doi: 10.1002/lno.10291. [DOI] [Google Scholar]
- 77.Lorenzen CJ. A method for the continuous measurement of in vivo chlorophyll concentration. Deep Sea Res Oceanogr Abstr. 1966;13:223–7. doi: 10.1016/0011-7471(66)91102-8. [DOI] [Google Scholar]
- 78.Lorenzen CJ. Determination of cholophyll and pheo-pigments: spectrophotometric equations. Limnol Oceanogr. 1967;12:343–6. doi: 10.4319/lo.1967.12.2.0343. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


