Abstract
In the surface ocean, light fuels photosynthetic carbon fixation of phytoplankton, playing a critical role in ecosystem processes including carbon export to the deep sea. In oligotrophic oceans, diatom–diazotroph associations (DDAs) play a keystone role in ecosystem function because diazotrophs can provide otherwise scarce biologically available nitrogen to the diatom host, fueling growth and subsequent carbon sequestration. Despite their importance, relatively little is known about the nature of these associations in situ. Here we used metatranscriptomic sequencing of surface samples from the North Pacific Subtropical Gyre (NPSG) to reconstruct patterns of gene expression for the diazotrophic symbiont Richelia and we examined how these patterns were integrated with those of the diatom host over day–night transitions. Richelia exhibited significant diel signals for genes related to photosynthesis, N2 fixation, and resource acquisition, among other processes. N2 fixation genes were significantly co-expressed with host nitrogen uptake and metabolism, as well as potential genes involved in carbon transport, which may underpin the exchange of nitrogen and carbon within this association. Patterns of expression suggested cell division was integrated between the host and symbiont across the diel cycle. Collectively these data suggest that symbiont–host physiological ecology is strongly interconnected in the NPSG.
Subject terms: Water microbiology, Microbial ecology
Introduction
Oligotrophic waters comprise an estimated 30% of the global surface ocean [1]. However, these seeming deserts of primary production are estimated to contribute up to 90% of photosynthetic carbon fixation in the marine environment, of which <15% is exported out of the euphotic zone [2]. In the North Pacific Subtropical Gyre (NPSG), much of this carbon export has been linked to annual summer increases in biomass of diatom–diazotroph associations (DDAs) [2, 3]. Composition of NPSG DDA blooms varies by year, but blooms are often dominated by Rhizosolenia sp. with the symbiotic cyanobacterium Richelia intracellularis [3–5]. Fixed N2 from Richelia is transferred to the host and is estimated to provide much of the diatom nitrogen (N) quota in similar associations such as with Hemiaulus [6]. Estimates of daytime N2 fixation rates by Richelia associated with Rhizosolenia are near 1 pmol heterocyst−1 h−1 [7] and have been shown to provide substantial inputs of new N into the systems where they occur [8, 9].
In oligotrophic waters, access to fixed N2 provided by a symbiotic partner likely provides distinct advantages to the diatom host. Currently, only a few genera of diatoms are known to form symbiotic relationships with diazotrophs. Further, Rhizosolenia is known to exist without Richelia [10] suggesting this partnership is facultative. Although the transfer of N from symbionts to diatom hosts has been demonstrated [6, 11], there are no long-term cultures of this symbiosis to study and our current understanding of this association in situ is limited. Fundamental questions remain about how the symbiosis is maintained, how resources beyond N might be modulated within the association, and how cell division might be synchronized between host and symbiont in order to propagate this facultative association. To better understand this keystone symbiosis, we reconstructed patterns of gene expression for the diazotroph Richelia and examined how these patterns were co-expressed with those of the diatom host over day–night transitions in the NPSG.
Methods
Sample collection
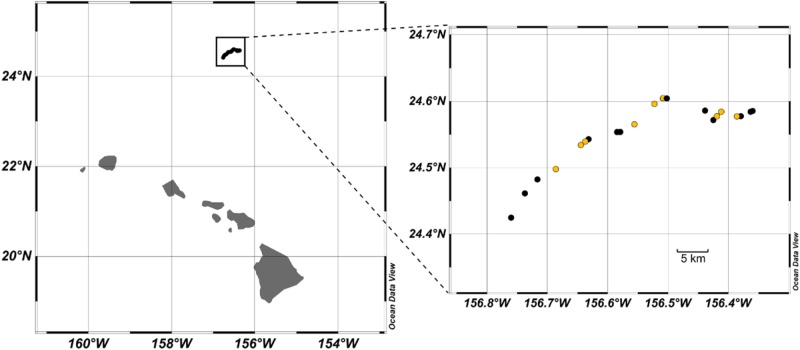
Seawater for diel in situ metatranscriptomes was collected aboard the R/V Kilo Moana as part of the 2015 Simons Collaboration on Ocean Processes and Ecology (SCOPE) research collaboration HOE-Legacy II Research Cruise near Station ALOHA (Fig. 1; http://scope.soest.hawaii.edu/data/hoelegacy/). During this cruise, samples were collected from 15 m depth within an anticyclonic eddy tracking a Lagrangian drifter. Detailed information regarding the sampling regime on the cruise has been described previously [12]. Hydrocasts for sampling were performed using a 24-bottle conductivity–temperature–depth (CTD) rosette sampler at a depth of 15 m every 4 h from 10 p.m. (local time) July 26, 2015 to 6 a.m. (local time) on July 30, 2015 for a total of 21 time points (Fig. 1). Water was collected in acid-washed 20 L carboys and seawater was prescreened through 200 µm nylon mesh and then filtered onto two 5 µm polycarbonate filters (47 mm) by way of a peristaltic pump, passing ~10 L through each filter. This size fraction was targeted to sample large eukaryotic phytoplankton and associated particle-attached and symbiotic prokaryotes. Samples collected at night were processed in the laboratory under red light. Filters were placed into cryovials and stored in liquid nitrogen until extraction. Total length of time from collection to placement into liquid nitrogen did not exceed 40 min.
Fig. 1.
Map of Lagrangian sampling stations on a July 2015 cruise in the North Pacific Subtropical Gyre. Left panel indicates location relative to the Hawaiian Islands and the right panel displays an expanded view, with stations (21 in total spanning 4.5 days) sampled during night hours in black and stations sampled during daylight hours in yellow
RNA extraction and sequencing
Total RNA was extracted from individual filters using a Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) modifying the lysis step with the addition of ~500 μL of 0.5 mm zirconia/silica beads (Biospec, Bartlesville, OK, USA). Briefly, lysis buffer and beads were added to each filter set (n = 2) for each time point and vortexed for 1 min, placed on ice for 30 s, and vortexed again for 1 min. Lysate from each filter set (n = 2) was removed with a pipette and pooled into a single 5 mL microcentrifuge tube. The rest of the Qiagen RNeasy Mini Kit protocol was then followed according to the manufacturer’s instructions, adjusting volumes accordingly and incorporating the on-column DNase digestion step, using a Qiagen RNase-free DNase kit. Resulting total RNA was eluted with RNase-free water and then purified and concentrated with a Qiagen RNeasy MinElute kit according to the manufacturer’s instructions. Quantity and quality of extracted total RNA was assessed on an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
To identify both eukaryotic and prokaryotic gene expression signals, extracted samples were split into two equal volumes of total RNA where one volume was sequenced after a polyA pull-down step (Illumina Truseq library preparation kit; Illumina, San Diego, CA, USA) to enrich for eukaryotic mRNA (herein referred to as “selected reads”) and one volume was sequenced directly (herein referred to as “unselected reads”). Illumina Truseq libraries were sequenced with an Illumina HiSeq 2000 at the JP Sulzberger Genome Center (CUGC) of Columbia University following Center protocols. Samples targeting eukaryotic signals (selected) were sequenced to produce 90 million 100 bp, paired-end reads while samples targeting prokaryotic signals (unselected) were sequenced to produce 60 million 100 bp, paired-end reads. These environmental sequence data are deposited in the Sequence Read Archive through the National Center for Biotechnology Information under accession no. SRP136571. Raw sequence quality was visualized using FastQC [13] and then cleaned and trimmed using Trimmomatic [14] version 0.27 (paired-end mode, 4 bp-wide sliding window for quality below 15, minimum length of 25 bp).
Mapping and periodic expression analysis
Reads were mapped to a reference database after Alexander et al. [15]. The transcriptome Rhizosolenia setigera CCMP 1694 (MMETSP0789; https://www.imicrobe.us/#/projects/104) was used as a reference to elucidate transcriptional patterns of selected reads for Rhizosolenia sp. and combined with transcriptomes of Chaetoceros sp. (MMETSP1336, 200, 149, 751, 752, 753, 754, 1429, 1447, 90, 91, 92, 717, and 718), a species commonly associated with diazotroph endosymbionts [16], to create a dataset large enough to minimize false positive read alignments. For Richelia sp. analyses, unselected reads were mapped to the genome of Richelia intracellularis RC01 (https://www.patricbrc.org/view/Genome/1164990.3), which was isolated from Rhizosolenia clevei via flow cytometry sorting and included both vegetative cells as well as heterocysts (http://www.ebi.ac.uk/ena/data/view/GCA_000613065; [17]). As with Rhizosolenia, a reference database of common diazotrophs was created from genomes of Richelia intracellularis RC01, Trichodesmium erythraeum IMS101, Calothrix rhizosoleniae SC01, Crocosphaera watsonii WH8501, UCYN-A1, and UCYN-A2. Mapping of both selected and unselected reads was conducted with the Burrows–Wheeler Aligner (BWA-MEM, parameters –k 10 –aM; [18]) and counted using the HTSeq 0.6.1 package (options –a 0, --m intersection-strict, -s no; [19]). Sequencing results can be found in Table S1. Alignment of trimmed reads to diatom and diazotroph reference databases resulted in a total of 4,229 (55% of total, Tables S2, S3) Richelia and 21,705 (92% of total, Table S2, S3) Rhizosolenia features (gene or contig) that were detected at sufficient coverage (≥3 read counts for both species) for subsequent analyses.
Annotations for Rhizosolenia were taken from the MMETSP assignments using the most abundant annotation hit for each contig (Pfam) while Richelia annotations were obtained from Patric assignments (https://www.patricbrc.org/view/Genome/1164990.3). Kyoto Encyclopedia of Genes and Genomes (KEGG) biochemical pathways for each genetic feature (contig or gene) were identified with KEGG Automatic Annotation Server (KAAS) using the partial genome single-directional best-hit method [20, 21]. Features (gene or contig) receiving KEGG assignments were binned by KEGG module. Analysis of periodic expression of count data was performed using the R package RAIN [22] by first filtering low abundance reads (see above) after Wilson et al. [12]. and then normalizing with the DESeq2 “varianceStabilizingTransformation” command [23]. Resulting p-values from this analysis were corrected for multiple testing using the false-discovery rate method [24], with corrected p-values ≤ 0.05 considered to have significant diel periodicity. Peak times were calculated with a harmonic regression, fitting the expression data to a sine curve, as has been used in several recent studies [25, 26]. These estimates of peak expression time were congruent with the normalized expression patterns across the time series and were used as a means of estimating at what time the highest relative expression was occurring.
Network analysis
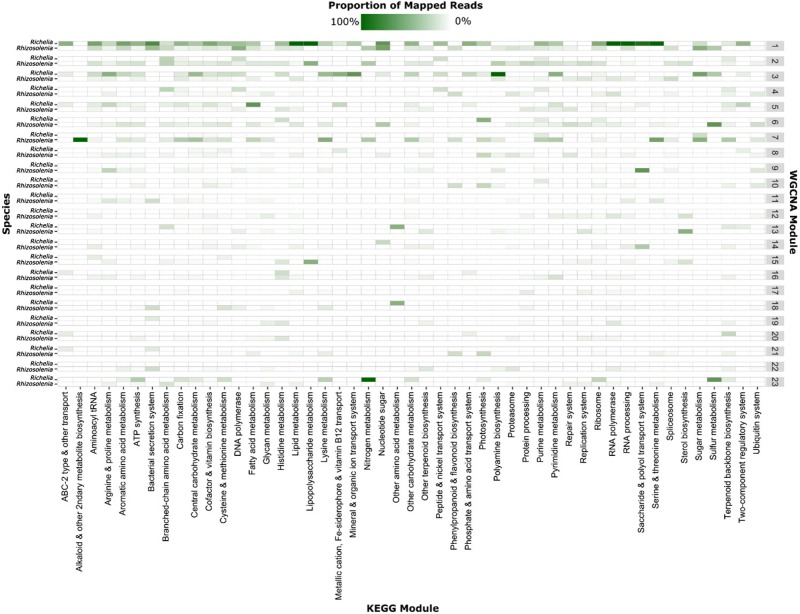
To examine co-expression patterns between Rhizosolenia and Richelia, a weighted gene co-expression network analysis was conducted using the R package WGCNA [27] on normalized counts (DESeq2 “varianceStabilizingTransformation” command; [23]) using methods described previously [25]. WGCNA uses a correlation network to identify clusters (modules) of highly correlated genes, which can have a co-expression pattern that is either synchronous or orthogonal. This type of analysis has been used in previous studies comparing host–symbiont dynamics [28–30]. Briefly, a soft-threshold of 7 was chosen based upon a scale-free network topology test (Figure S1) and transcriptional modules were identified using the “blockwiseModules” command (minModuleSize = 30, mergeCutHeight = 0.25). The TOMsimilarityFromExpr command was used to calculate topological overlap between transcripts. To evaluate variability in core metabolic processes for both Rhizosolenia and Richelia in each WGCNA module, normalized read counts for features (gene or contig) in each species with KEGG assignments were summed by KEGG module for WGCNA modules containing >200 genes or contigs (nodes). The relative abundance by species was then determined to view species-specific distribution of KEGG modules within each WGCNA module.
Results and discussion
The oscillation from day to night is one of the strongest and most predictable perturbations imposed on organisms, selecting for pronounced diel patterns in both metabolism and growth [31]. Although it is expected that the diazotroph Richelia exhibits daily oscillations in N2 fixation rates based upon diel patterns of nifH gene transcript copy abundance [32], little is known about diel patterns across other aspects of metabolism for Richelia and its diatom host. Here we mined high frequency (21 time points over 4.5 days) environmental metatranscriptomes for signals of a common bloom forming DDA Rhizosolenia–Richelia in order to investigate the diel rhythms in Richelia and identify significantly co-expressed transcripts between the diazotroph symbiont and its diatom host.
The Rhizosolenia–Richelia DDA has been observed in a wide variety of environments and in the NPSG it is often the dominant organism in summer phytoplankton blooms [3]. During the sampling period, the presence of Richelia (associated with Rhizosolenia) was observed in micrographs taken from seawater filtered from a shipboard uncontaminated seawater line [33]. The presence of the DDA was further confirmed with quantitative PCR (qPCR) of nifH gene copy abundance [12], being the second most abundant diazotroph at 15 m after UCYN-B (Figure S2). Although Richelia was not the most abundant diazotroph, these results are consistent with the importance of the Rhizosolenia–Richelia DDA in the NPSG during the summer export period [2, 3].
Patterns of diel gene expression in Richelia from the NPSG
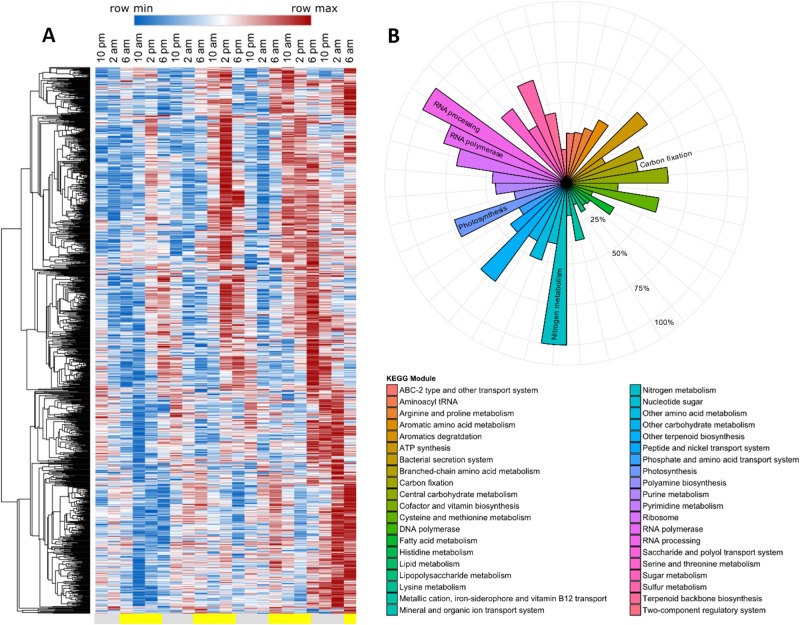
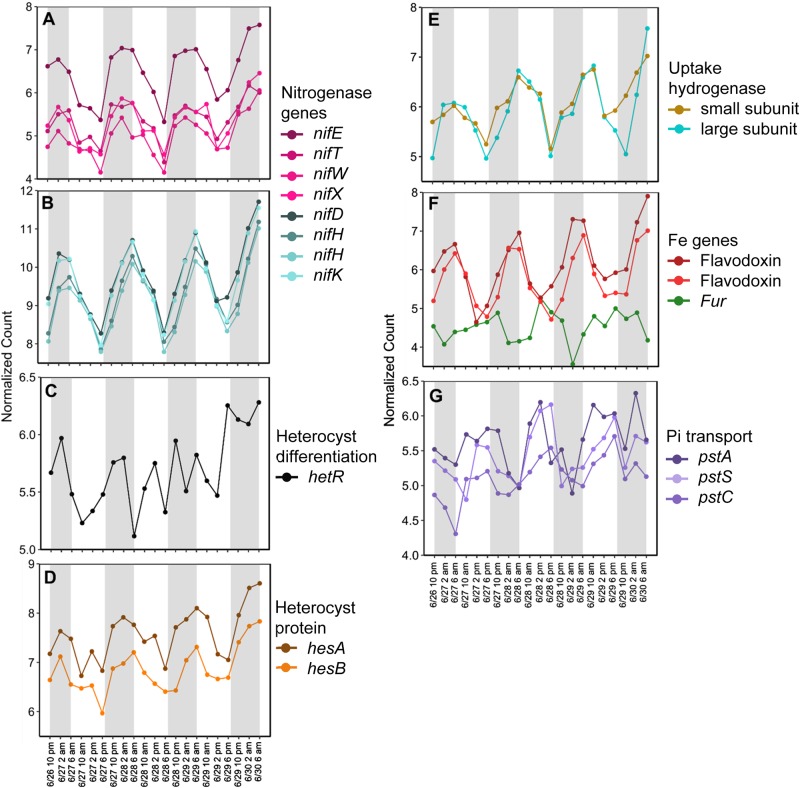
RAIN analysis identified 1,053 genes with significant periodicity in Richelia, representing 25% of the analyzed transcriptome (Fig. 2a, Table S2). A similar magnitude of the transcriptome (34%) was identified as significantly periodic in NPSG field populations of the free-living diazotroph Crocosphaera [12] suggesting that although Richelia typically occurs in association with other organisms, it may (at least when in association with Rhizosolenia) be equally subject to diel forcing as other free-living diazotrophs. As with free-living diazotrophs, N2 and photosynthetic carbon fixation processes exhibited significant periodicity [12, 34], in addition to RNA synthesis and processing (Fig. 2b). Diel patterns in nitrogenase (nif) gene expression are also common in diazotrophs [34–36] and here a total of 16 genes were identified as encoding nitrogenase components in Richelia, of which 12 had significant periodicity (Table S2). The KEGG module for N metabolism contained the nifHDK nitrogenase genes with significantly periodic expression (Fig. 2b). Nitrogenase genes involved in assembly and incorporation of iron and molybdenum into nitrogenase (nifE, nifT, nifW, and nifX) [37] had peak expression around 4 a.m. (Fig. 3a), followed by expression of the core components of nitrogenase (the heterotetrameric core encoded by nifD and nifK, as well as the subunits encoded by nifH) [38], with peak times at 6 a.m. (Fig. 3b). This cascade in gene expression is consistent with a coordinated assembly of the functional enzyme components, and the timing mirrored prior investigations in the region focused solely on Richelia nifH gene expression [32].
Fig. 2.
Significantly periodic gene expression and metabolic profiles for Richelia. a Hierarchically clustered heat map of Richelia genes that were significantly periodic (RAIN, FDR < 0.05) where red indicates the maximum normalized expression (row max) and blue indicates the minimum normalized expression (row min) for that row (gene). Gray bars below the figure indicate dark periods (7 p.m. to 6 a.m.) while yellow indicates light periods (6 a.m. to 7 p.m.). b The percent of significantly periodic Richelia genes with KEGG annotations clustered by KEGG module. KEGG modules mentioned in the text are labeled in the circle plot as a guide to the color legend. Each ring is labeled as percent increases from the center of the plot
Fig. 3.
Normalized expression patterns for genes in Richelia with significant periodic expression (RAIN, FDR < 0.05). a Nitrogen fixation genes with peak expression ~4 a.m. b Nitrogen fixation genes with peak expression ~6 a.m. c The heterocyst differentiation regulator hetR. d Heterocyst-associated genes with peak expression ~4 a.m. e Uptake hydrogenase subunits with peak expression ~6 a.m. f Genes related to iron (Fe) homeostasis, and g) genes involved in phosphorus (Pi) acquisition. Gray shading highlights the dark periods (7 p.m. to 6 a.m.) while light periods are white (6 a.m. to 7 p.m.)
In Richelia, N2 fixation occurs in heterocysts and the heterocyst differentiation regulator, hetR (Rich_326, peak expression at 1 a.m.), heterocyst-associated genes hesA (Rich_5163, peak expression at 4 a.m.) and hesB, (Rich_5165, peak expression at 4 a.m.), as well as genes encoding an uptake hydrogenase system (Rich_4671 and Rich_4672, peak expression 7 a.m. and 6 a.m., respectively) were significantly periodic (Fig. 3c–e). Although the function of hesA and hesB are uncharacterized, they are thought to be linked to N2 fixation, as insertional inactivation of hesA impairs N2 fixation [39] and both are upregulated during N depletion [40]. Here, peak expression of these genes occurred prior to dawn (Fig. 3d), mirroring the pattern that is observed in other heterocystous cyanobacteria [41] and the pattern observed for the Richelia nif genes (Fig. 3a, b). Uptake hydrogenases, located in the heterocysts [42], are thought to efficiently recycle H2 (consuming O2 in the process) which is produced during N2 fixation [43]. They may also act to provide ATP via the oxyhydrogen (knallgas) reaction or provide reducing equivalents to nitrogenase, further enhancing N2 fixation [44–46]. In prokaryotes, transcription and translation are closely coupled in time and space with many genes translated shortly after transcription [47]. The diel oscillations in gene expression for N2 fixation genes in Richelia therefore likely represent the biosynthesis and assembly of nitrogenase in the early morning in preparation for daytime N2 fixation.
In the NPSG, seasonal fluctuations in phosphorus (P), iron (Fe), and light are thought to drive annual blooms of DDAs in the region [48], and Fe and P in diazotrophs is important for fueling maximal N2 fixation [49]. Given the Fe demand associated with N2 fixation, Fe homeostasis may be tightly controlled over the diel cycle. Consistent with this hypothesis a ferric uptake regulatory (Fur) protein, typically involved in maintaining Fe homeostasis [50, 51], was significantly periodic in Richelia with peak expression at 4 p.m. (Fig. 3f, Table S2). Fur typically acts as a negative regulator of Fe stress genes, however, it can both induce and repress a suite of genes in other bacteria [52, 53]. When Fe is plentiful, Fur binds to Fe2+ acquiring a configuration that enables it to bind to target Fur boxes on the genome, inhibiting transcription. When iron is scarce, Fe2+ is released from Fur and transcription of Fe-related functions commences [54]. The expression pattern suggests that Richelia Fe quota and Fe demand are tightly modulated over the diel cycle.
Non-Fe requiring flavodoxins can substitute for ferredoxin to help mitigate Fe demand, and their induction can be an indicator of Fe stress in cyanobacteria [55]. Two flavodoxin transcripts (Rich_2552, Rich_7201) were significantly periodic (Fig. 3f), with peak expression near dawn (4 and 5 a.m., respectively), mirroring N2 fixation genes. Flavodoxin genes have been shown to track nif gene expression patterns in other diazotrophs [34], consistent with the patterns observed here. Flavodoxins in cyanobacteria can be under the transcriptional control of Fur [50, 51]. In other bacteria, Fur regulates a cascade of genes starting with Fe transport, siderophore production, and then genes related to Fe substitution and sparing-related functions such as flavodoxin [53]. This potential cascade may explain the offset in peak time observed for Fur and the flavodoxin genes (Fig. 3f). Regardless of the expression pattern, these day–night patterns in Fur and flavodoxin expression suggest oscillations in Fe demand across the 24 h cycle. These apparent oscillations in Fe demand have also been observed in free-living diazotrophs like Crocosphaera [34, 56] and Trichodesmium [36] and may be a common feature of these genera.
As with the apparent diel oscillations in demand for Fe, P demand appeared to also fluctuate over a diel cycle. Peak expression of genes within the inorganic phosphate (Pi) transport system (pstACS; Rich_4810, 4809, and 1959) occurred between 1 and 3 p.m. and were significantly periodic (Fig. 3g), suggesting variation in P demand and homeostasis over the diel cycle. Similar diel modulations of pstS and other genes involved in P acquisition were reported in free-living diazotrophs [12, 57], suggesting this apparent modulation of P acquisition may be a common feature. Unlike the choreography observed between Fe demand and nifH expression (Fig. 3b), peak expression time for the pst genes (Fig. 3g) was offset relative to that of nifH (Fig. 3b). Given the direct role (e.g. enzyme co-factors) Fe has in N2 fixation and photosynthesis, these apparent fluctuations in Fe demand may be more tightly linked to these processes than apparent fluctuations in P demand. The increased expression of pstS is often used as a marker of P stress [58–60]. In studies of P physiology, the diel modulations of pstS observed here for Richelia further emphasizes the importance of comparing samples from the same time of day to avoid diel variability which may confound interpretation of P stress signals observed in field populations [57].
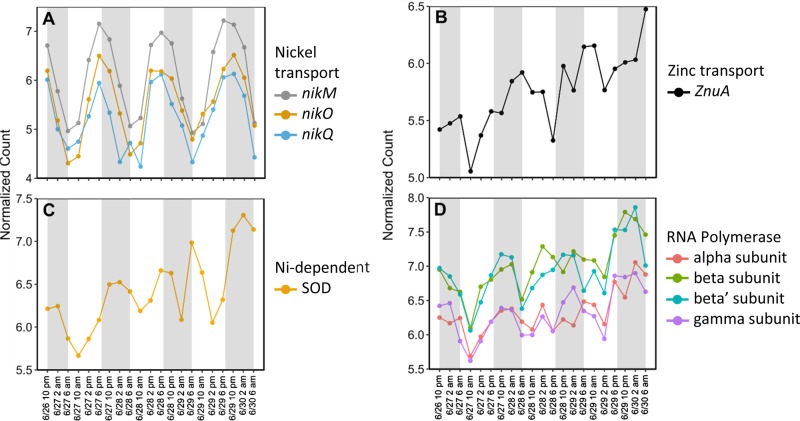
Diel oscillations were apparent for additional resources in Richelia. For example, a set of Nickel (Ni) transport genes (Rich_1479,1480,1481, nikMOQ, 7–8 p.m. peak time) exhibited significant periodicity (Fig. 4a). Ni is an essential transition metal, present at low levels in oceanic environments [61] and is a co-factor in metalloenzymes like uptake hydrogenase and Ni-superoxide dismutase (Ni-SOD) [62]. Ni transport appears to be linked to the periodic expression of metalloenzymes as the Ni-dependent SOD (Rich_4190, 2 a.m. peak time, Fig. 4b) and Ni-dependent uptake hydrogenase subunits (Rich_4671, 4672, 6–7 a.m. peak time, Fig. 3e) were also significantly periodic (Table S2). In the diazotroph Trichodesmium, increased Ni availability is correlated with increased N2 fixation where Ni-dependent enzymes like SOD and uptake hydrogenase reduce oxidative stress by removing oxygen radicals and utilizing released H2 [63]. As such, daily modulation of Ni demand might be common in both symbiotic and free-living diazotrophs.
Fig. 4.
Normalized expression patterns for genes in Richelia with significant periodic expression (RAIN, FDR < 0.05). a Nickel transport genes, b Ni-dependent superoxide dismutase (SOD), c zinc transport, and d RNA polymerase subunits. Gray shading highlights the dark periods (7 p.m. to 6 a.m.) while light periods are white (6 a.m. to 7 p.m.)
Coincident with oscillations in Ni was a significantly periodic Richelia Zn ABC-transporter (Rich_242, znuA), peaking in expression at 5 a.m. (Table S2, Fig. 4c). The modulation of this Zn transporter suggests diel oscillations in internal Zn inventory. In the NPSG, zinc (Zn) is depleted in surface waters [64] but, like Ni, is required for many metalloenzymes critical to processes, such as carbon fixation (carbonic anhydrase), nucleic acid replication (RNA polymerase, reverse transcriptase), and phosphorus metabolism (alkaline phosphatase) [65]. Although the Zn-dependent alkaline phosphatase (phoA) and carbonic anhydrases were not significantly periodic, a group of alpha, beta, and gamma RNA polymerases (RNAP) were significantly periodic with peak expression after midnight (Rich_1744, 3195, 3196, 3197; Fig. 4d). Zinc appears to play a structural role in promoting RNAP assembly and additions of Zn promote production of RNAP [66, 67]. As RNA polymerase is transcribed, Zn inventories may become depleted prompting the cell to express genes for Zn transport.
Taken together, these diel patterns in resource-related gene targets suggests that both macro and micronutrient demand is tightly modulated in Richelia over diel cycles. These diel patterns could influence the cycling of these resources within the host–symbiont association, and the surrounding water column. More diel studies of field populations are required to know to what extent patterns in resource demand are consistent across other diazotroph associations and phytoplankton in general, and to what extent synchronous competition for resources might drive competitive outcomes and resulting bloom dynamics in oligotrophic regions.
Patterns of diel gene expression in Rhizosolenia from the NPSG
The analysis of selected reads from the same pool of RNA allowed us to also identify diel patterns in the diatom host Rhizosolenia and evaluate coordinated patterns in gene expression within the symbiosis. RAIN identified 398 Rhizosolenia contigs with significant periodicity in the NPSG, representing 2% of the analyzed transcriptome (Figure S3, Table S2). Many of these periodically expressed genes were related to photosynthesis, carbon fixation and related processes (Figure S4, Table S2). For example, 43 chlorophyll a/b binding proteins were periodically expressed (Figure S4A) and significant periodicity was also observed in violaxanthin de-epoxidase (VDE), an enzyme that converts diadinoxanthin into diatoxanthin, protecting against photooxidative damage [68]. Peak expression of VDE in Rhizosolenia (1 p.m.; Figure S4B) was later than that observed in cultures of Thalassiosira pseudonana (dawn) [69] perhaps suggesting differing strategies or responses to this ubiquitous problem in photosynthetic organisms. Lastly, phosphoglycerate kinase, which catalyzes an essential step in glycolysis was found to have peak expression in Rhizosolenia at 8 a.m. (Figure S4C) while in T. pseudonana this gene peaked in expression near dusk [69]. It is clear that further investigation is warranted to determine if differences between Rhizosolenia and Thalassiosira are intrinsic to different diatom genera or due to inherent differences between culture studies and observations of field populations.
Significantly coordinated patterns of expression in the Rhizosolenia–Richelia association
Chronobiological observations of symbiotic partnerships have revealed metabolic interdependence between host and symbiont, largely centered around the exchange of resources and cell division processes [70, 71]. To explore this interdependence in the Rhizosolenia–Richelia partnership, a weighted gene co-expression network analysis (WGCNA) was used to examine patterns of co-expression between the partners over the time series. This analysis was independent of the RAIN analysis and was used to identify clusters (modules) of transcripts that were significantly correlated with either a synchronous or orthogonal pattern. Of the 126 modules constructed, Richelia genes were represented in all but five (Figure S5), suggesting that many cellular processes between the host and symbiont were significantly co-expressed. A number of WGCNA modules had patterns of day–night periodicity and contained features with significantly periodic expression (Figure S6). Some of this co-expression was due in part to the coordinated day–night expression of overlapping metabolic pathways like carbon fixation and nucleotide sugar metabolism (e.g. WGCNA module 1) (Fig. 5) and may reflect the shared response that phytoplankton have to light. However, the majority of the co-expression modules contained different KEGG pathways for each species (Fig. 5), and this variability may reflect each organism being individually influenced by day–night transitions. For example, N metabolism was observed in WGCNA modules 1, 2, and 7 for Rhizosolenia but only in module 23 for Richelia (Fig. 5). As another example, Richelia serine and threonine metabolism was predominately in WGCNA module 1, whereas Rhizosolenia serine and threonine metabolism was predominately in WGCNA module 7, reflecting different expression patterns for the host and symbiont (Fig. 5). The unique distribution of metabolic processes in the co-expression network analysis suggests that these organisms operate on different clocks in the same environment, where shared aspects of metabolism are offset. As with many host–symbiont relationships, it is assumed resource exchange is the primary function of this DDA partnership, and this metabolic offset may reflect this. However, it may also reflect inherent differences between diatoms and cyanobacteria. Additional surveys of diel patterns in field populations of diatoms and cyanobacteria would help resolve the extent to which these patterns reflect intrinsic differences between different photosynthetic taxa, versus patterns driven by the two organisms living in association. Despite these uncertainties, each species clearly has distinct metabolic patterns, and their metabolisms also appear to be tightly coordinated with each other. This significant coordination is likely important to maintaining the symbiosis and driving the exchange of resources between host and symbiont.
Fig. 5.
The species-specific proportion of mapped reads per KEGG module and their distribution across WGCNA modules containing >200 nodes (Richelia genes or Rhizosolenia contigs). Color corresponds to the proportion of mapped reads in each KEGG module (column) for each species visualized for WGCNA modules
Potential exchange of resources between host and symbiont over light–dark cycles
The transfer of N between symbiont and host has been shown in DDA partnerships [6, 11] and to explore this function we searched for WGCNA modules containing nif genes from Richelia. Module 23 of the WGCNA analysis contained 10 of the 12 significantly periodic nitrogenase genes (Fig. 6, Figure S8A) and these were significantly co-expressed with a Rhizosolenia ammonium transporter (Rhizo_7354), an amino acid permease (Rhizo_10488), and two major facilitator superfamily (MFS) proteins (Rhizo_156 and Rhizo_18969); a large gene family involved in the transport of amino acids among other potential substrates [72] (Fig. 6, Figure S8A). Although there are many factors regulating expression of N transport functions [73], these significantly co-expressed genes between the host and symbiont may, in part, underpin the known transfer of newly fixed nitrogen from Richelia to the diatom [11].
Fig. 6.
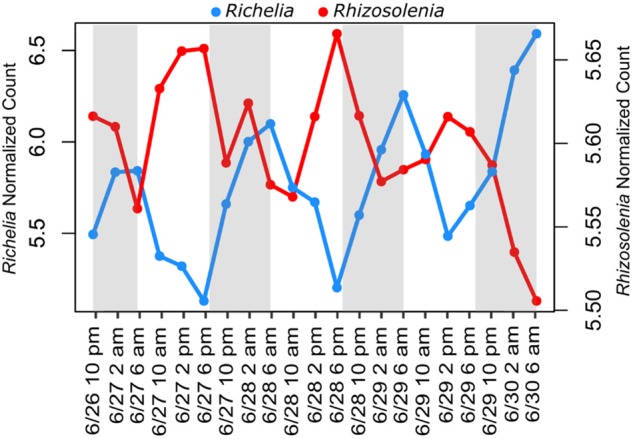
Average normalized expression patterns of significantly co-expressed features within the WGCNA module 23 for Richelia (103 genes, SE ± 0.21) and Rhizosolenia (105 contigs, SE ± 0.15). Key features within this module relating to potential exchange of resources are a TRAP = tripartite ATP-independent periplasmic and nitrogenase gene (nifHDKETWX) in Richelia and transporters for ammonium, amino acids, major facilitator superfamily (MFS) and efflux and EAMA-like transporters in Rhizosolenia. Gray shading highlights dark hours (7 p.m. to 6 a.m.) while light periods are white (6 a.m. to 7 p.m.)
Diatoms in the NPSG are limited by N [74] and Richelia is thought to provide a distinct advantage to the diatom host by providing N via N2 fixation. What Richelia gets back in return remains an open question. In legumes (as well as other plant/bacteria symbiosis), reduced carbon such as dicarboxylates, sugars, and amino acids are thought to be exchanged for fixed N2 [75, 76]. Although Richelia expresses genes for photosynthesis (Fig. 2, Table S2), it may still import carbon substrates from the diatom host. In Module 23, Rhizosolenia genes for the transport of carbon substrates, including an EAMA-like transporter (Rhizo_1833) and an efflux transporter (Rhizo_14884), were significantly co-expressed with the Richelia nitrogenase genes (Fig. 6, Figure S8A). In the diatom Phaeodactylum tricornutum, EAMA-like transporters (named after the O-acetyl-serine/cysteine export gene in E. coli) were associated with triose-phosphate translocators (TPTs), which are thought to export photosynthesis-derived carbohydrates [77]. These transporters may be involved in the export of substrates to Richelia in exchange for fixed N2. In support of this hypothesis, a Richelia tripartite ATP-independent periplasmic (TRAP) solute receptor gene (Rich_1143) was also co-expressed in the same module (Fig. 6, Figure S8B). TRAP transporters are unique to bacteria and archaea and are involved in the uptake of a number of organic substrates, including sugars [78], and in this case their co-expression may be indicative of Richelia actively transporting substrates produced by the host.
In addition to N, diatoms can also be limited by trace nutrients such as vitamins, especially during bloom conditions [74]. As they are not able to synthesize vitamin B12, diatoms in DDA partnerships may benefit from their cyanobacteria symbiont as a source for required vitamins such as B12 [79]. Like other cyanobacteria, Richelia likely produces a form of B12 (pseudocobalamin) that is unusable by most eukaryotic phytoplankton [80]. Notably, Rhizosolenia has genetic machinery to alter pseudocobalamin to a useable form (cobST; Rhizo_5562; Table S2), by replacing the axial ligand adenine with exogenous sources of 5,6-dimethylbenzimidazole (DMB) [80]. It appears both genes are present within the assembled contig. These genes are expressed in the NPSG but are not significantly diel (Table S2). In Richelia, a gene for cobalamin (B12) synthase (Rich_4005; Table S3) was expressed at too low a level to pass our stringent analysis threshold and portions of the cobalamin biosynthesis pathway cbiX (Rich_4021,3078), cobF (Rich_5476,5860), and cobN (Rich_155) were not significantly periodic (Table S2), suggesting perhaps constitutive low-level exchange of this essential resource. It may be that other vitamins are also exchanged within the association. For example, Richelia biotin synthase (Rich_1540) expression is significantly periodic, as are two Rhizosolenia biotin ligases (Rhizo_7013 and Rhizo_9114) (Table S2), the latter of which activates biotin and transfers biotin to biotin-accepting proteins [81]. Diatoms are known to produce biotin, but may rely on exogenous sources of biotin in Fe-limited conditions [82]. Although the Richelia and Rhizosolenia biotin genes do not fall into the same WGCNA module, these diel patterns may still underpin host–symbiont biotin exchange in the low Fe NPSG. Taken together, these data are consistent with the exchange of vitamins within the association. If this is confirmed with further study, then the vitamins from Richelia may free the host from competition for vitamins like B12 in addition to N.
Potential coordination of Rhizosolenia–Richelia growth
In Rhizosolenia, Richelia intracellularis is an extracellular endosymbiont located in the periplasmic space between the plasmalemma and silica wall [83, 84]. Early culture work suggested asynchronous division frequency between host and symbiont despite similar growth rates in N-limited media [85]. However, little is known regarding natural population division or how the host and symbiont maintain this extracellular association. RAIN analysis identified a putative ftsH cell division marker with significant periodicity for both the Rhizoselenia host (Rhizo_8174) and the Richelia symbiont (Rich_842) with both having peak expression at 7 a.m. (Fig. 7a). Although the genes did not fall into the same WGCNA module, their peak times are consistent, suggesting similar division timing. Uptake and incorporation of silicic acid appears to be linked to cell division as well [86, 87] and so the timing in expression of these transporters in Rhizosolenia may be an effective proxy for cell division processes in natural populations of Rhizosolenia. Significantly periodic Rhizosolenia silicic acid transporters (Rhizo_8532 and Rhizo_9964) had peak expression at ~2 a.m. (Fig. 7a, Figure S7) slightly before the peak expression of the fstH gene cell division markers observed here (Fig. 7a). Taken together, these expression patterns are consistent with a pattern of early morning division that is coordinated between the host and symbiont. If this pattern is supported with additional observations, then the association may be maintained, at least in part, by a coordinated pattern of cell division which is synchronized over the diel cycle in field populations.
Fig. 7.
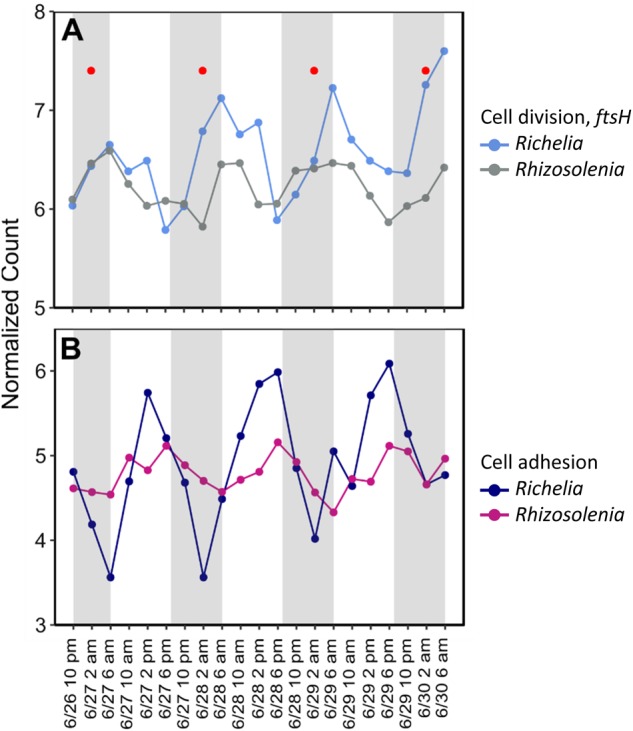
Normalized expression patterns of a the putative cell division marker ftsH for both Richelia and Rhizosolenia and b cell adhesion gene forming pili in Richelia and a fasciclin domain contig in Rhizosolenia. All genes/contigs were significantly periodic (RAIN, FDR < 0.05). Red dots in plot A correspond to peak expression timing of silicic acid transporters (Figure S7) in Rhizosolenia. Gray shading highlights dark hours (7 p.m. to 6 a.m.) while light periods are white (6 a.m. to 7 p.m.)
Fasciclin domain proteins in symbioses are thought to be related to adhesion between host and symbiont and they have been found in a wide variety of symbiotic associations [88], where increased expression of fasciclin domain proteins in the host has been linked to the presence of the symbiont [89]. WGCNA Module 3 contained a significantly periodic Rhizosolenia fasciclin domain protein (Rhizo_11521) with peak expression at 5 p.m. (Fig. 7b). This fasciclin domain protein was significantly co-expressed with a Richelia gene related to pilus formation (Rich_7610) (Fig. 7b). Filamentous surface appendages called pili mediate pilus-dependent adhesion in other associations, such as those found in N2-fixing bacteria associated with plant roots [90]. Given these coordinated expression patterns, we hypothesize that pili formation and related processes in the diatom host may help maintain the symbiosis over the day–night cycle as each species divides. Collectively, these gene expression patterns suggest that host and symbiont ecology is tightly integrated over the diel cycle and these data provide gene targets with which to further evaluate the ecology of this DDA.
Conclusions
DDAs such as the Rhizosolenia–Richelia symbiosis are known to exert profound control over the cycling of carbon and nitrogen, particularly in oligotrophic regions like the NPSG, yet there are still major knowledge gaps regarding DDA physiological ecology. In surface ocean prokaryotic communities, diel patterns of synchronous gene expression suggest there is cross-species coordination driving carbon metabolism and cycling in the community [25]. Here, we observed offsets in the expression of core metabolic processes in a photosynthetic host–symbiont pair. These differences may underpin the ability of these two species to live together by providing a temporal compartmentalization of metabolism. Temporal compartmentalization could minimize competition for key resources, while facilitating exchange of resources, like N, and maintain the processes that coordinate growth and stabilize the association over light–dark transitions. Given that oligotrophic waters are expanding due to climate change, with the NPSG expanding most rapidly [91, 92], DDAs may be increasingly important in the future ocean, underscoring the importance of understanding the physiological ecology of this keystone symbiosis.
Electronic supplementary material
Acknowledgements
For assistance with field work, the authors thank the operational staff of the Simons Collaboration on Ocean Processes and Ecology (SCOPE) and the captain and crew of the R/V Kilo Moana.
Funding
Funding was provided by the Simons Foundation (SCOPE award ID 329108 to STD and JPZ). Computational support was provided by the National Science Foundation under Grant Nos. DBI-1458641 and ABI-1062432 to Indiana University.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0262-2) contains supplementary material, which is available to authorized users.
References
- 1.Carr ME, Friedrichs MAM, Schmeltz M, Noguchi Aita M, Antoine D, Arrigo KR, et al. A comparison of global estimates of marine primary production from ocean color. Deep Sea Res Part 2. 2006;53:741–70. [Google Scholar]
- 2.Karl DM, Church MJ, Dore JE, Letelier RM, Mahaffey C. Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc Natl Acad Sci USA. 2012;109:1842–9. doi: 10.1073/pnas.1120312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dore JE, Letelier RM, Church MJ, Lukas R, Karl DM. Summer phytoplankton blooms in the oligotrophic North Pacific Subtropical Gyre: historical perspective and recent observations. Prog Oceanogr. 2008;76:2–38. [Google Scholar]
- 4.Villareal TA, Adornato L, Wilson C, Schoenbaechler CA. Summer blooms of diatom-diazotroph assemblages and surface chlorophyll in the North Pacific gyre: a disconnect. J Geophys Res Oceans. 2011; 116: 10.1029/2010JC006268
- 5.Wilson C Late Summer chlorophyll blooms in the oligotrophic North Pacific Subtropical Gyre. Geophys Res Lett. 2003; 30: 10.1029/2003GL017770
- 6.Foster RA, Kuypers MM, Vagner T, Paerl RW, Musat N, Zehr JP. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 2011;5:1484–93. doi: 10.1038/ismej.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitajima S, Furuya K, Hashihama F, Takeda S, Kanda J. Latitudinal distribution of diazotrophs and their nitrogen fixation in the tropical and subtropical western North Pacific. Limnol Oceanogr. 2009;54:537–47. [Google Scholar]
- 8.Carpenter EJ, Montoya JP, Burns J, Mulholland MR, Subramaniam A, Capone DG. Extensive bloom of a N2-fixing diatom/cyanobacterial association in the tropical Atlantic Ocean. Mar Ecol Prog Ser. 1999;185:273–83. [Google Scholar]
- 9.Böttjer D, Dore JE, Karl DM, Letelier RM, Mahaffey C, Wilson ST, et al. Temporal variability of nitrogen fixation and particulate nitrogen export at Station ALOHA. Limnol Oceanogr. 2017;62:200–16. [Google Scholar]
- 10.Villareal TA, Pilskaln CH, Montoya JP, Dennett M. Upward nitrate transport by phytoplankton in oceanic waters: balancing nutrient budgets in oligotrophic seas. PeerJ. 2014;2:e302. doi: 10.7717/peerj.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villareal TA. Laboratory culture and preliminary characterization of the nitrogen-fixing Rhizosolenia-Richelia symbiosis. Mar Ecol. 1990;11:117–32. [Google Scholar]
- 12.Wilson ST, Aylward FO, Ribalet F, Barone B, Casey JR, Connell PE, et al. Coordinated regulation of growth, activity and transcription in natural populations of the unicellular nitrogen-fixing cyanobacterium Crocosphaera. Nat Microbiol. 2017;2:17118. doi: 10.1038/nmicrobiol.2017.118. [DOI] [PubMed] [Google Scholar]
- 13.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 14.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander H, Jenkins BD, Rynearson TA, Dyhrman ST. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc Natl Acad Sci USA. 2015;112:E2182–90. doi: 10.1073/pnas.1421993112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster RA, Goebel NL, Zehr JP. Isolation of Calothrix rhizosoleniae (Cyanobacteria) Strain Sc01 from Chaetoceros (Bacillariophyta) Spp. diatoms of the Subtropical North Pacific Ocean. J Phycol. 2010;46:1028–37. [Google Scholar]
- 17.Hilton JA. Ecology and evolution of diatom-associated cyanobacteria through genetic analyses. Dissertation. University of California: Santa Cruz, CA; 2014.
- 18.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–7. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 25.Aylward FO, Eppley JM, Smith JM, Chavez FP, Scholin CA, DeLong EF. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc Natl Acad Sci USA. 2015;112:5443–8. doi: 10.1073/pnas.1502883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, 3rd, Schuster SC, et al. Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–12. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenkel CD, Matz MV. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat Ecol Evol. 2016;1:14. doi: 10.1038/s41559-016-0014. [DOI] [PubMed] [Google Scholar]
- 29.Rose NH, Seneca FO, Palumbi SR. Gene networks in the wild: identifying transcriptional modules that mediate coral resistance to experimental heat stress. Genome Biol Evol. 2015;8:243–52. doi: 10.1093/gbe/evv258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grote A, Voronin D, Ding T, Twaddle A, Unnasch TR, Lustigman S, et al. Defining Brugia malayi and Wolbachia symbiosis by stage-specific dual RNA-seq. PLoS Negl Trop Dis. 2017;11:e0005357. doi: 10.1371/journal.pntd.0005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One. 2012;7:e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2005;71:5362–70. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Follett Christopher L., White Angelicque E., Wilson Samuel T., Follows Michael J. Nitrogen fixation rates diagnosed from diurnal changes in elemental stoichiometry. Limnology and Oceanography. 2018;63(5):1911–1923. [Google Scholar]
- 34.Shi T, Ilikchyan I, Rabouille S, Zehr JP. Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J. 2010;4:621–32. doi: 10.1038/ismej.2009.148. [DOI] [PubMed] [Google Scholar]
- 35.Stockel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc Natl Acad Sci USA. 2008;105:6156–61. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frischkorn KR, Haley ST, Dyhrman ST. Coordinated gene expression between Trichodesmium and its microbiome over day-night cycles in the North Pacific Subtropical Gyre. ISME J. 2018;12:997–1007. doi: 10.1038/s41396-017-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiig JA, Rebelein JG, Hu Y. Nitrogenase complex. eLS: John Wiley & Sons, Ltd, Chichester. 2014 http://www.els.net [10.1002/9780470015902.a0001386.pub2]
- 38.Zehr JP, Turner PJ. Nitrogen fixation: Nitrogenase genes and gene expression. Methods Microbiol. 2001;30:271–86. [Google Scholar]
- 39.Borthakur D, Basche M, Buikema WJ, Borthakur PB, Haselkorn R. Expression, nucleotide sequence and mutational analysis of two open reading frames in the nif gene region of Anabaena sp. strain PCC 7120. Mol General Genet Manage. 1990;221:227–34. doi: 10.1007/BF00261725. [DOI] [PubMed] [Google Scholar]
- 40.Flaherty BL, Nieuwerburgh FV, Head SR, Golden JW. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combinded-nitrogen deprivation. BMC Genom. 2011;12:332. doi: 10.1186/1471-2164-12-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kushige H, Kugenuma H, Matsuoka M, Ehira S, Ohmori M, Iwasaki H. Genome-wide and heterocyst-specific circadian gene expression in the filamentous cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2013;195:1276–84. doi: 10.1128/JB.02067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson RB, Wolk CP. Localization of an upatke hydrogenase in Anabaena. Plant Physiol. 1978;61:688–91. doi: 10.1104/pp.61.4.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamagnini P, Axelsson R, Lindberg P, Oxelfelt F, Wunschiers R, Lindblad P. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol Mol Biol Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Sherman DM, Sherman LA. The uptake hydrogenase in the unicellular diazotrophic cyanobacterium Cyanothece sp. strain PCC 7822 protects nitrogenase from oxygen toxicity. J Bacteriol. 2014;196:840–9. doi: 10.1128/JB.01248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bothe H, Schmitz O, Yates MG, Newton WE. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol Mol Biol Rev. 2010;74:529–51. doi: 10.1128/MMBR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson ST, Tozzi S, Foster RA, Ilikchyan I, Kolber ZS, Zehr JP, et al. Hydrogen cycling by the unicellular marine diazotroph Crocosphaera watsonii strain WH8501. Appl Environ Microbiol. 2010;76:6797–803. doi: 10.1128/AEM.01202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGary K, Nudler E. RNA polymerase and the ribosome: the close relationship. Curr Opin Microbiol. 2013;16:112–7. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Follett CL, Dutkiewicz S, Karl DM, Inomura K, Follows MJ. Seasonal resource conditions favor a summertime increase in North Pacific diatom-diazotroph associations. ISME J. 2018;12:1543–57. doi: 10.1038/s41396-017-0012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills MM, Ridame C, Davey M, La Roche J, Geider RJ. Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature. 2004;429:292–4. doi: 10.1038/nature02550. [DOI] [PubMed] [Google Scholar]
- 50.Bes MT, Hernandez JA, Peleato ML, Fillat MF. Cloning, overexpression and interaction of recombinant Fur from the cyanobacterium Anabaena PCC 7119 with isiB and its own promoter. FEMS Microbiol Lett. 2001;194:187–92. doi: 10.1111/j.1574-6968.2001.tb09467.x. [DOI] [PubMed] [Google Scholar]
- 51.Ghassemian M, Straus NA. Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology. 1996;142:1469–76. doi: 10.1099/13500872-142-6-1469. [DOI] [PubMed] [Google Scholar]
- 52.Thompson DK, Beliaev AS, Giometti CS, Tollaksen SL, Khare T, Lies DP, et al. Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl Environ Microbiol. 2002;68:881–92. doi: 10.1128/AEM.68.2.881-892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pi H, Helmann JD. Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc Natl Acad Sci USA. 2017;114:12785–90. doi: 10.1073/pnas.1713008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–9. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fillat M.F., Peleato M.L., Razquin P., Gomez-Moreno C. Iron Nutrition in Soils and Plants. Dordrecht: Springer Netherlands; 1995. Effects of iron-deficiency in photosynthetic electron transport and nitrogen fixation in the cyanobacterium Anabaena: Flavodoxin induction as adaptative response; pp. 315–321. [Google Scholar]
- 56.Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, Monteiro FM, et al. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc Natl Acad Sci USA. 2011;108:2184–9. doi: 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereira N, Shilova IN, Zehr JP. Molecular markers define progressing stages of phosphorus limitation in the nitrogen-fixing cyanobacterium, Crocosphaera. J Phycol. 2016;52:274–82. doi: 10.1111/jpy.12396. [DOI] [PubMed] [Google Scholar]
- 58.Dyhrman ST, Haley ST. Phosphorus scavenging in the unicellular marine diazotroph Crocosphaera watsonii. Appl Environ Microbiol. 2006;72:1452–8. doi: 10.1128/AEM.72.2.1452-1458.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harke MJ, Berry DL, Ammerman JW, Gobler CJ. Molecular response of the bloom-forming cyanobacterium, Microcystis aeruginosa, to phosphorus limitation. Microb Ecol. 2012;63:188–98. doi: 10.1007/s00248-011-9894-8. [DOI] [PubMed] [Google Scholar]
- 60.Scanlan DJ, Silman NJ, Donald KM, Wilson WH, Carr NG, Joint I, et al. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol. 1997;63:2411–20. doi: 10.1128/aem.63.6.2411-2420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyle EA, Huested SS, Jones SP. On the distribution of copper, nickel, and cadmium in the surface waters of the North Atlantic and North Pacific Ocean. J Geophys Res Oceans. 1981; 86. 10.1029/JC086iC09p08048
- 62.Huertas MJ, Lopez-Maury L, Giner-Lamia J, Sanchez-Riego AM, Florencio FJ. Metals in cyanobacteria: analysis of the copper, nickel, cobalt and arsenic homeostasis mechanisms. Life (Basel) 2014;4:865–86. doi: 10.3390/life4040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez IB, Ho TY. Diel nitrogen fixation pattern of Trichodesmium: the interactive control of light and Ni. Sci Rep. 2014;4:4445. doi: 10.1038/srep04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruland KW. Oceanographic distributions of cadmium, zinc, nickel, and copper in the North Pacific. Earth Planet Sci Lett. 1980;47:176–98. [Google Scholar]
- 65.Twining BS, Baines SB. The trace metal composition of marine phytoplankton. Annu Rev Mar Sci. 2013;5:191–215. doi: 10.1146/annurev-marine-121211-172322. [DOI] [PubMed] [Google Scholar]
- 66.Markov D, Naryshkina T, Mustaev A, Severinov K. A zinc-binding site in the largest subunit of DNA-dependent RNA polymerase is involved in enzyme assembly. Genes Dev. 1999;13:2439–48. doi: 10.1101/gad.13.18.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr Purif. 1993;4:503–11. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 68.Lavaud J, Materna AC, Sturm S, Vugrinec S, Kroth PG. Silencing of the violaxanthin de-epoxidase gene in the diatom Phaeodactylum tricornutum reduces diatoxanthin synthesis and non-photochemical quenching. PLoS One. 2012;7:e36806. doi: 10.1371/journal.pone.0036806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashworth J, Coesel S, Lee A, Armbrust EV, Orellana MV, Baliga NS. Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana. Proc Natl Acad Sci USA. 2013;110:7518–23. doi: 10.1073/pnas.1300962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sorek M, Diaz-Almeyda EM, Medina M, Levy O. Circadian clocks in symbiotic corals: the duet between Symbiodinium algae and their coral host. Mar Genomics. 2014;14:47–57. doi: 10.1016/j.margen.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 71.Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–64. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pao SS, Paulsen IT, Saier MH., Jr. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collos Y, Berges JA. Nitrogen metabolism in phytoplankton. In: Duarte CM, editor. Marine ecology: Encyclopedia of Life Support Systems (EOLSS), Paris, France. 2002;1–18.
- 74.Alexander H, Rouco M, Haley ST, Wilson ST, Karl DM, Dyhrman ST. Functional group-specific traits drive phytoplankton dynamics in the oligotrophic ocean. Proc Natl Acad Sci USA. 2015;112:E5972–9. doi: 10.1073/pnas.1518165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Udvardi MK, Day DA. Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:493–523. doi: 10.1146/annurev.arplant.48.1.493. [DOI] [PubMed] [Google Scholar]
- 76.Mus F, Crook MB, Garcia K, Garcia Costas A, Geddes BA, Kouri ED, et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol. 2016;82:3698–710. doi: 10.1128/AEM.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moog D, Rensing SA, Archibald JM, Maier UG, Ullrich KK. Localization and evolution of putative triose phosphate translocators in the diatom Phaeodactylum tricornutum. Genome Biol Evol. 2015;7:2955–69. doi: 10.1093/gbe/evv190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mulligan C, Fischer M, Thomas GH. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol Rev. 2011;35:68–86. doi: 10.1111/j.1574-6976.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 79.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–3. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 80.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Krautler B, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol. 2016;26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slavoff SA, Chen I, Choi YA, Ting AY. Expanding the substrate tolerance of biotin ligase through exploration of enzymes from diverse species. J Am Chem Soc. 2008;130:1160–2. doi: 10.1021/ja076655i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen NR, Ellis KA, Burns WG, Lampe RH, Schuback N, Johnson Z, et al. Iron and vitamin interactions in marine diatom isolates and natural assemblages of the Northeast Pacific Ocean. Limnol Oceanogr. 2017;62:2076–96. [Google Scholar]
- 83.Sundström BG. Observations on Rhizosolenia clevei Ostenfeld (Bacillariophyceae) and Richelia intracellularis Schmidt (Cyanophyceae) Bot Mar. 1984;27:345–55. [Google Scholar]
- 84.Villareal Tracy A. Marine Pelagic Cyanobacteria: Trichodesmium and other Diazotrophs. Dordrecht: Springer Netherlands; 1992. Marine Nitrogen-Fixing Diatom-Cyanobacteria Symbioses; pp. 163–175. [Google Scholar]
- 85.Villareal TA. Division cycles in the nitrogen-fixing Rhizosolenia (Bacillariophyceae)-Richelia (Nostocaceae) symbiosis. Eur J Phycol. 1989;24:357–65. [Google Scholar]
- 86.Chisolm SW, Azam F, Eppley RW. Silicic acid incorportation in marine diatoms on light:dark cycles: use as an assay for phased cell division. Limnol Oceanogr. 1978;23:518–29. [Google Scholar]
- 87.Knight MJ, Senior L, Nancolas B, Ratcliffe S, Curnow P. Direct evidence of the molecular basis for biological silicon transport. Nat Commun. 2016;7:11926. doi: 10.1038/ncomms11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paulsrud P, Lindblad P. Fasciclin domain proteins are present in Nostoc symbionts of lichens. Appl Environ Microbiol. 2002;68:2036–9. doi: 10.1128/AEM.68.4.2036-2039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reynolds WS, Schwarz JA, Weis VM. Symbiosis-enhanced gene expression in cnidarian-algal associations: cloning and characterization of a cDNA, sym32, encoding a possible cell adhesion protein. Comp Biochem Physiol A. 2000;126:33–44. doi: 10.1016/s0742-8413(00)00099-2. [DOI] [PubMed] [Google Scholar]
- 90.Rodriguez-Navarro DN, Dardanelli MS, Ruiz-Sainz JE. Attachment of bacteria to the roots of higher plants. FEMS Microbiol Lett. 2007;272:127–36. doi: 10.1111/j.1574-6968.2007.00761.x. [DOI] [PubMed] [Google Scholar]
- 91.Polovina JJ, Howell EA, Abecassis M. Ocean’s least productive waters are expanding. Geophys Res Lett. 2008; 35. 10.1029/2007GL031745
- 92.Signorini SR, Franz BA, McClain CR. Chlorophyll variability in the oligotrophic gyres: mechanisms, seasonality and trends. Front Mar Sci. 2015;2:1. doi: 10.3389/fmars.2015.00001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.