Abstract
3′ untranslated regions (3′ UTRs) and particularly long 3′ UTRs have been shown to act as a new class of post-transcriptional regulatory element. We previously reported that hmsT mRNA stability is negatively regulated by the 3′ UTR of hmsT in Yersinia pestis. To investigate more general effects of 3′ UTRs in Y. pestis, we selected 15 genes potentially possessing long 3′ UTRs with different AU content and constructed their 3′ UTR deletion mutants. Deletion of AU-rich 3′ UTRs increased mRNA levels, whereas deletion of 3′ UTRs with normal AU content resulted in slight or no changes in the mRNA level. In addition, we found that PNPase was important for 3′ UTR-mediated mRNA decay when the transcriptional terminator was Rho-dependent. Finally, we showed that ribosomes promote mRNA stability when bound to a 3′ UTR. Our findings suggest that functional 3′ UTRs might be broadly distributed in bacteria and their novel regulatory mechanisms require further investigation.
Keywords: 3′ UTR, AU-rich region, post-transcriptional regulation, mRNA stability, Yersinia pestis
Introduction
To adapt to changes in the environment, an intricate regulatory network that accurately modulates gene expression has evolved in bacteria. Gene expression in bacteria is primarily regulated at the transcriptional level. However, unlike transcriptional regulation, post-transcriptional regulation of gene expression allows bacteria to adjust rapidly to the changing environment. One way in which post-transcriptional regulation is achieved is via changes in mRNA stability (Laalami et al., 2014).
5′ and 3′ untranslated regions (UTRs) contain many elements that assist in the regulation of gene expression (Pesole et al., 2001). 5′ UTR mediated gene regulation has been extensively studied in bacteria (Oliva et al., 2015). The 5′ UTR reportedly folds into a specific secondary structure, such as an RNA thermometer or riboswitch (Henkin, 2008; Breaker, 2011; Kortmann and Narberhaus, 2012; Serganov and Patel, 2012; Krajewski and Narberhaus, 2014). In addition to secondary structures, ribosome binding to the Shine-Dalgarno sequence increases the stability of the downstream mRNA (Agaisse and Lereclus, 1996; Sharp and Bechhofer, 2003; Daou-Chabo et al., 2009). Traditionally, bacterial 3′ UTRs were thought to comprise mainly transcriptional terminators. However, transcriptional terminators rarely exceed a size of 40–50 nucleotides; thus 3′ UTRs of greater length (>100 nt) have been hypothesized to contain other regulatory elements (Ruiz de los Mozos et al., 2013). Recent work has shown that 3′ UTRs are involved in regulating gene expression in bacteria (Ren et al., 2017). In particular, 3′ UTRs regulate the decay rate and translational initiation of mRNAs(Selinger et al., 2003; Maeda and Wachi, 2012; Lopez-Garrido et al., 2014; Liu et al., 2016; Zhu et al., 2016). Regulatory small RNAs (sRNAs) target 3′ UTRs, which in turn regulates the expression of the UTR-containing mRNAs (Opdyke et al., 2004; Silvaggi et al., 2005). In addition, 3′ UTRs provide a rich reservoir of sRNAs for the targeted regulation of gene expression (Chao et al., 2012; Gossringer and Hartmann, 2012; Kim et al., 2014; Tree et al., 2014; Miyakoshi et al., 2015; Chao and Vogel, 2016; Peng et al., 2016). The RNA chaperone Hfq binds to the 3′ ends of mRNAs (Holmqvist et al., 2016) and protects RNA from 3′ to 5′ exonuclease activity (Hajnsdorf and Regnier, 2000; Le Derout et al., 2003).
Several 3′ UTRs are reportedly attacked by ribonucleases to initiate mRNA degradation. One early example is the 3′ UTR of aceA in Corynebacterium glutamicum, which contains an AU-rich region cleaved by RNase E/G (Maeda and Wachi, 2012). Although the aceA 3′ UTR is only 63 nt in length, other 3′ UTRs involved in mRNA decay are much longer. For example, the 3′ UTR of hilD mRNA is 310 nt and is degraded by RNase E and polynucleotide phosphorylase (PNPase) in Salmonella enterica (Lopez-Garrido et al., 2014). Another example is the 3′ UTR of hmsT, which is 283 nt in length and cleaved by PNPase in Yersinia pestis (Zhu et al., 2016). The 3′ UTRs of hilD and aceA possess a specific AU-rich regulatory region (Maeda and Wachi, 2012; Lopez-Garrido et al., 2014). Although the hmsT 3′ UTR does not contain a specific AU-rich regulatory element, it is rich in AU nucleotides (70%). Thus, AU-rich 3′ UTRs appear to be susceptible to RNase attack, which in turn initiates mRNA decay.
Y. pestis causes plague and has two hosts: mammals and fleas. Y. pestis undergoes significant change when it is transmitted from the flea vector to the mammalian host. Recently, we found that the 3′ UTR increases hmsT mRNA decay at the body temperature of the mammalian host but not at the body temperature of the flea vector (room temperature) (Zhu et al., 2016). In the present study, to investigate the functions of 3′ UTRs in Y. pestis in greater detail, we analyzed 15 Y. pestis genes potentially possessing long 3′ UTRs but with differences in AU-content. The results showed that AU-rich 3′ UTRs can negatively regulate the expression of their own genes. Our findings suggest that functional 3′ UTRs are widespread in bacteria and warrant further investigations to uncover their regulatory mechanisms.
Results
Investigation of the General Effects of Long 3′ UTRs on Gene Expression in Y. pestis
Long 3′ UTRs play an important role in mRNA turnover (Selinger et al., 2003; Maeda and Wachi, 2012; Liu et al., 2016; Zhu et al., 2016). More interestingly, these long 3′ UTRs are usually rich in AU nucleotides (Maeda and Wachi, 2012; Lopez-Garrido et al., 2014; Zhu et al., 2016), indicating that AU-rich elements might be important for 3′ UTR-mediated mRNA decay. To investigate the role of AU-rich elements and the general effects of 3′ UTRs on mRNA turnover, 15 genes with long 3′ intergenic regions (IGRs > 250 bp) were randomly selected from the Y. pestis genome (Table 1). Seven genes had high (>68%) and eight genes had normal (45–60%) AT content in their 250 bp-3′ IGRs compared with the 52% AT content of the Y. pestis chromosome (Parkhill et al., 2001; Deng et al., 2002) (Table 1). Nucleotides 1–300 of the 3′ terminus (3′ T) of the 15 selected genes were deleted and replaced with a kanamycin (kan) cassette, and their mRNA levels were compared between wild-type (WT) and 3′ T1-300 deletion mutants using quantitative real-time PCR (qRT-PCR). As shown in Figure 1, for six of seven genes with high AT content (>68%) in their 3′ T regions, mRNA levels were significantly increased when the 3′ T region (potential 3′ UTR) was deleted. By contrast, for genes with normal AT content, only one gene (y2419, AT content = 49% in its 3′ T region) displayed a slightly increased mRNA expression when its 3′ T region (potential 3′ UTR) was deleted, whereas three genes (y2237, y1288, and nadB with AT content = 48%, 52%, and 46% in their 3′ T region, respectively) displayed slightly decreased mRNA expression.
Table 1.
Analysis of potential 3′ UTRs of 17 genes with long 3′ IGRs in Y. pestis.
| Gene name | IGR length (bp) | AT% in 3′ terminus (1–250 bp) | TTS (bp)∗ | AT% in potential 3′ UTR |
|---|---|---|---|---|
| hmsT | 540 | 73% | 283∗ | 70% |
| dsbA | 491 | 70% | 48∗∗ | 69% |
| y0624 | 314 | 72% | Unknown | – |
| y2025 | 837 | 68% | 63∗∗ | 59% |
| y1235 | 371 | 70% | 250 | 70% |
| lrhA | 677 | 70% | 60∗∗ | 72% |
| y4098 | 349 | 69% | 284 | 68% |
| y0961 | 459 | 70% | Unknown | – |
| cafA | 825 | 45% | Unknown | – |
| y2237 | 592 | 48% | 223∗∗ | 45% |
| y2419 | 661 | 49% | Unknown | – |
| y3757 | 540 | 57% | Unknown | – |
| hmsP | 639 | 60% | Unknown | – |
| amn | 424 | 55% | Unknown | – |
| y1288 | 282 | 52% | Unknown | – |
| nadB | 282 | 46% | 243∗∗ | 46% |
IGR, intergenic region; TTS, transcriptional termination site. ∗TTS of hmsT mRNA was determined by the 3′ RACE analysis as described in our previous study (Zhu et al., 2016). ∗∗Predicted Rho-independent TTS was determined based on the ARNold program. “Unknown” indicates the TTS is unknown.
FIGURE 1.
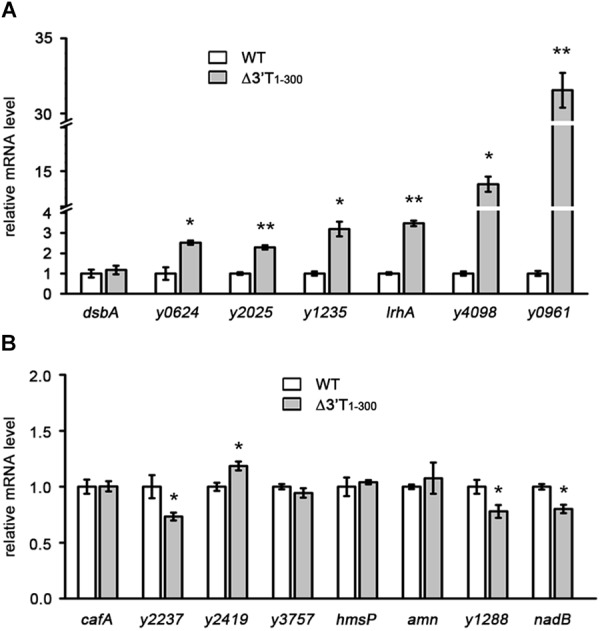
Characterization of the general effects of 3′ UTRs on mRNA expression in Yersinia pestis. (A,B) qRT-PCR analysis of transcripts of genes containing AT-rich (A) or normal (B) 3′ IGRs in the wild type (WT) and nucleotides 1–300 of the 3′ terminus (3′ T1-300) mutant strains (Δ3′T1-300), in which the 3′ T1-300 is disrupted by insertion of a kanamycin (kan) cassette. The Y. pestis WT strain was used as a control, and the fold change in relative mRNA level for each strain was compared with that of the WT (set as 1). Student’s t-test was used to calculate p-values. ∗p < 0.05, ∗∗p < 0.001. Means ± standard deviation (SD) from three independent experiments are shown.
To determine the role of these 3′ UTRs, we investigated their influence on the expression of a heterologous green fluorescent protein (gfp) gene. The 3′ T1-300 fragments of eight genes (six AT-rich and two normal) were cloned immediately into the downstream region of the gfp gene and the constructs were transformed into Escherichia coli cells, along with the vector control pAcGFP1 and a 3′ terminal control rrnB-TT (Figure 2A). As shown in Figure 2B, five 3′ terminal regions with high AT content caused a significant decrease in GFP fluorescence compared with the two controls (pAcGFP1 and rrnB-TT), whereas the y2237 and nadB 3′ terminal regions, which have moderate AT content, did not significantly affect gfp expression. Taken together, the results indicate that five out of seven selected AU-rich 3′ UTRs acted as independent units for the regulation of heterogonous gene expression. These results imply that the effects of 3′ UTRs on mRNA turnover might be broadly distributed in Y. pestis, and long AU-rich 3′ UTRs may be intimately involved in mRNA turnover.
FIGURE 2.
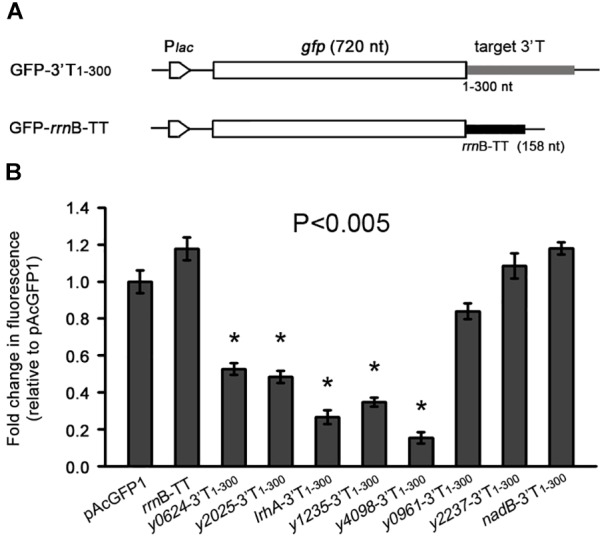
The effects of 3′ UTRs on expression of a heterologous green fluorescent (GFP) reporter. (A) Schematic representation of GFP reporter with different 3′ UTRs and an rrnB transcriptional terminator. (B) Relative GFP fluorescence of E. coli cells expressing GFP with different 3′ UTRs (GFP-3′T1-300) and the rrnB terminator, and without an additional 3′ UTR (pAcGFP1). Relative fluorescence intensities were normalized against the cell density (OD600 value) of the bacterial culture, and the fold change in fluorescence was compared with that of the vector control (set as 1). Student’s t-tests were used to calculate p-values. ∗p < 0.01. Means ± SD from three independent experiments are indicated.
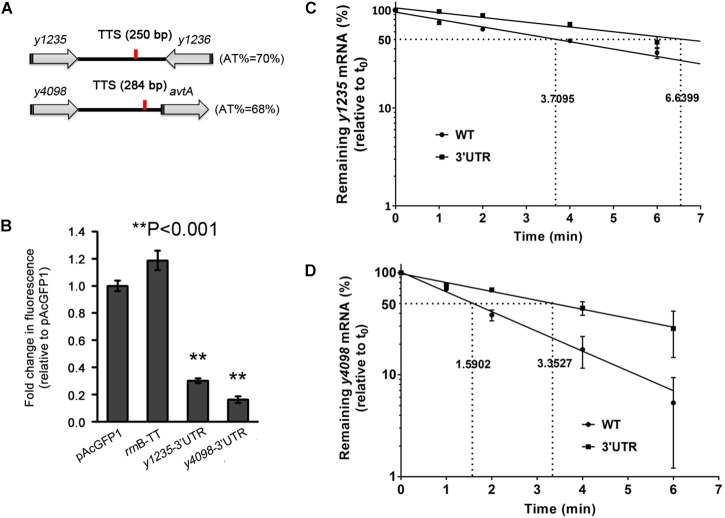
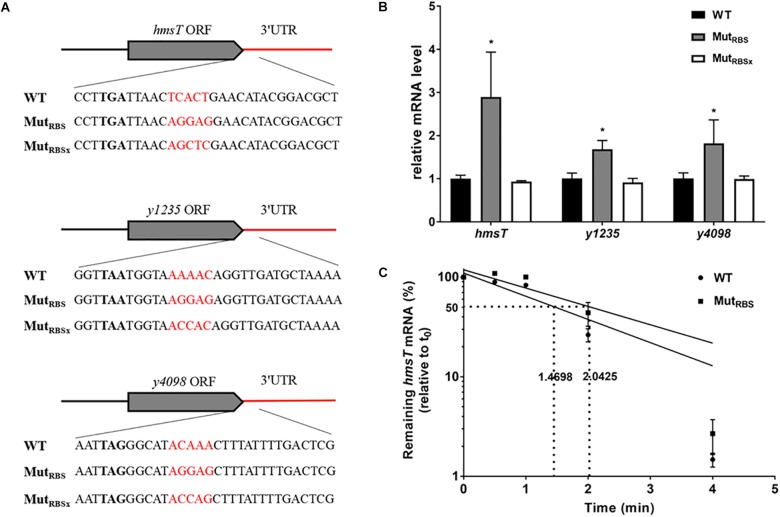
The 3′ UTRs of y1235 and y4098 Negatively Regulate mRNA Stability
To explore further the regulatory functions of the 3′ UTRs, those of y1235 and y4098, which significantly affected gfp expression (Figure 2B), were selected for further study. Sequence analysis using the ARNold program (Naville et al., 2011) revealed that no Rho-independent transcriptional terminator (intrinsic terminator) was present downstream of y1235 or y4098. 3′ rapid amplification of cDNA ends (RACE) data showed that the transcriptional termination sites (TTS) of y1235 and y4098 mRNAs are located 250- and 284-nt downstream of the stop codon, respectively (Figure 3A and Supplementary Figure S1). To further verify the role of these two 3′ UTRs, 250 and 284 bp fragments corresponding to the 3′ UTRs of y1235 and y4098 were cloned downstream of the gfp coding sequence, respectively. GFP reporter analysis showed that these constructs performed a similar regulatory role compared with their 3′ T1-300 regions (Figures 2B, 3B), further suggesting that the 3′ UTRs of y1235 and y4098 are involved in regulation of mRNA decay.
FIGURE 3.
The 3′ UTRs of y1235 and y4098 negatively regulate mRNA stability. (A) Schematic representation of y1235, y4098, and their transcriptional terminator (TTS, red line). (B) Relative GFP fluorescence of E. coli cells expressing GFP with the 3′ UTR of y1235 or y4098, or the rrnB terminator, or without an additional 3′ UTR (pAcGFP1). Relative fluorescence intensities were normalized against the OD600 value of the bacterial culture, and the fold change in fluorescence was compared with that of the vector control (set as 1). Student’s t-tests were used to calculate p-values. ∗∗p < 0.001. Means ± SD from three independent experiments are indicated. (C,D) The half-life of y1235 and y4098 mRNAs in WT and Δ3′ UTR mutant strains. Cells were grown at 26°C to an OD600 of 0.8, and rifampicin was added at time 0 to block RNA transcription. Samples were removed at time 0, 1 min, 2 min, 4 min, and 6 min. The percentage of residual mRNA at each time-point was measured by qRT-PCR and compared with that at time 0. Dashed lines indicate the time at which half of the detected mRNA remained. mRNA half-life was determined using linear regression analysis and is shown on the dashed lines. Error bars indicate SD of the mean.
To determine whether the 3′ UTR affects the expression of y1235 and y4098 at the post-transcriptional level, the half-lives of y1235 and y4098 mRNAs were measured by qRT-PCR in WT and 3′ UTR deletion mutant Y. pestis strains. The y1235 transcript half-life in the WT strain was 3.71 min, while the y1235 transcript half-life in the y1235 3′ UTR mutant was 6.64 min (Figure 3C). Deletion of the 3′ UTR of the y4098 transcript increased its half-life from 1.59 min to 3.35 min (Figure 3D). These results suggest that y1235 and y4098 3′ UTRs regulate mRNA decay at the post-transcriptional level.
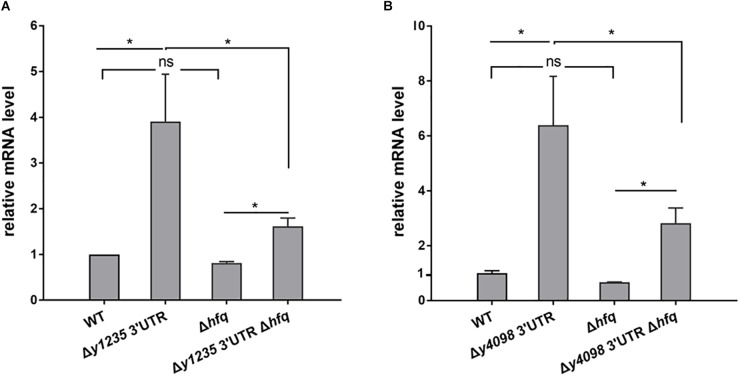
Examination of the Role of Hfq in 3′ UTR-Mediated y1235 and y4098 mRNA Decay
Hfq commonly binds to 3′ ends of mRNA (Holmqvist et al., 2016) and regulates mRNA stability by protecting RNA from degradation (Zhang et al., 2003; Chao et al., 2012). To investigate whether Hfq is involved in the regulation of 3′ UTR-mediated y1235 and y4098 mRNA decay, we used an hfq single deletion strain (Zhu et al., 2016), and constructed an hfq and y1235 or y4098 3′ UTR double deletion strain, and analyzed y1235 and y4098 mRNA levels in these strains by qRT-PCR. Deletion of hfq resulted in slightly decreased y1235 and y4098 mRNA levels relative to the WT strain (Figure 4). However, deletion of hfq in the 3′ UTR mutants caused a much stronger decrease in y1235 and y4098 mRNA levels relative to those in the 3′ UTR mutants (Figure 4), suggesting that Hfq protects their mRNA stability in a 3′UTR-dependent manner. In addition, y1235 and y4098 mRNA levels were much higher in the Δhfq Δ3′ UTR strain than in the Δhfq strains (Figure 4), suggesting that 3′ UTRs regulate y1235’ and y4098’ mRNA decay in an Hfq-independent manner.
FIGURE 4.
Examination of the roles of Hfq in 3′ UTR-mediated decay of y1235 and y4098 mRNA. Relative amounts of y1235 (A) and y4098 (B) mRNA produced by the WT, Δ3′ UTR, Δhfq, and Δ3′ UTR Δhfq strains. Student’s t-test was used to calculate p-values. ∗p < 0.01. Means ± standard deviation (SD) from three independent experiments are shown.
The Roles of RNases in the Expression of y1235 and y4098
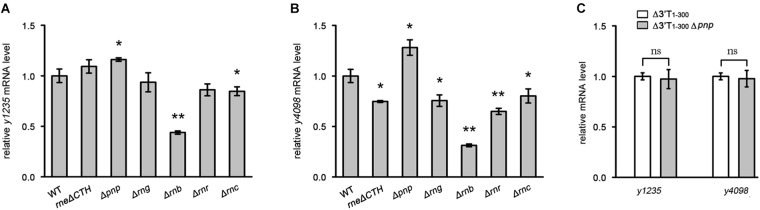
3′ UTRs may be targeted by RNases to initiate mRNA decay (Maeda and Wachi, 2012; Lopez-Garrido et al., 2014). To investigate which RNases contribute to 3′ UTR-mediated mRNA decay, we analyzed y1235 and y4098 mRNA abundance in cells in which RNase E, RNase G, RNase II, RNase R, RNase III, or PNPase was mutated (Zhu et al., 2016). RNase E is essential, but deletion of its C-terminal CTH region impairs RNase E-mediated RNA degradation without causing cell death (Bouvier and Carpousis, 2011; Ow et al., 2000). Thus, we used a Y. pestis C-terminal truncated RNase E mutant (Zhu et al., 2016) to determine whether it is involved in 3′ UTR-mediated mRNA decay. Mutation of RNase E, RNase G, RNase II, RNase R, or RNase III did not increase the mRNA levels of y1235 and y4098. However, mutation of PNPase caused increased y1235 and y4098 mRNA levels (Figures 5A,B), suggesting that PNPase might target these mRNAs for degradation. To determine whether the 3′ UTRs of y1235 and y4098 are targeted by PNPase, a PNPase mutation was generated in Y. pestis y1235 3′ UTR and y4098 3′ UTR deletion mutants. The levels of y1235 and y4098 mRNAs were similar in PNPase+ and PNPase- backgrounds (Figure 5C), indicating that PNPase is involved in 3′ UTR-mediated degradation of y1235 and y4098 mRNAs.
FIGURE 5.
The effects of the major RNases on 3′ UTR-mediated mRNA turnover. The major RNases coding genes, CTH of rne [the carboxyl terminal half (CTH) region of RNase E], pnp (PNPase), rng (RNase G), rnb (RNase II), rnr (RNase R), and rnc (RNase III) were deleted, respectively. (A,B) Relative y1235 (A) and y4098 (B) mRNA levels in Y. pestis RNases mutants. (C) Relative y1235 and y4098 mRNA levels in Y. pestis Δ3′ UTR (white bars) and PNPase-Δ3′ UTR (gray bars) isogenic strains. Student’s t-test was used to calculate p-values. ∗p < 0.05, ∗∗p < 0.001. Means ± SD from three independent experiments are indicated.
The y1235 and y4098 3′ UTRs Regulate Gene Expression in Response to Temperature Change
The regulation of gene expression in a precise manner is critical for bacterial adaptation to changing environments. Recently, we reported that 3′ UTR-mediated hmsT mRNA decay in Y. pestis is affected by changes in temperature (Zhu et al., 2016). To test whether temperature change affects 3′ UTR-dependent stability of y1235 and y4098 mRNAs, we compared y1235 and y4098 mRNA levels in WT and 3′ UTR mutant strains at 21°C, 26°C, and 37°C. Figure 6A shows that the level of the y1235 transcript was moderately high at 21°C (2.8-fold), but much higher at 26°C (3.8-fold) and 37°C (4.3-fold). To our surprise, the level of the y4098 transcript was highest at 26°C (13.8-fold), although its level was also high at 21°C (5.8-fold) and 37°C (5.7-fold) (Figure 6B). The results suggest that the 3′ UTRs of y1235 and y4098 function as thermosensors and differentially regulate gene expression in response to temperature changes.
FIGURE 6.
The effects of temperature on 3′ UTR-mediated mRNA turnover. qRT-PCR analysis of relative y1235 (A) and y4098 (B) mRNA levels in the WT and Δ3′ UTR mutant strain at 21°C, 26°C, and 37°C. Student’s t-test was used to calculate p-values. ∗p < 0.01, ∗∗p < 0.005. Means ± SD from three independent experiments are indicated.
The Effects of Ribosome Binding to 3′ UTRs
In general, 5′ UTRs are targeted by RNases to induce mRNA decay, while ribosome binding to a strong Shine-Dalgarno-like sequence (ribosome binding site, RBS) promotes mRNA stability (Agaisse and Lereclus, 1996; Sharp and Bechhofer, 2003; Daou-Chabo et al., 2009). Since 3′ UTRs may also be attacked by RNases to initiate mRNA decay, we wondered whether protein binding to 3′ UTR could affect mRNA stability. To answer this question, we introduced an RBS region into 5′ end of the 3′ UTR of hmsT, y1235, and y4098 genes (Figure 7A). Introduction of an RBS region in the 3′ UTR resulted in a significant increase in the mRNA levels of all three genes (Figure 7B). However, introduction of a sequence (RBSx) not recognized by ribosomes did not affect mRNA levels (Figures 7A,B). To determine whether introduction of an RBS in the 3′ UTR affects post-transcriptional gene expression, the mRNA half-life of hmsT was measured by qRT-PCR. Consistent with the above results, introduction of an RBS region but not a non-specific sequence into the 3′ UTR increased the hmsT mRNA half-life (Figure 7C). Taken together, these results suggest that binding of some proteins, such as ribosomes, to the 3′ UTR can protect it from degradation, which in turn promotes the stability of upstream mRNA.
FIGURE 7.
The effects of ribosome binding to 3′ UTR on mRNA decay. (A) Schematic representation of mutations introduced in 3′ UTRs of hmsT, y1235, and y4098. (B) qRT-PCR analysis of relative hmsT, y1235, and y4098 mRNA levels in Y. pestis WT strain, in the strain with a 3′ UTR containing a ribosome binding site (RBS), and in the strain containing the 3′ UTR with a sequence not recognized by the ribosome (RBSx). Student’s t-test was used to calculate p-values. ∗p < 0.05. Means ± SD from three independent experiments are presented. (C) hmsT mRNA half-lives in the WT and 3′ UTR with RBS strains. Cells were grown at 26°C to an OD600 of ∼0.8, and rifampicin was added at time 0 to block RNA transcription. Samples were removed at time 0, 0.5 min, 1 min, 2 min, and 4 min. The percentage of mRNA remaining at each time-point was measured by qRT-PCR and compared with that at time 0. Dashed lines indicate the time at which half of the detected mRNA remained. The half-life of mRNAs was determined by linear regression analysis and is shown on the dashed lines. Error bars indicate the SD of the mean.
Discussion
Degradation of mRNA is dependent on a rate-determining initial step in bacteria (Kushner, 2002; Bechhofer, 2009). mRNA is initially degraded by endonucleolytic enzymes. After endonucleolytic cleavage, exoribonucleases degrade the mRNA fragments, but ribosomes binding prevent initial mRNA endonucleolytic degradation of mRNA (Deneke et al., 2013). Thus, ribonucleases tend to degrade more readily the untranslatable regions of mRNA. Although 3′ UTRs are generally more stable than 5′ UTRs, subgenic-resolution oligonucleotide microarrays analysis of the positional patterns of transcript degradation in E. coli showed that some mRNAs have unstable 3′ UTRs, indicating that 3′ UTRs might also initiate mRNA decay (Selinger et al., 2003). 3′ UTRs have recently been shown to affect gene expression in bacteria by altering mRNA stability and translation in trans and cis, and thus have been classified as new post-transcriptional regulatory elements (Novick et al., 1993; Balaban and Novick, 1995; Felden et al., 2011; Ruiz de los Mozos et al., 2013; Laalami et al., 2014). Although a number of functional 3′ UTRs and their regulatory functions have been identified in bacteria, further work will be required to identify additional functional 3′ UTRs and their regulatory mechanisms.
We identified additional functional 3′ UTRs by focusing on long 3′ UTRs and found that among 3′ UTRs, those with long AU-rich regions are probably more important for mRNA expression regulation. Interestingly, mRNA levels were higher when six out of seven potential AU-rich 3′ UTRs were deleted, whereas mRNA levels were only slightly affected in the absence of 3′ UTRs with normal AU content (Figure 1). Together with the results of GFP reporter assays showing that these 3′ UTRs regulate heterologous gene expression (Figure 2), these results suggest that AU-rich long 3′ UTRs play a prominent role in regulating gene expression.
Why are AU-rich 3′ UTRs more likely to be involved in the regulation of mRNA decay? There are at least three possible reasons: (1) AU-rich sequences are preferentially cleaved by endoribonucleases such as RNase E and G; (2) AU-rich sequences do not tend to form stable secondary structures and thus can be readily degraded by exoribonucleases; (3) the structures formed by AU-rich sequences are more likely to undergo secondary structural changes in response to temperature change, which would facilitate the fine-tuning of mRNA degradation to changes in temperature. In addition, 3′ UTRs may contain other elements that regulate gene expression. For example, icaR expression is controlled by base pair interactions between 5′ and 3′ UTRs in Staphylococcus aureus (Ruiz de los Mozos et al., 2013). The 5′/3′-UTR interaction controls ribonuclease activity to regulate hbs mRNA stability in Bacillus subtilis (Braun et al., 2017). In addition, RNA binding proteins such as Hfq prefer to bind to AU-rich sequences and regulate mRNA stability (Zhang et al., 2003; Chao et al., 2012; Holmqvist et al., 2016). Taken together, it appears that mRNA turnover is regulated by different types of 3′ UTRs in bacteria.
Similar to hmsT and slrA (Liu et al., 2016; Zhu et al., 2016), y1235 and y4098, for which PNPase is involved in 3′ UTR-mediated mRNA decay, contain Rho-dependent terminators. PNPase cannot degrade strong stem-loop structures and its function relies on the presence of PNPase-susceptible 3′ ends (Liu et al., 2016). Rho-independent terminators but not Rho-dependent terminators usually form a strong stem-loop structure on the end of 3′ UTRs. Thus, PNPase is more likely to cleave from the 3′ ends of mRNAs with Rho-dependent terminators. This could explain why PNPase plays an important role in mRNA decay when 3′ UTRs contain Rho-dependent terminators. However, y1235 and y4098 mRNA levels were increased by only 1.2-fold and 1.3-fold in the absence of PNPase, respectively, suggesting that other ribonucleases may contribute to 3′ UTR-mediated mRNA decay. In addition, 3′ UTRs containing a Rho-independent terminator such as y2025 and lrhA might also regulate mRNA decay (Figures 1, 2). More than 21 ribonucleases are known in E. coli and most have Y. pestis homologs (Mackie, 2013). The ribonucleases involved in 3′ UTR-mediated mRNA degradation remain to be identified.
SmAP2 binds to 3′ UTRs and regulates mRNA stability in the crenarchaeum Sulfolobus solfataricus (Martens et al., 2017). Our results showed that Hfq protects mRNA from degradation in a 3′ UTR-dependent manner (Figure 4) and ribosome binding to the 3′ UTR promotes mRNA stability in Y. pestis (Figure 7C), suggesting that other RNA-binding proteins in bacteria might also regulate gene expression by binding to 3′ UTRs. RNA-binding proteins might mask endoribonucleolytic sites in 3′ UTRs or protect mRNA from degradation from 3′ ends by exoribonucleases. Several different mechanisms of 3′ UTR-regulated gene expression have been reported, but others remain to be elucidated (Ren et al., 2017). Further work should be directed toward identifying new functional 3′ UTRs and novel 3′ UTR-dependent regulatory mechanisms. This will increase our knowledge of how bacteria use 3′ UTR-dependent mRNA degradation to regulate gene expression in response to environmental changes.
Materials and Methods
Strains and Plasmids
Strains and plasmids used in this study are listed in Supplementary Table S1. Y. pestis KIM6+ was the WT strain. Y. pestis with deletions in 3′ intergenic regions and hfq was constructed by inserting a kanamycin (kan) and chloramphenicol (cat) resistance cassette, respectively, using the Lambda red protocol as previously described (Datsenko and Wanner, 2000; Sun et al., 2008). The 3′ UTR mutants of hmsT, y1235, and y4098 genes were generated using the CRISPR-Cas12a-assisted genome editing tool (Yan et al., 2017). Overlapping PCR was used to insert 300 bp 3′ T regions of selected genes immediately downstream of the pAcGFP1 gfp stop codon, resulting in plasmids pAcGFP1-3′ T1-300. Plasmids encoding gfp fusion with 3′ UTR of y1235 or y4098 were constructed by reverse PCR using the corresponding pAcGFP1-3′ T1-300 as template. Strains and plasmids were constructed using the oligonucleotides listed in Supplementary Table S2. Cells were grown at 26°C in LB medium unless otherwise specified.
Quantitative RT-PCR
qRT-PCR was performed as described previously with minor modifications (Sun et al., 2011). Briefly, cells in LB broth were grown overnight at 26°C with shaking at 250 rpm, diluted to an OD600 of ∼0.02, and then grown at 26°C in LB broth to an OD600 of ∼0.8. The RNeasy mini kit (Qiagen) was used to isolate total RNA from cells and DNase I (Invitrogen) was used to remove residual DNA. The purified RNA was used for quantitative PCR using the TaqMan® RNA -to-CTTM 1-Step Kit (ABI) and CFX96TM Real-Time System (Bio-Rad) according to the user guide. mRNA quantity was normalized with respect to the 16S rRNA gene, and ratios of normalized mRNA quantities in different strains to the normalized quantities in the WT strain were calculated. The data presented represents the results of three independent experiments performed in triplicate. Probes and primer sets are listed in Supplementary Table S2.
mRNA Half-Life Determination
mRNA half-life was determined as described previously with minor modifications (Bellows et al., 2012). WT Y. pestis (KIM6+) and 3′ UTR mutant strains in LB broth were grown overnight at 26°C with shaking at 250 rpm, diluted to an OD600 of ∼0.02, and then grown at 26°C to an OD600 of approximately ∼0.8. Rifampicin was added at a final concentration of 250 μg/ml to prevent initiation of transcription. Samples were collected at times 0, 0.5 min, 1 min, 2 min, and 4 min. RNA was extracted and analyzed by qRT-PCR, and the mRNA half-life was determined by linear regression analysis.
3′ RACE
3′ RACE was used to determine the transcription termination sites of y1235 and y4098 genes, as described previously (Zhu et al., 2016), using the 3′ RACE System for Rapid Amplification of cDNA Ends Kit (Invitrogen) according to the manufacturer’s instructions. RT-PCR products were separated using agarose gel electrophoresis, gel-purified, cloned into the pGEM-T vector, and transformed into E. coli cells. Transformants were picked for DNA sequencing and analysis.
GFP Assay
Green fluorescent protein reporter analysis was performed as described previously (Zhu et al., 2016). Briefly, overnight cultures of E. coli strains harboring GFP reporters were diluted 1:1000 in triplicate and grown in 5 ml LB at 37°C to an OD600 of ∼0.35. Aliquots (100 μl) were then placed in 96-well Opti-Plates and GFP fluorescence was detected using a Multi label Reader (PerkinElmer) with an excitation wave length of 475 nm and an emission wavelength of 505 nm. The fluorescence of a control strain not expressing GFP was subtracted from the fluorescence of the samples, and relative fluorescence intensities were normalized against cell density (OD600 value).
Author Contributions
Experiments were designed and manuscript was written by J-PZ, HZ, and Y-CS. Experiments were carried out by J-PZ, HZ, and X-PG. Manuscript review and modification were performed by J-PZ, HZ, X-PG, and Y-CS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was funded by National Natural Science Foundation of China (31870067), the National Basic Research Program of China (973 Program) (2015CB554200), the Youth Fund of PUMC (3332016084), and the CAMS Initiative for Innovative Medicine (2016-I2M-1-013).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03080/full#supplementary-material
Mapping of the transcription termination site of y1235 (A) and y4098 (B) mRNA by 3′ rapid amplification of cDNA ends (RACE). 3′ RACE products were analyzed by agarose gel electrophoresis. Fragments of upper bands (red arrows) obtained by PCR amplification were purified, cloned into pUC19, and transformed into E. coli cells. The resulting clones were picked for sequencing. The corresponding regions of 3′ UTRs are underlined, and the specific primers used for 3′ RACE are labeled.
Strains and plasmids employed in this study.
Primers and probes used in this work.
References
- Agaisse H., Lereclus D. (1996). STAB-SD: a shine-dalgarno sequence in the 5’ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20 633–643. 10.1046/j.1365-2958.1996.5401046.x [DOI] [PubMed] [Google Scholar]
- Balaban N., Novick R. P. (1995). Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3’-end deletion. FEMS Microbiol. Lett. 133 155–161. 10.1016/0378-1097(95)00356-A [DOI] [PubMed] [Google Scholar]
- Bechhofer D. H. (2009). Messenger RNA decay and maturation in Bacillus subtilis. Prog. Mol. Biol. Transl. Sci. 85 231–273. 10.1016/S0079-6603(08)00806-4 [DOI] [PubMed] [Google Scholar]
- Bellows L. E., Koestler B. J., Karaba S. M., Waters C. M., Lathem W. W. (2012). Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86 661–674. 10.1111/mmi.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M., Carpousis A. J. (2011). A tale of two mRNA degradation pathways mediated by RNase E. Mol. Microbiol. 82 1305–1310. 10.1111/j.1365-2958.2011.07894.x [DOI] [PubMed] [Google Scholar]
- Braun F., Durand S., Condon C. (2017). Initiating ribosomes and a 5’/3’-UTR interaction control ribonuclease action to tightly couple B. subtilis hbs mRNA stability with translation. Nucleic Acids Res. 45 11386–11400. 10.1093/nar/gkx793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. R. (2011). Prospects for riboswitch discovery and analysis. Mol. Cell 43 867–879. 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y., Papenfort K., Reinhardt R., Sharma C. M., Vogel J. (2012). An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31 4005–4019. 10.1038/emboj.2012.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y., Vogel J. (2016). A 3’ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell 61 352–363. 10.1016/j.molcel.2015.12.023 [DOI] [PubMed] [Google Scholar]
- Daou-Chabo R., Mathy N., Benard L., Condon C. (2009). Ribosomes initiating translation of the hbs mRNA protect it from 5’-to-3’ exoribonucleolytic degradation by RNase J1. Mol. Microbiol. 71 1538–1550. 10.1111/j.1365-2958.2009.06620.x [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneke C., Lipowsky R., Valleriani A. (2013). Effect of ribosome shielding on mRNA stability. Phys. Biol. 10:046008. 10.1088/1478-3975/10/4/046008 [DOI] [PubMed] [Google Scholar]
- Deng W., Burland V., Plunkett G., III, Boutin A., Mayhew G. F., Liss P., et al. (2002). Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184 4601–4611. 10.1128/JB.184.16.4601-4611.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B., Vandenesch F., Bouloc P., Romby P. (2011). The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 7:e1002006. 10.1371/journal.ppat.1002006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossringer M., Hartmann R. K. (2012). 3’-UTRs as a source of regulatory RNAs in bacteria. EMBO J. 31 3958–3960. 10.1038/emboj.2012.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E., Regnier P. (2000). Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. U.S.A. 97 1501–1505. 10.1073/pnas.040549897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T. M. (2008). Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 22 3383–3390. 10.1101/gad.1747308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E., Wright P. R., Li L., Bischler T., Barquist L., Reinhardt R., et al. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35 991–1011. 10.15252/embj.201593360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Shin J. H., Cho Y. B., Roe J. H. (2014). Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3’UTR of the sodF mRNA. Nucleic Acids Res. 42 2003–2014. 10.1093/nar/gkt1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J., Narberhaus F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10 255–265. 10.1038/nrmicro2730 [DOI] [PubMed] [Google Scholar]
- Krajewski S. S., Narberhaus F. (2014). Temperature-driven differential gene expression by RNA thermosensors. Biochim. Biophys. Acta 1839 978–988. 10.1016/j.bbagrm.2014.03.006 [DOI] [PubMed] [Google Scholar]
- Kushner S. R. (2002). mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184 4658–4665; discussion 4657. 10.1128/JB.184.17.4658-4665.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laalami S., Zig L., Putzer H. (2014). Initiation of mRNA decay in bacteria. Cell Mol. Life Sci. 71 1799–1828. 10.1007/s00018-013-1472-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Derout J., Folichon M., Briani F., Deho G., Regnier P., Hajnsdorf E. (2003). Hfq affects the length and the frequency of short oligo(A) tails at the 3’ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 31 4017–4023. 10.1093/nar/gkg456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Kearns D. B., Bechhofer D. H. (2016). Expression of multiple Bacillus subtilis genes is controlled by decay of slrA mRNA from Rho-dependent 3’ ends. Nucleic Acids Res. 44 3364–3372. 10.1093/nar/gkw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garrido J., Puerta-Fernandez E., Casadesus J. (2014). A eukaryotic-like 3’ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res. 42 5894–5906. 10.1093/nar/gku222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. (2013). RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11 45–57. 10.1038/nrmicro2930 [DOI] [PubMed] [Google Scholar]
- Maeda T., Wachi M. (2012). 3’ Untranslated region-dependent degradation of the aceA mRNA, encoding the glyoxylate cycle enzyme isocitrate lyase, by RNase E/G in Corynebacterium glutamicum. Appl. Environ. Microbiol. 78 8753–8761. 10.1128/AEM.02304-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens B., Sharma K., Urlaub H., Blasi U. (2017). The SmAP2 RNA binding motif in the 3’UTR affects mRNA stability in the crenarchaeum Sulfolobus solfataricus. Nucleic Acids Res. 45 8957–8967. 10.1093/nar/gkx581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi M., Chao Y., Vogel J. (2015). Regulatory small RNAs from the 3’ regions of bacterial mRNAs. Curr. Opin. Microbiol. 24 132–139. 10.1016/j.mib.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D. (2011). ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8 11–13. 10.4161/rna.8.1.13346 [DOI] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12 3967–3975. 10.1002/j.1460-2075.1993.tb06074.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva G., Sahr T., Buchrieser C. (2015). Small RNAs, 5’ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 39 331–349. 10.1093/femsre/fuv022 [DOI] [PubMed] [Google Scholar]
- Opdyke J. A., Kang J. G., Storz G. (2004). GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186 6698–6705. 10.1128/JB.186.20.6698-6705.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M. C., Liu Q., Kushner S. R. (2000). Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38 854–866. 10.1046/j.1365-2958.2000.02186.x [DOI] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Thomson N. R., Titball R. W., Holden M. T., Prentice M. B., et al. (2001). Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413 523–527. 10.1038/35097083 [DOI] [PubMed] [Google Scholar]
- Peng T., Berghoff B. A., Oh J. I., Weber L., Schirmer J., Schwarz J., et al. (2016). Regulation of a polyamine transporter by the conserved 3’ UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides. RNA Biol. 13 988–999. 10.1080/15476286.2016.1212152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G., Mignone F., Gissi C., Grillo G., Licciulli F., Liuni S. (2001). Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276 73–81. 10.1016/S0378-1119(01)00674-6 [DOI] [PubMed] [Google Scholar]
- Ren G. X., Guo X. P., Sun Y. C. (2017). Regulatory 3’ untranslated regions of bacterial mRNAs. Front. Microbiol. 8:1276 10.3389/fmicb.2017.01276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de los Mozos I., Vergara-Irigaray M., Segura V., Villanueva M., Bitarte N., Saramago M., et al. (2013). Base pairing interaction between 5’- and 3’-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 9:e1004001. 10.1371/journal.pgen.1004001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D. W., Saxena R. M., Cheung K. J., Church G. M., Rosenow C. (2003). Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13 216–223. 10.1101/gr.912603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A., Patel D. J. (2012). Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu. Rev. Biophys. 41 343–370. 10.1146/annurev-biophys-101211-113224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. S., Bechhofer D. H. (2003). Effect of translational signals on mRNA decay in Bacillus subtilis. J. Bacteriol. 185 5372–5379. 10.1128/JB.185.18.5372-5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaggi J. M., Perkins J. B., Losick R. (2005). Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 187 6641–6650. 10.1128/JB.187.19.6641-6650.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Hinnebusch B. J., Darby C. (2008). Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U.S.A. 105 8097–8101. 10.1073/pnas.0803525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Koumoutsi A., Jarrett C., Lawrence K., Gherardini F. C., Darby C., et al. (2011). Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6:e19267. 10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree J. J., Granneman S., McAteer S. P., Tollervey D., Gally D. L. (2014). Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell 55 199–213. 10.1016/j.molcel.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M. Y., Yan H. Q., Ren G. X., Zhao J. P., Guo X. P., Sun Y. C. (2017). CRISPR-Cas12a-assisted recombineering in bacteria. Appl. Environ. Microbiol. 83:e00947-17. 10.1128/aem.00947-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman K. M., Rosenow C., Tjaden B. C., Storz G., Gottesman S. (2003). Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50 1111–1124. 10.1046/j.1365-2958.2003.03734.x [DOI] [PubMed] [Google Scholar]
- Zhu H., Mao X. J., Guo X. P., Sun Y. C. (2016). The hmsT 3’ untranslated region mediates c-di-GMP metabolism and biofilm formation in Yersinia pestis. Mol. Microbiol. 99 1167–1178. 10.1111/mmi.13301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mapping of the transcription termination site of y1235 (A) and y4098 (B) mRNA by 3′ rapid amplification of cDNA ends (RACE). 3′ RACE products were analyzed by agarose gel electrophoresis. Fragments of upper bands (red arrows) obtained by PCR amplification were purified, cloned into pUC19, and transformed into E. coli cells. The resulting clones were picked for sequencing. The corresponding regions of 3′ UTRs are underlined, and the specific primers used for 3′ RACE are labeled.
Strains and plasmids employed in this study.
Primers and probes used in this work.