Abstract
We investigated the pH dependence of the fluorescence spectra of ADIFAB (FFA Sciences), a probe used for the quantification of free fatty acids (FFA). Data reports the change in the emission peak of ADIFAB and in the affinity for FFA as a function of pH. An algorithm based on spectral deconvolution allowed to correct ADIFAB fluorescence spectra for the spectroscopic effect caused by pH. Kd values were calculated at each pH based on a calibration with oleic acid. This method allows estimating FFA concentration by ADIFAB in media at different pH. The current data are related to the research article “Phospholipid components of the synthetic pulmonary surfactant CHF5633 probed by fluorescence spectroscopy” (Faggiano et al., 2018) [1].
Specifications table
| Subject area | Biochemistry, Spectroscopy, Pharmaceutical Science |
| More specific subject area | Free fatty acid quantification, fluorescence |
| Type of data | Figures |
| How data was acquired | Spectrofluorometer |
| Data format | Analyzed |
| Experimental factors | Samples equilibrated at 20 °C |
| Experimental features | ADIFAB fluorescence spectra in buffer phosphate at different pH and oleic acid concentrations were acquired and analyzed. |
| Data source location | Parma, Italy |
| Data accessibility | All data are with this article |
| Related research article | “Phospholipid components of the synthetic pulmonary surfactant CHF5633 probed by fluorescence spectroscopy” |
| Serena Faggiano, Luca Ronda, Samanta Raboni, Franco Sartor, Valeria Cavatorta, Elisa Sgarbi, Grazia Caivano, Marisa Pertile, Andrea Mozzarelli. International Journal of Pharmaceutics [1] |
Value of the data
-
•
ADIFAB is a fluorescent probe that can be used for free fatty acid quantification in a standard buffer at pH 7.4.
-
•
ADIFAB fluorescence spectrum depends on pH, limiting probe applications.
-
•
The data can be used to show how dependence of free fatty acids affinity for ADIFAB upon pH affects their quantification.
-
•
An algorithm allows correcting pH effect on ADIFAB spectra and on the affinity for free fatty acids.
-
•
This method enables the direct use of ADIFAB in media at different pH, allowing the determination of free fatty acids concentrations in liposomal preparations without prior dilution.
1. Data
1.1. Fluorescence spectra of ADIFAB at different pH and correction of the effect of pH
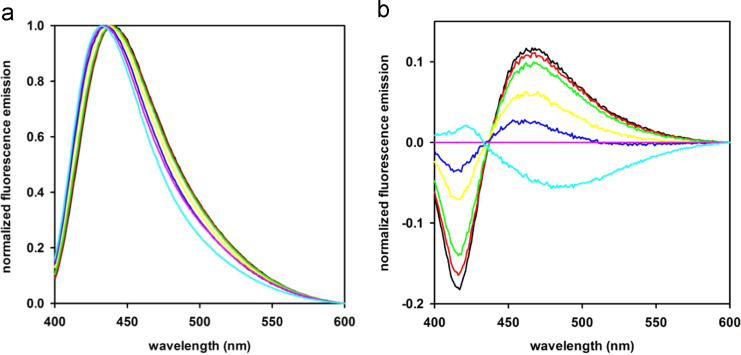
We recorded fluorescence spectra of ADIFAB [2] in 50 mM Na phosphate, 140 mM NaCl, prepared at pH between 6.2 and 7.6 by mixing NaH2PO4 and Na2HPO4 solutions (Fig. 1, a). Acquired spectra were normalized from 0 to 1.
Fig. 1.
a) Normalized spectra of the fluorescence of ADIFAB in measuring buffer at different pH: 6.2 (black line), 6.4 (red), 6.6 (green), 7.0 (yellow), 7.2 (blue), 7.4 (purple), 7.6 (cyan). All spectra were acquired at 20 °C. b) Difference spectra obtained by subtraction of the spectrum at reference pH (7.4, pH of the standard measuring buffer for ADIFAB). Color code as in a).
Difference spectra were obtained by subtracting the normalized spectrum at the reference pH (7.4) to all other spectra (Fig. 1, b). Then, by using MATLAB, a Singular Value Decomposition (SVD) matrix factorization was carried out on the difference spectra dataset [3], [4], [5]. In SVD analysis, the difference spectra data matrix A (m × n), where m is the number of wavelengths and n is the number of collected spectra, is resolved into a product of three matrices, named U, S, and VT:
| (1) |
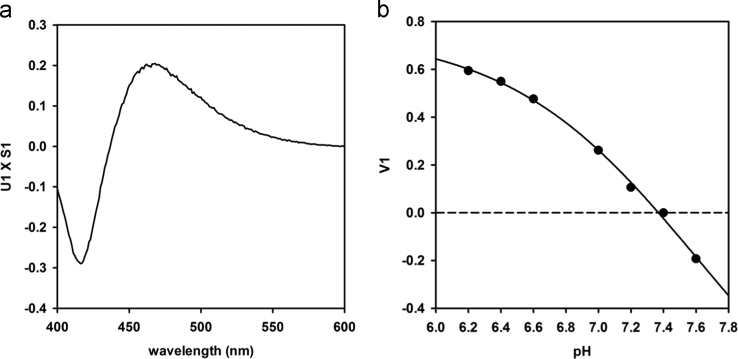
The U matrix consists of n orthonormal eigenvectors which are the spectral components, S is a square diagonal matrix with non-negative values on the diagonal, called singular values, and VT is a matrix containing eigenvalues, in which rows are also orthonormal, which correspond to the relative weight of each spectral component. The first singular value S1 accounts for 82% of the total of the singular values. The product U1 X S1 (Fig. 2, a) resulted to have the same spectral shape of difference spectra (Fig. 1, b).
Fig. 2.
a) First spectral component (U1) multiplied by S1 from SVD analysis. b) pH dependence of V1 (closed circles) fitted to Eq. (2) (solid line).
The dependence of the first V column on pH (Fig. 2, b) was fitted to the sigmoidal equation:
| (2) |
The fitting gave the following parameters: A = 1.91 ± 1.36, b = -0.59 ± 0.30, pH0 = 7.62 ± 0.65, V10 = -1.15 ± 1.22. These parameters were used to determine the V1 at the pH of the media. V1 was then multiplied by the vector U1 and the first singular value S1. The resulting spectrum is the correction curve to be subtracted to the experimental spectra in order to eliminate the effect of pH dependence.
1.2. Fluorescence spectra of ADIFAB in the presence of 0.2 μM oleic acid and extrapolation of corrected spectra
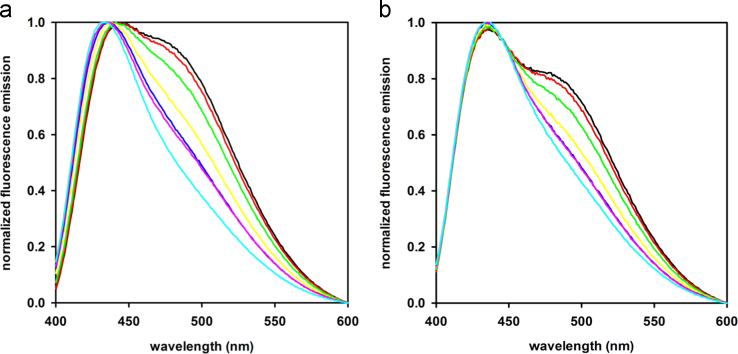
We acquired fluorescence spectra of ADIFAB at different pH in the presence of 0.2 μM oleic acid (OA) in a buffer containing 50 mM Na phosphate, 140 mM NaCl. (Fig. 3, a). The spectrum at each pH in the absence of OA was subtracted to the spectrum with 0.2 μM OA. Then, the spectrum without OA at pH 7.4 was summed. This procedure permitted to obtain a set of spectra in which the contribution of the change in fluorescence purely due to pH change was eliminated (Fig. 3, b).
Fig. 3.
a) Normalized spectra of the fluorescence of ADIFAB in measuring buffer at different pH the presence of 0.2 μM OA: 6.2 (black line), 6.4 (red), 6.6 (green), 7.0 (yellow), 7.2 (blue), 7.4 (purple), 7.6 (cyan). b) Normalized spectra of the fluorescence of ADIFAB in measuring buffer upon pH effect correction based on SVD analysis, same color code as in a).
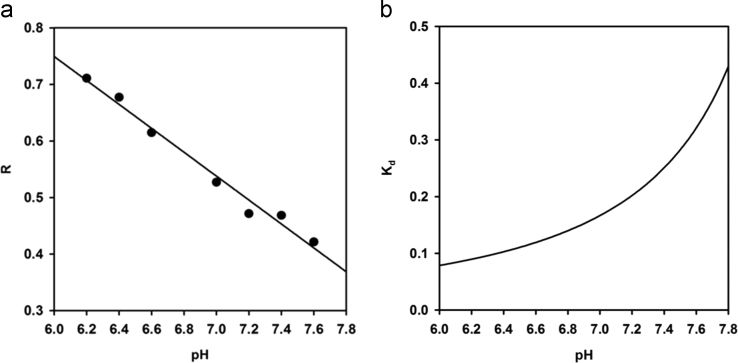
The R value (E505/E432, corresponding to the ratio of the fluorescence emission at 505 nm and at 432 nm upon excitation at 386 nm), necessary to measure the concentration of FFA according to ADIFAB protocol [2], was extrapolated at each experimental pH (Fig. 4, a) and fitted to a linear equation:
| (3) |
Fig. 4.
a) R (E505/E432 nm fluorescence emission ratio) dependence upon pH (closed circles) fitted to a linear equation (solid line). b) pH dependence of the Kd of ADIFAB for OA.
Fitting gave the following parameters: y0 = 2.02 ± 0.08 and a = -0.21 ± 0.01.
The correlation between R and Kd (Eq. (7)) was derived from Eqs. (4), (5), (6) (FFA Sciences protocol), in which Rmax (ratio of the fluorescence intensity at 505 to 432 nm when ADIFAB is saturated with FFA) was set to 9.9, and R0 (ratio of the fluorescence intensity at 505 to 432 nm in the absence of FFA) was set to 0.2. Concentrations were expressed in micromolar units:
| (4) |
| (5) |
| (6) |
| (7) |
Putting together Eqs. (3), (7), we obtained:
| (8) |
where FFAtotal = 0.2 μM, ADIFABtotal = 0.2 μM, R0 = 0.2, Rmax = 9.9, y0 = 2.02 and a = -0.21. We therefore extrapolated the Kd dependence on pH, as reported in Fig. 4, b.
The value of Kd can be then used together with the R value of the spectra corrected for pH effect to obtain FFA concentration applying Eqs. (4), (5), (6).
2. Experimental design, materials and methods
2.1. Materials
ADIFAB was purchased from FFA Sciences (San Diego, CA). Buffers were purchased from Sigma Aldrich (St. Louis, MO, USA).
2.2. Spectra acquisition
The fluorescence spectra of ADIFAB were acquired with a FluoroMax-3 fluorometer (HORIBA Jobin Yvon, Longjumeau Cedex, France) using a quartz cuvette of 0.5 × 0.5 mm. ADIFAB concentration was 0.2 μM. All measurements were performed at 20 °C. ADIFAB was excited at 386 nm and fluorescence emission spectra were recorded in the range 400–600 nm. A spectrum of the buffer was subtracted to ADIFAB spectra as a reference. OA was added from a 50 μM stock.
2.3. Data analysis
The SVD analysis, spectral subtraction and data fitting were performed using MATLAB. Graphical representations were carried out using the software Sigmaplot.
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.12.006.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Faggiano S., Ronda L., Raboni S., Sartor F., Cavatorta V., Sgarbi E., Caivano G., Pertile M., Mozzarelli A. Phospholipid components of the synthetic pulmonary surfactant CHF5633 probed by fluorescence spectroscopy. Int. J. Pharm. 2018;553:290–297. doi: 10.1016/j.ijpharm.2018.10.045. [DOI] [PubMed] [Google Scholar]
- 2.Richieri G.V., Ogata R.T., Kleinfeld A.M. The measurement of free fatty acid concentration with the fluorescent probe ADIFAB: a practical guide for the use of the ADIFAB probe. Mol. Cell. Biochem. 1999;192:87–94. [PubMed] [Google Scholar]
- 3.Henry E.R. The use of matrix methods in the modeling of spectroscopic data sets. Biophys. J. 1997;72:652–673. doi: 10.1016/s0006-3495(97)78703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry E.R., Hofrichter I. Singular value decomposition: application to analysis of experimental data. Methods Enzymol. 1992:129–192. [Google Scholar]
- 5.Ronda L., Pioselli B., Catinella S., Salomone F., Marchetti M., Bettati S. Quenching of tryptophan fluorescence in a highly scattering solution: insights on protein localization in a lung surfactant formulation. PLoS One. 2018;13:e0201926. doi: 10.1371/journal.pone.0201926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material