Cryptococcus is a fungal pathogen that kills an estimated quarter of a million individuals yearly and is the most common cause of nonviral meningitis in the United States. The fungus is carried in about 10% of the adult population and, after reactivation, causes disease in a wide variety of immunosuppressed individuals, including the HIV infected and patients receiving transplant conditioning, cancer therapy, or corticosteroid therapy for autoimmune diseases. The fungus is widely carried in the soil but can also cause infections in plants and mammals. However, the mechanisms for this widespread ability to infect a variety of hosts are poorly understood. The present study identified adaptation to low nutrients as a key property that allows the fungus to inhabit these diverse environments. Further studies identified a nutrient transporter gene, STL1, to be upregulated under low nutrients and to be associated with early fungicidal activity, a marker of poor clinical outcome in a cohort of HIV/AIDS patients. Understanding molecular mechanisms involved in adaptation to the human host may help to design better methods of control and treatment of widely dispersed fungal pathogens such as Cryptococcus.
KEYWORDS: CAC2, CAF-1, Cryptococcus neoformans, STL1, TOR pathway, VAD1
ABSTRACT
Human infection with Cryptococcus causes up to a quarter of a million AIDS-related deaths annually and is the most common cause of nonviral meningitis in the United States. As an opportunistic fungal pathogen, Cryptococcus neoformans is distinguished by its ability to adapt to diverse host environments, including plants, amoebae, and mammals. In the present study, comparative transcriptomics of the fungus within human cerebrospinal fluid identified expression profiles representative of low-nutrient adaptive responses. Transcriptomics of fungal isolates from a cohort of HIV/AIDS patients identified high expression levels of an alternative carbon nutrient transporter gene, STL1, to be associated with poor early fungicidal activity, an important clinical prognostic marker. Mouse modeling and pathway analysis demonstrated a role for STL1 in mammalian pathogenesis and revealed that STL1 expression is regulated by a novel multigene regulatory mechanism involving the CAC2 subunit of the chromatin assembly complex 1, CAF-1. In this pathway, the global regulator of virulence gene VAD1 was found to transcriptionally regulate a cryptococcal homolog of a cytosolic protein, Ecm15, in turn required for nuclear transport of the Cac2 protein. Derepression of STL1 by the CAC2-containing CAF-1 complex was mediated by Cac2 and modulated binding and suppression of the STL1 enhancer element. Derepression of STL1 resulted in enhanced survival and growth of the fungus in the presence of low-nutrient, alternative carbon sources, facilitating virulence in mice. This study underscores the utility of ex vivo expression profiling of fungal clinical isolates and provides fundamental genetic understanding of saprophyte adaption to the human host.
INTRODUCTION
Cryptococcus neoformans is a major fungal pathogen causing a highly lethal meningoencephalitis (CM), primarily in individuals with impaired host cell immunity, such as those infected with HIV/AIDS, causing a quarter of a million deaths annually (1, 2). Mortality exceeds 20 to 50% despite therapy (3), and recent failed attempts to improve outcomes (4) highlight our profound lack of understanding of the pathophysiology of human infections. While studies in host models such as mice or invertebrates have generated essential insights into fungal virulence, evolutionary differences in host response and pathogen environment suggest a need to model studies using human disease correlates (5).
Peculiar to Cryptococcus is the ability to live for extended periods both within the environment (6) and within the mammalian host in a dormant state (7). It can also cause disease in a wide variety of plants (8), free-living amoebae (9), and humans (10). Similarities between mammalian host environments such as the macrophage phagolysosome and those more primitive organisms such as amoebae have been implicated as exerting evolutionary pressure on facultative intracellular pathogens, such as C. neoformans (11), and gene expression studies suggest adaptation to nutrient deprivation is important within this environment (12). Indeed, requirements for the gluconeogenesis enzyme Pck1 (13) and a high-affinity glucose transporter (14) suggest roles for nutrient homeostasis in cryptococcal virulence. Phagocytosis also stimulates a starvation response in other pathogenic fungi, such as Candida albicans, that induce a shift to fatty acids as a carbon source by upregulating the glyoxylate cycle, requiring the enzyme isocitrate lyase (15). However, the dispensability of isocitrate lyase by C. neoformans during infection suggests that species-specific pathways for starvation tolerance are also important within the mammalian host (16). In addition, induction of the nutrient-recycling autophagy pathway is particularly important for C. neoformans but less important for other fungal pathogens such as Aspergillus and Candida (17). For this important stress response, global nutrient regulators such as the target of rapamycin (TOR) (18) play an important link between cryptococcal starvation response, macrophage survival, and pathogen fitness (19). In addition, downstream of TOR the virulence-associated dead box protein, Vad1, has been shown to act as a global posttranscriptional regulator of TOR-dependent processes, such as autophagy (19). Similarly, the role of the cAMP nutrient-sensing pathway has featured prominently in cryptococcal pathogenesis (20). In this pathway, adenylyl cyclase is activated by a Gα subunit (Gpa1), resulting in the production of cAMP that binds to regulatory subunit Pkr1 of the protein kinase A (PKA) complex to release an active form of the catalytic subunit Pka1, which activates downstream proteins (21). However, little is known regarding regulatory relationships that link these nutrient master regulators and gene targets responsible for mammalian virulence, as well as possible commonalities between the various infective environments, including humans.
Previous studies have not always found robust quantitative relationships between mammalian infectious outcome and classical virulence factors in vitro (22, 23), implying that other mechanisms of virulence remain to be identified. Thus, in the present work, to characterize virulence-associated starvation responses most relevant to human infections, transcriptional profiling of the fungus inoculated into human cerebrospinal fluid (CSF) was first compared to that from a starvation medium previously used to identify a clinical association between expression levels of a CTR4 copper transporter and dissemination to the brain in a cohort of human solid organ transplant patients (24). Comparison of the expression profiles of fungus incubated in human cerebrospinal fluid or under nutrient deficiency identified a large set of 855 genes upregulated under both conditions. To identify genes within this subgroup whose expression might be most relevant to human infections, we compared transcription profiles of isolates from those who died versus those surviving 10 weeks obtained from a previously described cohort of HIV/AIDS-infected patients with CM (23). The second most highly differentially expressed annotated gene (locus tag CNAG_01683 [accession no. CP022331.1]) between patient isolates showed highest homology to a sugar transporter-like gene (STL1) from Saccharomyces cerevisiae implicated previously in glycerol metabolism. STL1 expression levels showed a significant correlation with poor early fungicidal activity (EFA), a clinical prognostic marker of patient outcome (25), and in situ hybridization demonstrated STL1 expression in a brain of an HIV-infected patient with CM. Furthermore, mouse modeling was used to demonstrate attenuated virulence of an stl1Δ strain, confirming its role in mammalian virulence. STL1 was also found to be a target gene of TOR, regulated by a novel regulatory pathway involving mRNA decay and a multicomponent Cac2-Ecm15 nuclear shuttle regulatory complex. These studies thus provide a direct link between STL1, TOR signaling, starvation response, and CAF1-mediated regulation of microbial pathogenesis relevant to human fungal infections.
RESULTS
Transcriptional profiling of C. neoformans suggests similarity in adaptive stress responses between human CSF and starvation medium at 37°C.
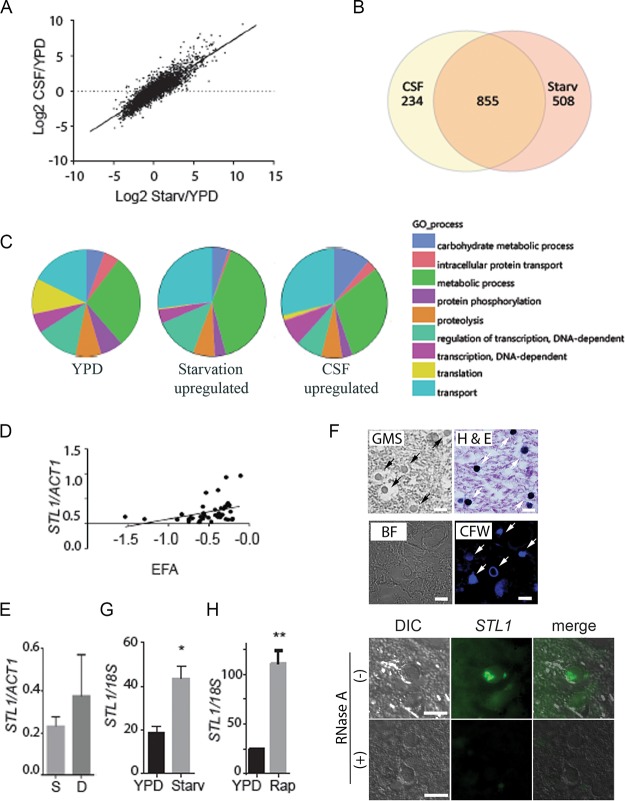
Previous studies utilized nutrient-deficient media to simulate the environment of the host CSF and successfully correlated expression levels of a CTR4 copper transporter with dissemination to the brain in a cohort of solid organ transplant patients (24). These conditions were utilized to compare transcriptional profiles of a pathogenic C. neoformans strain (H99, serotype A, molecular types VNI and VNII) (26) between nutrient-rich and nutrient-poor environments and to compare the starvation adaptation profiles to that obtained after transfer to pooled human cerebrospinal fluid from patients with cryptococcal meningitis (see Tables S1 and S2 in the supplemental material). Serotype A strains are the predominant strains that cause HIV-associated CM and were selected to reduce potential genetic confounding. A strong correlation was demonstrated between expression-level changes between YPD and either CSF versus starvation medium at 37°C (Fig. 1A) (R2 = 0.79; slope non-zero P value of <0.0001). Transcriptional profiles after transfer to each respective condition demonstrated not only common identities of upregulated genes, but also the comparatively similar magnitudes of transcriptional changes. Of 1,363 genes upregulated in starvation and 1,089 in CSF, 855 were upregulated under both conditions, of which 332 were annotated sufficiently to allow Gene Ontology (GO) term assignments (Fig. 1B). Compared to the overall annotated transcriptome (Fig. 1C, left panel), transition of C. neoformans to either starvation conditions or CSF resulted in an expansion of gene expression associated with the GO biological term “carbohydrate metabolic processes,” including key enzymes involved in β-oxidation, the glyoxylate cycle, and gluconeogenesis (see Fig. S1 in the supplemental material). In contrast, genes in the category “translation” were underrepresented in both media, consistent with the slow growth anticipated under these conditions. Using GO functional terms, transition of fungal cells to either starvation or CSF resulted in differential expression of genes involved in the transport function (see Fig. S2 in the supplemental material). These data thus emphasize similarities in gene expression profiles of C. neoformans between nutrient deprivation and CSF, as well as a predominance of genes involved in carbohydrate homeostasis and transport function.
FIG 1.
Expression profiling demonstrates a concordance of cryptococcal genes upregulated in fungus-infected human cerebrospinal fluid and starvation. (A) C. neoformans strain H99 was grown to the mid-log phase in YPD and then subjected to starvation (Starv) in asparagine salts without glucose or sterile cerebrospinal fluid (CSF) at 37°C for 3 h. RNA was recovered and subjected to microarray analysis, and values were expressed as log2 ratios of expression in CSF/YPD. n = 2 for each condition. (B) Venn diagram of genes upregulated >2-fold and adj P < 0.05 after incubation in fungus-infected CSF or under starvation conditions. (C) GO biological process term occurrence among genes upregulated under the indicated conditions. (D) Transcriptional ratio of STL1/ACT1 determined by qRT-PCR of patient isolates compared to EFA (n = 46; regression analysis, n − 2 degrees of freedom [df], P < 0.05). (E) Expression ratios of STL1/ACT1 in survivors at 10 weeks (S [n = 35]) versus those who died (D [n = 11]). (F) Sections of a brain autopsy specimen were obtained from a 42-year-old man with HIV/AIDS who died of severe and diffuse C. neoformans infection and were visualized by bright-field microscopy (BF) or differential interference contrast microscopy (DIC) or stained with Gomori methenamine silver stain (GMS), hematoxylin and eosin (H&E stain), calcofluor white (CFW), or C3-fluorescein-labeled oligonucleotide probes for STL1 transcripts (STL1) by fluorescent in situ hybridization in the presence (+) or absence (−) of RNase A treatment. Arrows point to stained yeast cells. Bar = 10 µm. (G and H) Expression levels of STL1/18S rRNA under mid-log growth in YPD or 3 h of incubation under starvation conditions or in the presence of 5 µM rapamycin (Rap). n = 3 independent experiments. *, P < 0.05, and **, P < 0.01, by Student’s t test. Error bars represent standard deviation.
Metabolic reprogramming of carbohydrate metabolism in C. neoformans in response to incubation in fungus-infected human CSF or starvation. Pathway schemes demonstrate differential expression ratios of C. neoformans after incubation in human fungus-infected CSF or under starvation conditions (starv) relative to mid-log-phase cells grown in YPD of genes expressing enzymes active in β-oxidation, glyoxylate cycle, or gluconeogenesis. Adapted from Derengowski et al. (12). Download FIG S1, PDF file, 0.1 MB (91KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression profiling demonstrates transcriptional expansion of genes related to transporter activity (GO function). Shown is transcriptional profiling of C. neoformans cells as in Fig. 1A displayed by the most common GO function terms. Shown are the percentage of annotated GO terms relative to total count of gene annotations (left upper panel), starvation (right upper panel), CSF alone (right lower panel), or both CSF and starvation (right lower panel). Download FIG S2, PDF file, 0.1 MB (95.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome-wide expression-level changes after transition from YPD to CSF. Download Table S1, PDF file, 0.3 MB (345.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome-wide expression-level changes after transition from YPD to starvation medium. Download Table S2, PDF file, 0.4 MB (417.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression profiling of isolates from a cohort of AIDS-related CM identifies a putative sugar transporter-like gene, STL1, associated with clinical outcome marker EFA.
Further studies were then conducted to determine whether starvation-induced genes may provide associations with clinical outcome that may help to understand human-related pathogenicity of the fungus. Transcriptional profiling under starvation conditions was conducted as described below, which previously identified the copper transporter gene CTR4 as a potential biomarker of brain dissemination in solid organ transplant recipients (27) using a set of serotype A C. neoformans isolates (molecular types VNI and VNII) from a cohort of HIV/AIDS patients described previously (28). A demographic table of the cohort from which isolates were obtained is shown in Table 1. Median age was 36 years, and all patients were treated with amphotericin B-containing regimens. Despite therapy, 5 died by 2 weeks, and an additional 6 died by 10 weeks, generating a 10-week mortality of 11/45 (24%), typical of CM in developing countries (3). Pretreatment covariables were significantly different for mental status, as measured by Glasgow coma score (P = 0.02), but parameters including CSF initial fungal burden, EFA, VN genotype, CSF opening pressure, or white blood cell (WBC) or CD4 cell count did not differ significantly. Expression profiles were compared using analysis of variance (ANOVA) with adjustment, grouping patient isolates associated with mortality at 10 weeks versus survivors. Ninety-one genes (28 annotated) showed significantly different expression in patients who died versus those who lived at least 10 weeks after therapy (see Table S3 in the supplemental material) (adjusted P value [adj P] of <0.02). The first highly expressed gene was the nitroreductase family protein gene (CNAG_02692 [CP022323.1]). The second most highly differentially expressed gene showed highest homology to a sugar transporter-like gene, STL1, from S. cerevisiae previously implicated in glycerol transport (29, 30). STL1 was selected for further study because of its putative role in alternative carbon metabolism and transport function exemplifying the aggregate transcriptional changes in human CSF. STL1 expression levels by quantitative reverse transcription-PCR (qRT-PCR) of the clinical isolates showed a significant correlation with poor early fungicidal activity (EFA [P = 0.018]) (Fig. 1D (25), as well as a trend with clinical outcome (Fig. 1E) (P = 0.07); however, the latter was not statistically significant, possibly due to nonmicrobiological confounders, such as immune responses or delays in medical care. EFA is the rate of fungal clearance in each of the HIV patient’s CSF during therapy and is a microbiological prognostic marker of clinical outcome (25, 31). We did not find a relationship between STL1 expression and molecular genoype (P = 0.73).In addition, STL1 expression was demonstrated in a brain autopsy specimen from an HIV/AIDS patient with CM using fluorescent in situ hybridization (FISH) (66/66 yeast cells with positive STL1 signal versus 9/78 in RNase-treated negative-control slides; Fisher’s exact test, P < 0.0001) (Fig. 1F) (19). Furthermore, qRT-PCR demonstrated that STL1 expression was induced during starvation, as well as in the presence of the TOR inhibitor rapamycin (Fig. 1G and H). TOR is a particularly important pathway as the TOR inhibitors sirolimus and everolimus are in widespread clinical use as immunosuppressants and may play a risk factor in cryptococcosis (32). In summary, these data identify a starvation-induced putative sugar transporter gene, STL1, associated with poor microbiological clearance (EFA) during HIV-associated human infections.
TABLE 1.
Baseline clinical and laboratory characteristics
| Parametera | Result for patients |
||
|---|---|---|---|
| All | Survived | Died | |
| n | 45 | 34 | 11 |
| Age, yrb | 36 (21–62) | 37 (21–62) | 33 (23–51) |
| Male, % | 53 | 53 | 55 |
| Abnormal mental status, % | 9 | 3 | 27 |
| Fungal burden, log10 CFU/ml | 5.9 | 5.9 | 5.8 |
| EFA | −0.51 | −0.55 | −0.37 |
| CSF opening pressure, cm H2Ob | 26 (7–76) | 26 (10–76) | 15 (7–42) |
| Genotype, % VNI | 68 | 58 | 42 |
CFU, colony-forming unit; CSF, cerebrospinal fluid; EFA, early fungicidal activity.
The median is shown with range in parentheses.
Genome-wide expression changes between isolates of patients who died versus survived at 10 weeks. Download Table S3, PDF file, 0.1 MB (54.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
STL1 plays a role in cryptococcal growth on alternative carbon sources and is required for full virulence in mice.
Further characterization demonstrated a role for STL1 in expression of the virulence factors capsule (Fig. 2A) and laccase (Fig. 2B), but STL1 was dispensable for mating (see Fig. S3A in the supplemental material). The induction of capsule under the nutrient-deficient (Glu−) conditions was decreased from a small rim of capsule in the wild type (WT) but markedly increased by overexpression under both YPD (yeast extract-peptone-dextrose) and nutrient-deficient conditions. Because of its putative role in alternative carbon homeostasis and poor fungal clearance and a trend toward worse survival in clinical strains showing increased expression, we tested to see if STL1 overexpression could facilitate growth on alternative carbon sources and play a role in mammalian virulence using a mouse model. Interestingly, STL1 overexpression (Fig. S3B) facilitated increased growth in nutrient limitation medium containing 0.03% of either pyruvate, lactate or acetate—intermediates in gluconeogenesis and previously identified substrates within mammalian brains during CM infections—as well as glycerol, which has been previously described for an STL1 homolog from Candida (Fig. 2C) (29, 33). In addition, STL1 overexpression facilitated increased growth in nutrient limitation medium containing either 0.03% ribose or citrate but not equivalent concentrations of glucose versus an identical strain transformed with empty vector alone in identical copy number (Fig. 2D). These data extend the spectrum of alternative carbon substrates related to fungal STL1. However, STL1 did not facilitate growth on other sugars, including galactose, glucosamine, and rhamnose, suggesting carbon substrate specificity (Fig. S3C). In addition, the stl1Δ strain exhibited moderately reduced virulence in an intravenous mouse brain dissemination model compared to the wild-type strain (Fig. 2E, left panel). However, inoculation of C. neoformans strains overexpressing STL1 resulted in no additional increase in virulence in the highly virulent strain H99, compared with identical strains transformed with empty vector alone (Fig. 2E, right panel). Lack of increased virulence could be because the H99 strain used is already highly virulent (34). Taken together, these data suggest that STL1 is associated with alternative carbon acquisition and plays a role in capsule, laccase activity, and virulence.
FIG 2.
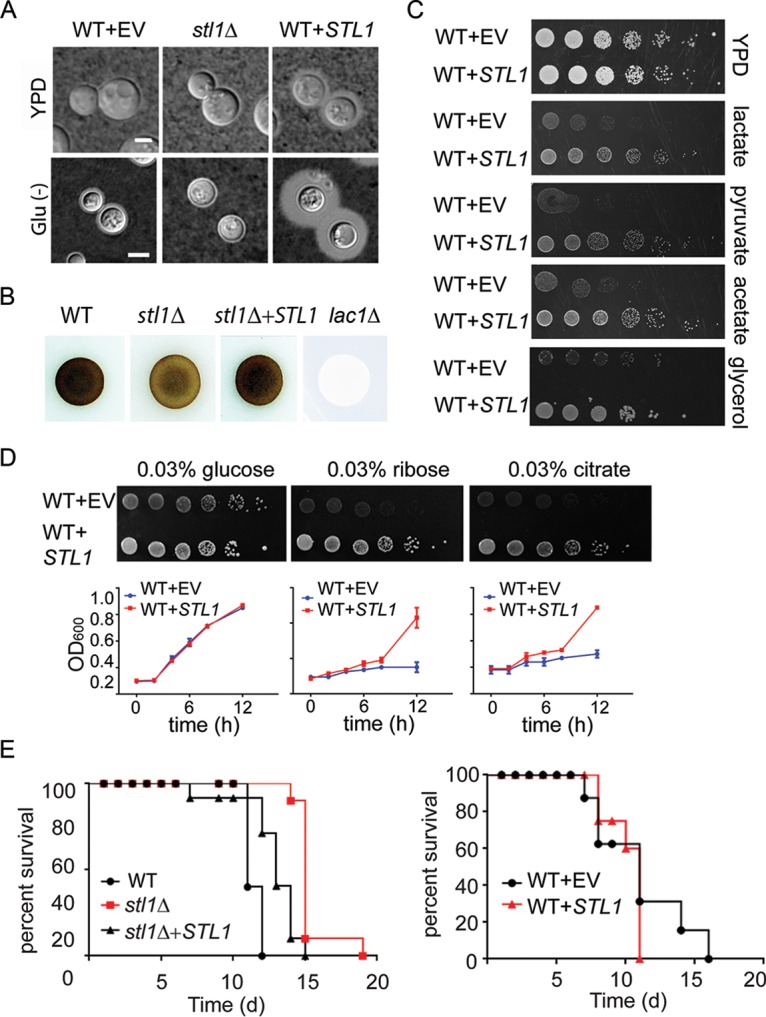
STL1 is required for growth on alternative carbon substrates, capsule formation, laccase activity, and mammalian virulence. (A) Indicated strains were incubated on YPD or ASN without glucose (Glu−) agar for 3 days at 30°C and examined by India ink microscopy. Bar = 5 µm. (B) The indicated cells were inoculated on ASN containing 100 mg/ml norepinephrine to assay production of melanin pigment by laccase. (C) The indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on YPD or ASN containing the indicated substrates (0.03%) and incubated at 30°C for 5 days. (D) In the upper panels, the indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on ASN containing the indicated substrates (0.03%) and incubated at 30°C for 7 days. In the lower panels, the indicated strains were cultured with ASN broth containing the indicated substrates (0.03%), and optical density at 600 nm (OD600) was calculated at the indicated time points. (E) Mice were inoculated by tail vein (106 of the indicated cells), and progress was followed until the mice were moribund. n = 10 mice per group. P < 0.0001 (left panel) and P = 0.5 (right panel) by log rank test.
Mating and spotting assay. (A) The indicated MATα strains were coincubated with a MATa mating partner (strain KN99) on nitrogen-limiting mating medium (V8 agar) for 3 weeks at room temperature. The edges of the mating mixtures were photographed (40×). Bar = 500 µm. (B) qRT-PCR of STL1 of indicated strains grown to the mid-log phase in ASN containing 0.03% acetate (without glucose). n = 3 independent experiments. ***, P < 0.001 by Student’s t test. (C) The indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on ASN containing the indicated substrates (0.03%) and incubated at 30°C for 5 to 10 days. Download FIG S3, PDF file, 0.2 MB (259.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The nuclear regulator CAC2 is a suppressor of STL1 and plays a role in capsule and virulence in mice.
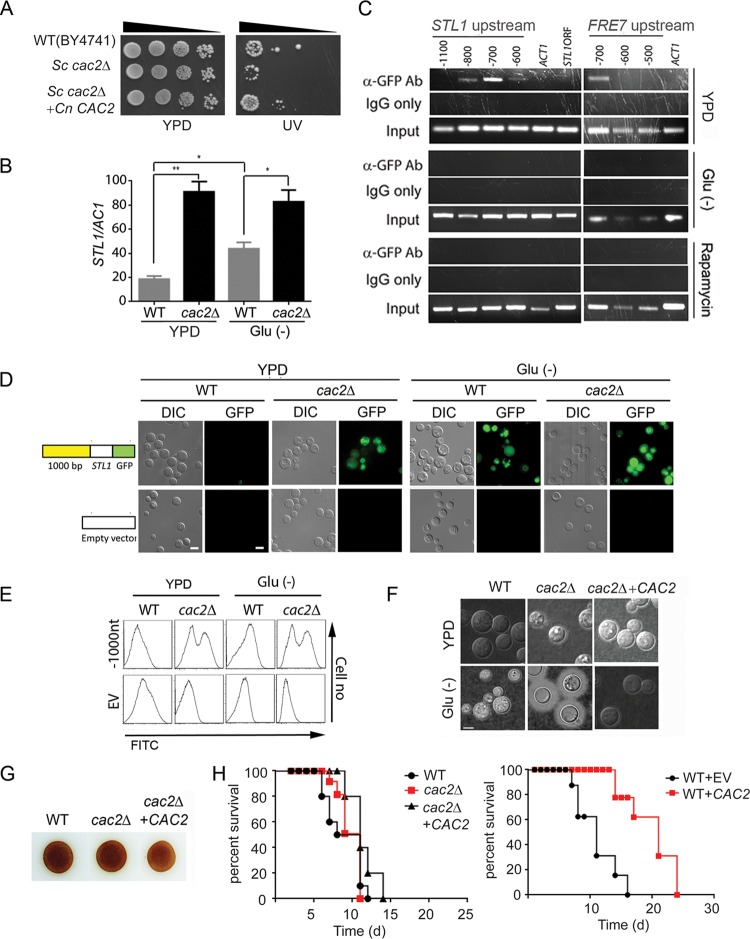
We next sought to investigate mechanisms of starvation-associated STL1 regulation. Previous work had identified ECM15 mRNA as a regulatory target of the starvation/TOR-associated global virulence-associated deadbox (Vad1) protein (35). Although ECM15 is not known to directly regulate gene expression, a whole-proteome interaction study previously suggested that Ecm15 from the yeast Saccharomyces cerevisiae interacts with a second S. cerevisiae protein, ScCac2 (36). Cac2 is a key constituent of the CAF-1 chromatin assembly factor, which assembles histones H3 and H4 and mediates chromatin suppression of genes at subtelomeric locations and tolerance to UV irradiation in both S. cerevisiae and C. neoformans (37, 38). However, its precise role as a potential regulator remains unexplored in eukaryotes. Thus, we sought to determine if a putative VAD1-ECM15-CAC2 regulatory pathway could play a role in the starvation/TOR-dependent regulation of STL1 demonstrated in Fig. 1G and H.
We first identified a cryptococcal CAC2 putative protein sequence demonstrating 33% identity to that of the S. cerevisiae. The identity of C. neoformans CAC2 (CnCAC2) was confirmed by restoring UV tolerance to an Sccac2Δ mutant strain (Fig. 3A). To directly evaluate CAC2 in C. neoformans, the cryptococcal cac2Δ strain was generated and demonstrated increased expression of STL1 by qRT-PCR (Fig. 3B). Early studies suggested that orthologs of CAF-1 members such as Cac2 are associated with suppression of genes in the subtelomeric region (37), which include cryptococcal genes such as FRE7 (ferric-chelate reductase: CNAG_00876 [CP022325.1]), CNAG_05333 (XM_012198174.1), and the HpcH/Hpa1 family protein gene (CNAG_06874 [XM_012194223.1]) (see Table S4 in the supplemental material); however, the STL1 gene does not reside within this region (location in chromosome 11 [CP003830.1], positions 603867 to 606604). Thus, to identify a possible direct STL1 Cac2-specific DNA binding region(s), chromatin immunoprecipitation (ChIP) was performed using an anti-green fluorescent protein (anti-GFP) antibody directed against a Cac2-GFP fusion protein. These studies demonstrated Cac2 binding within a 100-bp fragment centered at −700 bp from the transcriptional start site of STL1, as well as that of FRE7 under the nutrient condition (Fig. 3C, top panel). In addition, STL1 Cac2-specific DNA binding efficiency was reduced under both starvation (Fig. 3C, middle panel) and rapamycin (Fig. 3C, bottom panel) conditions. To determine the functional significance of the binding, we tested expression levels of an Stl1-GFP fusion having serial deletions of the STL1 promoter (positions −1,000, −500, and −50 from the transcript start site) on basal transcriptional activity in C. neoformans (see Fig. S4A in the supplemental material). The plasmid containing an STL1 open reading frame (ORF) with various lengths of the STL1 5′-promoter–GFP fusion gene were transformed into wild-type or cac2Δ C. neoformans strains. Empty vector was used as a control. These results indicated the presence of basal transcription originating from the region from −1000 to −500 of STL1 under both nutrient-replete (YPD) and nutrient-poor (Glu−) conditions, corresponding to the region of Cac2 binding by ChIP. However, under the nutrient-rich condition, a subpopulation of WT cells displayed suppression using episomal constructs which replicate autonomously, which could suggest a requirement for chromatin-dependent promoter-binding factor(s) for more effective CAC2-dependent chromatin silencing (39). Thus, linear PCR fragments containing the indicated promoter, STL1 coding region, and selection marker were transformed into WT and cac2Δ strains as integrated fragments, demonstrated comigration with genomic DNA on Southern blot analysis. As shown in Fig. 3D and E, the integrated construct exhibited more effective repressed STL1 expression under nutrient-rich conditions (YPD), which was derepressed under either nutrient-poor conditions (Glu−) or in the cac2Δ mutant strain. Interestingly there was heterogeneity in Stl1-GFP expression in the cac2Δ strain, which may suggest posttranslational modification of the fusion protein.
FIG 3.
Cac2 is a repressor of STL1 and capsule and overexpression reduces virulence in mice. (A) The indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on YPD or on YPD with UV irradiation (UV) for 1 min and incubated at 30°C for 3 days. (B) qRT-PCR of STL1 of indicated strains grown to the mid-log phase in YPD and ASN without glucose (Glu−) media. n = 3 independent experiments. *, P < 0.05, and **, P < 0.01, by Student’s t test. (C) Chromatin immunoprecipitation (ChIP) of a Cac2-promoter complex. Nuclear extract from induced cryptococcal cells expressing a Cac2-GFP fusion protein was immunoprecipitated using an anti-GFP antibody (α-GFP Ab) or control IgG (IgG only) and assayed by PCR for the presence of the indicated regions of STL1, FRE7 promoter, or control promoter sequences of ACT1. (D and E) Effects of serial deletion of the STL1 5′-promoter (−1000 from the transcript start site) on basal transcriptional activity in C. neoformans. Linear constructs containing STL1 ORF with indicated regions of the STL1 5′-promoter–GFP fusion gene were transformed into WT or cac2Δ strains of C. neoformans, integration was confirmed by Southern blotting, and cells were grown to the mid-log phase in YPD or ASN salts without glucose (Glu−), and the population was subjected flow cytometry or microscopy (DIC or GFP fluorescence microscopy). Cells transformed with empty vector were used as control. Bar = 5 μm. (F) The indicated strains were inoculated onto the indicated media, recovered, and visualized by India ink microscopy. (G) Indicated cells were inoculated on ASN containing 100 mg/ml norepinephrine to assay production of melanin pigment by laccase. (H) Mice were inoculated by tail vein (106 cells of the indicated strains), and progress was followed until mice were moribund. n = 10 mice per group. P = 0.7 (left panel) and P < 0.0001 (right panel) by log rank test.
Effects of serial deletion of the STL1 5′-promoter (−1000 from the transcript start site) on basal transcriptional activity in C. neoformans. A plasmid containing the STL1 ORF with indicated regions of the STL1 5′-promoter–GFP fusion gene was transformed into the WT strain of C. neoformans and grown to mid-log phase in YPD (Glu+) and ASN salts without glucose (Glu−), and the population was subjected to microscopy (DIC or GFP fluorescence) (A) or flow cytometry (FITC) (B). Empty vector was used as a control. Bar = 5μm. (C) Capsule thickness across mutant strains in C. neoformans. The indicated strains were incubated on YPD or ASN without glucose (Glu−) agar for 3 days at 30°C and examined by India ink microscopy. Bar = 5 µm. Download FIG S4, PDF file, 0.1 MB (110.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of genes suppressed by both ECM15 and CAC2 determined by genome-wide expression analysis. Download Table S4, PDF file, 0.1 MB (82.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Further characterization of CAC2 demonstrated that, similar to the STL1 overexpression strain, the CAC2 deletion strain showed increased capsule expression under nutrient-poor conditions (Fig. 3F), but there was little effect on melanin formation (Fig. 3G), mating (Fig. S3A), or virulence (Fig. 3H, left panel). However, overexpression of CAC2 resulted in markedly reduced virulence in mice (Fig. 3H, right panel), compared to an empty vector control strain, suggestive of a suppression of genes involved in virulence, such as STL1. The cac2Δ strain was thus a phenocopy of the STL1 overexpressor strain, though epistatic experiments demonstrated that STL1 overexpression in the CAC2 overexpressor strain could not restore capsule induction, suggesting that CAC2 may suppress additional genes involved in capsule synthesis besides STL1 (Fig. S4B). However, the data do suggest that CAC2 is a transcriptional repressor of STL1 and is associated with capsule expression and murine virulence.
Nuclear localization of CAC2 is nutrient dependent and requires ECM15.
Saccharomyces interactome studies (36) suggested that ECM15 may regulate CAC2 by a posttranslational interactive mechanism. ECM15 in S. cerevisiae is a nonessential poorly understood protein, proposed to be involved in cell wall competency (36, 40). Interestingly, deletion of ECM15 resulted in at least a 2-fold increased transcription (Fig. 4A) (adj P < 0.05) of 106 genes, while transcription of only 8 genes increased after CAC2 deletion, 4 of which were shared with ECM15. Interestingly, the gene showing highest suppression by both ECM15 and CAC2 was STL1 (Table S4). In addition, examination of protein sequences using cNLS Mapper (41) identified putative nuclear localization sequences in Ecm15 in the region D63 to A91 and Cac2 in the region E778 to V802 (see Fig. S5 in the supplemental material). Thus, to assess for possible regulatory interactions, as well as common nuclear localizations, in Cryptococcus and whether this could change in response to nutrient starvation or rapamycin, both proteins were tagged with GFP. These studies demonstrated colocalization of Cac2-GFP (Pearson correlation coefficient [PCC] = 0.77) and Ecm15-mCherry (PCC = 0.87) with the nuclear dye 4′,6-diamidino-2-phenylindole (DAPI) (Fig. 4B) and with each other (PCC = 0.77) under nutrient-rich conditions (YPD) with cytoplasm localization under starvation or in the present of rapamycin, consistent with a possible role as nuclear suppressors under nutrient-rich conditions and derepression under starvation conditions. Interestingly, rapamycin resulted in cytoplasmic localization of EMC15-mCherry with an additional suppression of visible fluorescence in starvation, which could be due to transcriptional suppression by VAD1, as alluded to above. Epistatic studies were next conducted, which demonstrated that the increased production of capsule in the ecm15Δ mutation could be readily suppressed by CAC2 overexpression, but ECM15 overexpression was not as effective at suppressing the increased capsule phenotype of the cac2Δ mutant (Fig. 4C), suggesting that ECM15 is hypostatic to CAC2. Additional studies demonstrated that deletion of ECM15 resulted in mislocalization of the Cac2-GFP fusion protein under nutrient-replete conditions (WT, CAC2-GFP-DAPI; PCC = 0.77) to the cytoplasm (ecm15Δ, CAC2-GFP-DAPI; PCC = 0.57), similar to that during starvation (WT, CAC2-GFP-DAPI; PCC = 0.53) or in rapamycin-treated cells (WT, CAC2-GFP-DAPI; PCC = 0.24) (Fig. 4D). These results suggest that Ecm15 is required for Cac2 nuclear targeting, leading to downstream repression of virulence-associated phenotypes and genes, including STL1.
FIG 4.
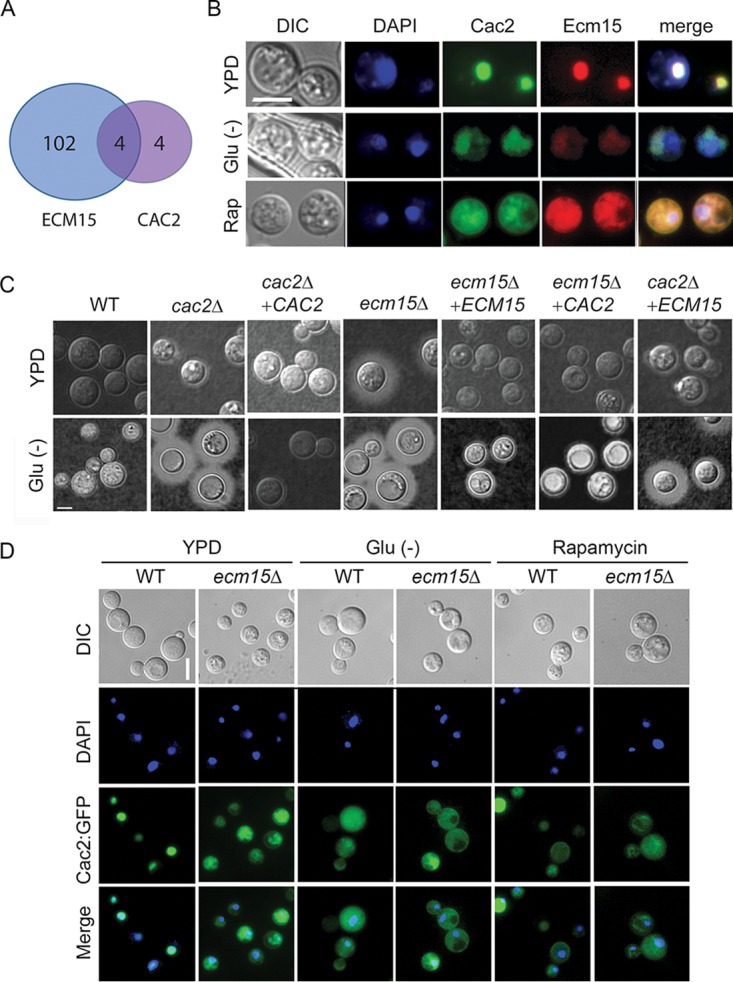
Cac2 and Ecm15 shares overlapping expression profiles and undergoes nutrient- and TOR-dependent nuclear localization. (A) Venn diagram of genes showing >2× increased transcription relative to WT and adj P < 0.05 after deletion of ECM15 or CAC2. (B) C. neoformans cells expressing Cac2-GFP or Ecm15-mCherry were incubated in YPD, ASN without glucose (Glu−), or YPD containing rapamycin and observed by fluorescence (DAPI, Cac2, or Ecm15) or differential interference contrast (DIC) microscopy. Bar = 5 µm. (C) Indicated strains were incubated on YPD or ASN without glucose (Glu−), recovered, and observed by India ink microsocopy. (D) Cells expressing Cac2-GFP in wild-type (WT) or ecm15Δ strains were incubated in YPD ASN without glucose (Glu−) or YPD containing rapamycin and observed by fluorescence or by DIC microscopy. Bar = 5 µm.
Prediction of putative nuclear localization signals. Ecm15 (A) and Cac2 (B) putative nuclear localization signals were determined by cNLS Mapper. Download FIG S5, PDF file, 0.1 MB (120.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ECM15 plays a role in capsule, mating, and cell wall stability and is regulated by VAD1.
Further studies were then conducted to further characterize the cryptococcal ECM15 gene and characterize its relationship with starvation regulatory pathways that could play a role in regulation of genes such as STL1. The deduced amino acid sequence of the cryptococcal Ecm15 showed 66.8% identity to its respective homolog in S. cerevisiae. Susceptibility of an Scecm15Δ mutant to the cell wall active agent calcofluor white as previously reported (42) was complemented by the cryptococcal ECM15 gene (Fig. 5A). These results suggest that CnEcm15 is a functional homolog of ScEcm15p. Phenotypic studies shown in the preceding figure (Fig. 4B) demonstrated a role for ECM15 in extracellular capsule as well as mating, with a subtle effect on melanin formation (Fig. 5B). These studies thus implicate a role for ECM15 in the regulation of several virulence-associated phenotypes as well as mating of C. neoformans.
FIG 5.
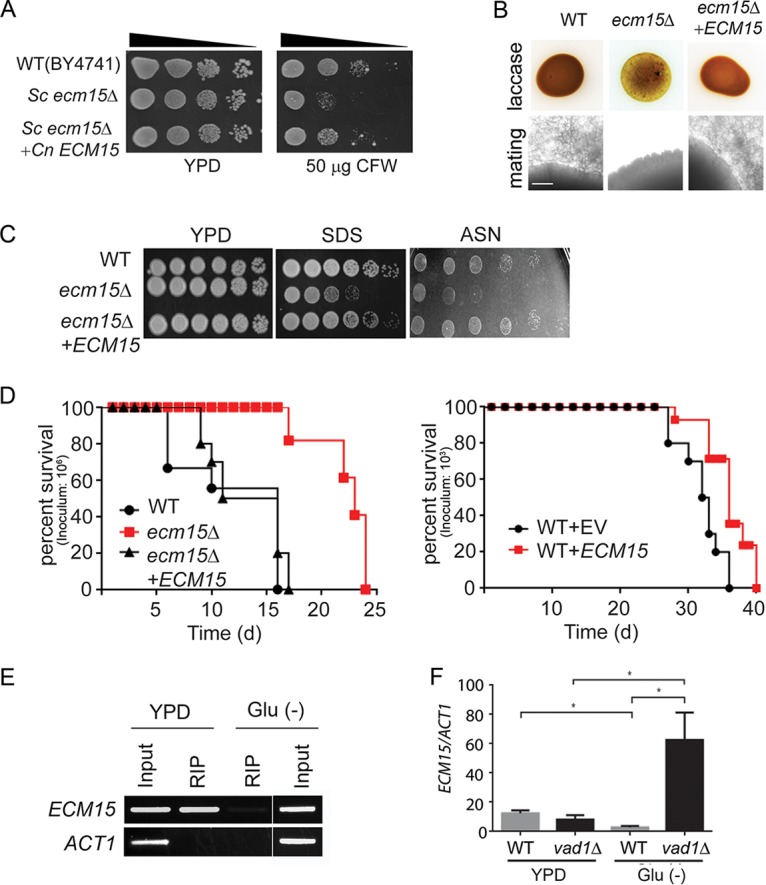
VAD1 is a repressor of ECM15, a regulator of capsule, laccase activity, and virulence. (A) The indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on YPD medium or YPD containing 50 μg/ml of calcofluor white (CFW) and incubated at 30°C for 3 days. (B) The indicated strains were inoculated on ASN containing 100 mg/ml norepinephrine and observed after 2 days at 37°C (upper panel). Indicated MATα strains were coincubated with a MATa mating partner (strain KN99) on nitrogen-limiting mating medium (V8 agar) for 3 weeks at room temperature. The edges of the mating mixtures were photographed (40×). Bar = 500 µm (lower panel). (C) The indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on YPD, ASN, or YPD containing SDS and incubated at 30°C for 3 to 7 days. (D) Mice were inoculated by tail vein (106 [left] or 103 [right] of the indicated strains), and progress was followed until they were moribund. n = 10 mice per group. P< 0.0001 (left panel) and P < 0.01 (right panel) by log rank test. (E) Lysates from cells expressing a c-myc-tagged Vad1 fusion protein incubated under the indicated conditions were immunoprecipitated followed by RT-PCR/gel electrophoresis using primers for the indicated gene transcripts. (F) Steady-state transcript levels of ECM15 from indicated strains grown to the mid-log phase in YPD or ASN salts without glucose (Glu−). n = 3 independent experiments. *; P < 0.05 by Student’s t test.
Extending these results to cell wall phenotypes associated with virulence, as suggested by studies in S. cerevisiae, the cryptococcal ecm15Δ mutants were found to exhibit increased susceptibility to the ionic detergent SDS (Fig. 5C). Interestingly the ecm15Δ mutant also displayed poor growth on nutrient-poor agar consisting of asparagine salts (ASN) without glucose (Fig. 5C), similar to that of the stl1Δ mutant. In addition, the Cnecm15Δ strain exhibited attenuated virulence in a mouse model (Fig. 5D, left panel). Interestingly attenuated virulence was also exhibited by an ECM15-overexpressing strain (Fig. 5D, right panel). ECM15 was overexpressed using an ACT1 constitutive promoter and compared with strains expressing empty vector alone in equivalent copy number, as previously described (43). As alluded to above, the cryptococcal RNA chaperone Vad1 regulates gene expression in a starvation-dependent fashion by recruiting mRNA to a decapping complex protein, Dcp2, leading to transcript degradation (19), and previous studies demonstrated that VAD1 binds ECM15 mRNA (35). In the present study, ECM15 mRNA binding to Vad1 was confirmed by qRT-PCR after radioimmunoprecipitation of a c-myc-Vad1 fusion protein (RIP-qRT-PCR) compared to that of an equivalent precipitation using an untagged strain (Fig. 5E). In addition, a vad1Δ mutant showed accumulation of ECM15 transcripts compared with the wild type from cells incubated under starvation conditions (Fig. 5F), suggesting that VAD1 is a repressor of ECM15. Taken together, these data suggest that deletion of cryptococcal VAD1 increases transcript abundance of ECM15, a regulator of capsule, mating, laccase, cell wall stability, starvation tolerance, and mammalian virulence.
In summary, these data identify a starvation-dependent epistatic regulatory pathway involving VAD1, ECM15, and CAC2 controlling growth on alternative carbon sources, expression of the virulence factors capsule and laccase, and mammalian virulence (Fig. 6).
FIG 6.
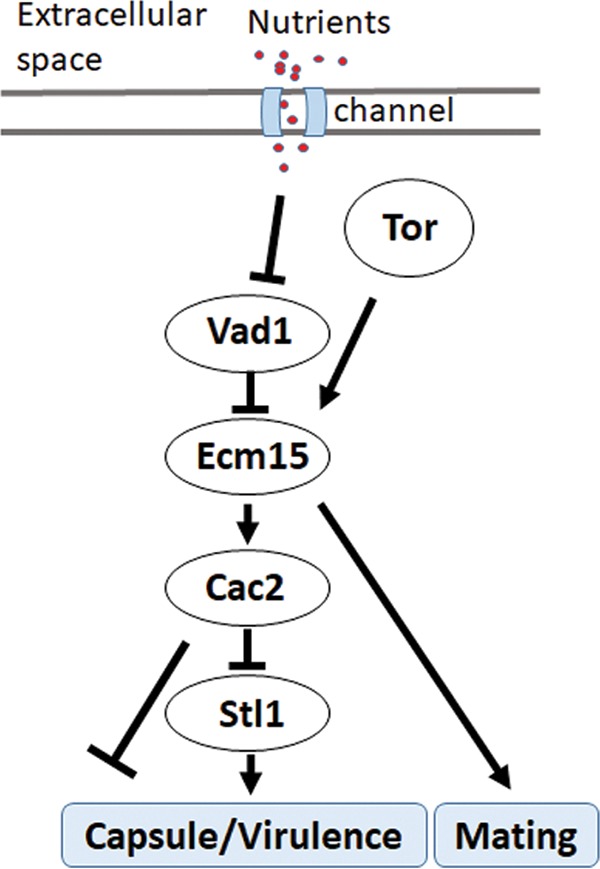
Scheme of starvation regulation of STL1 related to capsule formation and mammalian virulence.
DISCUSSION
Pathogen survival and virulence entail selection for traits necessary within the infective niche. For opportunist pathogens such as C. neoformans, evolutionary pressures exerted during residence in nutrient-poor soil or free-living amoebae (9), or in the presence of diphenolic toxins within plants (8), have likely shaped expression patterns necessary for effective mammalian infection. After infection, C. neoformans is also thought to reside in the human host for a prolonged time as a latent, growth-arrested organism (7). Thus, nutrient limitation could provide a common condition that shapes adaptation and pathogen optimization within diverse environments and was the subject of the present study.
In this study, expression patterns of C. neoformans within human cerebrospinal fluid from actively infected patients was highly correlated with that induced under starvation conditions at 37°C in both identity and levels of transcription. In addition, of the 222 genes previously reported to be upregulated after incubation in the macrophage cell line J774.1 (12), 147 were also found to be upregulated in CSF in the present studies, and of the 323 genes upregulated in free-living amoeba reported in the same paper, 164 were also upregulated in CSF and similar numbers in starvation, suggesting metabolic response commonality between these infective niches. Brain infections with C. neoformans are typically associated with low CSF glucose levels (3), and the pooled patient CSF used in the present studies reflected this. Interestingly, several studies suggest that the macrophage phagolysosome is also a nutrient-deprived environment (4, 43), implicating starvation response pathways in cryptococcal virulence in a number of human infective niches. Specifically, CSF incubation resulted in reduced expression of genes related to protein translation typical of growth limitations from reduced nutrients (44). In contrast, exposure to infected human CSF resulted in increased gene expression of processes related to sugar transport and alternative substrate utilization exemplified by genes involved in β-oxidation, the glyoxylate cycle, and gluconeogenesis. This metabolic reprogramming toward more efficient utilization of alternative substrates is shared by other organisms, such as Candida albicans (45). However, these expression patterns differed from that reported from RNA isolated directly from patient CSF of two CM patients, which more closely resembled that from nutrient-replete media and could have reflected an unusually vigorous growth condition or the higher levels of CSF glucose concentrations typical of some HIV/AIDS patients (46). However, such disparate results caution that expression of diverse strains from individuals with varied clinical presentations may be required to understand heterogeneity within the human infective niche.
Thus, the present studies utilized multiple patient isolates from a cohort of previously described HIV/AIDS patients presenting with cryptococcal meningoencephalitis and treated uniformly with amphotericin-based regimens to study gene expression under the same nutrient limitation conditions described above. These studies identified an STL1 sugar transporter whose expression levels were negatively associated with low rates of clearance from the CSF and showed a trend toward increased 10-week mortality. STL1 is named as a sugar transporter gene but has also been implicated in glycerol acquisition (29, 47), as was found for C. neoformans in the present studies with additional roles in lactate, pyruvate, and acetate metabolism. Interestingly, glycerol is elevated during cell membrane degradation and brain injury (48), and acetate has been demonstrated in large amounts by magnetic imaging spectroscopy during human cryptococcal infections (49, 50), and thus, STL1 may play a role in acquisition of these metabolites during infection. Early fungicidal activity (EFA) is a research measurement of CSF microbiological clearance in patients and is an important clinical prognostic marker of mortality. It reflects both microbiological virulence and drug responses, as well as host aspects of the infection (51). Notable in the present studies was reduced expression of the virulence factor laccase in the stl1Δ mutant. High laccase expression has previously been shown to be correlated with low EFA and worse patient survival, and higher expression could have contributed to microbiological retention in the CSF (23). While elevated STL1 expression tended to suggest poor survival, large variability in expression levels between wild-type C. neoformans strains could have impacted the significance of the relationship. Such variability was previously demonstrated by other cryptococcal biomarkers, such as CTR4, which demonstrated over 100-fold difference among clinical isolates from a cohort of solid organ transport patients (24). There also exist a number of potential nonmicrobiological factors that affect mortality. For example, initial patient mental status was more frequently altered in those who died and is an important risk factor for death (3), as was found in our cohort. Other previously reported clinical risk factors such as age and opening pressure on lumbar puncture did not differ between groups in our study. The relative immune response of the host could also be an important contributor to death in primary infections with C. neoformans. However, previous studies have shown only minor associations between mortality and markers such as CD4 count, peripheral white blood cell count, or CSF white cell count (3). The present cohort did not include patients with cryptococcal immune reconstitution syndrome (cIRIS), where immune responses may be more predominant (52), or other inflammatory syndromes associated with non-HIV-infected individuals (53). In addition, increased expression alone does not necessarily identify genes important to virulence, exemplified by the virulence factor laccase, which is downregulated at elevated temperatures (54), or the highly upregulated PTP1 sugar transporter, which also demonstrated no such role in virulence (12). Other upregulated genes such as PCK1 have had variable roles in virulence, with mutants attenuated in mice (13) but not in rabbits (14). All of these considerations led us to attempt to provide additional validation of the human cohort fungal expression studies by utilizing a model with genetically defined C. neoformans isolates. These studies did show differences in virulence between wild-type and stl1Δ strains, further implicating STL1 in fungal virulence. The intravenous model was tailored to the clinical cohort that all had brain infections and focuses on pathogen-related outcomes of murine brain infection, rather than a pulmonary (intratracheal or posterior pharyngeal inoculation) model, in which survival is more related to altered lung pathology (55). However, additional cohorts reflecting different clinical scenarios may be required to fully understand the complexity of the cryptococcus-human host interaction.
The present studies also identified an important molecular pathway in starvation response and its relation to human infections by examining STL1 as a target gene of the starvation/TOR stress pathway. TOR is an important mediator of the starvation/stress response, as well as cryptococcal survival (56), and inhibitors of TOR such as sirolimus and everolimus are in widespread use in patient populations at risk for CM, including transplant recipients (57). More recent work has shown that many TOR-dependent starvation processes are regulated via mRNA stability in yeast (58), and an important TOR-dependent regulator of mRNA stability, VAD1, is also a major virulence determinant in C. neoformans (19). The present studies identified a role for TOR/VAD1-dependent regulation of a novel CAC2/ECM15 regulatory pair of nuclear factors that demonstrated starvation-dependent regulation of STL1. In C. neoformans, a deletion mutant of CAC2 was previously found to exhibit normal growth characteristics and a slight susceptibility to UV irradiation (59). In S. cerevisiae, deletion of CAF-1 subunits such as CAC2 results in increased UV sensitivity and defects in subtelomeric gene silencing (37), suggesting a role in stress response, but the present studies are the first describing the role of Cac2 as a dynamic regulator of nutrient levels. As expected of an STL1 repressor, CAC2 overexpression resulted in reduced virulence in mice. The virulence phenotype was more evident in the CAC2 overexpressor than the STL1 knockout mutant and suggests a combined effect on virulence of multiple CAC2-regulated genes, some of which may not have reached significance in the patient cohort. Another ECM15/CAC2-dependent gene, CNAG_00876 (CP022325.1), has been previously identified as a ferric-chelate reductase gene (FRE7), regulating the important iron acquisition pathway in C. neoformans (60). The FRE7 gene was also upregulated in the patient cohort, although this did not approach significance and was not further characterized. The STL1 regulatory pathway (Fig. 6) has been simplified into a linear chain for study purposes but is likely much more complex due to the presence of other known virulence pathways related to alternative carbon acquisition/glycerol acquisition and metabolism, including HOG1, calcineurin, PI3K, PLC1, and PKA1 (19, 61 – 66). Repression by CAC2 may be chromatin dependent as promoter studies demonstrated greater CAC2-dependent suppression in glucose when the STL1 promoter constructs were integrated. Indeed, previous studies suggested that the Cac2-Caf1 complex binds preferentially to modified histones that have not been reported in plasmid DNA (67) and may thus be required as part of the chromatin repressor complex (68). The related factor Cac1 has recently been shown by single-particle electron microscopy to act as a histone binding platform, linking Cac2 within the Caf1-histone assembly complex implicated in maintenance of molecular architecture (69); however, the present studies extend the role of Cac2 within the Caf1 complex from merely a housekeeping function to that of a dynamic regulator of TOR-mediated nutrient stress response. Future studies may provide more detailed structural mechanistic insight for this complex regulation in eukaryotes and highlight the utility of C. neoformans as a model eukaryote. Nevertheless, the present data demonstrate that the VAD1/ECM15/CAC2 regulatory pathway is integral to the connection of the starvation/TOR virulence response through the sugar transporter gene STL1 (Fig. 6).
MATERIALS AND METHODS
Ethics statement.
Written informed consent was obtained, and the study was approved by the Research Ethics Committee of the University of Cape Town, the Medicines Control Council of South Africa, and the London-Surrey Borders Research Ethics Committee on behalf of St. George’s University of London and by an institutional review board (IRB)-approved protocol from the National Institute of Allergy and Infectious Diseases. All experimental procedures were conducted under a protocol approved by the Institutional Animal Care and Use Committee of the Intramural Research Program of the NIAID, NIH (protocol no. LCIM12E). All experimental studies were approved by the relevant NIAID Animal Care and Use Committee, as per the Guide for the Care and Use of Laboratory Animals (70) from the National Research Council of the National Academies, Washington, DC.
Study subjects.
Subjects providing isolates for expression analysis were control participants in a randomized trial of adjunctive gamma interferon (IFN-γ) in HIV-infected patients and a randomized trial examining alternative amphotericin B combinations, both described previously (71). We attempted to standardize the antifungal drug factor impact on EFA by only selecting for inclusion patients from a single clinical trial site treated using similar study protocols with amphotericin B-based induction regimens (i.e., no fluconazole and no adjunctive IFN-γ, both of which we know lead to significantly slower/faster clearance). CSF from 2 pooled donors was provided from stored specimens under an observational protocol previously described (53). As shown in Table 1, the 45 patients had their age, CD4 count, etc., recorded. All strains were previously serotyped to be serotype A and were genotyped by multilocus sequence typing (MLST) (72).
Strains and media.
Experiments were conducted in a genetic background of C. neoformans WT strain serotype A H99 (MATα [ATCC 208821]), which was the kind gift of J. Perfect. A complete list of strains used in this study is described in Table S5A in the supplemental material. Escherichia coli DH10B (Invitrogen) was the host strain for recovery and amplification of plasmids. The fungal strains were grown in YPD (2% glucose, 1% yeast extract, 2% Bacto-peptone) or YPD agar (YPD and 2% agar). Asparagine minimum selective medium (ASN) for transformant selection and incubation of YPD-grown mid-log cells for transcriptional studies or for detection of laccase production were described previously (73). V8 juice medium was used for mating assays as described previously (74).
Strains, primers, and probes used in this study. (A) Strains used in this study. (B) Primers used in this study. (C) FISH probes used in this study. Download Table S5, PDF file, 0.1 MB (88.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Microarray experiments.
Isolates from patients in Table 1 or the H99 lab strain were grown to mid-log phase and then transferred to asparagine medium without glucose (1 g/liter asparagine, 10 mM sodium phosphate [pH 7.4], and 0.25 g/liter MgSO4) or human CSF pooled from 4 patients with cryptococcal meningitis (having glucose concentrations from 10 to 25 mg/dl [0.55 to 1.4 mmol/liter]) and incubated for 3 h at 37°C, and RNA was recovered as described previously (24). Serotype A H99-based microarrays (hybridization probe sequences described previously in reference 27) with two unique probes (3 replicate features per probe) for each of 6,969 transcripts (1 per locus) were used for expression profiling. Two-color cohybridizations were performed with a common reference sample in the Cy5 channel on each array. Agilent Feature Extraction software (v.11.5.1.1, protocol GE2_107_Sep09) estimated the median pixel intensity of each feature, which was then log2 transformed. Replicate RNA samples from CSF or starvation buffer conditions were generated in two independent experiments. The reference sample was grown in YPD and used both for loess normalization and as a comparison condition in ANOVA. Each feature signal was normalized by loess (locally weighted scatterplot smoothing) and averaged per locus (3 replicates of two probes for 1 transcript per locus). A mixed-effects ANOVA model (fixed effect of growth medium, random effect of array identification) was computed, and expression difference estimates for each gene were calculated for CSF or starvation medium versus YPD. For the patient isolates (Table 1), the reference pool was created from 12 of the samples. Probe signals were summarized as the median of the 3 replicate features, then loess-normalized ratios against the cognate reference signal were averaged for the two probes per locus. A mixed-effects ANOVA model (fixed effect of survival or mortality time, random effect of study group, two levels) was computed, and expression difference estimates for each gene were calculated for week 2 mortality versus survived, week 10 mortality versus survived, or both week 2 and week 10 mortality versus survived. For both experiments, the false-discovery rate (FDR) was estimated from raw ANOVA P values to compensate for multiple testing of 6,969 genes. SAS and JMP/Genomics software (SAS, Cary, NC) was used for statistical analysis.
Overexpression, disruption, and complementation of ECM15, CAC2, and STL1 in C. neoformans.
Standard methods were used for overexpression, disruption, and complementation of the ECM15, CAC2, and STL1 genes in strain H99 (MATα), as described previously using two PCR-amplified fragments and a 1.3-kb PCR fragment of the URA5 gene previously described to effect a deletion within the target coding regions and was complemented using a 1-kb of up- and downstream genomic fragment of the target genes (13, 75, 76). Complementation in all cases retained the original deletion construct. The primers used in this study are listed in Table S5B.
Ecm15-mCherry and Cac2-GFP fusion proteins.
The cryptococcal shuttle vector pORA-YP142 (77) was used to express a fusion between the Ecm15 protein and a synthetic mCherry protein (Cneo-mCherry), utilizing C. neoformans codon usage produced by standard methods (75). The plasmid was digested with NdeI and PstI, and a PCR-amplified fragment of the H99 ECM15 gene-containing promoter region was digested with NdeI and PstI and ligated into compatible sites to produce YP148. The plasmids were recovered, the sequences were verified, and the plasmids were linearized with SceI and transformed into C. neoformans H99 MATα FOA cells by electroporation using standard methods (78). All cell preparations grown on nonselective medium were assayed at the end of each experiment by simultaneous inoculation of selective and nonselective plates to verify >90% retention of plasmid. Colocalization was observed using a Leica DMI 6000B microscope with a Hamamatsu camera using LAS AF6000 v.2.1.2 software (Leica). Pearson’s correlation coefficients of colocalization were calculated on a minimum of 20 cells using Leica LASX software. Predicted nuclear localization sequences were identified in the Ecm15 and Cac2 proteins using cNLS Mapper (41).
qRT-PCR experiments.
C. neoformans H99 strains were grown on YPD or ASN without glucose. Real-time PCR was performed using primer sets as described in Table S5B. Reverse transcription was performed on DNase-treated RNA using the iScript kit (Bio-Rad Laboratories) according to the manufacturer's protocol. PCRs were set up using iQ SYBR green supermix (Bio-Rad Laboratories), according to the manufacturer's protocol. qRT-PCR was performed using a Bio-Rad iCycler (MyiQ2).
FISH.
Sections of a brain autopsy specimen were obtained from a 42-year-old male who died of severe and diffuse C. neoformans infection as reported previously (13). Fluorescent in situ hybridization (FISH) was performed as previously described (19). Briefly, cells were washed once with 1× phosphate-buffered saline (PBS) and fixed for 4 h with 4% wt/vol paraformaldehyde in PBS at 4°C. Probes were labeled at the STL1 ORF with C3-fluorescein (LGC Bioresearch [Table S5C]). For the negative control, samples were treated with RNase A (50 µg/ml) for 1 h at 37°C, prior to the hybridization step. Fixed cells were hybridized in 20 μl of hybridization buffer (0.9 M NaCl, 0.01% wt/vol SDS, 20 mM Tris-HCl [pH 7.2], 20% formamide) with 5 ng of C3 fluorescein-labeled probe and incubated at 46°C for 16 h. After incubation, cells were pelleted by centrifugation and resuspended in 1 ml of prewarmed washing buffer (20 mM Tris-HCl [pH 8.0], 0.01% wt/vol SDS, 5 mM EDTA, 225 mM NaCl) for 30 min at 46°C. The slides were then mounted in ProLong Gold antifade reagent (Invitrogen) and observed using a Leica DMI 6000B microscope with a Hamamatsu camera and LAS AF6000 v.2.1.2 software (Leica). Cells were scored as positive by a blind observer and analyzed by Fisher’s exact test.
Measurement of capsular size and virulence studies.
To induce capsule, yeast cells were grown on ASN at 30°C for 4 days. Capsule was measured by microscopy after the fungal cells were suspended in India ink (79) and laccase by melanin production on norepinephrine agar. Virulence studies were conducted according to a previously described intravenous mouse meningoencephalitis model (80) using 10 CBA/J mice for each C. neoformans strain. All experimental procedures were conducted under a protocol approved by the Institutional Animal Care and Use Committee of the Intramural Research Program of the NIAID, NIH.
ChIP assay.
The ChIP assay was adapted and modified from a previously described protocol (81) for C. neoformans. PCR detection of the STL1 gene was performed using a primer set described in Table S5B. Input DNA was used as a positive control, consisting of unprecipitated genomic DNA as a loading control and to show intact function of each primer set and PCR.
Promoter deletion studies.
The 5′-truncated promoter sequences were obtained by PCR using a single reverse primer and different forward primers (positions −1000, −500, and −20 from the transcript start site) carrying BglII and NdeI restriction sites (Table S5B). The amplified fragments were inserted upstream of the GFP gene coding region of the plasmid, producing a series of STL1 promoter-STL1-GFP vectors. After verification by sequence analysis, the confirmed constructs or linear amplified fragments containing the indicated upstream region, STL1 reading frame, and GFP marker and terminator as indicated were transformed into WT or cac2Δ strains of C. neoformans. Integrated constructs were confirmed by uncut Southern blots (81). These strains were subjected to microscopy (differential interference contrast [DIC] or GFP fluorescence) or flow cytometry (fluorescein isothiocyanate [FITC]) to determine the promoter activity of STL1 under glucose or starvation conditions.
Statistics.
Errors were expressed as standard error of the mean (SEM). Fluorescence-positive cells (Fig. 1) were scored in a blind fashion and analyzed by Fisher’s exact test.
Calculations involving statistics available from the ANOVA output (JMP/Genomics version 8.0) include mean square error (MSE), error degrees of freedom (DDF), model degrees of freedom (NDF), and the proportion of the variance accounted for by the model (R2). From these we derive the following: SSE (sum of squares error) = MSE × DDF, SSM (sum of squares model) = SSE × R2/(1 − R2), MSM (mean square model) = SSM/NDF, and F ratio = MSM/MSE.
Raw P values are retrieved from the cumulative probability distribution of the F ratio using the parameters F ratio, NDF, and DDF (e.g., with the SAS function PROBF), where n (total samples) = NDF + DDF + 1 and C (number of groups) = NDF + 1.
A hypothetical increase of n samples per group by iteration i (steps) leads to an increase in DDF, DDF(ni) = n + 2 × n × i – C, which propagates to recalculations of SSE, SSM, F ratio, and P value for i = 1,2,3… iterations.
Data availability.
Data from the microarray experiments have been deposited in the National Library of Medicine Gene Expression Ontology (GEO) database.
ACKNOWLEDGMENTS
We acknowledge all members of JMP Technical Support team, SAS Institute, for assistance with power calculation formulas. We thank J. Powell (Bioinformatics and Molecular Analysis Section, CIT, NIH) for assistance with microarray data management and informatics.
This work was supported, in part, by the intramural research program of the NIAID, NIH. T.B. was funded by a Wellcome Trust Intermediate Clinical Fellowship (WT 089966). J.N.J. was supported by the Wellcome Trust, London, United Kingdom (WT081794). This publication was made possible through core services and support from the Penn Center for AIDS Research, a National Institutes of Health-funded program (grant no. P30 AI 045008 to J.N.J.).
Footnotes
Citation Park Y-D, Jarvis JN, Hu G, Davis SE, Qiu J, Zhang N, Hollingsworth C, Loyse A, Gardina PJ, Valyi-Nagy T, Myers TG, Harrison TS, Bicanic T, Williamson PR. 2018. Transcriptional profiling of patient isolates identifies a novel TOR/starvation regulatory pathway in cryptococcal virulence. mBio 9:e02353-18. https://doi.org/10.1128/mBio.02353-18.
Contributor Information
Joseph Heitman, Duke University.
John Perfect, Duke University.
Bettina Fries, Stony Brook University.
REFERENCES
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Rajasingham R, Smith R, Park B, Jarvis J, Govender NP, Chiller T, Denning D, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarvis JN, Bicanic T, Loyse A, Namarika D, Jackson A, Nussbaum JC, Longley N, Muzoora C, Phulusa J, Taseera K, Kanyembe C, Wilson D, Hosseinipour MC, Brouwer AE, Limmathurotsakul D, White N, van der Horst C, Wood R, Meintjes G, Bradley J, Jaffar S, Harrison T. 2014. Determinants of mortality in a combined cohort of 501 patients with HIV-associated cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis 58:736–745. doi: 10.1093/cid/cit794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beardsley J, Wolbers M, Kibengo FM, Ggayi AB, Kamali A, Cuc NT, Binh TQ, Chau NV, Farrar J, Merson L, Phuong L, Thwaites G, Van Kinh N, Thuy PT, Chierakul W, Siriboon S, Thiansukhon E, Onsanit S, Supphamongkholchaikul W, Chan AK, Heyderman R, Mwinjiwa E, van Oosterhout JJ, Imran D, Basri H, Mayxay M, Dance D, Phimmasone P, Rattanavong S, Lalloo DG, Day JN, CryptoDex Investigators. 2016. Adjunctive dexamethasone in HIV-associated cryptococcal meningitis. N Engl J Med 374:542–554. doi: 10.1056/NEJMoa1509024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson PR. 2013. Advancing translational immunology in HIV-associated cryptococcal meningitis. J Infect Dis 207:1793–1795. doi: 10.1093/infdis/jit102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazera MS, Pires FD, Camillo-Coura L, Nishikawa MM, Bezerra CC, Trilles L, Wanke B. 1996. Natural habitat of Cryptococcus neoformans var. neoformans in decaying wood forming hollows in living trees. Med Mycol 34:127–131. doi: 10.1080/02681219680000191. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Hermoso D, Janbon G, Dromer F. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol 37:3204–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warpeha KM, Park YD, Williamson PR. 2013. Susceptibility of intact germinating Arabidopsis thaliana to human fungal pathogens Cryptococcus neoformans and C. gattii. Appl Environ Microbiol 79:2979–2988. doi: 10.1128/AEM.03697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steenbergen JN, Shuman HA, Casadevall A. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A 98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson PR, Jarvis JN, Panackal AA, Fisher MC, Molloy SF, Loyse A, Harrison TS. 2017. Cryptococcal meningitis: epidemiology, immunology, diagnosis and therapy. Nat Rev Neurol 13:13–24. doi: 10.1038/nrneurol.2016.167. [DOI] [PubMed] [Google Scholar]
- 11.Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68:4225–4237. doi: 10.1128/IAI.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derengowski LS, Paes HC, Albuquerque P, Tavares AH, Fernandes L, Silva-Pereira I, Casadevall A. 2013. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell 12:761–774. doi: 10.1128/EC.00073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panepinto J, Liu L, Ramos J, Zhu X, Valyi-Nagy T, Eksi S, Fu J, Jaffe H, Wickes B, Williamson P. 2005. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J Clin Invest 115:632–641. doi: 10.1172/JCI200523048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR. 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2:e00103-11. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Arenas E, Cabezon V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, Gil C. 2007. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics 6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Rude T, Toffaletti D, Cox G, Perfect J. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun 70:5684–5694. doi: 10.1128/IAI.70.10.5684-5694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer GE, Askew DS, Williamson PR. 2008. The diverse roles of autophagy in medically important fungi. Autophagy 4:982–988. doi: 10.4161/auto.7075. [DOI] [PubMed] [Google Scholar]
- 18.Tatebe H, Shiozaki K. 2017. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 7:E77. doi: 10.3390/biom7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu G, McQuiston T, Bernard A, Park YD, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, Maraia RJ, Kehrl JH, Uzel G, Klionsky DJ, Williamson PR. 2015. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol 17:930–942. doi: 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza C, Heitman J. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev 25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 21.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. 2002. Adenylylcyclase functions downstream of the G-alpha protein GPA1 and controls mating and pathogenicity. Eukaryot Cell 1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClelland EE, Perrine WT, Potts WK, Casadevall A. 2005. Relationship of virulence factor expression to evolved virulence in mouse-passaged Cryptococcus neoformans lines. Infect Immun 73:7047–7050. doi: 10.1128/IAI.73.10.7047-7050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabiiti W, Robertson E, Beale MA, Johnston SA, Brouwer AE, Loyse A, Jarvis JN, Gilbert AS, Fisher MC, Harrison TS, May RC, Bicanic T. 2014. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J Clin Invest 124:2000–2008. doi: 10.1172/JCI72950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterman S, Hacham M, Hu G, Zhu X, Park Y, Shin S, Panepinto J, Valyi-Nagy T, Beam C, Husain S, Singh N, Williamson P. 2007. Role of a CUF1-CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J Clin Invest 117:794–802. doi: 10.1172/JCI30006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bicanic T, Meintjes G, Wood R, Hayes M, Rebe K, Bekker L, Harrison T. 2007. Fungal burden, early fungicidal activity, and outcome in cryptococcal meningitis in antiretroviral-naive or antiretroviral-experienced patients treated with amphotericin B or fluconazole. Clin Infect Dis 45:76–80. doi: 10.1086/518607. [DOI] [PubMed] [Google Scholar]
- 26.Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, Bahn Y-S, Chen W, Chen Y, Chow EWL, Coppée J-Y, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh Y-P, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuvéglise C, Newlon CS, Perfect JR, Lodge JK, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, Cuomo CA, Dietrich FS. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adler A, Park YD, Larsen P, Nagarajan V, Wollenberg K, Qiu J, Myers TG, Williamson PR. 2011. A novel specificity protein 1 (SP1)-like gene regulating protein kinase C-1 (Pkc1)-dependent cell wall integrity and virulence factors in Cryptococcus neoformans. J Biol Chem 286:20977–20990. doi: 10.1074/jbc.M111.230268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loyse A, Wilson D, Meintjes G, Jarvis JN, Bicanic T, Bishop L, Rebe K, Williams A, Jaffar S, Bekker LG, Wood R, Harrison TS. 2012. Comparison of the early fungicidal activity of high-dose fluconazole, voriconazole, and flucytosine as second-line drugs given in combination with amphotericin B for the treatment of HIV-associated cryptococcal meningitis. Clin Infect Dis 54:121–128. doi: 10.1093/cid/cir745. [DOI] [PubMed] [Google Scholar]
- 29.Kayingo G, Martins A, Andrie R, Neves L, Lucas C, Wong B. 2009. A permease encoded by STL1 is required for active glycerol uptake by Candida albicans. Microbiology 155:1547–1557. doi: 10.1099/mic.0.023457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Lu X, Zong H, Zhuge B. 2018. Functional and expression studies of two novel STL1 genes of the osmotolerant and glycerol utilization yeast Candida glycerinogenes. J Gen Appl Microbiol 64:121–126. doi: 10.2323/jgam.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Montezuma-Rusca JM, Powers JH, Follmann D, Wang J, Sullivan B, Williamson PR. 2016. Early fungicidal activity as a candidate surrogate endpoint for all-cause mortality in cryptococcal meningitis: a systematic review of the evidence. PLoS One 11:e0159727. doi: 10.1371/journal.pone.0159727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klawitter J, Nashan B, Christians U. 2015. Everolimus and sirolimus in transplantation—related but different. Expert Opin Drug Saf 14:1055–1070. doi: 10.1517/14740338.2015.1040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himmelreich U, Malik R, Kuhn T, Daniel HM, Somorjai RL, Dolenko B, Sorrell TC. 2009. Rapid etiological classification of meningitis by NMR spectroscopy based on metabolite profiles and host response. PLoS One 4:e5328. doi: 10.1371/journal.pone.0005328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Movahed E, Munusamy K, Tan GMY, Looi CY, Tay ST, Wong WF. 2015. Genome-wide transcription study of Cryptococcus neoformans H99 clinical strain versus environmental strains. PLoS One 10:e0137457. doi: 10.1371/journal.pone.0137457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park YD, Panepinto J, Shin S, Larsen P, Giles S, Williamson PR. 2010. Mating pheromone in Cryptococcus neoformans is regulated by a transcriptional/degradative “futile” cycle. J Biol Chem 285:34746–34756. doi: 10.1074/jbc.M110.136812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CS, Drubin D, Barnes G, Hazbun T. 2007. A protein interaction map of the mitotic spindle. Mol Biol Cell 18:3800–3809. doi: 10.1091/mbc.e07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman PD, Kobayashi R, Stillman B. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev 11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 38.Yang DH, Maeng S, Bahn YS. 2013. Msi1-like (MSIL) proteins in fungi. Mycobiology 41:1–12. doi: 10.5941/MYCO.2013.41.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi X. 2014. Heterochromatin structure: lessons from the budding yeast. IUBMB Life 66:657–666. doi: 10.1002/iub.1322. [DOI] [PubMed] [Google Scholar]
- 40.Galat A. 2003. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity—targets-functions. Curr Top Med Chem 3:1315–1347. doi: 10.2174/1568026033451862. [DOI] [PubMed] [Google Scholar]
- 41.Kosugi S, Hasebe M, Tomita M, Yanagawa H. 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lussier M, White AM, Sheraton J, di Paolo T, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, Roemer TW, Vo DH, Bondoc DC, Hall J, Zhong WW, Sdicu AM, Davies J, Klis FM, Robbins PW, Bussey H. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu G, Chen SH, Qiu J, Bennett JE, Myers TG, Williamson PR. 2014. Microevolution during serial mouse passage demonstrates FRE3 as a virulence adaptation gene in Cryptococcus neoformans. mBio 5:e00941-14. doi: 10.1128/mBio.00941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coller J, Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenz M, Bender J, Fink G. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Toffaletti D, Tenor J, Litvintseva AP, Fang C, Mitchell TG, McDonald T, Nielsen K, Boulware D, Bicanic T, Perfect JR. 2014. The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio 5:e01087-13. doi: 10.1128/mBio.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao S, Douglas NW, Heine MJ, Williams GM, Winther-Larsen HC, Meaden PG. 1994. The STL1 gene of Saccharomyces cerevisiae is predicted to encode a sugar transporter-like protein. Gene 146:215–219. doi: 10.1016/0378-1119(94)90295-X. [DOI] [PubMed] [Google Scholar]
- 48.Clausen T, Alves OL, Reinert M, Doppenberg E, Zauner A, Bullock R. 2005. Association between elevated brain tissue glycerol levels and poor outcome following severe traumatic brain injury. J Neurosurg 103:233–238. doi: 10.3171/jns.2005.103.2.0233. [DOI] [PubMed] [Google Scholar]
- 49.Himmelreich U, Allen C, Dowd S, Malik R, Shehan B, Mountford C, Sorrell T. 2003. Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes Infect 5:285–290. doi: 10.1016/S1286-4579(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 50.Himmelreich U, Dzendrowskyj TE, Allen C, Dowd S, Malik R, Shehan BP, Russell P, Mountford CE, Sorrell TC. 2001. Cryptococcomas distinguished from gliomas with MR spectroscopy: an experimental rat and cell culture study. Radiology 220:122–128. doi: 10.1148/radiology.220.1.r01jl25122. [DOI] [PubMed] [Google Scholar]
- 51.Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, Taseera K, Rebe K, Loyse A, Jarvis J, Bekker LG, Wood R, Limmathurotsakul D, Chierakul W, Stepniewska K, White NJ, Jaffar S, Harrison TS. 2009. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis 49:702–709. doi: 10.1086/604716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarvis JN, Meintjes G, Bicanic T, Buffa V, Hogan L, Mo S, Tomlinson G, Kropf P, Noursadeghi M, Harrison TS. 2015. Cerebrospinal fluid cytokine profiles predict risk of early mortality and immune reconstitution inflammatory syndrome in HIV-associated cryptococcal meningitis. PLoS Pathog 11:e1004754. doi: 10.1371/journal.ppat.1004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panackal AA, Wuest SC, Lin YC, Wu T, Zhang N, Kosa P, Komori M, Blake A, Browne SK, Rosen LB, Hagen F, Meis J, Levitz SM, Quezado M, Hammoud D, Bennett JE, Bielekova B, Williamson PR. 2015. Paradoxical immune responses in non-HIV cryptococcal meningitis. PLoS Pathog 11:e1004884. doi: 10.1371/journal.ppat.1004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu X, Williamson P. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res 5:1–10. doi: 10.1016/j.femsyr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Huffnagle GB, Traynor TR, McDonald RA, Olszewski MA, Lindell DM, Herring AC, Toews GB. 2000. Leukocyte recruitment during pulmonary Cryptococcus neoformans infection. Immunopharmacology 48:231–236. doi: 10.1016/S0162-3109(00)00222-8. [DOI] [PubMed] [Google Scholar]
- 56.Cruz MC, Cavallo LM, Gorlach JM, Cox G, Perfect JR, Cardenas ME, Heitman J. 1999. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol 19:4101–4112. doi: 10.1128/MCB.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aguiar PV, Campistol JM, Diekmann F. 2015. Safety of mTOR inhibitors in adult solid organ transplantation. Expert Opin Drug Saf 15:303–319. doi: 10.1517/14740338.2016.1132698. [DOI] [PubMed] [Google Scholar]
- 58.Messier V, Zenklusen D, Michnick SW. 2013. A nutrient-responsive pathway that determines M phase timing through control of B-cyclin mRNA stability. Cell 153:1080–1093. doi: 10.1016/j.cell.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 59.Yang DH, Maeng S, Strain AK, Floyd A, Nielsen K, Heitman J, Bahn YS. 2012. Pleiotropic roles of the Msi1-like protein Msl1 in Cryptococcus neoformans. Eukaryot Cell 11:1482–1495. doi: 10.1128/EC.00261-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saikia S, Oliveira D, Hu G, Kronstad J. 2014. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect Immun 82:839–850. doi: 10.1128/IAI.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strucko T, Zirngibl K, Pereira F, Kafkia E, Mohamed ET, Rettel M, Stein F, Feist AM, Jouhten P, Patil KR, Forster J. 2018. Laboratory evolution reveals regulatory and metabolic trade-offs of glycerol utilization in Saccharomyces cerevisiae. Metab Eng 47:73–82. doi: 10.1016/j.ymben.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Banerjee D, Bloom AL, Panepinto JC. 2016. Opposing PKA and Hog1 signals control the post-transcriptional response to glucose availability in Cryptococcus neoformans. Mol Microbiol 102:306–320. doi: 10.1111/mmi.13461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bahn YS, Kojima K, Cox GM, Heitman J. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell 16:2285–2300. doi: 10.1091/mbc.e04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Souza C, Alspaugh A, Yue C, Harashima T, Cox G, Perfect J, Heitman J. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kontoyiannis DP, Lewis RE, Alexander BD, Lortholary O, Dromer F, Gupta KL, John GT, Del Busto R, Klintmalm GB, Somani J, Lyon GM, Pursell K, Stosor V, Munoz P, Limaye AP, Kalil AC, Pruett TL, Garcia-Diaz J, Humar A, Houston S, House AA, Wray D, Orloff S, Dowdy LA, Fisher RA, Heitman J, Albert ND, Wagener MM, Singh N. 2008. Calcineurin inhibitor agents interact synergistically with antifungal agents in vitro against Cryptococcus neoformans isolates: correlation with outcome in solid organ transplant recipients with cryptococcosis. Antimicrob Agents Chemother 52:735–738. doi: 10.1128/AAC.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park YD, Williamson PR. 2012. 'Popping the clutch': novel mechanisms regulating sexual development in Cryptococcus neoformans. Mycopathologia 173:359–366. doi: 10.1007/s11046-011-9464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winkler DD, Zhou H, Dar MA, Zhang Z, Luger K. 2012. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res 40:10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Mondon P, Chang YC, Varma A, Kwon-Chung KJ. 2000. A novel episomal shuttle vector for transformation of Cryptococcus neoformans with the ccdB gene as a positive selection marker in bacteria. FEMS Microbiol Lett 187:41–45. doi: 10.1111/j.1574-6968.2000.tb09134.x. [DOI] [PubMed] [Google Scholar]
- 69.Kim D, Setiaputra D, Jung T, Chung J, Leitner A, Yoon J, Aebersold R, Hebert H, Yip CK, Song JJ. 2016. Molecular architecture of yeast chromatin assembly factor 1. Sci Rep 6:26702. doi: 10.1038/srep26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 71.Jarvis JN, Meintjes G, Rebe K, Williams GN, Bicanic T, Williams A, Schutz C, Bekker LG, Wood R, Harrison TS. 2012. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS 26:1105–1113. doi: 10.1097/QAD.0b013e3283536a93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beale MA, Sabiiti W, Robertson EJ, Fuentes-Cabrejo KM, O’Hanlon SJ, Jarvis JN, Loyse A, Meintjes G, Harrison TS, May RC, Fisher MC, Bicanic T. 2015. Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across southern Africa. PLoS Negl Trop Dis 9:e0003847. doi: 10.1371/journal.pntd.0003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X, Williamson PR. 2003. A CLC-type chloride channel gene is required for laccase activity and virulence in Cryptococcus neoformans. Mol Microbiol 50:1271–1281. doi: 10.1046/j.1365-2958.2003.03752.x. [DOI] [PubMed] [Google Scholar]
- 74.Xue C, Tada Y, Dong X, Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 75.Liu X, Hu G, Panepinto J, Williamson P. 2006. Role of a VPS41 homolog in starvation response and virulence of Cryptococcus neoformans. Mol Microbiol 61:1132–1146. doi: 10.1111/j.1365-2958.2006.05299.x. [DOI] [PubMed] [Google Scholar]
- 76.Panepinto JC, Misener AL, Oliver BG, Hu G, Park YD, Shin S, White TC, Williamson PR. 2010. Overexpression of TUF1 restores respiratory growth and fluconazole sensitivity to a Cryptococcus neoformans vad1Delta mutant. Microbiology 156:2558–2565. doi: 10.1099/mic.0.035923-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park Y-D, Shin S, Panepinto J, Ramos J, Qiu J, Frases S, Albuquerque P, Cordero RJB, Zhang N, Himmelreich U, Beenhouwer D, Bennett JE, Casadevall A, Williamson PR. 2014. A role for LHC1 in higher order structure and complement binding of the Cryptococcus neoformans capsule. PLoS Pathog 10:e1004037. doi: 10.1371/journal.ppat.1004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Panepinto J, Komperda K, Frases S, Park Y, Djordjevic J, Casadevall A, Williamson P. 2009. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol. 71:1165–1176. doi: 10.1111/j.1365-2958.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- 79.Erickson T, Liu L, Gueyikian A, Zhu X, Gibbons J, Williamson P. 2001. Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Mol Microbiol 42:1121–1131. doi: 10.1046/j.1365-2958.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 80.Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. 1996. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med 184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang S, Hacham M, Panepinto J, Hu G, Shin S, Zhu X, Williamson P. 2006. The Hsp70 member, Ssa1 acts as a DNA-binding transcriptional co-activator in Cryptococcus neoformans. Mol Microbiol 62:1090–1101. doi: 10.1111/j.1365-2958.2006.05422.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolic reprogramming of carbohydrate metabolism in C. neoformans in response to incubation in fungus-infected human CSF or starvation. Pathway schemes demonstrate differential expression ratios of C. neoformans after incubation in human fungus-infected CSF or under starvation conditions (starv) relative to mid-log-phase cells grown in YPD of genes expressing enzymes active in β-oxidation, glyoxylate cycle, or gluconeogenesis. Adapted from Derengowski et al. (12). Download FIG S1, PDF file, 0.1 MB (91KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression profiling demonstrates transcriptional expansion of genes related to transporter activity (GO function). Shown is transcriptional profiling of C. neoformans cells as in Fig. 1A displayed by the most common GO function terms. Shown are the percentage of annotated GO terms relative to total count of gene annotations (left upper panel), starvation (right upper panel), CSF alone (right lower panel), or both CSF and starvation (right lower panel). Download FIG S2, PDF file, 0.1 MB (95.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome-wide expression-level changes after transition from YPD to CSF. Download Table S1, PDF file, 0.3 MB (345.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome-wide expression-level changes after transition from YPD to starvation medium. Download Table S2, PDF file, 0.4 MB (417.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genome-wide expression changes between isolates of patients who died versus survived at 10 weeks. Download Table S3, PDF file, 0.1 MB (54.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Mating and spotting assay. (A) The indicated MATα strains were coincubated with a MATa mating partner (strain KN99) on nitrogen-limiting mating medium (V8 agar) for 3 weeks at room temperature. The edges of the mating mixtures were photographed (40×). Bar = 500 µm. (B) qRT-PCR of STL1 of indicated strains grown to the mid-log phase in ASN containing 0.03% acetate (without glucose). n = 3 independent experiments. ***, P < 0.001 by Student’s t test. (C) The indicated strains were diluted to an A600 of 1.0, and 1:5 serial dilutions (5 μl) were plated on ASN containing the indicated substrates (0.03%) and incubated at 30°C for 5 to 10 days. Download FIG S3, PDF file, 0.2 MB (259.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effects of serial deletion of the STL1 5′-promoter (−1000 from the transcript start site) on basal transcriptional activity in C. neoformans. A plasmid containing the STL1 ORF with indicated regions of the STL1 5′-promoter–GFP fusion gene was transformed into the WT strain of C. neoformans and grown to mid-log phase in YPD (Glu+) and ASN salts without glucose (Glu−), and the population was subjected to microscopy (DIC or GFP fluorescence) (A) or flow cytometry (FITC) (B). Empty vector was used as a control. Bar = 5μm. (C) Capsule thickness across mutant strains in C. neoformans. The indicated strains were incubated on YPD or ASN without glucose (Glu−) agar for 3 days at 30°C and examined by India ink microscopy. Bar = 5 µm. Download FIG S4, PDF file, 0.1 MB (110.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of genes suppressed by both ECM15 and CAC2 determined by genome-wide expression analysis. Download Table S4, PDF file, 0.1 MB (82.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Prediction of putative nuclear localization signals. Ecm15 (A) and Cac2 (B) putative nuclear localization signals were determined by cNLS Mapper. Download FIG S5, PDF file, 0.1 MB (120.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Strains, primers, and probes used in this study. (A) Strains used in this study. (B) Primers used in this study. (C) FISH probes used in this study. Download Table S5, PDF file, 0.1 MB (88.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Data from the microarray experiments have been deposited in the National Library of Medicine Gene Expression Ontology (GEO) database.