Abstract
Bacterial adaptive responses to biotic and abiotic stresses often involve large-scale reprogramming of the transcriptome. Since nitrogen is an essential component of the bacterial cell, the transcriptional basis of the adaptive response to nitrogen starvation has been well studied. The adaptive response to N starvation in Escherichia coli is primarily a ‘scavenging response’, which results in the transcription of genes required for the transport and catabolism of nitrogenous compounds. However, recent genome-scale studies have begun to uncover and expand some of the intricate regulatory complexities that underpin the adaptive transcriptional response to nitrogen starvation in E. coli. The purpose of this review is to highlight some of these new developments.
Keywords: Escherichia coli, nitrogen metabolism, RNA polymerase, transcriptomics
Introduction
Conditions that sustain constant bacterial growth are seldom found in nature. Bacterial growth is often limited by the availability of nutrients: soil, water, and even host environments such as macrophages can lack essential nutrients to support growth. Hence, many bacteria spend the majority of their time in states of little or no growth because they are starved of essential nutrients. The nutrient-starved and growth-attenuated state is widely considered an important physiological condition in bacterial pathogenesis and survival and is thus an area of intense research. Nitrogen (N) is an essential element of most macromolecules in the bacterial cell (including proteins, nucleic acids, and cell wall components). Many bacterial pathogens experience nitrogen limitation in host environments such as the urinary tract (e.g. uropathogenic Escherichia coli [1]) or macrophages (e.g. Salmonella Typhimurium [2]) and respond by activating specific adaptive processes. Emerging evidence also indicates that bacterial nitrogen metabolism and nitrogen stress responses are important for the gut microbiota to be properly established and maintained [3]. Bacteria can assimilate a variety of N sources where ammonia typically supports the fastest growth, serving as the preferred N source for many bacteria including E. coli [4]. The adaptive responses that allow bacteria to cope with, and survive, N starvation primarily manifest themselves through large-scale changes in the transcriptome. This review summarises the transcriptional basis underpinning how E. coli adapts to N starvation, specifically highlighting two new branches in this important adaptive process.
A sensing mechanism for N starvation
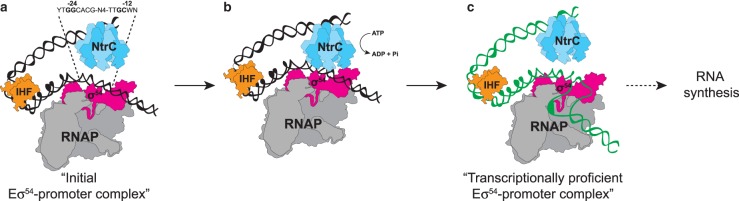
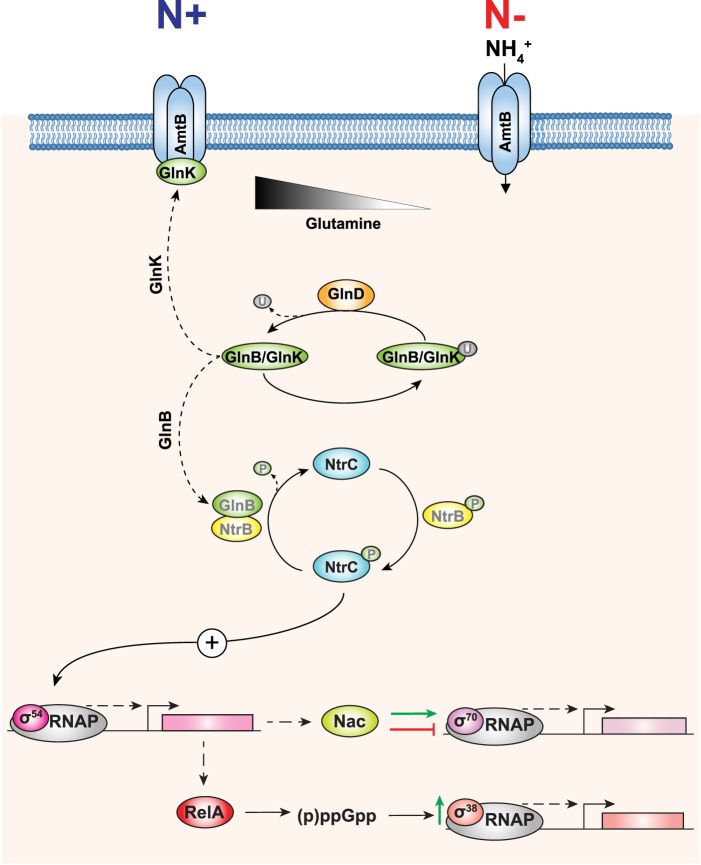
The initial transcriptional response to nitrogen starvation is dependent on the RNA polymerase (RNAP; E) containing the major variant promoter-specificity sigma (σ) factor subunit σ54 (reviewed in ref. [5]). In contrast with the major E. coli σ70 family of σ factors involved in adaptive responses to diverse stress cues, including housekeeping functions, σ54 directs its RNAP (Eσ54) to promoters uniquely characterised by consensus dinucleotide sequences positioned at −24 and −12 relative to the transcription start site at +1. Promoter complexes formed by Eσ54 initially exist in a transcriptionally inactive state until activated by a specialised activator protein, which binds to enhancer-like sequences positioned ∼100–150 base pairs upstream of the +1 site and interact with the Eσ54 at the promoter by looping (assisted by the DNA-bending protein integration host factor) out the intervening DNA. The transcriptionally proficient promoter complex is only formed when the activator protein remodels the initial promoter complex in a reaction consuming ATP (Figure 1) [6]. In Enterobacteria, the master transcription regulator of the adaptive response to N starvation, the Nitrogen regulation (Ntr) stress response, is NtrC of the NtrBC two-component system, where NtrB is the cognate histidine kinase of NtrC. Under N replete growth, ammonia is converted to glutamine and glutamate, which are the primary nitrogen donors in the cell. The intracellular concentration of glutamine is thought to be the main intracellular signal for N availability in E. coli and its levels are detected by the uridylyltransferase/uridylyl-removing enzyme, GlnD (Figure 2). We refer readers to reviews by Dixon and Kahn [7], Larry Reitzer [4] and Huergo et al. [8] for a detailed description of the mechanism by which glutamine is sensed by UT/UR (uridylyltransferase/uridylyl-removing). At low glutamine concentrations, GlnD deuridylylates the PII homologues, GlnB and GlnK. The deuridylylated form of GlnB binds to NtrB to activate its phosphatase activity and consequently dephosphorylates NtrC. The deuridylylated form of GlnK interacts with the ammonium transporter, AmtB, to inhibit ammonium uptake. Conversely, under N sufficiency, when the intracellular concentration of glutamine is high, GlnB and GlnK become deuridylylated, which results in (i) the phosphorylation of NtrC (by NtrB) leading to expression of the NtrC regulon and (ii) the inhibition of the interaction of GlnK with AmtB, thus enabling the uptake of ammonium (Figure 2).
Figure 1. Steps involved in transcription initiation by the major variant bacterial RNAP, Eσ54, which is required for the expression of the NtrC regulon in N-starved E. coli.
In (a), Eσ54 forms the initial promoter complex, which is transcriptionally silent. The enhancer DNA-bound NtrC interacts with the initial Eσ54 promoter complex by looping out the intervening DNA in an ATP hydrolysis-dependent manner (b), leading to the formation of the transcriptionally proficient Eσ54 promoter complex (c).
Figure 2. The Ntr response in E. coli.
The signalling cascade E. coli uses to sense and respond to N starvation, which results in reprogramming of the transcriptome by least three different forms of RNAP.
The expanded regulon of NtrC
Previous studies from Sydney Kustu and colleagues dubbed the adaptive response to N starvation as a scavenging response because many of the genes that are activated by NtrC encode transport systems for nitrogenous compounds [9]. One of the NtrC-activated genes encodes the N assimilation control (Nac) protein, which is a DNA-binding dual transcription regulator. Nac serves to both activate and repress Eσ70-dependent genes required for adaptation to N starvation. Nac-regulated genes are involved in diverse physiological processes ([10], and our unpublished data). We refer readers interested in the structure–function–regulation of Nac to a detailed review by Bob Bender [11]. Recently, a study by Brown et al. [12] highlighted the need for E. coli to integrate the specific adaptive response to N starvation with bacterial stringent response-mediated changes in gene expression to allow the cell to optimally cope with low N availability. By combining global gene expression analysis with genome-wide profiling of NtrC-binding sites in N-starved E. coli, Brown et al. discovered that NtrC also activates the transcription of relA; the product of which is responsible for the synthesis of the major stress signalling nucleotide, (p)ppGpp [13,14]. Accumulation of (p)ppGpp has far-reaching consequences for major cellular processes, including transcription, translation and DNA replication, which collectively form the cell's stringent response. The stringent response leads to down-regulation of stable RNA synthesis (rRNA and tRNA), required in high abundance for fast-growing cells, while biosynthetic operons are up-regulated to promote survival until growth conditions improve. (p)ppGpp also induces intracellular accumulation of σ38 (the predominant σ factor active in stationary phase E. coli) and thus transcription from Eσ38-dependent promoters [15]. In essence, in its simplest form, the adaptive transcriptional response to N starvation, initiated by the transcription regulators, NtrC and Nac, can be considered to represent the integration of at least three distinct regulons (however, see later), which are dependent on the three different forms of the bacterial RNAP, namely Eσ54, Eσ70 and Eσ38 (Figure 2). Consequently, the initial transcriptional response to N starvation results in the global reprogramming of transcription, involving ∼40% of all E. coli genes, to allow E. coli cope with N starvation.
A role for a kinase in the adaptive transcriptional response to N starvation
Until recently, the roles of all but one — the yeaGH operon — of the transcription units within the NtrC regulon during adaptation to N starvation were well understood. The transcription of the yeaGH operon is driven by two adjacently positioned promoters, which are dependent on Eσ54 and Eσ38, respectively. Thus, unsurprisingly, yeaGH is one of the most highly expressed operons during the initial adaptive transcriptional response to N starvation [9,12]. Intriguingly, the yeaGH operon is also highly expressed in E. coli and Salmonella in response to diverse stresses including low pH [16], hyperosmotic conditions [17], entry into stationary phase [16], nitrogen starvation [12], sulfur limitation [18], in biofilms [19] and exposure to antimicrobial peptide Polymyxin B [20], clearly suggesting that the products of yeaG and yeaH have a role in how bacteria adapt to these stresses. The yeaGH operon is highly conserved across several bacterial species, particularly Enterobacteria, where both genes are present in numerous pathogenic species, including Salmonella, Shigella, Yersinia and Klebsiella, while prkC, the homologue of yeaG, is found in some Gram-positive bacteria such as Bacillus or Clostridium species. The product of yeaH, YeaH, is a 49 kDa uncharacterised protein with very little sequence or structural similarity to any protein described to date. However, intriguingly, the product of yeaG, YeaG, is a 75 kDa protein, which contains a carboxyl-terminal domain that shows strong amino acid sequence homology to Hank's type kinase domains [21] and an amino-terminal domain that resembles an ATPase Associated with diverse cellular Activities (AAA+) domain [22]. Indeed, an earlier study by Tagourti et al. [23] had demonstrated that recombinant YeaG had weak kinase activity in vitro. Interestingly, the comparison of protein sequences of bacterial kinases available in the public databases indicates that the domain organisation of YeaG is atypical among Hank's type kinases, potentially suggesting that YeaG might be subject to a novel mode of regulation.
A study by Figueira et al. [24] investigated the role of the yeaGH operon in the adaptive response to N starvation in E. coli. This work revealed that the viability of ΔyeaG and ΔyeaH E. coli became considerably compromised under sustained N starvation when compared with the wild-type strain. The viability of a ΔyeaGH E. coli strain was comparable to that of the respective single mutant strains, implying that YeaG and YeaH functionally interact in the adaptive response to N starvation. Intriguingly, Figueira et al. discovered that the intracellular concentration of σ38 was lower in N-starved ΔyeaG E. coli than in the wild-type strain. This was primarily due to the transcriptional dysregulation of two toxin–antitoxin gene pairs (mqsR/mqsA and dinJ/yafQ) in ΔyeaG E. coli, which likely antagonised a regulatory cascade that was responsible for the accumulation of σ38. The study concluded that YeaG acts upstream of σ38 and that its absence results in a perturbed adaptive transcriptional response to N starvation due to compromised intracellular σ38 levels.
In expanding, the knowledge of how yeaG contributes to adaptation to sustained nitrogen starvation, a recent study by Switzer et al. [25] used global gene expression analysis to compare the transcriptomes of wild-type and ΔyeaG E. coli growing in batch cultures under N replete conditions (N+), and short- (N-starved for 20 min; N−) and long-term (N-starved for 24 h; N-24) N-starved conditions. Surprisingly, the present study uncovered that, despite the intracellular levels of σ38 being different in wild-type and ΔyeaG E. coli (see above), the σ38 regulon was not dysregulated at N+ or N− and only modestly dysregulated at N-24 in mutant bacteria. A possible explanation for this could be that E. coli has evolved ‘failsafe’ mechanisms to — at least partly — compensate for the transcription of σ38-dependent genes by utilising Eσ70 under conditions where Eσ38 availability or activity is compromised. The fact that many σ70- and σ38-dependent promoters are difficult to distinguish at the level of activity (despite moderate differences at sequence level) might be consistent with such a view. Alternatively, the reduction in the intracellular levels of σ38 in ΔyeaG E. coli may not be sufficiently low to have a detectable impact on transcriptional output from its promoters [26].
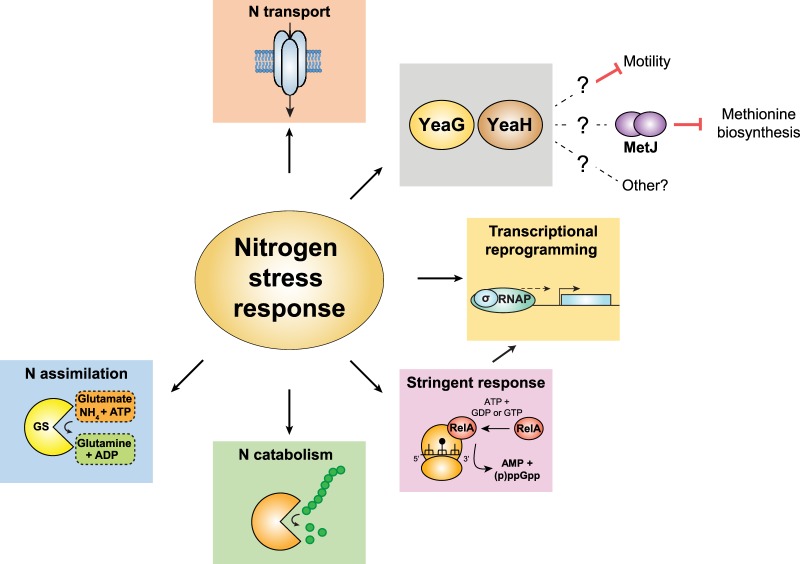
Intriguingly, Switzer et al. also discovered that the ΔyeaG E. coli displayed perturbed phenotypic and metabolic properties, which point to a dysregulation of the adaptive response to N starvation at the transcription level. Firstly, although the adaptive response to N starvation is a scavenging response, it does not involve chemotactic-like behaviour [27]. At N−, shortly after N run-out, several σ28-dependent genes, which are associated with the biosynthesis of flagella and motility, become up-regulated in ΔyeaG E. coli. Consequently, the mutant bacteria were more motile at N− than their wild-type counterparts. However, at N-24, following a sustained period of time under N starvation, the mutant bacteria ceased being motile. Secondly, the study also uncovered that genes associated with methionine biosynthesis (which are primarily under negative control by the transcription repressor, MetJ) become up-regulated at N-24 in ΔyeaG E. coli, consequently resulting in the aberrant biosynthesis of methionine in growth-attenuated and N-starved ΔyeaG E. coli cells. Although subsequent analysis revealed that the ability of MetJ to repress transcription (but not to bind to its cognate sites on the DNA) was compromised in mutant bacteria at N-24, the molecular basis by which YeaG affects MetJ activity remains to be elucidated. Since YeaG is a Hank's type kinase, it is tempting to speculate that regulation of MetJ activity during sustained N starvation involves it, or an effector protein, is phosphorylated by YeaG. Similarly, how YeaG influences Eσ28 activity at N− warrants further investigation. Clearly, YeaG appears to be a novel regulatory factor affecting the transcriptional regulation of (at least) two distinct genetic networks in N-starved E. coli. Thus, we propose that the yeaGH operon, alongside relA, constitutes a new branch in the Ntr response in E. coli (Figure 3).
Figure 3. The expanded regulon of NtrC.
Summary of the processes affected by the adaptive response to N starvation in E. coli, highlighting the role(s) played by the kinase, YeaG.
Adaptation to N starvation — a dynamic and temporally regulated transcriptional response?
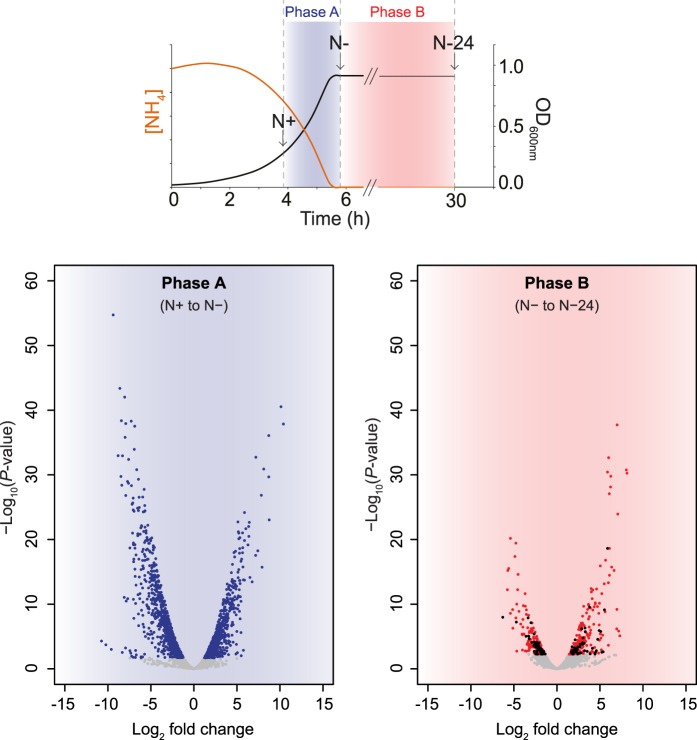
Perhaps, the most striking observation made by Switzer et al. [25] is that the adaptive transcriptional response in E. coli grown in batch cultures is dynamic and subject to temporal regulation (our unpublished observations). For example, upon transition from N+ to N− (phase A), 1787 genes (∼40% of E. coli genes) become differentially expressed (Figure 4). However, upon transition from N− to N-24 (phase B), 451 genes (10% of E. coli genes) are differentially expressed, of which 105 genes (2% of E. coli genes), that were not differentially expressed in phase A, become differentially expressed in phase B (Figure 4). Functional categorisation of the genes specifically differentially expressed in phase B shows that they are involved in diverse processes: transcription, amino acid transport, metabolism and several genes of unknown function. It is therefore tempting to speculate that these 105 genes exclusively contribute to the adaptive response to sustained N starvation. It remains to be investigated whether any of the genes that specifically become differentially expressed in phase B are subject to control by NtrC or Nac. The fact that intracellular NtrC levels remain unchanged at N− and N-24 would be consistent with the view that the role of NtrC (and possibly Nac) extends beyond the initial transcriptional response to N starvation. Although the observations made by Switzer et al. do not take the changes in gene expression associated with the massive change in the growth rate following N run-out into consideration, a previous study by Hua et al. [28] studied the transcriptome of N-starved E. coli under steady-state conditions (i.e. where the E. coli cells are growing at the same specified low growth rate). Their results revealed that only a subset of genes (235) become differentially expressed; just 5% of the E. coli genome compared with 40% identified by Switzer et al. that become differentially expressed in response to nitrogen run-out. Most of these genes belonged to the NtrC and Nac regulons. This observation further underscores the need to study the changes the transcriptome of E. coli undergoes as N starvation ensues for a longer period of time under steady-state conditions to uncouple changes in gene expression directly associated with adaptation to sustained N starvation from the indirect changes starved, and thus growth-impaired, E. coli cells undergo. The study by Switzer et al. ultimately indicates that the adaptive transcriptional response to N starvation occurs in phases to likely allow the cells to both optimally cope with N starvation, and be prepared — at least for a certain period of time — to resume growth when N becomes replenished. The role of YeaG in the adaptive transcriptional response to N starvation, based on the present study, can be considered as ‘molecular brake’, which functions to dampen transcription of ATP-consuming pathways in a temporally co-ordinated manner as the progression of N starvation ensues.
Figure 4. The dynamic adaptive transcriptome of N-starved E. coli.
Volcano plots illustrating significant differential gene expression shortly (20 min; N−) after N run-out (with respect to N+; phase A; in blue) and following sustained (24 h; N-24) N starvation (with respect to N−; phase B; in red). Significantly differentially expressed genes were defined as those with expression levels changed ≥ twofold with a false discovery rate-adjusted P-value < 0.05. The genes that are exclusively differentially expressed in phase B are shown in black.
Perspectives
Although our current understanding of the adaptive response to N starvation in E. coli and related bacteria are based on pioneering work from the laboratories of primarily Bob Bender, Martin Buck, Ray Dixon, Sydney Kustu, Boris Magasanik, Mike Merrick, Alexander Ninfa and many others, the recent genome-scale studies point to a complex and temporally co-ordinated and integrated reprogramming of the transcriptome in N-starved E. coli. Many details of the newly discovered features of the adaptive transcriptional response to N starvation are still unclear and many more features are likely to be discovered still. Unsurprisingly, the new findings open new questions and provide a springboard for further investigation. In our (perhaps biased) view, we consider the following as priorities:
YeaG is Hank's type kinase. What are the phosphorylation targets of YeaG in N-starved E. coli?
NtrC and Nac are the master transcriptional regulators of the initial adaptive transcriptional response to N starvation. What is the transcriptional regulatory basis of genes that constitute the transcriptional changes as sustained N starvation ensues?
Many of the genes of the NtrC and Nac regulons are expressed in pathogenic bacteria during the infection process. To what extent do pathogenic bacteria experience N starvation in the host? To what extent does the adaptive transcriptional response to N starvation differ in the context of the host and that seen in the laboratory setting? How does the dysregulation of the adaptive response to N starvation affect bacterial pathogenesis and infection outcome?
The transcriptome of N-starved and growth-attenuated bacteria is dynamic. What is the regulatory basis of the temporal changes that the transcriptome undergoes in nutrient-starved growth-attenuated bacteria and what are the phenotypic consequences of its dysregulation?
Acknowledgements
We thank members of the Wigneshweraraj group for helpful comments.
Abbreviations
- N
Nitrogen
- Nac
nitrogen assimilation control
- Ntr
nitrogen regulation
- RNAP (E)
RNA polymerase
Funding
This work was supported by a Wellcome Trust Investigator award WT100958MA, a Leverhulme Trust project grant [RPG-2017-431] and an MRC doctoral studentship.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Hagan E.C., Lloyd A.L., Rasko D.A., Faerber G.J. and Mobley H.L.T. (2010) Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6, e1001187 10.1371/journal.ppat.1001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose K.E. and Mekalanos J.J. (1997) Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect. Immun. 65, 587–596 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni J., Shen T.D., Chen E.Z., Bittinger K., Bailey A., Roggiani M. et al. (2017) A role for bacterial urease in gut dysbiosis and Crohn's disease. Sci. Transl. Med. 9, eaah6888 10.1126/scitranslmed.aah6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reitzer L. (2003) Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57, 155–176 10.1146/annurev.micro.57.030502.090820 [DOI] [PubMed] [Google Scholar]
- 5.Zhang N., Darbari V.C., Glyde R., Zhang X. and Buck M. (2016) The bacterial enhancer-dependent RNA polymerase. Biochem. J. 473, 3741–3753 10.1042/BCJ20160741C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glyde R., Ye F., Jovanovic M., Kotta-Loizou I., Buck M. and Zhang X. (2018) Structures of bacterial RNA polymerase complexes reveal the mechanism of DNA loading and transcription initiation. Mol. Cell 70, 1111–1120.e3 10.1016/j.molcel.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon R. and Kahn D. (2004) Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631 10.1038/nrmicro954 [DOI] [PubMed] [Google Scholar]
- 8.Huergo L.F., Chandra G. and Merrick M. (2013) PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. 37, 251–283 10.1111/j.1574-6976.2012.00351.x [DOI] [PubMed] [Google Scholar]
- 9.Zimmer D.P., Soupene E., Lee H.L., Wendisch V.F., Khodursky A.B., Peter B.J. et al. (2000) Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl Acad. Sci. U.S.A. 97, 14674–14679 10.1073/pnas.97.26.14674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aquino P., Honda B., Jaini S., Lyubetskaya A., Hosur K., Chiu J.G. et al. (2017) Coordinated regulation of acid resistance in Escherichia coli. BMC Syst. Biol. 11, 1 10.1186/s12918-016-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender R.A. (2010) A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae. J. Bacteriol. 192, 4801–4811 10.1128/JB.00266-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown D.R., Barton G., Pan Z., Buck M. and Wigneshweraraj S. (2014) Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 5, 4115 10.1038/ncomms5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauryliuk V., Atkinson G.C., Murakami K.S., Tenson T. and Gerdes K. (2015) Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalebroux Z.D. and Swanson M.S. (2012) Ppgpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212 10.1038/nrmicro2720 [DOI] [PubMed] [Google Scholar]
- 15.Kvint K., Farewell A. and Nyström T. (2000) RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of ςs. J. Biol. Chem. 275, 14795–14798 10.1074/jbc.C000128200 [DOI] [PubMed] [Google Scholar]
- 16.Weber H., Polen T., Heuveling J., Wendisch V.F. and Hengge R. (2005) Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603 10.1128/JB.187.5.1591-1603.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozen Y. and Belkin S. (2001) Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25, 513–529 10.1111/j.1574-6976.2001.tb00589.x [DOI] [PubMed] [Google Scholar]
- 18.Gyaneshwar P., Paliy O., McAuliffe J., Popham D.L., Jordan M.I. and Kustu S. (2005) Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 187, 1074–1090 10.1128/JB.187.3.1074-1090.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schembri M.A., Kjaergaard K. and Klemm P. (2003) Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267 10.1046/j.1365-2958.2003.03432.x [DOI] [PubMed] [Google Scholar]
- 20.Erickson K.D. and Detweiler C.S. (2006) The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol. Microbiol. 62, 883–894 10.1111/j.1365-2958.2006.05420.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stancik I.A., Šestak M.S., Ji B., Axelson-Fisk M., Franjevic D., Jers C. et al. (2018) Serine/threonine protein kinases from bacteria, archaea and eukarya share a common evolutionary origin deeply rooted in the tree of life. J. Mol. Biol. 430, 27–32 10.1016/j.jmb.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 22.Hanson P.I. and Whiteheart S.W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- 23.Tagourti J., Landoulsi A. and Richarme G. (2008) Cloning, expression, purification and characterization of the stress kinase YeaG from Escherichia coli. Protein Expr. Purif. 59, 79–85 10.1016/j.pep.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 24.Figueira R., Brown D.R., Ferreira D., Eldridge M.J.G., Burchell L., Pan Z. et al. (2015) Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci. Rep. 5, 17524 10.1038/srep17524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switzer A., Evangelopoulos D., Figueira R., de Carvalho L.P.S., Brown D.R. and Wigneshweraraj S. (2018) A novel regulatory factor affecting the transcription of methionine biosynthesis genes in Escherichia coli experiencing sustained nitrogen starvation. Microbiology 164, 1457–1470 10.1099/mic.0.000683 [DOI] [PubMed] [Google Scholar]
- 26.Wong G.T., Bonocora R.P., Schep A.N., Beeler S.M., Lee Fong A.J., Shull L.M. et al. (2017) Genome-wide transcriptional response to varying RpoS levels in Escherichia coli K-12. J. Bacteriol. 199, e00755-16 10.1128/JB.00755-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyaneshwar P., Paliy O., McAuliffe J., Jones A., Jordan M.I. and Kustu S. (2005) Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc. Natl Acad. Sci. U.S.A. 102, 3453–3458 10.1073/pnas.0500141102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hua Q., Yang C., Oshima T., Mori H. and Shimizu K. (2004) Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 70, 2354–2366 10.1128/AEM.70.4.2354-2366.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]