Abstract
Epidemiological studies have revealed that offspring conceived by in vitro fertilization (IVF) have an elevated risk of cardiovascular malformations at birth, and are more predisposed to cardiovascular diseases. The renin-angiotensin system (RAS) plays an essential role in both the pathogenesis of congenital heart disease in fetuses and cardiovascular dysfunction in adults. This study aimed to assess the relative expression levels of genes in the RAS pathway in mice conceived using IVF, compared to natural mating with superovulation. Results demonstrated that expression of the angiotensin II receptor type 1 (AGTR1), connective tissue growth factor (CTGF), and collagen 3 (COL3), in the myocardial tissue of IVF-conceived mice, was elevated at 3 weeks, 10 weeks, and 1.5 years of age, when compared to their non-IVF counterparts. These data were supported by microRNA microarray analysis of the myocardial tissue of aged IVF-conceived mice, where miR-100, miR-297, and miR-758, which interact with COL3, AGTR1, and COL1 respectively, were upregulated when compared to naturally mated mice of the same age. Interestingly, bisulfite sequencing data indicated that IVF-conceived mice exhibited decreased methylation of CpG sites in Col1. In support of our in vivo investigations, miR-297 overexpression was shown to upregulate AGTR1 and CTGF, and increased cell proliferation in cultured H9c2 cardiomyocytes. These findings indicate that the altered expression of RAS in myocardial tissue might contribute to cardiovascular malformation and/or dysfunction in IVF-conceived offspring. Furthermore, these cardiovascular abnormalities might be the result of altered DNA methylation and abnormal regulation of microRNAs.
Keywords: DNA methylation, in vitro fertilization, mouse, myocardium, renin-angiotensin system, microRNA
Altered expression of the renin-angiotensin system in myocardial tissue might contribute to an increased risk of cardiovascular malformations and dysfunction in IVF offspring, and is involved in abnormal regulations of DNA methylation and microRNAs.
Introduction
Since the birth of Louise Brown, the first baby conceived via in vitro fertilization and embryo transfer (IVF-ET) in 1978, more than 5 million newborns have been conceived through assisted reproductive technology (ART) [1, 2]. ART includes controlled ovarian hyperstimulation, IVF-ET, intracytoplasmic sperm injection (ICSI), preimplantation genetic diagnosis, and gamete and embryo cryopreservation. The clinical implementation of ART has made a significant contribution to the treatment of infertility [3]. However, ART intervenes several key points during the process of human conception and early development, specifically gametogenesis, fertilization, and early embryonic maturation. Data has shown that during the periconceptional period, the oocyte and embryo are highly sensitive to signals from their surrounding environment and as such, are particularly susceptible to damage which might manifest in an altered adult phenotype [4]. Follow-up studies on the growth and development of ART-conceived human offspring have aroused widespread concern as their findings indicate that when compared with naturally conceived newborns those conceived by IVF are at greater risk of low birth weight, increasing their susceptibility in adulthood to cardiovascular disease and diabetes [5–7]. These data are of particular concern given the extensive clinical application of ART and a large number of resultant ART-conceived offspring [8]. Multicenter epidemiological studies have shown that the incidence of cardiovascular malformations in ART babies is markedly increased compared with that of naturally conceived offspring [9–11]. Furthermore, follow-up surveys of ART-conceived offspring have shown an increased risk of dysfunction [12] and remodeling in the cardiovascular system [13], including higher arterial blood pressure, altered vascular intima-media thickness, and compromised myocardial regulatory function [14]. Investigations using mouse models have further illustrated that ART-conceived offspring have abnormal vascular endothelial function and changes in lipid metabolism [15, 16]. While it is widely accepted that IVF-conceived offspring have an increased risk of poor cardiovascular health, the precise mechanisms underlying cardiovascular malformation and dysfunction remain unclear.
The renin-angiotensin system (RAS) is a major physiological system that regulates blood pressure and the balance of fluids and electrolytes within the body. In addition, the RAS is known to play an essential role in the pathogenesis of several types of cardiovascular disease as well as in cardiac remodeling [17, 18]. Growing evidence reveals that in addition to the classic systemic RAS, a local RAS operates, in organs such as the heart, large arteries, kidneys, and even in single cells to further modulate cardiovascular regulation [19, 20]. In the RAS, the precursor protein angiotensinogen (AGT) is first converted into angiotensin I (Ang I) by renin (Ren). Then, Ang I is hydrolyzed to Ang II-IX by angiotensin-converting enzymes (ACE). Ang II is a biological key hormone, and its function is mediated through the receptors of Ang II type 1 (AGTR1) or Ang II type 2 (AGTR2) [21]. Excessive activation of AGTR1 results in increased expression levels of the extracellular matrix components, such as collagen 1 (COL1) and collagen 3 (COL3) [22], which are distributed in several layers of the heart wall to provide rigidity and elasticity [23]. During the embryonic period, abnormal expression of COL1 and COL3 can affect the normal development of the cardiovascular structures [24, 25]. In adulthood, overstimulation of the RAS can contribute to the pathogenesis of a number of cardiovascular diseases, affecting the normal function of cardiovascular tissue cells and increasing the risk of hypertension, heart arrhythmias, and cardiac hypertrophy [26]. Connective tissue growth factor (CTGF) promotes fibrosis therefore playing a further key role in AT1-mediated intracellular signal transmission that results in myocardial remodeling and vascular fibrosis [27].
During gametogenesis and early embryogenesis, parental genomes undergo demethylation and remethylation for epigenetic reprogramming. The first round of genome reprogramming occurs in the germline, while the second round of genome reprogramming occurs after fertilization when somatic methylation patterns are generated [28]. Evidence suggests that the early embryo, and later the fetus, is highly sensitive to its in utero environment, and that a suboptimal uterine milieu, such as is experienced by the fetus during maternal nutritional restriction, could increase the incidence of chronic diseases in adulthood via epigenetic modifications in the embryo or fetus [4, 29, 30]. Since IVF-ET overlaps with the epigenetic reprogramming period of the embryo, the risk of imprinting defects that cause imprinted disorders such as Prader-Willi syndrome and Angelman syndrome may increase when fertilization and early embryonic development occur in vitro [31]. Recent experiments in humans and mice have revealed differences in the methylation levels of CpG islands in imprinting genes and developmentally important genes between ART-conceived offspring and control groups [32, 33]. A previous study from our research group showed alterations in Igf2/H19 expression and DNA methylation in both the liver and the skeletal muscle of IVF mice [34]. Interestingly, ICSI-conceived fetuses also exhibit a decreased rate of CpG island methylation, accompanied by increased expression of insulin-induced gene 1 (INSIG1) and resulting in significantly increased umbilical cord blood lipid levels [35]. In addition to ART-associated DNA methylation and chromatin acetylation modifications, the role of noncoding RNAs in regulating gene expression has become a concern in IVF-ET offspring [36]. It is known that multiple microRNAs (miRNAs) are involved in the regulation of hypertension, coronary heart disease, and glycolipid metabolic disorders [37, 38]. However, few studies have focused on whether ART affects the expression of miRNAs. Therefore, a miRNA array was performed in this present study to identify associations between miRNA expression and the use of IVF-ET, and also to explore possible miRNA mediated mechanisms influencing the increased incidence of cardiovascular disease in ART offspring.
Variations in the health status diseases, fertility, genetic background, and living environment of human patients make it difficult to clarify a causal link between the use of ART and the development of cardiovascular aberrations in offspring. In addition, it is currently almost impossible to assess the long-term health of a population conceived through ART, since most IVF-conceived offspring are less than 30 years old. Therefore, a mouse model was used in this study to evaluate the expression levels of RAS components in the myocardial tissue of IVF and naturally conceived mice, from 3 weeks to 1.5 years. In addition, corresponding DNA methylation levels in the RAS, and miRNA expression profiles were evaluated, with the view to investigate the presence of underlying epigenetic regulatory mechanisms that might influence cardiovascular dysfunction in ART-conceived offspring.
Materials and methods
Experimental animals
All treatment protocols were approved by the Animal Care Ethics Committee of Zhejiang University (No. ZJU2009101007Y). The methods used in these investigations were performed in accordance with the approved guidelines. C57BL/6J female mice (6–8 weeks old) and male mice (10–12 weeks old) were used in this study. All animals were provided with nesting materials and housed with a constant 12-h light/12-h dark cycle at 25°C ± 0.5°C with 50%–60% humidity. Mice were fed standard chow and water.
Experimental design
All female mice were superovulated and randomly divided into the IVF group and the in vivo naturally mated group.
Superovulation
C57BL/6J female mice were injected intraperitoneally with 7.5 IU (0.15 ml) of pregnant mare serum gonadotropin (PMSG) (GEN’s, Hangzhou, China). After 48 h, the female mice were injected with 7.5 IU (0.15 ml) of human chorionic gonadotropin (hCG) (GEN’s).
In vitro fertilization
Human tubal fluid (HTF) (Irvine Scientific, CA, USA) medium containing 10% synthetic serum substitute (SSS; Irvin Scientific) was equilibrated at 37°C with 5% CO2 and 95% humidity overnight before IVF. In the IVF group, sperms were collected from the cauda epididymis of C57BL/6J male mice in warmed modified HTF (mHTF) (containing 10% SSS) (Irvine Scientific) and then transferred to freshly warmed HTF for capacitation. Capacitation of the spermatozoa was performed in a 37°C environment with 5% CO2 and 95% humidity for 1–2 h. C57BL/6J female mice were euthanized by cervical dislocation at 15 h after hCG injection. The oviducts were separated and placed in warmed mHTF, and the ampulla of the fallopian tube was punctured. Cumulus-oocyte complexes were washed twice in mHTF and then transferred to HTF droplets. The capacitated sperm suspension was gently added to the HTF medium with metaphase II oocytes in a final sperm concentration of 1–2 × 106/ml. Sperm and oocytes were co-cultured for 6 h in the insemination medium. Then, the presumptive embryos were gently washed twice for further culture in fresh HTF. The two-cell-stage embryos of the IVF group were obtained 24 h later.
In the in vivo group, C57BL/6J female mice were caged with male mice at a ratio of 1:1 after hCG injection. The next day, female mice with a vaginal plug were separated from the males. After 44 h of hCG injection, the female mice were euthanized by cervical dislocation and two-cell embryos were obtained from the oviducts. The operating conditions were the same as those of the IVF group.
Embryo transfer
Female mice at least 8 weeks of age from the Institute of Cancer Research (ICR) were used as pseudopregnant recipients. Vaginal plugs were detected the morning after mating with vasectomized males from the ICR, and the animals were considered to be at day 0.5 of pseudopregnancy. Twelve morphologically normal two-cell-stage embryos in each group were transferred to the oviducts of pseudopregnant recipients.
Postnatal growth
The birth outcomes of mice were described previously [34]. Fifty-one mice were born in each of the IVF and in vivo groups. No significant differences were observed in pregnancy rate, birth rate, mortality, or sex ratio. The wet weights of mice were recorded weekly from birth to 10 weeks of age, and biweekly from 11 weeks to 1.5 years of age. The animals were euthanized, and the myocardial tissue was dissected. A part of the tissue was snap frozen in liquid nitrogen and stored at 80°C for RNA, DNA, and protein detection. The remaining tissue was fixed overnight with 4% paraformaldehyde in phosphate-buffered saline (PBS) for immunohistochemistry.
Cell culture
The H9c2 (2–1) cell line was purchased from American Type Culture Collection (ATCC, MD, USA) and cultured in Dulbecco modified Eagle's medium (DMEM) (Life Sciences, UT, USA) containing 10% fetal bovine serum (FBS) (Gibco, NY, USA) and 1% penicillin/streptomycin solution (Gibco) in 100-cm2 flasks at 37°C in a humidified atmosphere of 5% CO2.
MicroRNA transfection
The H9c2 cells in the exponential phase of growth (2 × 105/well) were cultured overnight in six-well plates to overexpress miR-297. The cells were transfected with a miR-297 mimic (100 nM) or negative control RNA (100 nM) using the riboFECT CP reagent and buffer (RiboBio) according to the manufacturer's protocol. First, 10 μl of miRNAs was diluted with 120 μl of riboFECT CP buffer for 10 min at room temperature. This diluent was mixed with 12 μl of riboFECT CP reagent and incubated for 10 min at room temperature. Finally, the total mixture was added to each well of the plate together with 1.8 ml of DMEM and incubated at 37°C. RNA and protein was extracted from the H9c2 cells after 24 and 48 h, respectively.
RNA extraction
Total RNA was extracted from the myocardial tissue of mice and H9c2 cells after transfection according to the manufacturer's protocol using the RNAiso Plus kit (TaKaRa, Tokyo, Japan).
Reverse transcription
For mRNAs (Ren1 Ace Ace2 Agt Agtr1a Agtr1b Agtr2a Col1a1 Col1a2 Col3 Ctgf), 0.5-μg RNA samples were reverse-transcribed with PrimeScript RT Reagent Kit (TaKaRa). For miRNAs (miR-100 miR-297 miR-758), reverse transcription was performed with Mir-X microRNA FirstStrand Synthesis Kit (TaKaRa). U6 snRNA controls served as a reference gene for analyzing microRNA expression levels.
Real-time quantitative polymerase chain reaction
Real-time quantitative polymerase chain reaction (PCR) analysis was performed using SYBR Premix Ex Taq (TaKaRa) for mRNAs and SYBR Premix Ex Taq II for miRNAs in an ABI 7900 thermocycler (Applied Biosystems, CA, USA). Gapdh served as a reference gene for analyzing gene expression levels. The real-time PCR data were analyzed using the 2−ΔΔCT method. The primers were synthesized by Sangon (Shanghai, China). The primer sequences for real-time PCR are shown in Supplementary Tables S1 and S2.
Western blotting
Myocardial tissues and H9c2 cells were homogenized in radioimmunoprecipitation assay lysis buffer (CWBIO, Beijing, China), incubated on ice for 20 min, and centrifuged at 14,000 g for 10 min at 4°C. The supernatant was transferred to a tube, and concentrations were detected using the Bradford assay (Bio-Rad Laboratories, CA, USA). Then, 50 μg of protein was transferred to each lane on a 10% SDS-polyacrylamide gel. These samples were transferred to polyvinylidene fluoride membranes (Millipore, MA, USA) and incubated with the following primary antibodies (Supplementary Table S3): anti-renin antibody (sc-137252; Santa Cruz Biotechnology, California, USA); anti-angiotensin converting enzyme 1 antibody (ab11734; Abcam, Cambridge, UK); anti-angiotensin II type 1 receptor antibody (ab124505; Abcam, Cambridge, UK); anti-angiotensin II type 2 receptor antibody (ab92445; Abcam); anti-collagen 3 antibody (ab7778; Abcam); anti-CTGF antibody (ab6992; Abcam); and anti-glyceraldehyde 3 phosphate dehydrogenase (GAPDH) antibody (ab181602; Abcam) overnight at 4°C. They were incubated with fluorescent secondary antibodies (680 and 790 nm; Jackson, PA, USA) for 2 h during RT. The protein bands were visualized using the Odyssey Imager (Li-Cor Biosciences, NE, USA). The GAPDH protein levels were used as the control protein.
DNA methylation analysis
Targeted bisulfite sequencing
The process of targeted bisulfite sequencing was performed to analyze DNA methylation, as described in a previous study [39]. The genomic DNA of myocardial tissue was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Genome bisulfite modification was performed using the EpiTect Bisulfite Kit (Qiagen) according to the operation manual, and its products were used for subsequent PCR amplification. Targeted sequences of primers are shown in Supplementary Table S4. Furthermore, a 1-μl bisulfite-converted DNA template was performed using the PCR program. The PCR conditions were as follows: 3 min at 94°C, 5 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C (decrease of 0.5°C in each cycle); then, 30 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C; and finally 72°C for 5 min. The PCR products were performed with end-repair, A-tailing, adaptor ligation, and clean-up according to the recommendations of KAPA Biosystems (MA, USA). A PCR program using Kapa Hifi DNA Polymerase (KAPA Biosystems) was performed as follows: 3 min at 94°C, 9 cycles of 30 s at 94°C, 30 s at 62°C, and 30 s at 72°C. The sequencing of samples was performed on the MiSeq System (Illumina, CA, USA) using 100-bp paired-end reads.
The methylation levels of each CpG site were calculated using the Bismark's methylation extractor tool [40] and shown using R scripts. Average methylation levels of the target regions and other information were generated with the local Perl, Python, and Shell scripts to compile counts of methylated and unmethylated reads at each CpG site.
MicroRNA microarray and bioinformatic analysis
MicroRNA profiling was performed by RiboBio. The quality and quantity of RNA were assessed using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). A260/A280 ≥1.5 and A260/A230 ≥1 were acceptable RNA purity levels, whereas an acceptable RNA integrity number (RIN) value was ≥5 using the Agilent 2200 RNA assay (Agilent Technologies, Santa Clara, CA, USA). gDNA contamination was detected using gel electrophoresis; 1 μg of total RNA was labeled with cy3 using the (Universal Linkage System) ULS method and hybridization was performed using RiboArray miDETECT mouse array (RiboBio). After the data were acquired, microRNA targets were predicted using the following databases: TargetScan (http://www.targetscan.org), http://www.microRNA.org (http://www.microrna.org), miRDB (http://www.mirdb.org/), and CLIP (http://lulab.life.tsinghua.edu.cn/clipdb/).
Luciferase assay
The AGTR1 3΄-UTR or AGTR1 3΄-UTR mutant fragment was amplified by PCR and inserted into the pmiR-RB-Report vector (Guangzhou Ruibo Biotech Company, China) to generate a luciferase reporter vector. Co-transfection of 293T cells with the recombinant vectors alongside miR-297a-3p mimic or negative control (NC) was performed with lipofectamine 2000 transfection reagent (Invitrogen, USA), according to the manufacturer's instructions. The relative luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega, USA). The reporter vector used was hRluc, with hluc as the internal reference corrected fluorescence. All transfection assays were performed in three independent biological replicates.
Cell proliferation assay
H9c2 cells were transfected with miRNA mimics in six-well plates. Forty-eight hours after cell transfection, 5-ethynyl-20-deoxyuridine (EdU) (10 μM) was added to the cultures and cells were grown for an additional 2 h.
For imaging analysis of cell proliferation, the cells were fixed with 4% formaldehyde in PBS for 30 minutes. After labeling, H9c2 cells were assayed using the Cell-Light EdU Apollo 488 in vitro imaging kit (RiboBio) according to the manufacturer's protocol. The proliferation rate was calculated after normalizing the number of EdU-positive cells to that of DAPI-stained cells in five randomly selected high power fields. Images were acquired under an FV1200 laser confocal microscopy (Olympus, PA, USA).
For flow cytometry analysis of cell proliferation, EdU (10 μM) (Cell Light EdU Apollo 488 in vitro flow cytometry kit; RiboBio) was added and the cells were cultured for an additional 2 h according to the manufacturer's protocol. H9c2 cells were resuspended in PBS and centrifuged at 15,000 rpm for 5 min. H9c2 cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. H9c2 cells were resuspended in PBS cells. Then, they were immunocytochemically stained with antibodies. The following tests were performed using ACEA NovoCyte flow cytometry (ACEA, San Diego, CA, USA).
Statistical analysis
The experimental data were compared using the Student t-test (version 19.0; SPSS, NY, USA) to perform statistical analyses of the results of the two groups. The results were presented as mean ± standard error of the mean. For all results, statistically significant differences were defined as P < 0.05 and P < 0.01.
Results
IVF altered the expression of genes in the RAS components and those related to myocardial remodeling in myocardial tissue
The expression levels of genes in the RAS components and corresponding effect genes were detected using quantitative RT-PCR to investigate the effect of IVF on the expression of essential genes related to the health of the cardiovascular system (Figure 1). The genes in the RAS components examined in this study included Ren1 Ace Ace2 Agt Agtr1a Agtr1b, and Agtr2. The corresponding effect genes of the RAS included Col1a1 Col1a2 Col3, and Ctgf. At 3 weeks of age, the expression levels of Ren1 Ace Agtr1a Col3, and Ctgf were found to be significantly increased in the IVF group compared with those in the in vivo group (1.14-fold, P < 0.05; 2.15-fold, P < 0.05; 2.01-fold, P < 0.05; 1.62-fold, P < 0.05; and 2.13-fold, P < 0.05, respectively) (Figure 1A). No statistically significant differences were observed in the expression levels of Ace2 Agt Agtr1b Agtr2 Col1a1 and Col1a2 (P > 0.05) between the in vivo group and the IVF group (Figure 1A). At 10 weeks of age, the expression levels of Ace Agt Agtr1b Agtr2 Col1a1, and Col3 were also found to be significantly increased in the IVF group compared with those in the in vivo group (1.74-fold, P < 0.05; 2.04-fold, P < 0.05; 1.79-fold, P < 0.05; 2.08-fold, P < 0.05; and 1.62-fold, P < 0.05, respectively) (Figure 1B). But no statistically significant differences were found in the expression levels of Ren1 Ace2 Agtr1a Col1a2, and Ctgf (P > 0.05) between the in vivo group and the IVF group (Figure 1B). When the mice reached the age of 1.5 years, the expression levels of Ren1 Ace Agtr1b Col3, and Ctgf were still increased in the IVF group compared with those in the in vivo group (2.37-fold, P < 0.01; 2.38-fold, P < 0.05; 1.68-fold, P < 0.05; 1.86-fold, P < 0.05; and 1.81-fold, P < 0.05, respectively) (Figure 1C). However, no significant differences were found in the expression levels of Ace2 Agt Agtr1a Agtr2 Col1a1, and Col1a2 (P > 0.05) between the in vivo group and the IVF group (Figure 1C).
FIGURE 1.
The mRNA relative expression of genes in the RAS components and those related to myocardial remodeling in myocardial tissue in the in vivo and IVF groups at (A) 3 weeks, (B) 10 weeks, and (C) 1.5 years. Ren1 Ace Ace2 Agt Agtr1a Agtr1b, and Agtr2 were critical elements in the RAS. Col1a1 Col1a2, and Col3 were the major components of the extracellular matrix of the heart and also crucial in the development of heart and myocardial remodeling. Ctgf promotes the pathological process of fibrosis. The data are presented as mean ± standard error of the mean. The relative levels were calculated using the 2−ΔΔCT method. *P < 0.05 and **P < 0.01, compared with the in vivo group; Student t-test (N, in vivo = 6, IVF = 6 in 3-week-old mice; N, in vivo = 6, IVF = 6 in 10-week-old mice; and N, in vivo = 6, IVF = 6 in 1.5-year-old mice).
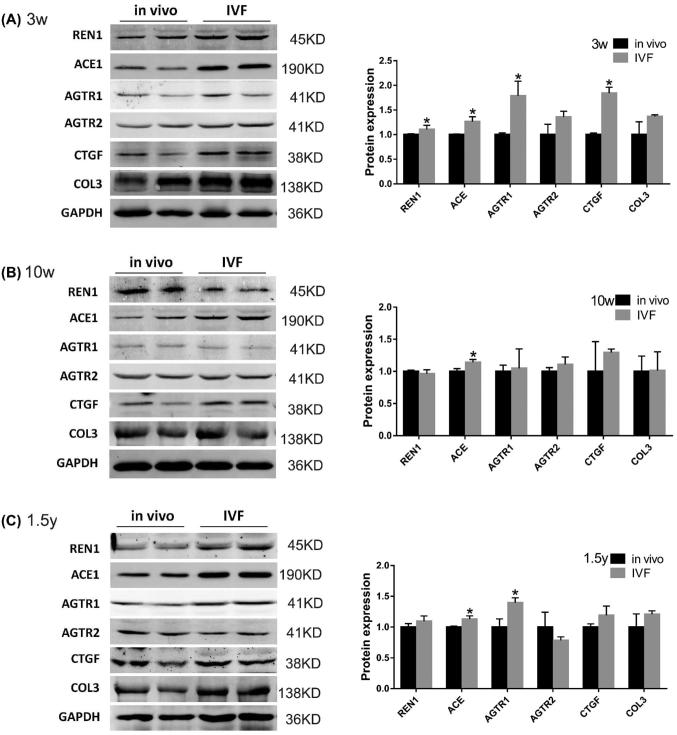
The protein expression levels of genes in the RAS components and corresponding effect genes were further assessed using quantitative western blotting (Figure 2). At 3 weeks of age, the expression levels of REN1, ACE, AGTR1, and CTGF were significantly increased in the IVF group compared with the in vivo group (1.11-fold, P < 0.05; 1.26-fold, P < 0.05; 1.79-fold, P < 0.05; 1.85-fold, P < 0.05, respectively) (Figure 2A). No significant differences were found in expression levels of AGTR2 and COL3 (P > 0.05) (Figure 2A). At 10 weeks of age, the expression levels of ACE were significantly increased in the IVF group compared with the in vivo group (1.14-fold, P < 0.05) (Figure 2B). The protein expression levels of AGTR1, AGTR2, and CTGF were not statistically different between the two groups at 10 weeks of age (P > 0.05) (Figure 2B). In old age, the protein expression of ACE and AGTR1 was upregulated in the IVF group (1.13-fold, P < 0.05; 1.40-fold, P < 0.05, respectively) (Figure 2C), but the protein expression levels of REN1, AGTR2, CTGF, and COL3 were not significantly different (P > 0.05) (Figure 2C).
FIGURE 2.
Western blot analysis of REN1, ACE1, AGTR1, AGTR2, CTGF, and COL3 in myocardial tissue in the in vivo and IVF groups at (A) 3 weeks, (B) 10 weeks, and (C) 1.5 years of age. GAPDH was used as the internal control. Data are presented as mean ± standard error of the mean. *P < 0.05 and **P < 0.01 compared with the in vivo group; Student t-test (N, in vivo = 4; IVF = 4 in 3-week-old mice; N, in vivo = 4; IVF = 4 in 10-week-old mice; and N, in vivo = 4; IVF = 4 in 1.5-year-old mice).
The altered methylation levels of CpG island regions of Col1a1 in IVF-conceived mice
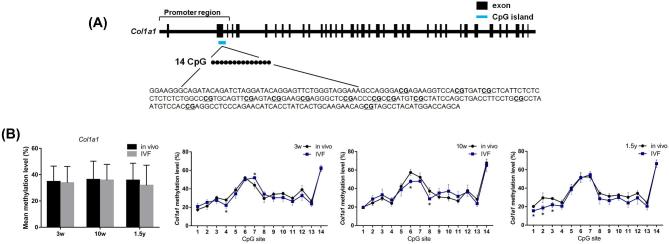
Among the differentially expressed genes, there are CpG islands near the promoters regions of Col1a1. Thus, Col1a1 were selected to explore whether the methylation levels of CpG island regions of Col1a1 would change in IVF-conceived mice (Figure 3). Targeted bisulfite sequencing analysis was used to detect the methylation level of each CpG site in the targeted gene. Using this method, we measured methylation levels of 14 CpG sites in Col1a1 target regions. The schematic overview of the CpG island regions of the Col1a1 is shown in Figure 3A. No statistically significant differences were observed in the mean DNA methylation level of Col1a1 between the IVF and in vivo groups (Figure 3B). At 3 weeks of age, the methylation level of the CpG site 4 of Col1a1 was significantly reduced in the IVF group (P < 0.05). The methylation levels of CpG sites 6 and 8 of Col1a1 at 10 weeks and of CpG sites 1, 2, and 3 of Col1a1 at 1.5 years of age were significantly reduced in the IVF group (P < 0.05). Only the methylation level of CpG site 7 at 3 weeks of age in the IVF group was increased (P < 0.05) (Figure 3B).
FIGURE 3.
Schematic overviews of the CpG island regions of Col1a1 (A). The target sequences examined by targeted bisulfite sequencing were shown, and CpG sites were underlined. Average DNA methylation levels for the whole regions of CpG islands and the mean methylation levels for the individual CpG site of Col1a1 (B) in the in vivo and IVF groups at 3 weeks, 10 weeks, and 1.5 years. Data are presented as mean ± standard error of the mean. *P < 0.05 and **P < 0.01 compared with the in vivo group; Student t-test (N, in vivo = 6; IVF = 6 in 3-week-old mice; N, in vivo = 6; IVF = 6 in 10-week-old mice; and N, in vivo = 6; IVF = 6 in 1.5-year-old mice).
MiRNAs were involved in the regulation of IVF-induced abnormal cardiac gene expression
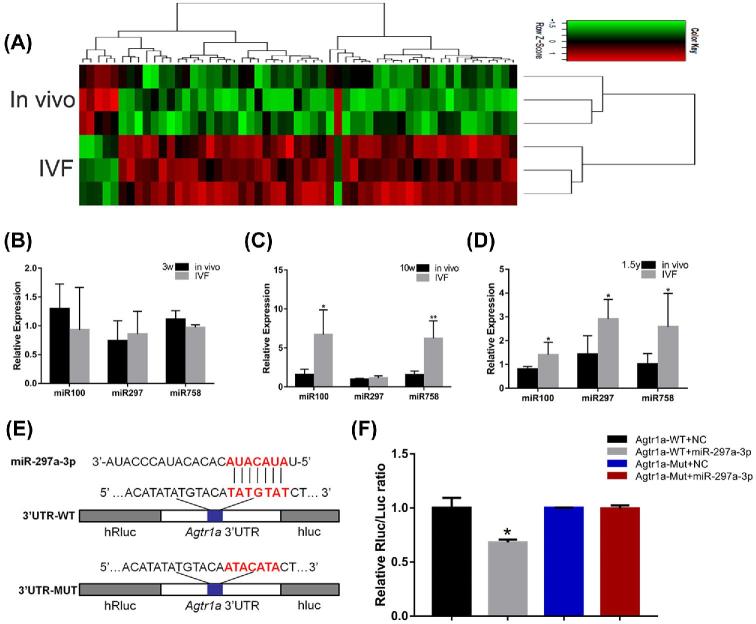
A microarray analysis was first used to detect the expression profile of miRNA in myocardial tissue in the IVF-conceived and in vivo groups at 1.5 years (Figure 4A). While some upregulated miRNAs were observed, the present study focused on significantly upregulated miR-100 miR-297, and miR-758. The miR-100 miR-297, and miR-758 were predicted to interact with Col3a1 Agtr1a, and Col1a2, respectively. Next, quantitative RT-PCR was used to detect the expression of miR-100 miR-297, and miR-758 in myocardium from the IVF-conceived and in vivo groups at 3 weeks, 10 weeks, and 1.5 years of age. At 3 weeks, the expression levels of miR-100 miR-297, and miR-758 were not found to be significantly altered between the two groups (P > 0.05) (Figure 4B), whereas at 10 weeks, a significant increase in miR-100 and miR-758 was observed in the IVF group compared with the in vivo group (4.24-fold, P < 0.05 and 4.03-fold, P < 0.01, respectively) (Figure 4C). A similar trend was found during old age; the expression levels of miR-100, miR-297 and miR-758 were found to be significantly increased in the IVF group (1.76-fold, P < 0.05; 2.04-fold, P < 0.05; and 2.52-fold, P < 0.05) (Figure 4D). To further verify the interaction between Agtr1 and miR-297, this study performed luciferase activity assay to confirm the prediction (Figure 4E and F). The results of the luciferase activity assay showed that miR-297a-3p mimic significantly modulated the luciferase activity of the pmiR-AGTR1-WT vector. However, the luciferase activity of the pmiR-AGTR1-Mut vector was not affected by the miR-297a-3p mimics. This evidence indicated that miR-297a-3p directly targeted AGTR1 and regulated its expression (Figure 4E and F).
FIGURE 4.
(A) Heat map showing the hierarchical clustering of miRNAs in the in vivo and IVF groups at 1.5 years. (B–D) Expression of miR-100, miR-297, and miR-758 was altered in the myocardial tissue of the IVF-conceived mice. The gene expression of miR-100 miR-297, and miR-758 at (B) 3 weeks, (C) 10 weeks, and (D) 1.5 years. (E and F) Correlation between AGTR1 and miR-297a-3p detected using the luciferase reporter gene assay. (E) The pmiR-AGTR1-WT plasmid was mutated in the miRNA-predicted target site and designated as pmiR-AGTR1-Mut. (F) Luciferase activity of AGTR1 3΄UTR luciferase reporters was detected after transfecting the recombinant vectors along with miR-297a-3p mimics or NC. Data concerning the relative concentrations of miR-100, miR-297, and miR-758were normalized to U6 snRNA; the relative levels were calculated using the 2−ΔΔCT method. The data are presented as mean ± standard error of the mean. *P < 0.05 and **P < 0.01, compared with the in vivo group; Student t-test (N, in vivo = 6, IVF = 6 in 3-week-old mice; N, in vivo = 6, IVF = 6 in 10-week-old mice; and N, in vivo = 6, IVF = 6 in 1.5-year-old mice). The data of luciferase activity are presented as mean ± standard error of triplicate cell cultures. *P < 0.05 and **P < 0.01.
Proliferation of H9c2 was promoted by miR-297 probably through regulating the expression of AGTR1 and CTGF
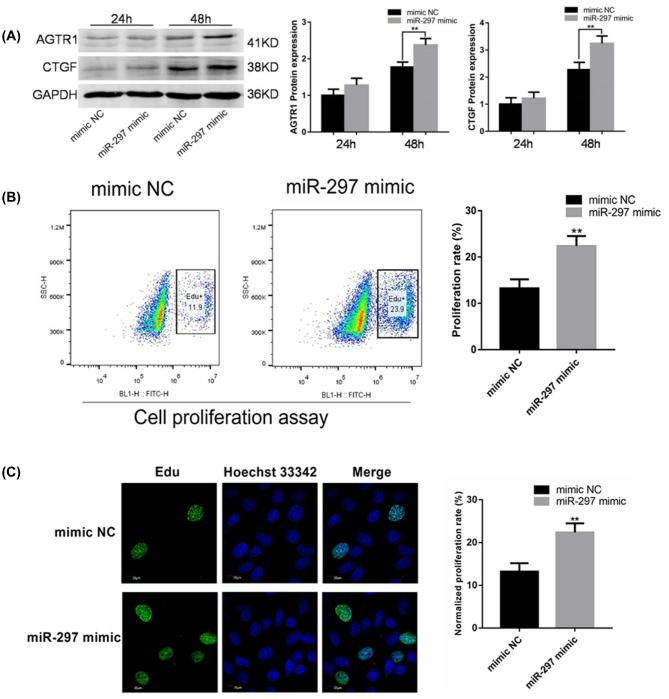
Because miR-297 was found to have an upregulation tendency in the myocardial tissue of IVF-conceived mice compared with the in vivo group, H9c2 cardiomyocytes were transfected with 100 nM miR-297 mimic to increase their expression level of miR-297 (Figure 5A). The expression of AGTR1 and CTGF did not change significantly after 24 h of overexpression of miR-297. However, the protein expression of AGTR1 and CTGF in the miR-297 overexpression group significantly increased after 48 h, indicating that miR-297 could increase the expression of AGTR1 and further promote the expression of CTGF protein.
FIGURE 5.
(A) The miR-297 promoted the expression of AGTR1 and CTGF in the proliferation of H9c2 cardiomyocytes. The H9c2 cells were transfected with negative control (NC) or miR-297 mimic for 24 or 48 h. The western blot analysis was used to analyze the expression of AGTR1 and CTGF proteins. GAPDH was the internal control. (B and C) miR-297 was involved in the regulation of the proliferation of cardiac H9c2 cells. The H9c2 cell proliferation assay using flow cytometry (B) and laser confocal microscopy (C). The cells were transfected with miR-297 and negative control (NC) after 48 h. The data are presented as mean ± standard error of the mean. *P < 0.05 and **P < 0.01, compared with the in vivo group; Student t-test.
To examine whether miR-297 affects cell proliferation in H9c2 cells, the H9c2 cells were treated with 100 nM miR-297 mimic for 48 h. Cell proliferation was examined to detect EdU-positive cells using flow cytometry and fluorescence microscopy. The flow cytometry results showed that the overexpression of miR-297 was able to promote cell proliferation (23.57% ± 0.30%) compared with the control group (13.23% ± 1.95%; Figure 5B). Significantly, more EdU-positive cells were observed in the miR-297 mimic group (25.47% ± 2.74%) compared with the control group (15.79% ± 1.43%; Figure 5C) using fluorescence microscope imaging.
Discussion
Since it was initially proposed that conception via ART might be linked to an increased risk of developing long-term cardiovascular health issues, several studies have confirmed an altered cardiovascular phenotype and elevated metabolic risk in children born following IVF [13, 14]. For example, investigations have shown that IVF-conceived children have significantly higher blood pressure compared with spontaneously conceived children [41–44], something that has recently been supported by a meta-analysis of 2112 children conceived by ART, where data showed that there was a slight, yet statistically significant, increase in blood pressure compared with naturally conceived offspring [45]. Furthermore, Brenda et al. reported that children conceived through ART showed cardiac and vascular remodeling during the fetal period that persisted in postnatal life [13]. Another meta-analysis of 25,856 children conceived using IVF/ICSI techniques showed an increased risk of developing congenital heart defects compared with those conceived spontaneously [46]. Using an IVF mouse model, this study goes some significant way to addressing the urgent need for us to better understand the underlying mechanisms influencing ART-induced cardiovascular dysfunction (Figure 6). Furthermore, we have clarified the long-term health effects of an ART-induced compromised cardiovascular system in mice from aged 3 weeks to 1.5 years.
FIGURE 6.
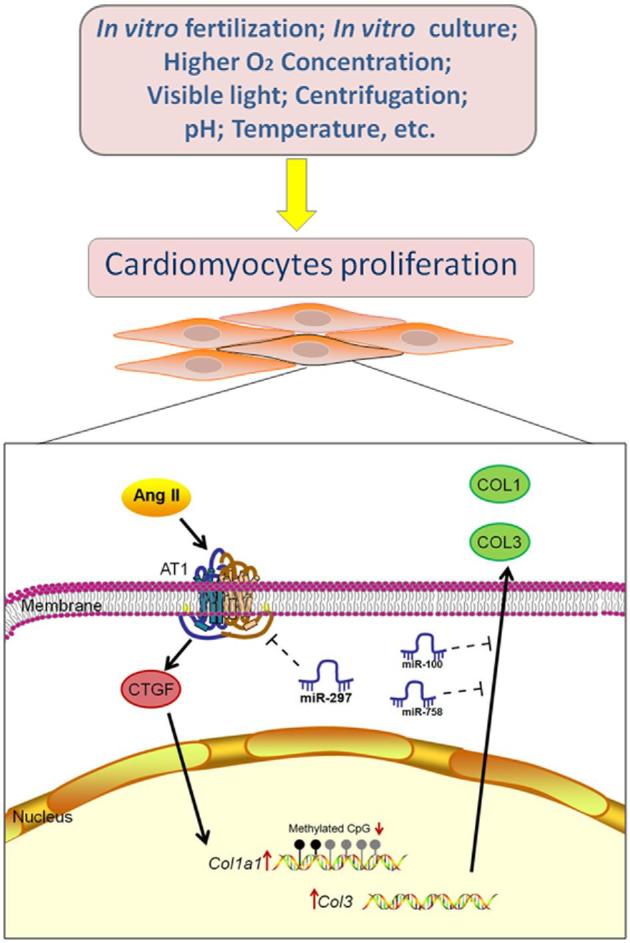
ART procedures affected the expression of RAS and epigenetic modifications in the myocardium. Oocytes and embryos were exposed to various nonphysiological factors during IVF, such as culture media, altered pH, visible light, higher oxygen pressure, and reproductive hormones. These factors might result in the increased expression of Agt Agtr1, and other genes in the RAS. CTGF is crucial in AT1-mediated intracellular signaling and can promote the expression of COL1 and COL3. Moreover, it was suggested that the changes in the expression of Col1a1 genes in IVF-conceived mice might have been caused by the decreased methylation of CpG sites in these genes. MiR-100, miR-297, and miR-758 might be involved in the regulation of the expression of Col3a1 Agtr1a, and Col1a2. In particular, miR-297 could promote the proliferation of cardiomyocytes by interacting with AGTR1.
Several studies have shown that structural and functional alterations of the heart are mediated through stimulation of a local cardiac RAS system in the cardiac tissue and large arteries. Data suggest that the cardiac RAS may act in an autocrine or paracrine manner by binding to AGTR1 [47, 48], an idea supported by evidence which suggests that the cardiac RAS is activated at advanced stages of heart failure in humans [47, 48]. Besides that, AGTR1 activation has an important role in the development of load-induced cardiac hypertrophy, even without Ang II involvement [49, 50]. In this present study, the mRNA expression level of Ace Agtr1, and Col3 was significantly increased in the myocardial tissue of IVF-conceived mice at 3 weeks and 1.5 years of age when compared to their naturally mated counterparts. These findings were reflected at the protein level, where AGTR1 was elevated in the myocardium of IVF-conceived mice at both 3 weeks and 1.5 years of age. These results might be applied to human physiology, providing a potential explanation for the cardiovascular alterations observed in ART-conceived children.
Based on the hypothesis of “developmental origins of health and disease” put forward by Barker, adverse events during the fetal development period can increase the risks of some adult diseases, including cardiovascular disorders [4]. Thus, it is not unreasonable to conclude that ART intervention during the periconception period might induce a health risk in the resulting offspring. During ART, specifically in vitro gamete manipulation and embryo system, spermatozoa, oocytes, and embryos are exposed to various nonphysiological factors, such as culture media, altered pH [51], light [52, 53], higher oxygen pressure [54], increased concentration of reproductive hormones [55], and mechanical injection or biopsy, all of which contribute to an altered periconception environment [56]. In addition to in vitro ART, a sub-optimal uterine environment following embryo transfer is thought to further influence the long-term viability of offspring. Interestingly, factors that create an adverse interuterine environment, including maternal high-salt diet, inflammation, and dietary restriction, have also been shown to alter the fetal and offspring RAS [57–60].
CTGF, an immediate response gene of the CCN (Cyr61 Ctgf Nov) family, is a critical cytokine in the pathophysiology of fibrosis that upregulates COL1 and COL3 in cardiomyocytes [61]. CTGF is essential in AGTR1-mediated intracellular signaling and can activate cytokines, such as transforming growth factor-β, resulting in myocardial remodeling, vascular fibrosis, endothelial dysfunction, and atherosclerosis progress [62]. Increased plasma levels of CTGF may be a marker of pulmonary arterial hypertension associated with congenital heart disease [63]. An abnormal expression of COL1 and COL3 in the embryonic period may affect the heart's structural development [25]. Sustained changes in the expression of COL1 and COL3 in adulthood would further increase the risk of cardiovascular diseases, such as hypertension, arrhythmia, and cardiac hypertrophy, and enhance the process of cardiovascular fibrosis and remodeling [26]. The effects of ART on genome-wide changes in DNA methylation levels have been confirmed by several studies [64–66]. For instance, ART-conceived mice showed arterial tissue endothelial dysfunction and increased DNA methylation of the promoter of the eNOS gene [15]. CpG islands are located in or near the promoter region of genes and hence, prevent transcription factors from binding, thus preventing transcription [67]. At 3 weeks and 1.5 years of age, the methylation levels of some CpG sites in Col1a1 were significantly decreased in the IVF-conceived group, which might contribute to the changes in genes expression of Col1a1.
In addition to DNA methylation and chromatin histone acetylation modification, accumulating studies have also shown noncoding RNA to be involved in the epigenetic modification of growth and development [68]. MiRNAs are small noncoding RNA molecules of approximately 22–24 nucleotides that participate in the transcriptional and posttranscriptional regulation of gene expression [69]. A variety of miRNAs have been shown to regulate hypertension, coronary heart disease, and glucose and lipid metabolism disorders [70, 71]. In this study, miR-100, miR-297 and miR-758 were chosen as candidate genes after miRNA database prediction indicated that they interact with Col3a1 Agtr1a, and Col1a2, respectively. Previous studies have shown that plasma miR-100 is positively associated with coronary plaque vulnerability [72] can inhibit angiogenesis [73] and is involved in the development and growth of pancreatic cancer [74], while miR-297 is involved in septic shock [75]. In this study, the luciferase assay was used to demonstrate that miR-297 targets AGTR1 and promotes the proliferation of cardiomyocytes by regulating the expression of AGTR1 and CTGF. Moreover, the expression of miR-297 was shown to promote the proliferation of H9c2 cells in culture, which might also indicate a role for miR-297 in cardiac structural and functional abnormalities. In accordance with the present results, previous studies have also demonstrated that upregulation of miR-297 promotes cardiomyocyte hypertrophy induced by Ang II [76]. The functions of the miR might be extensive due to its ability to bind multiple molecules [77]. The miR-297 could directly inhibit gene expression by targeting on the 3΄UTR region of mRNA [76]. In addition, miRs also indirectly affect the gene expression by regulating transcription factors, nuclear factor and binding proteins which are closely related to gene expression [77]. The miRs might also target important molecules acting on cellular signaling, indirectly affecting gene expression [78]. Therefore, we speculated that miR-297 may also interact with other regulatory factors in the signaling pathway regulated by AGRT1, and other mechanisms need to be explored in future studies.
Evidence presented in this manuscript indicates that the expressions of RAS-associated gene and protein expression in myocardial tissue have been changed in the myocardium of IVF-conceived mice. Furthermore, modifications to DNA methylation and abnormal patterns of expression of miRNAs may be involved in the regulations, and thus the cardiovascular health of IVF-conceived fetuses and offspring. The application of an IVF mouse model has enabled us to investigate the long-term health of ART offspring. Given that ART is now widely utilized, it is important to further explore the exact signaling pathways and genes involved in embryonic defects, associated with environmental factors, which manifest as clinical disorders in the adult. These findings provide new and important insights into the mechanisms which control the development of cardiovascular disease and will allow us to develop tools to minimize ART-induced cardiovascular dysfunction at the point of fetal origin and also as the offspring progress into adulthood [79].
Supplementary data
Supplementary data are available at BIOLRE online.
Supplementary Figure S1. The mRNA relative expression of genes in the RAS components and those related to myocardial remodeling in myocardial tissue in male and female mice respectively from the in vivo and IVF groups at (A) 3 weeks, (B) 10 weeks, and (C) 1.5 years. Ren1 Ace Ace2 Agt Agtr1a Agtr1b, and Agtr2 were critical elements in the RAS. Col1a1 Col1a2, and Col3 were the major components of the extracellular matrix of the heart and also crucial in the development of heart and myocardial remodeling. Ctgf promotes the pathological process of fibrosis. The data are presented as mean ± standard error of the mean. The relative levels were calculated using the 2−ΔΔCT method. *P < 0.05 and **P < 0.01, compared with the in vivo group; Student t-test (N, in vivo = 6, IVF = 6 in 3-week-old mice; N, in vivo = 6, IVF = 6 in 10-week-old mice; and N, in vivo = 6, IVF = 6 in 1.5-year-old mice).
Supplementary Figure S2. The mRNA relative expression of genes in the RAS components and those related to myocardial remodeling in myocardial tissue between males and females within the IVF and in vivo groups. Data are mean ± standard error of mean. *P < 0.05, compared with the in vivo group (Student t-test). (N = 6/group)
Supplementary Figure S3. Average DNA methylation levels for the whole region of CpG islands and mean methylation levels for individual CpG sites of DNA methyltransferases in heart samples from the in vivo and IVF groups. Gene methylation levels of Dnmt1 Dnmt3a, and Dnmt3b at (A) 3 weeks, (B) 10 weeks, and (C) 1.5 years of age. Data are mean ± standard error of mean. *P < 0.05 and **P < 0.01, compared with the in vivo group (Student t-test). (N = 6/group).
Supplementary Figure S4. The mRNA levels of imprinting genes in the myocardial tissue of in vivo and IVF groups at (A) 3 weeks, (B) 10 weeks, and (C) 1.5 years of age. Data are mean ± standard error of the mean; relative mRNA levels were calculated by the 2−ΔΔCT method. *P < 0.05 and **P < 0.01 compared with in vivo group; Student t-test (N = 6/group).
Supplementary Figure S5. Organ coefficient of the heart in the in vivo and IVF groups. *P < 0.05, compared with the in vivo group (Student t- test).
Supplementary Table S1. Sequence of RT-PCR primers
Supplementary Table S2. Sequence of RT-PCR primers for miRNAs
Supplementary Table S3. List of antibodies used
Supplementary Table S4. Primer sequence of targeted bisulfite sequencing
Supplementary Table S5. The methylation rates at each site.
Acknowledgments
The authors thank the valuable support and suggestions from other members of the laboratory.
Notes
Conference Presentation: Presented in part at the 32th Annual Meeting of the European Society of Human Reproduction and Embryology, July 3, 2016, Helsinki, Finland.
Edited by Dr. Sarah Kimmins, PhD, McGill University
Footnotes
Grant Support: The authors are supported by the National Basic Research Program of China (No. 2014CB943300), the National Natural Science Program of China (No. 81771652, 81571500, 81501321, and 81200475) and the Natural Science Foundation of Zhejiang Province of China (No. LY14H040009 and LQ17H040001).
References
- 1. Chronopoulou E, Harper JC. IVF culture media: past, present and future. Hum Reprod Update 2015; 21:39–55. [DOI] [PubMed] [Google Scholar]
- 2. Grafodatskaya D, Cytrynbaum C, Weksberg R. The health risks of ART. EMBO Rep 2013; 14:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiba H, Hiura H, Okae H, Miyauchi N, Sato F, Sato A, Arima T. DNA methylation errors in imprinting disorders and assisted reproductive technology. Pediatr Int 2013; 55:542–549. [DOI] [PubMed] [Google Scholar]
- 4. Lane M, Robker RL, Robertson SA. Parenting from before conception. Science 2014; 345:756–760. [DOI] [PubMed] [Google Scholar]
- 5. Whitelaw N, Bhattacharya S, Hoad G, Horgan GW, Hamilton M, Haggarty P. Epigenetic status in the offspring of spontaneous and assisted conception. Hum Reprod 2014; 29:1452–1458. [DOI] [PubMed] [Google Scholar]
- 6. Nwaru BI, McCleary N, Erkkola M, Kaila M, Virtanen SM, Sheikh A. Assisted reproductive technology and risk of asthma and allergy in the offspring: protocol for a systematic review and meta-analysis. BMJ Open 2016; 6:e010697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Massaro PA, MacLellan DL, Anderson PA, Romao RLP. Does intracytoplasmic sperm injection pose an increased risk of genitourinary congenital malformations in offspring compared to in vitro fertilization? A systematic review and Meta-Analysis. J Urol 2015; 193:1837–1842. [DOI] [PubMed] [Google Scholar]
- 8. Qin J, Liu X, Sheng X, Wang H, Gao S. Assisted reproductive technology and the risk of pregnancy-related complications and adverse pregnancy outcomes in singleton pregnancies: a meta-analysis of cohort studies. Fertil Steril 2016; 105:73–85.e1–6. [DOI] [PubMed] [Google Scholar]
- 9. Tararbit K, Houyel L, Bonnet D, De Vigan C Lelong N, Goffinet F, Khoshnood B. Risk of congenital heart defects associated with assisted reproductive technologies: a population-based evaluation. Eur Heart J 2011; 32:500–508. [DOI] [PubMed] [Google Scholar]
- 10. Davies MJ, Moore VM, Willson KJ, Van Essen P, Priest K, Scott H, Haan EA, Chan A. Reproductive technologies and the risk of birth defects. N Engl J Med 2012; 366:1803–1813. [DOI] [PubMed] [Google Scholar]
- 11. Tararbit K, Lelong N, Thieulin AC, Houyel L, Bonnet D, Goffinet F, Khoshnood B. The risk for four specific congenital heart defects associated with assisted reproductive techniques: a population-based evaluation. Hum Reprod 2013; 28:367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Padhee M, Zhang S, Lie S, Wang KC, Botting KJ, McMillen IC, MacLaughlin SM, Morrison JL. The periconceptional environment and cardiovascular disease: Does in vitro embryo culture and transfer influence cardiovascular development and health? Nutrients 2015; 7:1378–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Valenzuela-Alcaraz B, Crispi F, Bijnens B, Cruz-Lemini M, Creus M, Sitges M, Bartrons J, Civico S, Balasch J, Gratacos E. Assisted reproductive technologies are associated with cardiovascular remodeling in utero that persists postnatally. Circulation 2013; 128:1442–1450. [DOI] [PubMed] [Google Scholar]
- 14. Scherrer U, Rexhaj E, Allemann Y, Sartori C, Rimoldi SF. Cardiovascular dysfunction in children conceived by assisted reproductive technologies. Eur Heart J 2015; 36:1583–1589. [DOI] [PubMed] [Google Scholar]
- 15. Rexhaj E, Paoloni-Giacobino A, Rimoldi SF, Fuster DG, Anderegg M, Somm E, Bouillet E, Allemann Y, Sartori C, Scherrer U. Mice generated by in vitro fertilization exhibit vascular dysfunction and shortened life span. J Clin Invest 2013; 123:5052–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang LY, Wang N, Le F Li L, Lou HY, Liu XZ, Zheng YM, Qian YQ, Chen YL, Jiang XH, Huang HF, Jin F. Superovulation induced changes of lipid metabolism in ovaries and embryos and its probable mechanism. PLoS One 2015; 10:e0132638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramalingam L, Menikdiwela K, LeMieux M, Dufour JM, Kaur G, Kalupahana N, Moustaid-Moussa N. The renin angiotensin system, oxidative stress and mitochondrial function in obesity and insulin resistance. Biochim Biophys Acta 2017; 1863:1106–1114. [DOI] [PubMed] [Google Scholar]
- 18. van Thiel BS, van der Pluijm I, te Riet L, Essers J, Danser AH. The renin-angiotensin system and its involvement in vascular disease. Eur J Pharmacol 2015; 763:3–14. [DOI] [PubMed] [Google Scholar]
- 19. De Mello WC, Frohlich ED. Clinical perspectives and fundamental aspects of local cardiovascular and renal Renin-Angiotensin systems. Front Endocrinol (Lausanne) 2014; 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar R, Singh VP, Baker KM. The The intracellular renin-angiotensin system: a new paradigm. Trends Endocrinol Metab 2007; 18:208–214. [DOI] [PubMed] [Google Scholar]
- 21. Munoz-Durango N, Fuentes CA, Castillo AE, Gonzalez-Gomez LM, Vecchiola A, Fardella CE, Kalergis AM. Role of the Renin-Angiotensin-Aldosterone system beyond blood pressure regulation: Molecular and cellular mechanisms involved in End-Organ damage during arterial hypertension. Int J Mol Sci 2016; 17:E797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Billet S, Aguilar F, Baudry C, Clauser E. Role of angiotensin II AT1 receptor activation in cardiovascular diseases. Kidney Int 2008; 74:1379–1384. [DOI] [PubMed] [Google Scholar]
- 23. Carver W, Terracio L, Borg TK. Expression and accumulation of interstitial collagen in the neonatal rat heart. Anat Rec 1993; 236:511–520. [DOI] [PubMed] [Google Scholar]
- 24. Cooper WO, Hernandez-Diaz S, Arbogast PG, Dudley JA, Dyer S, Gideon PS, Hall K, Ray WA. Major congenital malformations after first-trimester exposure to ACE inhibitors. N Engl J Med 2006; 354:2443–2451. [DOI] [PubMed] [Google Scholar]
- 25. Tong W, Zhang L. Fetal hypoxia and programming of matrix metalloproteinases. Drug Discov Today 2012; 17:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferrario CM. Cardiac remodelling and RAS inhibition. Ther Adv Cardiovasc Dis 2016; 10:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khuman MW, Harikumar SK, Sadam A, Kesavan M, Susanth VS, Parida S, Singh KP, Sarkar SN. Candesartan ameliorates arsenic-induced hypertensive vascular remodeling by regularizing angiotensin II and TGF-beta signaling in rats. Toxicology 2016; 374:29–41. [DOI] [PubMed] [Google Scholar]
- 28. Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science 2010; 330:622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barker DJ. Sir Richard Doll Lecture. Developmental origins of chronic disease. Public Health 2012; 126:185–189. [DOI] [PubMed] [Google Scholar]
- 30. El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil Steril 2013; 99:632–641. [DOI] [PubMed] [Google Scholar]
- 31. Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril 2009; 91:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fauque P, Ripoche MA, Tost J, Journot L, Gabory A, Busato F, Le Digarcher A, Mondon F, Gut I, Jouannet P, Vaiman D, Dandolo L et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum Mol Genet 2010; 19:1779–1790. [DOI] [PubMed] [Google Scholar]
- 33. Zheng YM, Li L, Zhou LM, Le F Cai LY, Yu P, Zhu YR, Liu XZ, Wang LY, Li LJ, Lou YY, Xu XR et al. Alterations in the frequency of trinucleotide repeat dynamic mutations in offspring conceived through assisted reproductive technology. Hum Reprod 2013; 28:2570–2580. [DOI] [PubMed] [Google Scholar]
- 34. Le F, Wang LY, Wang N, Li L, Li le J, Zheng YM, Lou HY, Liu XZ, Xu XR, Sheng JZ, Huang HF, Jin F. In vitro fertilization alters growth and expression of Igf2/H19 and their epigenetic mechanisms in the liver and skeletal muscle of newborn and elder mice. Biol Reprod 2013; 88:75. [DOI] [PubMed] [Google Scholar]
- 35. Lou H, Le F Zheng Y, Li L, Wang L, Wang N, Zhu Y, Huang H, Jin F. Assisted reproductive technologies impair the expression and methylation of insulin-induced gene 1 and sterol regulatory element-binding factor 1 in the fetus and placenta. Fertil Steril 2014; 101:974–980.e2. [DOI] [PubMed] [Google Scholar]
- 36. Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development 2009; 136:3647–3655. [DOI] [PubMed] [Google Scholar]
- 37. Nemecz M, Alexandru N, Tanko G, Georgescu A. Role of MicroRNA in endothelial dysfunction and hypertension. Curr Hypertens Rep 2016; 18:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lew JK, Pearson JT, Schwenke DO, Katare R. Exercise mediated protection of diabetic heart through modulation of microRNA mediated molecular pathways. Cardiovasc Diabetol 2017; 16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kitamoto T, Kitamoto A, Ogawa Y, Honda Y, Imajo K, Saito S, Yoneda M, Nakamura T, Nakajima A, Hotta K. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol 2015; 63:494–502. [DOI] [PubMed] [Google Scholar]
- 40. Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakka SD, Loutradis D, Kanaka-Gantenbein C, Margeli A, Papastamataki M, Papassotiriou I, Chrousos GP. Absence of insulin resistance and low-grade inflammation despite early metabolic syndrome manifestations in children born after in vitro fertilization. Fertil Steril 2010; 94:1693–1699. [DOI] [PubMed] [Google Scholar]
- 42. Ceelen M, van Weissenbruch MM, Vermeiden JP, van Leeuwen FE, Delemarre-van de Waal HA. Cardiometabolic differences in children born after in vitro fertilization: follow-up study. J Clin Endocrinol Metab 2008; 93:1682–1688. [DOI] [PubMed] [Google Scholar]
- 43. Ceelen M, van Weissenbruch MM, Prein J, Smit JJ, Vermeiden JP, Spreeuwenberg M, van Leeuwen FE, Delemarre-van de Waal HA. Growth during infancy and early childhood in relation to blood pressure and body fat measures at age 8-18 years of IVF children and spontaneously conceived controls born to subfertile parents. Hum Reprod 2009; 24:2788–2795. [DOI] [PubMed] [Google Scholar]
- 44. Belva F, Roelants M, De Schepper J, Roseboom TJ, Bonduelle M, Devroey P, Painter RC. Blood pressure in ICSI-conceived adolescents. Hum Reprod 2012; 27:3100–3108. [DOI] [PubMed] [Google Scholar]
- 45. Guo XY, Liu XM, Jin L, Wang TT, Ullah K, Sheng JZ, Huang HF. Cardiovascular and metabolic profiles of offspring conceived by assisted reproductive technologies: a systematic review and meta-analysis. Fertil Steril 2017; 107:622–631.e5. [DOI] [PubMed] [Google Scholar]
- 46. Giorgione V, Parazzini F, Fesslova V, Cipriani S, Candiani M, Inversetti A, Sigismondi C, Tiberio F, Cavoretto P. Congenital heart defects in IVF/ICSI pregnancy: systematic review and meta-analysis. Ultrasound Obstet Gynecol 2018; 51:33–42. [DOI] [PubMed] [Google Scholar]
- 47. De Mello WC, Frohlich ED. On the local cardiac renin angiotensin system. Basic and clinical implications. Peptides 2011; 32:1774–1779. [DOI] [PubMed] [Google Scholar]
- 48. Singh VP, Le B, Bhat VB, Baker KM, Kumar R. High-glucose-induced regulation of intracellular ANG II synthesis and nuclear redistribution in cardiac myocytes. Am J Physiol Heart Circ Physiol 2007; 293:H939–H948. [DOI] [PubMed] [Google Scholar]
- 49. Yaras N, Bilginoglu A, Vassort G, Turan B. Restoration of diabetes-induced abnormal local Ca2+ release in cardiomyocytes by angiotensin II receptor blockade. Am J Physiol Heart Circ Physiol 2007; 292:H912–H920. [DOI] [PubMed] [Google Scholar]
- 50. Yasuda N, Miura S, Akazawa H, Tanaka T, Qin Y, Kiya Y, Imaizumi S, Fujino M, Ito K, Zou Y, Fukuhara S, Kunimoto S et al. Conformational switch of angiotensin II type 1 receptor underlying mechanical stress-induced activation. EMBO Rep 2008; 9:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Will MA, Clark NA, Swain JE. Biological pH buffers in IVF: help or hindrance to success. J Assist Reprod Genet 2011; 28:711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shahar S, Wiser A, Ickowicz D, Lubart R, Shulman A, Breitbart H. Light-mediated activation reveals a key role for protein kinase A and sarcoma protein kinase in the development of sperm hyper-activated motility. Hum Reprod 2011; 26:2274–2282. [DOI] [PubMed] [Google Scholar]
- 53. Li R, Liu Y, Pedersen HS, Callesen H. Effect of ambient light exposure of media and embryos on development and quality of porcine parthenogenetically activated embryos. Zygote 2015; 23:378–383. [DOI] [PubMed] [Google Scholar]
- 54. Ciray HN, Aksoy T, Yaramanci K, Karayaka I, Bahceci M. In vitro culture under physiologic oxygen concentration improves blastocyst yield and quality: a prospective randomized survey on sibling oocytes. Fertil Steril 2009; 91:1459–1461. [DOI] [PubMed] [Google Scholar]
- 55. La Bastide-Van Gemert S, Seggers J, Haadsma ML, Heineman MJ, Middelburg KJ, Roseboom TJ, Schendelaar P, Hadders-Algra M, Van den Heuvel ER. Is ovarian hyperstimulation associated with higher blood pressure in 4-year-old IVF offspring? Part II: an explorative causal inference approach. Hum Reprod 2014; 29:510–517. [DOI] [PubMed] [Google Scholar]
- 56. Dimitriadou E, Noutsopoulos D, Markopoulos G, Vlaikou AM, Mantziou S, Traeger-Synodinos J, Kanavakis E, Chrousos GP, Tzavaras T, Syrrou M. Abnormal DLK1/MEG3 imprinting correlates with decreased HERV-K methylation after assisted reproduction and preimplantation genetic diagnosis. Stress 2013; 16:689–697. [DOI] [PubMed] [Google Scholar]
- 57. Mao C, Liu R, Bo L, Chen N, Li S, Xia S, Chen J, Li D, Zhang L, Xu Z. High-salt diets during pregnancy affected fetal and offspring renal renin-angiotensin system. J Endocrinol 2013; 218:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang S, Morrison JL, Gill A, Rattanatray L, MacLaughlin SM, Kleemann D, Walker SK, McMillen IC. Dietary restriction in the periconceptional period in normal-weight or obese ewes results in increased abundance of angiotensin-converting enzyme (ACE) and angiotensin type 1 receptor (AT1R) in the absence of changes in ACE or AT1R methylation in the adrenal of the offspring. Reproduction 2013; 146:443–454. [DOI] [PubMed] [Google Scholar]
- 59. Deng Y, He X, Chu J, Zhou J, Zhang Q, Guo W, Huang P, Guan X, Tang Y, Wei Y, Zhao S, Zhang X et al. Prenatal inflammation-induced NF-kappaB dyshomeostasis contributes to renin-angiotensin system over-activity resulting in prenatally programmed hypertension in offspring. Sci Rep 2016; 6:21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Woods LL, Ingelfinger JR, Nyengaard JR, Rasch R. Maternal protein restriction suppresses the newborn renin-angiotensin system and programs adult hypertension in rats. Pediatr Res 2001; 49:460–467. [DOI] [PubMed] [Google Scholar]
- 61. Li Y, Jian Z, Yang ZY, Chen L, Wang XF, Ma RY, Xiao YB. Increased expression of connective tissue growth factor and transforming growth factor-beta-1 in atrial myocardium of patients with chronic atrial fibrillation. Cardiology 2013; 124:233–240. [DOI] [PubMed] [Google Scholar]
- 62. Daniels A, van Bilsen M, Goldschmeding R, van der Vusse GJ, van Nieuwenhoven FA. Connective tissue growth factor and cardiac fibrosis. Acta Physiol (Oxf) 2009; 195:321–338. [DOI] [PubMed] [Google Scholar]
- 63. Li G, Tang L, Jia P, Zhao J, Liu D, Liu B. Elevated plasma connective tissue growth factor levels in children with pulmonary arterial hypertension associated with congenital heart disease. Pediatr Cardiol 2016; 37:714–721. [DOI] [PubMed] [Google Scholar]
- 64. van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update 2012; 18:171–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nelissen EC, Dumoulin JC, Busato F, Ponger L, Eijssen LM, Evers JL, Tost J, van Montfoort AP. Altered gene expression in human placentas after IVF/ICSI. Hum Reprod 2014; 29:2821–2831. [DOI] [PubMed] [Google Scholar]
- 66. Ghosh J, Mainigi M, Coutifaris C, Sapienza C. Outlier DNA methylation levels as an indicator of environmental exposure and risk of undesirable birth outcome. Hum Mol Genet 2016; 25:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293:1089–1093. [DOI] [PubMed] [Google Scholar]
- 68. Bartel DP. MicroRNAs. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 69. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 2012; 13:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lew JKS, Pearson JT, Schwenke DO, Katare R. Exercise mediated protection of diabetic heart through modulation of microRNA mediated molecular pathways. Cardiovasc Diabetol 2017; 16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Soeki T, Yamaguchi K, Niki T, Uematsu E, Bando S, Matsuura T, Ise T, Kusunose K, Hotchi J, Tobiume T, Yagi S, Fukuda D et al. Plasma MicroRNA-100 is associated with coronary plaque vulnerability. Circ J 2015; 79:413–418. [DOI] [PubMed] [Google Scholar]
- 73. Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, Sluijter JP, Hoefer I, Pasterkamp G, Bode C, Moser M. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation 2011; 123:999–1009. [DOI] [PubMed] [Google Scholar]
- 74. Li ZP, Li X, Yu C, Wang M, Peng F, Xiao J, Tian R, Jiang JX, Sun CY. MicroRNA-100 regulates pancreatic cancer cells growth and sensitivity to chemotherapy through targeting FGFR3. Tumor Biol 2014; 35:11751–11759. [DOI] [PubMed] [Google Scholar]
- 75. Sari AN, Korkmaz B, Serin MS, Kacan M, Unsal D, Buharalioglu CK, Sahan Firat S, Manthati VL, Falck JR, Malik KU, Tunctan B. Effects of 5,14-HEDGE, a 20-HETE mimetic, on lipopolysaccharide-induced changes in MyD88/TAK1/IKKbeta/IkappaB-alpha/NF-kappaB pathway and circulating miR-150, miR-223, and miR-297 levels in a rat model of septic shock. Inflamm Res 2014; 63:741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bao Q, Zhao M, Chen L, Wang Y, Wu S, Wu W, Liu X. MicroRNA-297 promotes cardiomyocyte hypertrophy via targeting sigma-1 receptor. Life Sci 2017; 175:1–10. [DOI] [PubMed] [Google Scholar]
- 77. Wang H, Cai J. The role of microRNAs in heart failure. Biochim Biophys Acta 2017; 1863:2019–2030. [DOI] [PubMed] [Google Scholar]
- 78. von Haehling S, Ebner N, Dos Santos MR, Springer J, Anker SD. Role of microRNAs in wasting in heart failure. Nat Rev Cardiol 2017; 14:566–566. [DOI] [PubMed] [Google Scholar]
- 79. Barker DJP, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. N Engl J Med 2005; 353:1802–1809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.