Abstract
The receptor tyrosine kinase family of fibroblast growth factor receptors (FGFRs) play crucial roles in embryonic development, metabolism, tissue homeostasis and wound repair via stimulation of intracellular signalling cascades. As a consequence of FGFRs’ influence on cell growth, proliferation and differentiation, FGFR signalling is frequently dysregulated in a host of human cancers, variously by means of overexpression, somatic point mutations and gene fusion events. Dysregulation of FGFRs is also the underlying cause of many developmental dysplasias such as hypochondroplasia and achondroplasia. Accordingly, FGFRs are attractive pharmaceutical targets, and multiple clinical trials are in progress for the treatment of various FGFR aberrations. To effectively target dysregulated receptors, a structural and mechanistic understanding of FGFR activation and regulation is required. Here, we review some of the key research findings from the last couple of decades and summarise the strategies being explored for therapeutic intervention.
Keywords: drug discovery and design, fibroblast growth factor receptors, receptor tyrosine kinases, structural biology
Introduction
Through their role in signal transduction pathways, protein kinases mediate a plethora of cellular phenotypic changes such as cell growth, proliferation, differentiation and survival [1]. Receptor tyrosine kinases (RTKs) are an important kinase subfamily whose members span the cell surface and activate intracellular signalling cascades in response to exogenous growth signals via binding of family-specific extracellular ligands. Canonically, this is achieved through ligand-driven receptor dimerisation and subsequent trans-autophosphorylation of the cytosolic receptor tyrosine kinase domains, stimulating kinase activity [2]. Alternatively, in cases where receptors are believed to exist as constitutive dimers, activation can be achieved allosterically via ligand-induced conformational rearrangements of the receptors. Fibroblast growth factor receptors (FGFRs), the focus of this review, are one of these RTK subfamilies, responding to the binding of fibroblast growth factors (FGFs) (Figure 1) [2,3]. Through their activation, FGFRs have roles in embryonic development, tissue homeostasis and metabolism [4–7].
Figure 1. Stimulation of FGFRs.
FGFRs are composed of an extracellular domain comprising D1, acid box, and D2 and D3 domains, followed by a single helix TMD, the JMD, and an intracellular ‘split’ tyrosine KD. Two models describing receptor stimulation by FGF ligand and heparin/heparan sulfate cofactor have been described: the canonical ligand-induced receptor dimerisation model (left) and an allosteric ligand-induced conformational change model (right). Receptor activation leads to trans-autophosphorylation of the kinase domains and stimulation of intracellular signalling cascades. The boxed regions (A–C) correspond to those in Figure 2.
The FGFR family is composed of four separately encoded yet highly homologous receptors, FGFRs 1–4, sharing between 56 and 71% sequence identity [8]. Structurally, all members share the same architecture consisting of a large ligand-binding extracellular domain (ECD) that comprises three immunoglobulin (Ig)-like domains (D1–3); a single membrane-spanning helix; and an intracellular domain containing the catalytically active ‘split’ tyrosine kinase domain (Figure 1). Genetic, biochemical and structural studies have yielded extensive insights into understanding of FGFR activation.
Localisation of FGF ligand binding and receptor dimerisation
In mammals, there are 18 FGF ligands which can be subdivided into paracrine and endocrine families; all FGFs have a beta-trefoil fold with a heparan sulfate binding-site on its surface that facilitates sequestration of FGF ligands close to the cell surface for receptor binding [9,10]. FGF-ligand binding to FGFRs is localised to domains D2 and D3 of the extracellular domain [11], and in the case of paracrine FGFs, occurs in association with heparan sulfate proteoglycan cofactors [10] (Figure 1). While FGFs are able to bind independently to FGFRs in a 1:1 stoichiometry [12–14], heparan sulfate (or heparin) is necessary for receptor dimerisation and FGFR signalling [15–17]. Although dimerisation of a variety of 1:1 FGF:FGFR extracellular domain complexes was initially observed crystallographically in the absence of heparin [13,14,18], this dimerisation is likely to be a crystallisation artefact. However, these heparin-free crystallographically dimerised complexes were nonetheless useful for building models of heparin-mediated receptor dimerisation and are structurally similar to a 2:2:2 stoichiometry FGF2:FGFR1:heparin ternary complex structure solved at a later date (Figure 2) [19]. The stoichiometry and minimal heparan sulfate chain length required for receptor dimerisation have been disputed, resulting in different proposed models of FGFR activation [19–23]. However, regardless of the model, in all FGF:FGFR complex structures, FGF ligands make contacts with residues from D2, the D2–D3 linker and D3 domains of FGFRs, with the interfaces characterised by both hydrophobic and polar interactions. In the 2:2:2 stoichiometry ternary complex model, both FGF ligands and heparin make contacts with both FGFR molecules of the dimer (Figure 2). Unlike paracrine FGFs, endocrine FGFs such as FGF21 and FGF23 exhibit lower binding affinity for heparan sulfate [10] and require Klotho co-receptors to act as cofactors for activation of FGFRs [7,24,25].
Figure 2. Structures of FGFR extracellular, transmembrane, and kinase domains.
(A) Crystal structure of FGFR1 extracellular domains D2 and D3 (grey cartoon on transparent surface representation) in a 2:2:2 complex with FGF2 (light green, cartoon) and heparin (dark blue, sticks) (PDB: 1FQ9). Only one copy of FGF2 and FGFR1 are shown in cartoon representation for clarity. (B) An FGFR3 transmembrane domain dimer derived from NMR (PDB: 2LZL) in cartoon representation with the observed dimerisation interface and GxxxG-like motifs highlighted. (C) FGFR3 kinase domain crystal structure (PDB: 4K33) in cartoon representation on a transparent surface with the N- and C-lobes and structural elements, the αC helix (salmon), the P-loop (orange), the catalytic loop (blue), the A-loop (yellow), the kinase hinge (magenta) and the (incomplete) kinase insert (black) highlighted. Panels are not in scale with one another.
Though structurally homologous, FGFR family members exhibit different binding specificities to subsets of the 18 FGF ligands [8,10]. While some FGFs, namely FGF1 (acidic FGF) and FGF2 (basic FGF), show binding redundancy among the FGF receptors, others bind to sole members or only some of the family [8]. This variety in ligand-binding specificity is enhanced by tissue-specific alternative splicing of the third Ig-like domain D3 of FGFRs 1–3 [10,11,26,27] and can be attributed, in part, to consequent changes in FGF–FGFR contacts at the βC′-βE region of D3 [13].
The extracellular domain also displays receptor autoinhibition mechanisms, realised by the blocking of FGF binding by domain D1 and the D1–D2 ‘acid-box’ linker [23]; studies of alternative splicing of this region and biophysical analyses suggest that autoinhibition is mediated by competition between ‘acid box’ and heparan sulfate binding, and through back-binding of D1 to the FGF binding-site on domains D2 and D3 [28–31].
Dimerisation of FGFRs at the transmembrane domain and an alternative stimulation model
The dimerisation of the extracellular domains of FGFRs presumably positions the C-terminal ends of D3 domains such that the intracellular tyrosine kinase domains are arranged to perform trans-autophosphorylation; this would require the passing of spatial and conformational information across the plasma membrane. Studies of FGFR transmembrane domains (TMDs) are sparse; however, nuclear magnetic resonance studies of FGFR3 transmembrane domain in d38-dodecylphosphocholine/d29-sodium dodecyl sulfate (9/1) micelles revealed a symmetric left-handed dimer of helices with 310 and alpha-helical character (Figure 2) [32]. In this structure, the two helices cross one another with an angle of 23°, approximately at the midpoint of the helix length. However, this crossing does not occur at the GxxxG-like motifs of the helices which are observed at dimerisation interfaces of other receptor tyrosine kinases [33–35] and lie immediately upstream of the NMR-observed cross-point. The canonical model of receptor tyrosine kinase activation proceeds via ligand-induced dimerisation, but recent evidence suggests that FGFRs 1–3 may possess intrinsic dimerisation potential even when unliganded [36,37]. Consequently, an allosteric activation model featuring ligand-induced conformational change could be more appropriate for FGFRs (Figure 1), similar to that of insulin receptor tyrosine kinase [38]. Under this model, it is expected that there will be more than one dimerisation state of the transmembrane domain, reflecting different ligand binding at the extracellular domains. The extent of conformational change induced by ligand binding, perhaps manifested as the degree of C-terminal helix separation, could correlate to the level of kinase activation and signalling outcome. Altogether, these suggest that the NMR-observed symmetrical dimer may correspond to the basal dimerisation state of the receptor, while the alternative dimer interface at the GxxxG-like motifs may be that of the fully active state [32]. Additional intermediate activation states with alternative dimerisation interfaces cannot be ruled out. Further independent data lend support to the allosteric model: crystal structures of the extracellular region of FGFR2c bound to FGF8b and FGF2 show variation in the distances between D3 domain C-termini of ∼35 Å and ∼46 Å, respectively [10]. Furthermore, Förster resonance energy transfer (FRET)-based studies of FGF1 and FGF2 binding to FGFRs 1–3 show differences in ligand-induced helix separation, and at least in the case of FGFR3, this further correlates with the level of receptor (auto)phosphorylation [27,36].
The heart of the action: the FGFR tyrosine kinase domain
The intracellular ‘split’ tyrosine kinase domain of FGFRs shares the prototypical bi-lobed kinase fold (Figure 2) [39–42]. Binding of both adenosine triphosphate (ATP) and substrate occurs at the cleft between the two lobes. Nucleotide binding is facilitated by interactions with N-lobe residues of the hinge region, the glycine-rich P-loop (or nucleotide binding loop) which folds over and encloses ATP for phosphotransfer, and of conserved residue K514 (FGFR1) of helix αC (stabilised by salt-bridge formation with equally conserved E531 (FGFR1)). On the other hand, substrate binding is orchestrated by the C-lobe. Phosphorylation is catalysed by an invariant aspartate residue (D623 in FGFR1) of the His-Arg-Asp (HRD) motif, conserved among protein kinases and located on the C-lobe in the αE-β7 (catalytic) loop. Thus, to attain a catalytically competent state, the N- and C-lobes of the kinase require rotation towards one another during transition from the inactive state. During receptor activation, the kinase domains autophosphorylate one another in the dimer, firstly at Y653 (FGFR1) of the YYKK motif in the activation loop (A-loop) [43]. Seven phosphorylatable tyrosine residues have been identified in FGFR1 (Y463, Y583, Y585, Y653, Y654, Y730 and Y766), five of which are phosphorylated in an ordered fashion in vitro [43–46]. Tyrosine residues Y653 and Y654 are essential for kinase activity and their phosphorylation increases catalytic activity 50–100 and 500–1000 fold, respectively [43]. Other phospho-Tyr residues serve as docking sites for SH2 domain-containing adaptor proteins for the stimulation of downstream signalling cascades (Figure 1); for example, phospho-Y766 of FGFR1 serves as a binding-site for phospholipase Cγ (PLCγ) [47,48]. Likewise, Y724 of FGFR3 (equivalent to Y730 of FGFR1) appears to play a central role in FGFR3-mediated signalling, affecting activation of phosphoinositide 3-kinase (PI3K), signal transducer and activator of transcription protein (STAT) and mitogen-activated protein kinase (MAPK) pathways [49,50]. Immediately upstream of the kinase domain, the juxtamembrane domain (JMD) serves as a further site for coupling of receptor activation to downstream signalling cascades; here, the phosphotyrosine-binding domain of FGFR substrate 2 (FRS2α) binds constitutively to FGFR1 in a non-canonical, phosphotyrosine-independent manner and upon its own FGFR-dependent phosphorylation acts as a scaffold for Grb2 adaptor protein for MAPK signalling [51–53].
Activity regulation of the kinase domain
To trans-autophosphorylate in response to ligand binding, the FGFR kinase domain requires an intrinsic basal kinase activity. This requirement has led to a ‘two-state’ dynamic equilibrium model of kinase activation wherein a kinase can exhibit ensembles of a rigid, catalytically ‘inhibited’ state, or a dynamic, conformationally heterogeneous active state [54,55]. Under this model, kinase activity can be fine-tuned by shifting the kinase population between these two states. It is essential that kinases are tightly regulated, ensuring the ability for trans-autophosphorylation while preventing overstimulation of signalling cascades; dysregulation of FGFR kinases underpins a plethora of pathologies including developmental abnormalities such as achondroplasia, hypochondroplasia and lacrimo-auriculo-dento-digital (LADD) syndrome, and a host of human cancers [3,56,57].
In the basal, unstimulated state, the kinase domain is autoinhibited and has minimal kinase activity. Mechanisms of autoinhibition vary between kinases [2], and in FGFRs is achieved by the steric blocking of substrate Tyr binding by the protein tyrosine kinase-invariant P663 (FGFR1) at the C-terminal end of the A-loop [39]. Additionally, the so-called molecular brake of FGFR kinases, located at the kinase hinge, has a critical role in establishing autoinhibition and, via its release, activation (Figure 3) [40]. This structural motif is composed of a hydrogen bonding network between FGFR2 residues H544 and N549 (of the αC–β4 loop), E565 (of the kinase hinge) and K641 (of β8) in autoinhibited kinases (H541, N546, E562 and K638, respectively, in FGFR1), but is found to dissociate in activated kinase domain crystal structures (Figure 3) [40]. The release of the molecular brake occurs concomitantly with local A-loop conformational changes from an autoinhibited, substrate-blocked to an extended conformation, and the global conformational rotation of the αC helix (and N-lobe) towards the C-lobe, facilitating the generation of a catalytically competent state. The extended active A-loop conformation of the kinase is stabilised by salt-bridge interactions between phospho-Y657 (of FGFR2, equivalent to Y654 of FGFR1) and conserved residue R649 (of FGFR2), also of the A-loop (Figure 3) [40]. Residues of the molecular brake region are mutated in patients with dwarfism and glioblastoma, highlighting its importance in kinase activity regulation [40,58].
Figure 3. Comparison of active and inactive FGFR kinase domain states.
(A) Structural overlay (right) of non-phosphorylated, inactive FGFR2 kinase domain (light grey) (PDB: 2PSQ) and phosphorylated, active FGFR2 kinase domain (blue) (PDB: 2PVF), both in cartoon representation. Additionally, in the active kinase domain, the kinase regulatory spine and two participating residues, H624 of the HRD motif and F645 of the DFG motif, are highlighted in red sticks and surface representation. During kinase activation, the molecular brake hydrogen bonding network between H544, N549, E565, and K641 of FGFR2 is broken, as illustrated in the expanded sections (left). The same regions in the inactive state of FGFR1 kinase (PDB: 4V01) (dark grey) with the corresponding H541, N546, E562, and K638 residues are also presented (far left), illustrating the conservation of this feature among FGFRs. (B) Structural differences in A-loop conformation in active and inactive FGFR2 kinase domains where phosphorylation-dependent salt bridge interactions between R649 and phospho-Y657 (pY657) stabilise an extended conformation of the loop in the activated kinase.
Additional structural features of the kinase domain, including Asp-Phe-Gly (DFG)-motif conformation and kinase hydrophobic spines, are indicators of kinase activity status [59,60]. The DFG-motif, located at the start of the A-loop, classically exists in one of two states: the catalytically competent DFG-in and catalytically incompetent DFG-out state [61–63]. When in the DFG-in state, the Asp residue of the DFG-motif plays an essential role in ATP binding through coordination of all three phosphate groups of ATP, either directly or via magnesium ions; these interactions are not possible in the DFG-out state [62]. Furthermore, the ‘flipping’ of the DFG motif by ∼180° to its DFG-out position breaks the hydrophobic regulatory spine of the kinase. The regulatory spine is a structural entity that spans the N- and C-lobes of a kinase, composed of H624 (FGFR2) of the HRD motif, F645 (FGFR2) of the DFG motif and aliphatic side-chains of residues located on N-lobe elements αC and β4 (Figure 3). Conserved across kinases, its assembly is a hallmark of the active kinase state. A second hydrophobic spine, the catalytic spine, is assembled upon ATP binding, where the adenine base bridges further hydrophobic entities in the N- and C-lobes [59,64]. Recent analysis of inhibited, partially activated and fully activated (phosphorylated) FGFR kinase domains has revealed an interconnected allosteric network at the N- and C-lobe interface, permitting long-distance communication between the molecular brake and A-loop [65]. This allosteric network comprises the molecular brake, an ‘A-loop plug’ element (which holds the loop in a substrate-binding incompatible conformation), and hydrophobic patches of residues termed the DFG-latch and αC-tether. While these are distinct from those elements discussed previously, they are intimately associated with DFG-motif and αC conformational states. Strikingly, naturally found mutations within these elements alter the activity level of the kinase in vitro, with combined double mutations in different elements demonstrating additive effects, reflecting a population shift in the two-state dynamic equilibrium [65].
Dysregulation of FGFRs
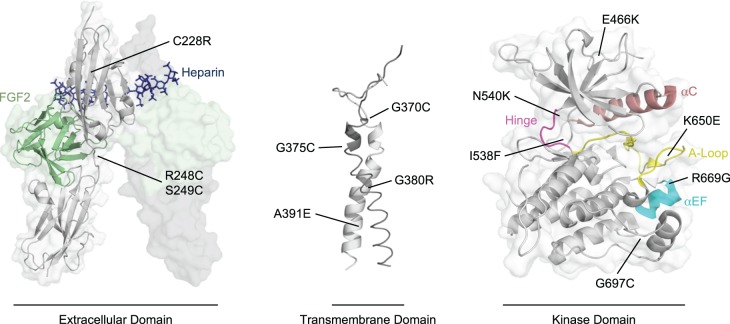
Subversion of FGFR kinase regulation and receptor hyperactivity is achieved in various ways, from overexpression of FGFRs and/or FGF ligands, to point mutations and gene fusion events [66]. Components of the FGF signalling pathways are the most frequently mutated kinases carrying non-synonymous somatic mutations in human cancers [67]. Though found in all FGFRs, point mutations occur most commonly in FGFR3 and are located in all three receptor domains (Figure 4). Consequently, here we focus on FGFR3 aberrations, though in many instances corresponding mutations can be found in FGFRs 1, 2 and 4 also. One can envisage that point mutations cause ligand-independent activity through either stabilisation of active conformational states or destabilisation of autoinhibitory states.
Figure 4. Point mutations of FGFR3.
The locations of a selection of developmental disease and cancer-associated point mutations of FGFR3 in the extracellular domain (left) (PDB: 1FQ9), the transmembrane domain (middle) (PDB: 2LZL), and the kinase domain (right) (PDB: 4K33), as discussed in the text. As no FGFR3 ligand-dimerised extracellular domain structure is available, the extracellular domain of FGFR1 in complex with FGF2 is shown, illustrating the localisation of FGFR3 point mutations to regions which could generate similar dimer structures in a ligand-independent manner. Similarly, in the kinase domain, the αC helix (salmon), the αEF helix (cyan), the hinge region (magenta), and the A-loop (yellow) are highlighted to illustrate the localisation of many point mutations to important regulatory elements of the kinase domain.
Point mutations in the extracellular domain of FGFR3, such as R248C/S249C (D2-D3 linker, thanatophoric dysplasia and keratosis [68,69]) and C228R (D2 domain, carcinoma [67,70]) are thought to recapitulate stimulation of the receptor in a ligand-independent manner through obligate receptor dimerisation. This is thought to be achieved by disulfide cross-linking of the extracellular domain and/or by induction of appropriate extracellular domain conformational arrangements for activation of the intracellular kinase domain. To this end, extracellular domain mutations are localised to regions where inter-receptor cross-linking may mimic ligand-bound states (Figure 4). Likewise, mutations in the transmembrane domain are believed to stabilise the active (or destabilise the basal) dimerisation interfaces of the helices, shifting the equilibrium towards a ligand-stimulated dimer arrangement. Indeed, cysteine-introducing point mutations such as G375C/G370C (N-terminal end of the transmembrane domain, achondroplasia and keratosis [69,71]) may act to cross-link the dimer at the extracellular domain/transmembrane domain interface and cause separation of the C-terminal ends of the transmembrane helices. Alternatively, substitutions of large, polar residues such as G380R (achondroplasia and hypochondroplasia [72,73]) and A391E (Crouzon syndrome with acanthosis nigricans [74]) in and around the transmembrane dimer interface may destabilise the basal dimerisation state by steric and Coulombic repulsion, or potentially stabilise the active state through hydrogen-bonding [32]. Supportively, energetic dimer stabilisation is observed following A391E substitution in the transmembrane domain [70].
The kinase domain is abundant in point mutations which are typically localised to regulatory elements such as the molecular brake, the A-loop, the kinase hinge, the DFG-latch and others. Recently, an extensive study on the prevalence and activating effect of point mutations in FGFR3 identified N540 (molecular brake) and K650 (A-loop) as mutational hotspots in FGFRs; these also elicit significant stimulation of FGFR3 kinase domain autophosphorylation [75]. Both mutation sites have been extensively characterised; molecular brake mutations are thought to overcome autoinhibitory mechanisms and facilitate transition to the active kinase state [40], while K650E substitution mimics A-loop Y648 (FGFR3) phosphorylation and stabilises the active, extended A-loop conformation [41]. Surprisingly, the study showed that clinical prevalence does not directly correlate with stimulatory effect. For example, R669G, the most activating mutation with respect to FGFR3 kinase domain autophosphorylation, is not a mutation hotspot, whereas G697C (of oral squamous cell carcinomas [76]), an identified FGFR3 mutation hotspot, has no stimulatory effect on kinase activity over wild-type kinase domain under the analysed conditions [75]. R669, due to its location at the C-terminal end of αEF in the C-lobe likely influences kinase activity by effect on the kinase A-loop (Figure 4); in fact, crystallographic evidence suggests that the corresponding residue R675 in FGFR1 contacts residues of the activation loop, stabilising the inactive kinase state. Upon R675 mutation of FGFR1, these contacts are lost and the kinase A-loop instead occupies an ‘open’ active conformation [75]. On the other hand, G697 is located within the αF-αG loop at the base of the C-lobe and does not appear to have any direct interactions with kinase regulatory elements (Figure 4). While the general frequency of G697C substitution in FGFR3 is disputed [77], the lack of stimulatory action by cancer-associated mutations is not unique to G697C; in truth, there are multiple mutations which are neutral-to-destabilising with respect to kinase activity, do not increase kinase domain autophosphorylation nor substrate phosphorylation, yet are nonetheless observed in tumours [75]. Furthermore, numerous deleterious point mutations in the FGFR kinase domain have been identified [75,78]. The role of deleterious, inhibitory and neutral mutations of FGFRs in pathologies is unclear but may be dependent on cellular context, and their purpose may become clearer when evaluated macroscopically with interaction partners and signalling networks in the cell [3]. For example, the destabilising and inactivating point mutations E466K and I538F of FGFR3, like kinase-activating N540K, enhance kinase domain binding to heat shock protein 90 (Hsp90) co-chaperone Cdc37 [79]. As Hsp90 has been implicated in the regulation and activation mechanisms of kinases [80], altered association with cellular chaperones is consistent with the notion that dysregulation of FGFRs by such mutations may play out at the level of the wider interactome of the kinase.
Although of relatively low clinical incidence, oncogenic gene fusions of FGFRs have recently come to light in a variety of cancer types [81,82]. Typically, these fuse self-associating elements of a second protein in frame with the C-terminal end (and less frequently the N-terminal end) of the receptor. Though supporting structural evidence is lacking to date, FGFR gene fusions are expected to cause ligand-independent constitutive kinase activity through fusion protein-induced dimerisation of the receptors, similar to that observed for the TPR-MET kinase fusion [83]. C-terminal fusions also lack exon 19 of the receptor, resulting in the inability to activate PLCγ signalling through loss of its phospho-Tyr binding-site [84]. While a variety of fusion partner genes have been identified for FGFR2 [66], gene fusions of FGFR3 almost exclusively occur with transforming acidic coiled-coil-containing protein 3 (TACC3) [66,84]. These fusions and the FGFR3-BAIAP2L1 (brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1) fusion are exquisitely sensitive to FGFR-selective inhibitors in urothelial cells, indicating that these aberrations are highly targetable [82,84].
Progress towards therapies for FGFR-driven diseases
The finding that aberrant FGFR signalling has driving roles in a plethora of cancers has spurred research interests in the development of anti-FGFR treatments, predominantly taking the form of small molecule kinase inhibitors (Figure 5). FGFR-targeting treatments under clinical development have been extensively reviewed previously [85–87] and will only be summarised here.
Figure 5. Binding modes of FGFR inhibitors.
Crystal structures of inhibitor-bound FGFR1 and FGFR4 kinase domains, illustrating the binding modes of reversible and irreversible (covalent) inhibitors. Reversible inhibitors can be classified into type I and type II inhibitors, differing in their binding modes (top). Type I inhibitors such as AZD4547 bind to active, DFG in-state kinases, whereas type II inhibitors such as ponatinib bind to inactive, DFG out-state kinases. In each instance, FGFR1 kinase domains are shown in full in cartoon representation with transparent surfaces (light grey) and inhibitors in stick representation (purple). Additionally, the inhibitor-binding site is expanded for each with FGFR1 (light grey) in cartoon representation alone, and F642 of the DFG motif (red) and the gatekeeper residue V550 (orange) shown in stick representation with transparent surfaces. In the ponatinib-bound structure, the asterisk (*) indicates the location of the ethynyl group attributed to the ability of ponatinib to accommodate gatekeeper residue mutations and to the multikinase selectivity profile of the inhibitor. The binding modes of three irreversible, covalent inhibitors to FGFR4 and FGFR4 surrogate kinase domain (FGFR1–Y563C) are presented in expanded panels in a similar manner (bottom). In these, where resolved in the crystal structures, the gatekeeper residue and Phe of the DFG motif is shown as above, and the Cys residues utilised in ligand conjugation are highlighted also (yellow). In the FIIN-2-bound structure, F631 (DFG motif, FGFR4) is observed in both the DFG in- and DFG out-states, marked with a double asterisk (**). The structures presented are: FGFR1 kinase domain bound to AZD4547 (PDB: 4V05) and ponatinib (PDB: 4V01); FGFR4 kinase domain bound to BLU-9931 (PDB: 4XCU) and FIIN-2 (PDB: 4QQC) and of FGFR4 surrogate kinase domain (FGFR1 harbouring a Y563C substitution) bound to H3B-6527 (PDB: 5VND).
Small molecule inhibitors of FGFRs can be classified into non-selective multi-kinase inhibitors and selective FGFR inhibitors. The first efforts to treat FGFR aberrations have made use of non-selective multi-kinase inhibitors such as dovitinib, ponatinib and lucitanib which show pan-FGFR inhibition with nanomolar IC50 values against FGFR family members (Table 1). With the notable exception of ponatinib, these compounds are type I inhibitors which bind to the active, DFG-in state of FGFR kinases in an ATP-competitive manner. Often, vascular endothelial growth factor receptors (VEGFRs) and platelet-derived growth factor receptors (PDGFRs) are also targeted by these non-selective FGFR inhibitors. While the ability to target multiple kinases with a single compound may be clinically beneficial under certain circumstances, non-selective multi-kinase FGFR inhibitors typically exhibit lower affinity binding to FGFRs than other targets. Consequently, the use of these non-selective inhibitors as FGFR-targeted therapies is associated with off-target-related toxicities [86]. The broad specificity of many kinase inhibitors has been attributed to the high degree of structural conservation between kinases rendering the development of selective inhibitors challenging, particularly against those in the active state. A second subclass of reversible kinase inhibitors, type II inhibitors, bind to kinases in a DFG-out, inactive state. To generate this kinase state, the Phe residue of the DFG-motif ‘flips’ outwards, breaking the regulatory spine and providing access to an additional hydrophobic pocket from the ATP binding-site (Figure 5) [63]. Type II inhibitors have proved to be generally more selective than their type I counterparts, while also exhibiting considerably slower dissociation kinetics [88,89]; however, the ability for a type II inhibitor to bind to its target is dependent on the propensity of the kinase to ‘visit’ the DFG-out state through conformational sampling, implying that some kinase classes may be innately more amenable to type II inhibition than others. Furthermore, a survey of many kinase inhibitors has established that not all type II inhibitors are necessarily more selective [90]. This is the case for the multi-kinase type II inhibitor ponatinib which was originally developed to target BCR-ABL aberrations harbouring the T315I ‘gatekeeper’ mutation in the ATP binding-site, conferring resistance to earlier generation BCR-ABL inhibitors [91]. While ponatinib is able to accommodate the Thr to Ile mutation of the gatekeeper residue in BCR-ABL through productive interactions with the unsaturated ethynyl bond of the inhibitor, this feature is also likely to be responsible for the relatively poor kinase selectivity of the inhibitor, which additionally exhibits potent pan-FGFR inhibition [91].
Table 1. Inhibitors of FGFR.
| Inhibitor name | Company | Measured IC50 (nM, in vitro) | Progress in clinical trials (with identifiers) | Ref. | |||
|---|---|---|---|---|---|---|---|
| Multi-kinase inhibitors | |||||||
| Ponatinib (AP24534) |
ARIAD Pharmaceuticals | FGFR1: 2.2 FGFR2: 1.6 FGFR3: 18.2 FGFR4: 7.7 |
Phase II NCT02272998 |
NCT02265341 |
|
[91] | |
| Dovitinib (CHIR258, TKI258) |
Novartis | FGFR1: 8 FGFR3: 9 |
Phase II NCT01732107† NCT01676714 NCT01379534 |
NCT01576380 NCT00790426 NCT01719549 |
NCT01058434 NCT01831726 NCT00958971 |
[92] | |
| Lucitanib (E-3810) |
Clovis Oncology | FGFR1: 17.5 FGFR2: 82.5 FGFR3: 237.5 FGFR4: >1000 |
Phase I NCT03117101 Phase I/II NCT01283945 Phase II NCT02202746 |
NCT02109016 |
NCT02053636 |
[93] | |
| Nintedanib (BIBF 1120) |
Boehringer Ingelheim | FGFR1: 69 FGFR2: 37 FGFR3: 108 FGFR4: 610 |
Phase II NCT01948141 |
[94] | |||
| ARQ 087 (Derazantinib) |
ArQule | FGFR1: 4.5 FGFR2: 1.8 FGFR3: 4.5 FGFR4: 34 |
Phase I/II NCT01752920 Phase II NCT03230318 |
[95] | |||
| FGFR-selective inhibitors | |||||||
| AZD4547 | AstraZeneca | FGFR1: 0.2 FGFR2: 2.5 FGFR3: 1.8 FGFR4: 165 |
Phase I NCT01213160 Phase I/II NCT01824901‡ NCT02824133§ Phase II/III NCT02965378‡ |
NCT00979134 NCT01202591‡ |
NCT01791985‡ |
[96] | |
| LYS2874455 | Eli Lilly | FGFR1: 2.8 FGFR2: 2.6 FGFR3: 6.4 FGFR4: 6 |
Phase I NCT01212107 |
NCT03125239‡ |
[97] | ||
| (NVP-)BGJ398 (Infigratinib) |
Novartis | FGFR1: 0.9 FGFR2: 1.4 FGFR3: 1 FGFR3K650E: 4.9 FGFR4: 60 |
Phase I NCT01697605 Phase II NCT02706691 NCT03510455 |
NCT01004224 NCT02150967 |
NCT01928459‡ NCT01975701 |
[98] | |
| Debio-1347 (CH5183284) |
Debiopharm International | FGFR1: 9.3 FGFR2: 7.6 FGFR3: 22 FGFR4: 290 |
Phase I NCT01948297 Phase I/II NCT03344536‡ |
[99] | |||
| Erdafitinib (JNJ-42756493) |
Janssen | FGFR1: 1.2 FGFR2: 2.5 FGFR3: 3.0 FGFR4: 5.7 |
Phase I NCT02421185 NCT03238196‡ Phase I/II NCT03473743‡ Phase II NCT03210714 NCT02952573‡ Phase III NCT03390504‡ |
NCT01962532 NCT02699606 |
NCT01703481 NCT02365597 |
[100] | |
| INCB054828 | Incyte Corporation | FGFR1: 3–50* |
Phase I NCT03235570 Phase I/II NCT02393248‡ Phase II NCT03011372 |
NCT02872714 |
NCT02924376 |
[101] | |
| Rogaratinib (BAY1163877) |
Bayer | FGFR1: 12–15 FGFR2: <1 FGFR3: 19 FGFR4: 33 |
Phase I NCT01976741 Phase I/II NCT03473756‡ Phase II/III NCT03410693 |
[102] | |||
| (NVP)FGF401 | Novartis | FGFR4: 1.1 |
Phase I/II NCT02325739‡ |
[103] | |||
| PD173074 | Pfizer | FGFR1: 22–25 FGFR3: 29 |
N/A | [104,105] | |||
| PD166866 | Pfizer | FGFR1: 52.4 | N/A | [106] | |||
| SSR128129E | N/A | FGFR1: 1900 | N/A | [107] | |||
A selection of small molecule multi-kinase and FGFR-selective reversible inhibitors, their measured in vitro IC50 values and clinical trial status.
Key: ‘Ref.’, reference.
*IC50 value measured using in cell assays.
Trial terminated due to funding.
Drug combination study.
Trial suspended.
To address the toxicity issues of multi-kinase inhibitors, efforts have been made to develop FGFR-selective kinase inhibitors, yielding numerous reversible type I inhibitor compounds with FGFR1–3 and pan-FGFR activities (Table 1). Of these, AZD4547, a potent inhibitor of FGFRs 1–3, has shown promising responses in preclinical and phase I clinical trials, particularly towards tumours with FGFR amplifications [108–110]. Several phase II clinical trials evaluating the efficacy of AZD4547 alone or in combination with other compounds are active or have completed. However, preclinical studies have also indicated that resistance can be conferred to AZD4547 via the gatekeeper mutation V555M in FGFR3 [111], much like that in BCR-ABL, highlighting the need for continued inhibitor development and the personalisation of FGFR-targeted therapies in the clinic. Towards this end, a second-generation FGFR-selective inhibitor Debio-1347 has been developed which has a different chemical scaffold to AZD4547, PD173074 and BGJ398, and has shown inhibition efficacy against Ba/F3 cells harbouring a FGFR2 fusion with V564F gatekeeper mutation [99]. Despite efforts, no FGFR-selective type II inhibitors in the vein of ponatinib have yet been reported, though several irreversible, covalent inhibitors of FGFRs have been developed. Unlike reversible inhibitors, covalent inhibition confers the advantage of partially circumventing high in vivo ATP concentrations [112]. Furthermore, covalent inhibition has facilitated the development of isoform-selective inhibitors; many these covalent inhibitors are highly selective for FGFR4 (Table 2). This FGFR isoform selectivity has been achieved in at least three cases (H3B-6527, BLU-9931 and BLU-554) through the use of the FGFR4-unique C552 residue of the hinge region which is occupied by a Tyr residue in the corresponding position in FGFRs 1–3 (Figure 5) [113–115]. Conversely, pan-FGFR covalent inhibition has been achieved through use of the FGFR-conserved C477 residue (FGFR4) in the cases of inhibitors FIIN-2 and FIIN-3 (Table 2), both of which also exhibit activities against FGFR2 harbouring gatekeeper mutations in cell-based assays [116]. Intriguingly, a crystal structure of FIIN-2 bound to FGFR4 indicates that the inhibitor can bind to both DFG-in and DFG-out states of the kinase, though the inhibitor does not occupy the additional hydrophobic pocket which is accessible in the DFG-out state (Figure 5). The significance, if any, of being able to bind to both states is unclear; however, FIIN-2 could form the foundation for development of next-generation type II-like covalent inhibitors. At the time of writing, four covalent FGFR inhibitors (PRN1371, TAS-120, H3B-6527 and BLU-554) are recruiting for phase I clinical trials.
Table 2. Irreversible, covalent FGFR-selective inhibitors under development.
| Inhibitor name | Company | Measured IC50 (nM, in vitro) |
Progress in clinical trials (with identifiers) |
Ref. |
|---|---|---|---|---|
| PRN1371 | Principia Biopharma | FGFR1: 0.6 FGFR2: 1.3 FGFR3: 4.1 FGFR4: 19.3 |
Phase I NCT02608125 |
[112] |
| TAS-120 | Taiho Oncology | FGFR1: 3.9 FGFR2: 1.3 FGFR3: 1.6 FGFR4: 8.3 |
Phase I/II NCT02052778 |
[117] |
| BLU-554 | Blueprint Medicines Corporation | FGFR1: 624 FGFR2: 1202 FGFR3: 2203 FGFR4: 5 |
Phase I NCT02508467 |
[118,119] |
| BLU-9931 | Blueprint Medicines Corporation | FGFR1: 591 FGFR2: 493 FGFR3: 150 FGFR4: 3 |
N/A | [115] |
| FIIN-2 | N/A | FGFR1: 3.09 FGFR2: 4.3 FGFR3: 27 FGFR4: 45.3 |
N/A | [116] |
| FIIN-3 | N/A | FGFR1: 13.1 FGFR2: 21 FGFR3: 31.4 FGFR4: 35.3 |
N/A | [116] |
| H3B-6527 | H3 Biomedicine, Eisai Incorporation |
FGFR1: 320 FGFR2: 1290 FGFR3: 1060 FGFR4: <1.2 |
Phase I NCT02834780 |
[114] |
FGFR-selective inhibitors that have an irreversible, covalent mode of action, their measured in vitro IC50 values and their clinical trial status. Key: ‘Ref.’, references.
FGFR-targeted therapies are not limited to the tyrosine kinase domain only; there have been multiple efforts to target the extracellular domains of FGFRs also, offering further opportunities for isoform-selective inhibition (Table 3). This is best exemplified by the development of anti-FGFR2 and anti-FGFR3 monoclonal antibodies/antibody-drug conjugates [120–124]. FP-1039, an FGF-ligand trap composed of an FGFR1 extracellular domain-IgG1 fusion which is able to inhibit tumour growth in xenograft models, has also been developed [125]. Furthermore, a novel small molecule inhibitor (SSR128129E) which binds to FGFR extracellular domains in a non-FGF competitive manner but induces selective, allosteric inhibition of receptor internalisation and ERK1/2 signalling has been described [107,126]. Lastly, there has also been exploration of the use of antisense therapy for targeting FGFR4 in obesity patients (Table 3) [127].
Table 3. Alternative therapies for FGFR aberrations under development.
| Molecule name | Company | Target | Progress in clinical trials (with identifiers) |
Ref. | |
|---|---|---|---|---|---|
| FGF ligand traps | |||||
| FP-1039 (GSK3052230) |
Five Prime Therapeutics | FGF2 and others |
Phase II NCT01244438* |
[125] | |
| Anti-FGFR monoclonal antibodies | |||||
| Bemarituzumab (FPA144) |
Five Prime Therapeutics | FGFR2b |
Phase I NCT02318329 |
NCT03343301† |
[120] |
| BAY1179470 | Bayer | FGFR2 |
Phase I NCT01881217 |
[122] | |
| LY3076226 | Eli Lilly | FGFR3 |
Phase I NCT02529553 |
[123] | |
| MFGR1877S | Genentech | FGFR3 |
Phase I NCT01122875 |
NCT01363024 |
[124] |
| Antisense therapy | |||||
| ISIS-FGFR4RX | ISIS Pharmaceuticals | FGFR4 |
Phase II NCT02476019 |
[127] | |
Additional non-kinase-domain inhibitor-based therapies under development, their targets and their clinical trial status.
Key: ‘Ref.’, references.
*Trial withdrawn.
Drug combination study.
While improved selectivity of therapies may be beneficial to target specific FGFR aberrations, it is important to recognise that clinical efficacy and selectivity are not necessarily related. Highly selective FGFR-targeted therapies may also be more prone to resistance development if not used in combination strategies. For example, in addition to the gain of gatekeeper mutations detailed above, resistance to FGFR inhibitors has also been acquired in cell lines harbouring FGFR3 amplification by switching to ErbB family signalling [128]. Moreover, while one aim of FGFR-selective inhibitor development is to overcome off-target effects of multi-kinase inhibitors, FGFR-selective therapies are not immune to side effects, exemplified by toxicity profiles associated with FGFR-selective inhibitors in the clinic [86]. FGFRs are still a relatively novel target and many anti-FGFR programmes are still in their early stages; however, with multiple clinical trials active or recruiting, in many cases with participants with specific FGFR aberrations, we should soon glean further insights that help to improve our approaches to treatment of FGFR dysregulation.
Conclusions and perspectives
Since their first description, it has been established that FGFRs play crucial roles in a host of physiological processes which when dysregulated result in a plethora of pathologies. From an extensive range of studies covering FGFR expression, structure and function, among others, mechanisms of FGFR regulation and activation have come to light, and good progress has been made in the development of anti-FGFR therapies. Despite this, due to the complexity of FGFR signalling inputs, outputs and FGFR interactomes, and difficulties faced with the biochemical and biophysical characterisation of full-length receptors, we are still far from an integrated understanding of FGFR biology. Crucially, mechanisms of activation in the context of the full-length receptor are unclear and will remain unresolved until structures of full-length FGFRs in autoinhibited, ligand-activated and pathogenically activated modes are solved. In fact, to date there are no high-resolution full-length structures of any receptor tyrosine kinase, severely limiting our understanding of this highly important class of kinases. Equally, while it is recognised that activation of FGFRs can lead to differential activation of intracellular signalling cascades, the underlying molecular basis of how this occurs and of how cellular context influences phenotypic outcome remain poorly understood. We anticipate that advances will be made in addressing these and other remaining questions in the coming decades, and with this, new and improved strategies for treatment of disorders arising due to aberrant FGFR signalling will develop.
Acknowledgements
We acknowledge all authors whose work is referred to in this review, along with those we have not cited for reasons of space.
Abbreviations
- ATP
adenosine triphosphate
- BAIAP2L1
brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1
- D1–3
immunoglobulin (Ig)-like domains 1–3
- DFG-motif
Asp-Phe-Gly motif
- ECD
extracellular domain
- ERK1/2
extracellular signal-regulated kinases 1/2
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- FRET
Förster resonance energy transfer
- FRS2α
FGFR substrate 2
- Grb2
growth factor receptor-bound protein 2
- HRD motif
His-Arg-Asp motif
- Hsp90
heat shock protein 90
- JMD
juxtamembrane domain
- KD
kinase domain
- LADD syndrome
lacrimo-auriculo-dento-digital syndrome
- MAPK
mitogen-activated protein kinase
- PDGFR
platelet-derived growth factor receptor
- PI3K
phosphoinositide 3-kinase
- PLCγ
phospholipase Cγ
- P-loop
nucleotide binding loop
- RTK
receptor tyrosine kinase
- SH2 domain
Src-homology 2 domain
- STAT
signal transducer and activator of transcription protein
- TACC3
transforming acidic coiled-coil-containing protein 3
- TMD
transmembrane domain
- VEGFR
vascular endothelial growth factor receptor
Author Contribution
B.F. wrote the manuscript with guidance and contributions from A.L.B.
Funding
B.F. is supported by a PhD studentship from the Wellcome Trust [109155/Z/15/Z]. Work in the A.L.B. laboratory is supported by funding from the UK Medical Research Council, the Wellcome Trust and the Pancreatic Cancer Research Fund.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Manning G., Whyte D.B., Martinez R., Hunter T. and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 2.Lemmon M.A. and Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner N. and Grose R. (2010) Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 10, 116–129 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- 4.Beenken A. and Mohammadi M. (2009) The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8, 235–253 10.1038/nrd2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart A.W., Baeza N., Apelqvist Å. and Edlund H. (2000) Attenuation of FGF signalling in mouse β-cells leads to diabetes. Nature 408, 864–868 10.1038/35048589 [DOI] [PubMed] [Google Scholar]
- 6.Dorey K., Amaya E., Harpal K., Yamaguchi T.P., Rossant J. and Papalopulu N. (2010) FGF signalling: diverse roles during early vertebrate embryogenesis. Development 137, 3731–3742 10.1242/dev.037689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Razzaque M.S. (2009) The FGF23–Klotho axis: endocrine regulation of phosphate homeostasis. Nat. Rev. Endocrinol. 5, 611–619 10.1038/nrendo.2009.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh N. and Ornitz D.M. (2004) Evolution of the Fgf and Fgfr gene families. Trends Genet. 20, 563–569 10.1016/j.tig.2004.08.007 [DOI] [PubMed] [Google Scholar]
- 9.Zhu X., Komiya H., Chirino A., Faham S., Fox G.M., Arakawa T. et al. (1991) Three-dimensional structures of acidic and basic fibroblast growth factors. Science 251, 90–93 PMID: [DOI] [PubMed] [Google Scholar]
- 10.Goetz R. and Mohammadi M. (2013) Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 14, 166–180 10.1038/nrm3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F., Kan M., Xu J., Yan G. and McKeehan W.L. (1995) Ligand-specific structural domains in the fibroblast growth factor receptor. J. Biol. Chem. 270, 10222–10230 10.1074/JBC.270.17.10222 [DOI] [PubMed] [Google Scholar]
- 12.Plotnikov A.N., Schlessinger J., Hubbard S.R. and Mohammadi M. (1999) Structural basis for FGF receptor dimerization and activation. Cell 98, 641–650 10.1016/S0092-8674(00)80051-3 [DOI] [PubMed] [Google Scholar]
- 13.Plotnikov A.N., Hubbard S.R., Schlessinger J. and Mohammadi M. (2000) Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101, 413–424 10.1016/S0092-8674(00)80851-X [DOI] [PubMed] [Google Scholar]
- 14.Stauber D.J., DiGabriele A.D. and Hendrickson W.A. (2000) Structural interactions of fibroblast growth factor receptor with its ligands. Proc. Natl Acad. Sci. U.S.A. 97, 49–54 10.1073/pnas.97.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapraeger A., Krufka A. and Olwin B. (1991) Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708 10.1126/science.1646484 [DOI] [PubMed] [Google Scholar]
- 16.Yayon A., Klagsbrun M., Esko J.D., Leder P. and Ornitz D.M. (1991) Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848 10.1016/0092-8674(91)90512-W [DOI] [PubMed] [Google Scholar]
- 17.Spivak-Kroizman T., Lemmon M.A., Dikic I., Ladbury J.E., Pinchasi D., Huang J. et al. (1994) Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell 79, 1015–1024 10.1016/0092-8674(94)90032-9 [DOI] [PubMed] [Google Scholar]
- 18.Olsen S.K., Li J.Y.H., Bromleigh C., Eliseenkova A.V., Ibrahimi O.A., Lao Z. et al. (2006) Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 20, 185–198 10.1101/gad.1365406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlessinger J., Plotnikov A.N., Ibrahimi O.A., Eliseenkova A.V., Yeh B.K., Yayon A. et al. (2000) Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 10.1016/S1097-2765(00)00073-3 [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini L. (2001) Role of heparan sulfate in fibroblast growth factor signalling: a structural view. Curr. Opin. Struct. Biol. 11, 629–634 10.1016/S0959-440X(00)00258-X [DOI] [PubMed] [Google Scholar]
- 21.Harmer N.J., Ilag L.L., Mulloy B., Pellegrini L., Robinson C V. and Blundell T.L. (2004) Towards a resolution of the stoichiometry of the fibroblast growth factor (FGF)–FGF receptor–heparin complex. J. Mol. Biol. 339, 821–834 10.1016/J.JMB.2004.04.031 [DOI] [PubMed] [Google Scholar]
- 22.Blundell T.L., Pellegrini L., Burke D.F., von Delft F. and Mulloy B. (2000) Crystal structure of fibroblast growth factor receptor ectodomain boundto ligand and heparin. Nature 407, 1029–1034 10.1038/35039551 [DOI] [PubMed] [Google Scholar]
- 23.Mohammadi M., Olsen S.K. and Ibrahimi O.A. (2005) Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 16, 107–137 10.1016/j.cytogfr.2005.01.008 [DOI] [PubMed] [Google Scholar]
- 24.Chen G., Liu Y., Goetz R., Fu L., Jayaraman S., Hu M.-C. et al. (2018) α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553, 461–466 10.1038/nature25451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S., Choi J., Mohanty J., Sousa L.P., Tome F., Pardon E. et al. (2018) Structures of β-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature 553, 501–505 10.1038/nature25010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beenken A., Eliseenkova A.V., Ibrahimi O.A., Olsen S.K. and Mohammadi M. (2012) Plasticity in interactions of fibroblast growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J. Biol. Chem. 287, 3067–3078 10.1074/jbc.M111.275891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F. and Hristova K. (2011) The physical basis of FGFR3 response to fgf1 and fgf2. Biochemistry 50, 8576–8582 10.1021/bi200986f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen S.K., Ibrahimi O.A., Raucci A., Zhang F., Eliseenkova A.V., Yayon A. et al. (2004) Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl Acad. Sci. U.S.A. 101, 935–940 10.1073/pnas.0307287101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalinina J., Dutta K., Ilghari D., Beenken A., Goetz R., Eliseenkova A.V. et al. (2012) The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure 20, 77–88 10.1016/j.str.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi E., Kan M., Xu J., Wang F., Hou J. and McKeehan W.L. (1993) Control of fibroblast growth factor receptor kinase signal transduction by heterodimerization of combinatorial splice variants. Mol. Cell. Biol. 13, 3907–3918 10.1128/MCB.13.7.3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F., Kan M., Yan G., Xu J. and McKeehan W.L. (1995) Alternately spliced NH2-terminal immunoglobulin-like loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 270, 10231–10235 10.1074/jbc.270.17.10231 [DOI] [PubMed] [Google Scholar]
- 32.Bocharov E V., Lesovoy D.M., Goncharuk S.A., Goncharuk M V., Hristova K. and Arseniev A.S. (2013) Structure of FGFR3 transmembrane domain dimer: implications for signaling and human pathologies. Structure 21, 2087–2093 10.1016/j.str.2013.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae J.H., Boggon T.J., Tomé F., Mandiyan V., Lax I. and Schlessinger J. (2010) Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proc. Natl Acad. Sci. U.S.A. 107, 2866–2871 10.1073/pnas.0914157107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cymer F. and Schneider D. (2010) Transmembrane helix-helix interactions involved in ErbB receptor signaling. Cell Adh. Migr. 4, 299–312 10.4161/cam.4.2.11191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sternberg M.J.E. and Gullick W.J. (1990) A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. Protein Eng. Des. Sel. 3, 245–248 10.1093/protein/3.4.245 [DOI] [PubMed] [Google Scholar]
- 36.Sarabipour S. and Hristova K. (2016) Mechanism of FGF receptor dimerization and activation. Nat. Commun. 7, 10262 10.1038/ncomms10262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comps-Agrar L., Dunshee D.R., Eaton D.L. and Sonoda J. (2015) Unliganded fibroblast growth factor receptor 1 forms density-independent dimers. J. Biol. Chem. 290, 24166–24177 10.1074/jbc.M115.681395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutmann T., Kim K.H., Grzybek M., Walz T. and Coskun Ü (2018) Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J. Cell Biol. 217, 1643–1649 10.1083/jcb.201711047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadi M., Schlessinger J. and Hubbard S.R. (1996) Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell 86, 577–587 10.1016/S0092-8674(00)80131-2 [DOI] [PubMed] [Google Scholar]
- 40.Chen H., Ma J., Li W., Eliseenkova A.V., Xu C., Neubert T.A. et al. (2007) A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell 27, 717–730 10.1016/j.molcel.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Z., Chen H., Blais S., Neubert T.A.A., Li X. and Mohammadi M. (2013) Structural mimicry of A-loop tyrosine phosphorylation by a pathogenic FGF receptor 3 mutation. Structure 21, 1889–1896 10.1016/j.str.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lesca E., Lammens A., Huber R. and Augustin M. (2014) Structural analysis of the human fibroblast growth factor receptor 4 kinase. J. Mol. Biol. 426, 3744–3756 10.1016/j.jmb.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 43.Furdui C.M., Lew E.D., Schlessinger J. and Anderson K.S. (2006) Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21, 711–717 10.1016/j.molcel.2006.01.022 [DOI] [PubMed] [Google Scholar]
- 44.Lew E.D., Furdui C.M., Anderson K.S. and Schlessinger J. (2009) The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci. Signal. 2, ra6 10.1126/scisignal.2000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohammadi M., Dikic I., Sorokin A., Burgess W.H., Jaye M. and Schlessinger J. (1996) Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol. Cell. Biol. 16, 977–989 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou J., Mckeehan K., Kan M., Crabb J.W., Mckeehan W.L., Carr S.A. et al. (2008) Identification of tyrosines 154 and 307 in the extracellular domain and 653 and 766 in the intracellular domain as phosphorylation sites in the heparin-binding fibroblast growth factor receptor tyrosine kinase (flg). Protein Sci. 2, 86–92 10.1002/pro.5560020109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohammadi M., Dionne C.A., Li W., Li N., Spivak T., Honegger A.M. et al. (1992) Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature 358, 681–684 10.1038/358681a0 [DOI] [PubMed] [Google Scholar]
- 48.Peters K.G., Marie J., Wilson E., Ives H.E., Escobedo J., Del R.M. et al. (1992) Point mutation of an FGF receptor abolishes phosphatidylinositol turnover and Ca2+ flux but not mitogenesis. Nature 358, 678–681 10.1038/358678a0 [DOI] [PubMed] [Google Scholar]
- 49.Hart K.C., Robertson S.C. and Donoghue D.J. (2001) Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, Stat activation, and phosphatidylinositol 3-kinase activation. Mol. Biol. Cell 12, 931–942 10.1091/mbc.12.4.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong M., Wang C.S. and Donoghue D.J. (2002) Interaction of fibroblast growth factor receptor 3 and the adapter protein SH2-B. A role in STAT5 activation. J. Biol. Chem. 277, 15962–15970 10.1074/jbc.M102777200 [DOI] [PubMed] [Google Scholar]
- 51.Dhalluin C., Yan K.S., Plotnikova O., Lee K.W., Zeng L., Kuti M. et al. (2000) Structural basis of SNT PTB domain interactions with distinct neurotrophic receptors. Mol. Cell 6, 921–929 10.1016/S1097-2765(05)00087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kouhara H., Hadari Y., Spivak-Kroizman T., Schilling J., Bar-Sagi D., Lax I. et al. (1997) A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89, 693–702 10.1016/S0092-8674(00)80252-4 [DOI] [PubMed] [Google Scholar]
- 53.Hadari Y.R., Gotoh N., Kouhara H., Lax I. and Schlessinger J. (2001) Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc. Natl Acad. Sci. U.S.A. 98, 8578–8583 10.1073/pnas.161259898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H., Huang Z., Dutta K., Blais S., Neubert T.A.A., Li X. et al. (2013) Cracking the molecular origin of intrinsic tyrosine kinase activity through analysis of pathogenic gain-of-function mutations. Cell Rep. 4, 376–384 10.1016/j.celrep.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karp J.M., Sparks S. and Cowburn D. (2017) Effects of FGFR2 kinase activation loop dynamics on catalytic activity. PLoS Comput. Biol. 13, e1005360 10.1371/journal.pcbi.1005360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou W.-Y., Zheng H., Du X. and Yang J. (2016) Characterization of FGFR signaling pathway as therapeutic targets for sarcoma patients. Cancer Biol. Med. 13, 260–268 10.20892/j.issn.2095-3941.2015.0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teven C.M., Farina E.M., Rivas J. and Reid R.R. (2014) Fibroblast growth factor (FGF) signaling in development and skeletal diseases. Genes Dis. 1, 199–213 10.1016/J.GENDIS.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkie A.O.M. (2005) Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 16, 187–203 10.1016/j.cytogfr.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 59.Kornev A.P., Taylor S.S. and Ten Eyck L.F. (2008) A helix scaffold for the assembly of active protein kinases. Proc. Natl Acad. Sci. U.S.A. 105, 14377–14382 10.1073/pnas.0807988105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu J., Ahuja L.G., Meharena H.S., Kannan N., Kornev A.P., Taylor S.S. et al. (2015) Kinase regulation by hydrophobic spine assembly in cancer. Mol. Cell. Biol. 35, 264–276 10.1128/MCB.00943-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hari S.B., Merritt E.A. and Maly D.J. (2013) Sequence determinants of a specific inactive protein kinase conformation. Chem. Biol. 20, 806–815 10.1016/j.chembiol.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kornev A.P., Haste N.M., Taylor S.S. and Ten Eyck L.F. (2006) Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl Acad. Sci. U.S.A. 103, 17783–17788 10.1073/pnas.0607656103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vijayan R.S.K., He P., Modi V., Duong-Ly K.C., Ma H., Peterson J.R. et al. (2015) Conformational analysis of the DFG-out kinase motif and biochemical profiling of structurally validated type II inhibitors. J. Med. Chem. 58, 466–479 10.1021/jm501603h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim J., Ahuja L.G., Chao F.-A., Xia Y., McClendon C.L., Kornev A.P. et al. (2017) A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci. Adv. 3, e1600663 10.1126/sciadv.1600663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H., Marsiglia W.M., Cho M.-K., Huang Z., Deng J., Blais S.P. et al. (2017) Elucidation of a four-site allosteric network in fibroblast growth factor receptor tyrosine kinases. eLife 6, 299–300 10.7554/eLife.21137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babina I.S. and Turner N.C. (2017) Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 17, 318–332 10.1038/nrc.2017.8 [DOI] [PubMed] [Google Scholar]
- 67.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G. et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 10.1038/nature05610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tavormina P.L., Rimoin D.L., Cohn D.H., Zhu Y.-Z., Shiang R. and Wasmuth J.J. (1995) Another mutation that results in the substitution of an unpaired cysteine residue in the extracellular domain of FGFR3 in thanatophoric dysplasia type I. Hum. Mol. Genet. 4, 2175–2177 10.1093/hmg/4.11.2175 [DOI] [PubMed] [Google Scholar]
- 69.Logié A., Dunois-Lardé C., Rosty C., Levrel O., Blanche M., Ribeiro A. et al. (2005) Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum. Mol. Genet. 14, 1153–1160 10.1093/hmg/ddi127 [DOI] [PubMed] [Google Scholar]
- 70.Sarabipour S. and Hristova K. (2015) FGFR3 unliganded dimer stabilization by the juxtamembrane domain. J. Mol. Biol. 427, 1705–1714 10.1016/j.jmb.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Superti-Furga A., Steinmann B., Gitzelmann R., Eich G., Giedion A., Bucher H.U. et al. (1995) A glycine 375-to-cysteine substitution in the transmembrane domain of the fibroblast growth factor receptor-3 in a newborn with achondroplasia. Eur. J. Pediatr. 154, 215–219 10.1007/BF01954274 [DOI] [PubMed] [Google Scholar]
- 72.Bellus G.A., McIntosh I., Smith E.A., Aylsworth A.S., Kaitila I., Horton W.A. et al. (1995) A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat. Genet. 10, 357–359 10.1038/ng0795-357 [DOI] [PubMed] [Google Scholar]
- 73.Bellus G.A., Hefferon T.W., Ortiz de Luna R.I., Hecht J.T., Horton W.A., Machado M. et al. (1995) Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 56, 368–373 PMID: [PMC free article] [PubMed] [Google Scholar]
- 74.Meyers G.A., Orlow S.J., Munro I.R., Przylepa K.A. and Jabs E.W. (1995) Fibroblast growth factor receptor 3 (FGFR3) transmembrane mutation in Crouzon syndrome with acanthosis nigricans. Nat. Genet. 11, 462–464 10.1038/ng1295-462 [DOI] [PubMed] [Google Scholar]
- 75.Patani H., Bunney T., Thiyagarajan N., Norman R., Ogg D., Breed J. et al. (2016) Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use. Oncotarget 7, 1949–2553 10.18632/oncotarget.8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y., Hiraishi Y., Wang H., Sumi K., Hayashido Y., Toratani S. et al. (2005) Constitutive activating mutation of the FGFR3b in oral squamous cell carcinomas. Int. J. Cancer 117, 166–168 10.1002/ijc.21145 [DOI] [PubMed] [Google Scholar]
- 77.Aubertin J., Tourpin S., Janot F., Ahomadegbe J.-C. and Radvanyi F. (2007) Analysis of fibroblast growth factor receptor 3 G697C mutation in oral squamous cell carcinomas. Int. J. Cancer 120, 2058–2059 10.1002/ijc.22285 [DOI] [PubMed] [Google Scholar]
- 78.George Priya Doss C., Rajith B. and Chakraborty C. (2013) Predicting the impact of deleterious mutations in the protein kinase domain of FGFR2 in the context of function, structure, and pathogenesis—a bioinformatics approach. Appl. Biochem. Biotechnol. 170, 1853–1870 10.1007/s12010-013-0315-y [DOI] [PubMed] [Google Scholar]
- 79.Bunney T.D., Inglis A.J., Sanfelice D., Farrell B., Kerr C.J., Thompson G.S. et al. (2018) Disease variants of FGFR3 reveal molecular basis for the recognition and additional roles for Cdc37 in Hsp90 chaperone system. Structure 26, 446–458.e8 10.1016/j.str.2018.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verba K.A. and Agard D.A. (2017) How Hsp90 and Cdc37 lubricate kinase molecular switches. Trends Biochem. Sci. 42, 799–811 10.1016/j.tibs.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A. et al. (2012) Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235 10.1126/science.1220834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu Y.-M., Su F., Kalyana-Sundaram S., Khazanov N., Ateeq B., Cao X. et al. (2013) Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 3, 636–647 10.1158/2159-8290.CD-13-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pal K., Bandyopadhyay A., Zhou X.E., Xu Q., Marciano D.P., Brunzelle J.S. et al. (2017) Structural basis of TPR-mediated oligomerization and activation of oncogenic fusion kinases. Structure 25, 867–877.e3 10.1016/j.str.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams S V., Hurst C.D. and Knowles M.A. (2013) Oncogenic FGFR3 gene fusions in bladder cancer. Hum. Mol. Genet. 22, 795–803 10.1093/hmg/dds486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Porta R., Borea R., Coelho A., Khan S., Araújo A., Reclusa P. et al. (2017) FGFR a promising druggable target in cancer: molecular biology and new drugs. Crit. Rev. Oncol. Hematol. 113, 256–267 10.1016/j.critrevonc.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 86.Chae Y.K., Ranganath K., Hammerman P.S., Vaklavas C., Mohindra N., Kalyan A. et al. (2017) Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget 8, 16052–16074 10.18632/oncotarget.14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Touat M., Ileana E., Postel-Vinay S., André F. and Soria J.-C. (2015) Targeting FGFR signaling in cancer. Clin. Cancer Res. 21, 2684–2694 10.1158/1078-0432.CCR-14-2329 [DOI] [PubMed] [Google Scholar]
- 88.Klein T., Vajpai N., Phillips J.J., Davies G., Holdgate G.A., Phillips C. et al. (2015) Structural and dynamic insights into the energetics of activation loop rearrangement in FGFR1 kinase. Nat. Commun. 6, 7877 10.1038/ncomms8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tucker J.A., Klein T., Breed J., Breeze A.L., Overman R., Phillips C. et al. (2014) Structural insights into FGFR kinase isoform selectivity: diverse binding modes of AZD4547 and ponatinib in complex with FGFR1 and FGFR4. Structure 22, 1764–1774 10.1016/j.str.2014.09.019 [DOI] [PubMed] [Google Scholar]
- 90.Davis M.I., Hunt J.P., Herrgard S., Ciceri P., Wodicka L.M., Pallares G. et al. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 10.1038/nbt.1990 [DOI] [PubMed] [Google Scholar]
- 91.O'Hare T., Shakespeare W.C., Zhu X., Eide C.A., Rivera V.M., Wang F. et al. (2009) AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell 16, 401–412 10.1016/j.ccr.2009.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trudel S., Li Z.H., Wei E., Wiesmann M., Chang H., Chen C. et al. (2005) CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood 105, 2941–2948 10.1182/blood-2004-10-3913 [DOI] [PubMed] [Google Scholar]
- 93.Bello E., Colella G., Scarlato V., Oliva P., Berndt A., Valbusa G. et al. (2011) E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 71, 1396–1405 10.1158/0008-5472.CAN-10-2700 [DOI] [PubMed] [Google Scholar]
- 94.Hilberg F., Roth G.J., Krssak M., Kautschitsch S., Sommergruber W., Tontsch-Grunt U. et al. (2008) BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res. 68, 4774–4782 10.1158/0008-5472.CAN-07-6307 [DOI] [PubMed] [Google Scholar]
- 95.Hall T.G., Yu Y., Eathiraj S., Wang Y., Savage R.E., Lapierre J.-M. et al. (2016) Preclinical activity of ARQ 087, a novel inhibitor targeting FGFR dysregulation. PLoS ONE 11, e0162594 10.1371/journal.pone.0162594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gavine P.R., Mooney L., Kilgour E., Thomas A.P., Al-Kadhimi K., Beck S. et al. (2012) AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 72, 2045–2056 10.1158/0008-5472.CAN-11-3034 [DOI] [PubMed] [Google Scholar]
- 97.Zhao G., Li W.-Y., Chen D., Henry J.R., Li H.-Y., Chen Z. et al. (2011) A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol. Cancer Ther. 10, 2200–2210 10.1158/1535-7163.MCT-11-0306 [DOI] [PubMed] [Google Scholar]
- 98.Guagnano V., Furet P., Spanka C., Bordas V., Le Douget M., Stamm C. et al. (2011) Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 54, 7066–7083 10.1021/jm2006222 [DOI] [PubMed] [Google Scholar]
- 99.Nakanishi Y., Akiyama N., Tsukaguchi T., Fujii T., Sakata K., Sase H. et al. (2014) The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Mol. Cancer Ther. 13, 2547–2558 10.1158/1535-7163.MCT-14-0248 [DOI] [PubMed] [Google Scholar]
- 100.Perera T.P.S., Jovcheva E., Mevellec L., Vialard J., De Lange D., Verhulst T. et al. (2017) Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Mol. Cancer Ther. 16, 1010–1020 10.1158/1535-7163.MCT-16-0589 [DOI] [PubMed] [Google Scholar]
- 101.Liu P.C., Wu L., Koblish H., Bowman K., Zhang Y., Klabe R. et al. (2015) Abstract 771: preclinical characterization of the selective FGFR inhibitor INCB054828. Cancer Res. 75, 771–771 10.1158/1538-7445.AM2015-771 [DOI] [Google Scholar]
- 102.Collin M.-P., Lobell M., Hübsch W., Brohm D., Schirok H., Jautelat R. et al. (2018) Discovery of rogaratinib (BAY 1163877): a pan-FGFR inhibitor. ChemMedChem 13, 437–445 10.1002/cmdc.201700718 [DOI] [PubMed] [Google Scholar]
- 103.Porta D.G., Weiss A., Fairhurst R.A., Wartmann M., Stamm C., Reimann F. et al. (2017) Abstract 2098: NVP-FGF401, a first-in-class highly selective and potent FGFR4 inhibitor for the treatment of HCC. Cancer Res. 77, 2098–2098 10.1158/1538-7445.AM2017-2098 [DOI] [Google Scholar]
- 104.Lamont F.R., Tomlinson D.C., Cooper P.A., Shnyder S.D., Chester J.D. and Knowles M.A. (2011) Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br. J. Cancer 104, 75–82 10.1038/sj.bjc.6606016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mohammadi M., Froum S., Hamby J.M., Schroeder M.C., Panek R.L., Lu G.H. et al. (1998) Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 17, 5896–5904 10.1093/emboj/17.20.5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Panek R.L., Lu G.H., Klutchko S.R., Batley B.L., Dahring T.K., Hamby J.M. et al. (1997) In vitro biological characterization and antiangiongenic effects of PD 1666866, a selective inhibitor of the FGF-1 receptor tyrosine kinase. J. Pharmacol. Exp. Ther. 283, 1433–1444 PMID: [PubMed] [Google Scholar]
- 107.Bono F., De Smet F., Herbert C., De Bock K., Georgiadou M., Fons P. et al. (2013) Inhibition of tumor angiogenesis and growth by a small-molecule multi-FGF receptor blocker with allosteric properties. Cancer Cell 23, 477–488 10.1016/j.ccr.2013.02.019 [DOI] [PubMed] [Google Scholar]
- 108.Andre, F., Ranson, M., Dean, E., Varga, A., van der Noll, R., Stockman, P.K et al. (2013) Abstract LB-145: results of a phase I study of AZD4547, an inhibitor of fibroblast growth factor receptor (FGFR), in patients with advanced solid tumors. Cancer Res. 73, LB-145-LB-145 10.1158/1538-7445.AM2013-LB-145 [DOI] [Google Scholar]
- 109.Xie L., Su X., Zhang L., Yin X., Tang L., Zhang X. et al. (2013) FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin. Cancer Res. 19, 2572–2583 10.1158/1078-0432.CCR-12-3898 [DOI] [PubMed] [Google Scholar]
- 110.Zhang J., Zhang L., Su X., Li M., Xie L., Malchers F. et al. (2012) Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin. Cancer Res. 18, 6658–6667 10.1158/1078-0432.CCR-12-2694 [DOI] [PubMed] [Google Scholar]
- 111.Chell V., Balmanno K., Little A.S., Wilson M., Andrews S., Blockley L. et al. (2013) Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene 32, 3059–3070 10.1038/onc.2012.319 [DOI] [PubMed] [Google Scholar]
- 112.Brameld K.A., Owens T.D., Verner E., Venetsanakos E., Bradshaw J.M., Phan V.T. et al. (2017) Discovery of the irreversible covalent FGFR inhibitor 8-(3-(4-Acryloylpiperazin-1-yl)propyl)-6-(2,6-dichloro-3,5-dimethoxyphenyl)-2-(methylamino)pyrido[2,3- d ]pyrimidin-7(8 H)-one (PRN1371) for the treatment of solid tumors. J. Med. Chem. 60, 6516–6527 10.1021/acs.jmedchem.7b00360 [DOI] [PubMed] [Google Scholar]
- 113.Katoh M. (2016) FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med. 38, 3–15 10.3892/ijmm.2016.2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joshi J.J., Coffey H., Corcoran E., Tsai J., Huang C.-L., Ichikawa K. et al. (2017) H3B-6527 is a potent and selective inhibitor of FGFR4 in FGF19-driven hepatocellular carcinoma. Cancer Res. 77, 6999–7013 10.1158/0008-5472.CAN-17-1865 [DOI] [PubMed] [Google Scholar]
- 115.Hagel M., Miduturu C., Sheets M., Rubin N., Weng W., Stransky N. et al. (2015) First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 5, 424–437 10.1158/2159-8290.CD-14-1029 [DOI] [PubMed] [Google Scholar]
- 116.Tan L., Wang J., Tanizaki J., Huang Z., Aref A.R., Rusan M. et al. (2014) Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc. Natl Acad. Sci. U.S.A. 111, E4869–E4877 10.1073/pnas.1403438111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meric-Bernstam, F., Arkenau, H.-T., Tran, B., Bahleda, R., Kelley, R.K, Hierro, C. et al. (2018) Efficacy of TAS-120, an irreversible fibroblast growth factor receptor (FGFR) inhibitor, in cholangiocarcinoma patients with FGFR pathway alterations who were previously treated with chemotherapy and other FGFR inhibitors [Internet]. Accessible at: http://www.taihoposters.com/ESMO-GI2018/ESMO-GI18_TPU-TAS-120_101_Meric-Bernstam.pdf [cited Jun 29, 2018]
- 118.Hoeflich, K. (2015) BLU-554, a novel, potent and selective inhibitor of FGFR4 for the treatment of liver cancer [Internet]. Accessible at: http://media.ilc-congress.eu/wp-content/uploads/2015/04/Dr-Klaus-Hoeflich_Friday-Press-Conference.pdf [cited Jun 29, 2018]
- 119.Kim R., Sharma S., Meyer T., Sarker D., Macarulla T., Sung M. et al. (2016) First-in-human study of BLU-554, a potent, highly selective FGFR4 inhibitor designed for hepatocellular carcinoma (HCC) with FGFR4 pathway activation. Eur J Cancer 69, S41 10.1016/S0959-8049(16)32704-6 [DOI] [Google Scholar]
- 120.Pierce K.L., Deshpande A.M., Stohr B.A., Gemo A.T., Patil N.S., Brennan T.J. et al. (2014) FPA144, a humanized monoclonal antibody for both FGFR2-amplified and nonamplified, FGFR2b-overexpressing gastric cancer patients. J. Clin. Oncol. 32, e15074 10.1200/jco.2014.32.15_suppl.e15074 [DOI] [Google Scholar]
- 121.Sommer A., Kopitz C., Schatz C.A., Nising C.F., Mahlert C., Lerchen H.-G. et al. (2016) Preclinical efficacy of the auristatin-based antibody–drug conjugate BAY 1187982 for the treatment of FGFR2-positive solid tumors. Cancer Res. 76, 6331–6339 10.1158/0008-5472.CAN-16-0180 [DOI] [PubMed] [Google Scholar]
- 122.Schatz C.A., Kopitz C., Wittemer-Rump S., Sommer A., Lindbom L., Osada M. et al. (2014) Abstract 4766: pharmacodynamic and stratification biomarker for the anti-FGFR2 antibody (BAY1179470) and the FGFR2-ADC. Cancer Res. 74, 4766–4766 10.1158/1538-7445.AM2014-4766 [DOI] [Google Scholar]
- 123.Eli Lilly and Company (2016) FGFR-3 Antibody-Drug Conjugate LY3076226 [Internet]. Accessible at: http://lillyoncologypipeline.com/assets/pdf/fgfr_3_antibody_drug_conjugate.pdf [cited Aug 8, 2018]
- 124.Trudel S., Bergsagel P.L., Singhal S., Niesvizky R., Comenzo R.L., Bensinger W.I. et al. (2012) A phase I study of the safety and pharmacokinetics of escalating doses of MFGR1877S, a fibroblast growth factor receptor 3 (FGFR3) antibody, in patients with relapsed or refractory t(4;14)-positive multiple myeloma. Blood 120, 4029 [Google Scholar]
- 125.Blackwell C., Sherk C., Fricko M., Ganji G., Barnette M., Hoang B. et al. (2016) Inhibition of FGF/FGFR autocrine signaling in mesothelioma with the FGF ligand trap, FP-1039/GSK3052230. Oncotarget 7, 39861–39871 10.18632/oncotarget.9515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Herbert C., Schieborr U., Saxena K., Juraszek J., De Smet F., Alcouffe C. et al. (2013) Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling. Cancer Cell 23, 489–501 10.1016/j.ccr.2013.02.018 [DOI] [PubMed] [Google Scholar]
- 127.Yu X.X., Watts L.M., Manchem V.P., Chakravarty K., Monia B.P., McCaleb M.L. et al. (2013) Peripheral reduction of FGFR4 with antisense oligonucleotides increases metabolic rate and lowers adiposity in diet-induced obese mice. PLoS ONE 8, e66923 10.1371/journal.pone.0066923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang J., Mikse O., Liao R.G., Li Y., Tan L., Janne P.A. et al. (2015) Ligand-associated ERBB2/3 activation confers acquired resistance to FGFR inhibition in FGFR3-dependent cancer cells. Oncogene 34, 2167–2177 10.1038/onc.2014.161 [DOI] [PMC free article] [PubMed] [Google Scholar]