Abstract
Streptococcus pneumoniae’s polysaccharide capsule is an important virulence factor; vaccine-induced immunity to specific capsular polysaccharide effectively prevents disease. Serotype 1 S. pneumoniae is rarely found in healthy persons, but is highly invasive and a common cause of meningitis outbreaks and invasive disease outside of the United States. Here we show that genes for polysaccharide capsule similar to those expressed by pneumococci were commonly detected by polymerase chain reaction among upper respiratory tract samples from older US adults not carrying pneumococci. Serotype 1-specific genes were predominantly detected. In five oropharyngeal samples tested, serotype 1 gene belonging to S. mitis expressed capsules immunologically indistinct from pneumococcal capsules. Whole genome sequencing revealed three distinct S. mitis clones, each representing a cps1 operon highly similar to the pneumococcal cps1 reference operon. These findings raise important questions about the contribution of commensal streptococci to natural immunity against pneumococci, a leading cause of mortality worldwide.
Introduction
Streptococcus pneumoniae’s polysaccharide capsule is believed to be its most important virulence factor, and natural or vaccine-induced immunity to the capsule is protective against disease. The capsular polysaccharide structure is highly variable, with over 90 different pneumococcal serotypes identified. In the early 20th century, serotype 1 was the most commonly identified serotype causing pneumococcal pneumonia and sepsis in the United States1. However, over the last few decades and well before the introduction of a 13-valent pneumococcal conjugate vaccine (PCV13) targeting serotype 1 into the national infant immunization program in 2010, serotype 1 became a rare cause of pneumococcal disease in the US population, with only sporadic increases being reported in Utah2,3. Among 42,059 cases of invasive pneumococcal disease (IPD) identified through Active Bacterial Core surveillance (ABCs), part of the Centers for Disease Control and Prevention’s Emerging Infections Program, from 1998 through 2009 (before PCV13 introduction), 609 (1.4%) were due to serotype 1 (CDC unpublished data).
At the same time, serotype 1 is a common cause of severe disease in several countries outside the United States. In African countries, serotype 1 remains a leading cause of IPD with the potential to cause large outbreaks and epidemics, even in countries that have already introduced PCV13, as recently reported in Ghana4–8. In England and Wales, serotype 1 was among the most common serotypes causing invasive disease in cases reported from 2008 to 20149. Serotype 1 pneumococci are significantly more resistant to opsonization and complement deposition compared to some other serotypes, which could explain the high degree of invasiveness of this serotype and its potential to cause epidemics10. Conversely, serotype 1 pneumococci rarely cause asymptomatic colonization of the upper respiratory tract in children or adults11,12. The reason that pneumococcal serotype 1 disease is now unusual in the United States but continues to commonly cause disease elsewhere is not understood; whether natural exposure to the serotype 1 capsule varies between populations is unknown.
In this report, we describe commensal streptococci – Streptococcus mitis – expressing pneumococcal serotype 1 capsule, which were discovered during a 2015–2016 study of pneumococcal colonization in US adults aged 65 years and older. Our results suggest that serotype 1-containing S. mitis may be common among US adults. In addition, we describe results of tests of genetic similarity and serologic cross-reactivity with serotype 1 pneumococci.
Methods
Study Design and Population
We conducted a cross-sectional survey from July 13, 2015 through March 31, 2016 among healthy adults 65 years of age and older enrolled from outpatient clinics, senior residential communities, senior day care centers and health fairs in four US states participating in the Centers for Disease Control and Prevention’s (CDC) Active Bacterial Core surveillance: Georgia, Maryland, New York and Tennessee. We excluded adults if they reported severe immunocompromising conditions (e.g., hematologic malignancies, transplant recipient, end-stage renal disease)13 or if they were residents in nursing homes or skilled nursing facilities.
For each eligible adult who consented to participate, we collected one nasopharyngeal (NP) and one oropharyngeal (OP) swab and conducted an interview to obtain demographic, clinical and vaccination data. Participants without paired NP and OP specimens were excluded from the study. Trained staff followed up with healthcare providers to confirm PCV13 receipt and dates of immunization.
The study was approved by the institutional review boards of the Centers for Disease Control and Prevention (CDC – Protocol #6725), Emory University, Georgia Department of Public Health, Maryland Department of Health, Johns Hopkins School of Public Health, University of Rochester, Vanderbilt University, and Tennessee Department of Health. Written informed consent was obtained from all study participants, and the research was performed according to federal regulations for protection of human subjects.
Specimen collection and processing
Trained staff collected NP and OP specimens using sterile nylon tipped flocked swabs (Copan Diagnostics INC, Murrieta, CA). NP and OP specimens were placed separately into cryovials containing 1.0 ml skim milk-tryptone-glucose-glycerol (STGG) medium, vortexed for 10 to 20 seconds to disperse the organisms from the swab, and frozen at −70 °C within 4 hours of collection.
Each swab was processed separately. For pneumococcal isolation and identification, 200 µl of either STGG-NP or STGG-OP inoculated medium was transferred into 5.0 ml Todd Hewitt broth containing 0.5% yeast extract (THY) and 1 ml of rabbit serum, which was then incubated at 37 °C in a CO2 incubator for 5–6 hours. After incubation, 10 µl of cultured broth was streaked on tryptone soy agar plates with 5% sheep blood (BAP) and incubated at 37 °C in CO2 for 18–24 hours14. Alpha-hemolytic colonies resembling pneumococci were tested for susceptibility to optochin and bile solubility, and serotyped by Quellung using CDC pneumococcal typing antisera15. Pneumococcal culture-negative NP and OP specimens underwent real-time PCR targeting lytA for detection of pneumococcal DNA16. DNA extraction was performed using a MagNA Pure Compact with an external lysis protocol using buffer #4 (Roche isolation kit III) or NucliSENS® EasyMag® automated system for total nucleic acid extraction, according to manufacture instructions. Specimens were considered to be lytA-positive if the cycle threshold (Ct) value was ≤35. Real-time multiplex PCR for detection of 37 serotypes (including PCV13 serotypes) was performed on culture-negative specimens from 53 and 395 adults with lytA-positive and lytA-negative specimens, respectively. For lytA-negative specimens, we assumed that 30% (+/−5%) would have a serotype detected by real-time PCR leading to a target sample size of 395; while for lytA-positive specimens almost all of them (71%) were tested. PCR for serotyping was performed using DNA extracted from 200 µl of each NP and OP swab-inoculated STGG media17. Because real-time PCR for serotyping is a multiplex reaction, Ct ≤35 for a specific serotype was considered positive, while Ct values between 36 and 40 were considered equivocal and repeated three times for confirmation.
Non-pneumococcal species isolation and characterization
The five specimens with the lowest PCR cycle threshold for serotype 1-specific wzy1 gene, which lies within the pneumococcal cps1 operon, were cultured using broth enrichment followed by plating on blood agar for isolation of alpha-hemolytic streptococcal species; these five were all OP specimens, two lytA positive and three lytA negative. Non-pneumococcal alpha-hemolytic streptococci colonies grown on the plates that were optochin resistant and bile insoluble were screened for wzy1 by real-time and conventional multiplex PCR. wzy1-positive non-pneumococcal isolates (N = 5, 1 from each specimen) were subjected to lytA screening by PCR, immunochromatographic testing (BinaxNow), and whole genome sequencing on Illumina MiSeq18. We compared the sequences of the 16,000 bp non-pneumococcal streptococci cps1 operons and flanking regions with the corresponding sequence of the 22,000 bp pneumococcal serotype 1 reference strain (GenBank accession CR931632) using the SSEARCH program19.
Species assignment was done by phylogenetic analysis of seven previously described concatenated housekeeping gene sequences (total length of 3063 bases)20. A selection of 3063 bp sequences20 that most closely matched the non-pneumococcal streptococcal strains were used to construct the phylogenetic tree. In addition, reference strain sequences were obtained from NCBI accessions for S. pseudopneumoniae (ATCCBAA960), S. mitis (B6 and NCTC 12261), S. infantis (ATCC 700779), and S. oralis (Uo5). Evolutionary distances were computed using the Maximum Composite Likelihood method21 and analyzed in MEGA722.
Capsule expression of non-pneumococcal streptococci isolates
Latex agglutination followed by the Quellung reaction using polyclonal serotype 1 S. pneumoniae typing antiserum prepared in rabbits was performed on the five isolates to assess capsule expression and compare capsule expression with that of S. pneumoniae serotype 1 (positive control). In addition, double immunodiffusion assays using the same antiserum assessed cross-reactivity of non-pneumococcal streptococci with serotype 1 pneumococcal antisera using the methods described by Sorensen et al.23. Briefly, both untreated (i.e. crude) bacterial suspension extracts and extracts treated with proteinase K were tested. The double immunodiffusion immunoprecipitation reaction was carried out on Pierce standard Ouchterlony agarose gel plates (Pierce #31111, Thermofisher Scientific) as previously described24,25. We applied 15 µl undiluted serotype 1 pneumococcal antiserum (AS) to the center well (2.8 mm) with 15 µl extract samples applied to the four surrounding wells, placed at a distance, edge to edge, of 3.2 mm. Two wells were left empty as controls. Plates were kept for 2 days at 5 °C and then photographed.
Opsonophagocytosis of pneumococcal and non-pneumococcal streptococci isolates
An opsonophagocytic killing assay26 was used to assess functional antibody activity against both the non-pneumococcal streptococci strains expressing serotype 1 capsular polysaccharide and the serotype 1 pneumococcus. S. mitis ATCC (NCTC 12261) strain (PCR negative for wzy1 gene) was used as a control. In a 96-well micro titer plate, 20 μl of S. mitis serotype 1 or serotype 1 pneumococcus was pre-opsonized with 10 μl anti-pneumococcal serotype 1 specific human polyclonal serum (8-dilutions, diluted 2-fold starting neat) or anti-S. mitis serotype 1 rabbit serum for 15 minutes at 37 °C with 5% CO2. After pre-opsonization, 10 μl baby rabbit complement was added followed by 40 μl of human promyelocyte (HL60) derived neutrophils (effector cells). Complement, neutrophil, and bacterial controls were maintained. After incubation for 45 minutes at 37 °C, 5 μl from each well was transferred onto air-dried Todd-Hewitt yeast extract agar. Opsonophagocytic titers were the reciprocal of the serum dilution with >50% killing compared with the growth in the complement control wells.
Data Analysis
Analyses were performed using SAS software, version 9.3. We calculated the prevalence of pneumococcal carriage as determined by culture. Among culture-negative patients, we calculated the prevalence of serotype genes detected by PCR among patients with lytA-positive or lytA-negative specimens. Comparison of the prevalence of pneumococcal vaccine-serotype homolog genes (serotypes 1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, 23F) by PCV13 status was done using the chi-square test. Patients were considered vaccinated if PCV13 was received at least 14 days before swab collection. Serotypes not distinguished by PCR (e.g. 9A/9V, 7A/7F, 18A/18B/18C/18F) were grouped.
The data that support the findings of this study are available from the corresponding authors upon reasonable request. The strains are stored in the CDC’s Streptococcus laboratory and can be available to investigators upon discussion and approval by the authors and site investigators. The method (composition of the strain with the purpose to induce immune response) is the subject of a U.S. Provisional Patent Application, No. 62/627,602 filed on February 7, 2018; non-exclusive royalty-free (NERF) academic research use licenses are available upon request.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Results
We enrolled 1,299 adults who had both NP and OP specimens successfully collected; of these, 67.9% were female, median age was 74 years (range: 65–102 years), and 73.9% were either recruited in Georgia or New York. Pneumococcus was isolated from specimens from 16 (1.2%) participants; 12 from NP, 2 from OP and 2 from both NP and OP. Of the 1,283 participants with culture-negative samples, 75 (5.8%) had a specimen that was lytA-positive: 1 from NP only, 71 from OP only, and 3 from both NP and OP (Supplementary Material, Figure).
PCR-based serotype deduction was performed on both NP and OP specimens from 448 culture-negative adults. Among the 53 adults with lytA-positive specimens from either NP or OP, 8 (15.1%) had wzy1 detected in one or both specimens; among the 395 adults with lytA-negative specimens from both NP and OP, 41 (10.4%) had wzy1 detected. Of the 49 adults with wzy1-positive, culture-negative specimens, 42 (85.6%) had wzy1 detected only in the OP specimen, 4 (8.2%) in both NP and OP specimens, and 3 (6.1%) in the NP specimen only; 23 (46.9%) of these 49 were enrolled in Georgia, 11 (22.5%) in Tennessee, 10 (20.4%) in New York and five (10.2%) in Maryland.
Of the 53 and 395 adults with lytA-positive and lytA-negative specimens, 27(50.9%) and 131(33.2%) were positive by PCR for genes for other pneumococcal serotypes; although overall wzy1 was most commonly detected (Table 1). Compared to NP specimens, OP specimens were more likely to be PCR-positive for serotype-specific pneumococcal genes in adults with lytA-positive (50.9% [27/53] vs. 3.8% [2/53]; P < 0.001) or lytA-negative (32.7% [129/395] vs. 2.3% [9/395]; P < 0.001) samples. From the 488 culture-negative adults with samples that underwent PCR for serotype genes, 424 had documented vaccination history and 251 received PCV13. Adults who were vaccinated with PCV13 were less likely to have pneumococci vaccine-type genes detected in their specimens compared to those who were unvaccinated (26.0% [45/173] vs. 35.8% [90/251]; P = 0.03).
Table 1.
PCR serotyping results from pneumococcal culture-negative specimens by lytA status, USA, 2015–2016.
| PCR Serotyping Results* | lytA positive specimens | lytA negative specimens |
|---|---|---|
| (N = 53) | (N = 395) | |
| n (%) | n (%) | |
| 001 | 8 (15.1) | 41 (10.4) |
| 004 | 9 (16.9) | 37 (9.4) |
| 9V/9A# | 6 (11.3) | 32 (8.1) |
| 005 | 1 (1.9) | 11 (2.8) |
| 18A/18B/18C/18F# | 7 (13.2) | 25 (6.3) |
| 23A | 2 (3.8) | 4 (1.0) |
| 19F | 1 (1.9) | 0 (0) |
| 14 | 0 | 2 (0.5) |
| 12F/12A/12B/44/46# | 1 (1.9) | 9 (2.3) |
| Others | 2 (3.8) | 13 (3.3) |
| Negative for the serotypes tested by PCR | 26 (49.1) | 264 (66.8) |
Specimens were nasopharyngeal and oropharyngeal samples from 448 adults aged ≥65 years; those from adults with lytA-positive PCR results from either specimen were classified as lytA positive; those with both specimens testing lytA negative were classified as lytA negative.
*Not mutually exclusive.
#PCR reaction groups these serotypes.
Isolation of non-pneumococcal strains and characterization
Streptococcus mitis was identified from culture from all 5 of the OP specimens, three lytA-negative (ID# L006, L115, L116) and two lytA-positive (ID# L121, L164), with the lowest cycle threshold values for wzy1 (between 22.0 and 27.5). The S. mitis isolates were positive for the serotype 1-specific wzy1 gene by both real-time and conventional PCR, and by latex agglutination for serotyping (Table 2). Two S. mitis isolates were Binax positive.
Table 2.
Results of characterization analyses for Streptoccocus mitis strains.
| Isolate ID | Type of Sample | lytA of OP specimen | Species isolated | Optochin | lytA of S. mitis isolate | Immunochro-matographic testing (BinaxNow) | Serotyping assays | Capsule visualization by Quellung using pneumococcal anti-serum | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Real-time PCR | Conventional PCR | Latex agglutination | ||||||||
| L006 | OP | Negative | S. mitis | Resistant | Negative | Negative | Serotype 1 | Serotype 1 | Serotype 1 | Weak visualization of capsule with serotype 1 anti-serum |
| L115 | OP | Negative | S. mitis | Resistant | Negative | Positive | Serotype 1 | Serotype 1 | Serotype 1 (weak reaction) | No visualization |
| L116 | OP | Negative | S. mitis | Resistant | Negative | Positive | Serotype 1 | Serotype 1 | Serotype 1 (weak reaction) | No visualization |
| L121 | OP | Positive (Ct = 31.19) | S. mitis | Resistant | Negative | Negative | Serotype 1 | Serotype 1 | Serotype 1 | Weak visualization of capsule with serotype 1 anti-serum |
| L164 | OP | Positive (Ct = 28.98) | S. mitis | Resistant | Negative | Negative | Serotype 1 | Serotype 1 | Serotype 1 (weak reaction) | Weak visualization of capsule with serotype 1 anti-serum |
Ct = cycle threshold.
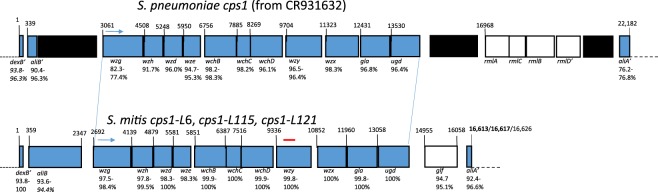
Whole genome sequence analysis of these five S. mitis isolates revealed three very similar, although distinct, capsule gene operons and adjacent genes. The lengths of the three loci, corresponding to the region between the flanking dexB and aliA segments of the 22,182 bp pneumococcal reference (GenBank accession CR931632), ranged between 16,613–16,626 bp and shared 98.0–98.2% sequence identity with one another. Each of the 11 genes within the polysaccharide synthetic cluster (wzg – ugd) were highly homologous to the S. pneumoniae serotype1 counterparts and shared the same order (Fig. 1).
Figure 1.
Genomic sequencing results showing alignment of cps1 regions between serotype 1 S. pneumoniae and S. mitis. Comparison of cps1-L polysaccharide synthetic cluster regions from S. mitis strains L006/L164 (16,613 bp), L115/L116 (16,617 bp) and L121 (16,626 bp) with the corresponding pneumococcal cps1 sequence. White rectangles represent open reading frames not shared between the species. Top diagram depicts ranges of sequence identity between pneumococcal cps1 genes and the S. mitis counterparts depicted on the bottom diagram. Ranges of sequence identity between the S. mitis alleles are given below S. mitis cps1 diagram. L006 and L164 shared identical 16613 sequence. L115 and L116 shared identical 16617 bp sequence. The connecting blue lines depict the alignment of the polysaccharide synthesis clusters. Black regions in the pneumococcal diagram depict remnants of transposase genes. Two inactive pneumococcal genes (aliB and rmlD, the latter renders the rhamnose biosynthesis cluster inactive) containing frameshift mutations are indicated, as well as the partial 5′ and 3′ ends of the dexB and aliA genes in both species (′). The region detected through use of PCR assays to first detect the presence of these strains in upper respiratory specimens is depicted by the red line above the wzy gene.
Three distinct S. mitis clones were found (Fig. 2). While 16 different pneumococcal strains representing 6 different serotypes formed a tight cluster based upon phylogenetic analysis of 3,063 bp concatenated housekeeping gene segments, the S. mitis strains formed a much more divergent cluster. At the level of direct DNA sequence comparison, the 3 serotype 1 S. mitis clones shared 97.1–97.3% sequence identity over the 3,063 bp, while the 16 pneumococcal strains shared 99.5–99.9% sequence identity. These S. mitis strains were all macrolide-resistant (all contained erm-methylase or mef genes) and cotrimoxazole-resistant (alterations within folA and/or folP genes). As in pneumococci, the cps loci in the S. mitis strains were situated between the penicillin binding protein genes pbp2x and pbp1a. All five S. mitis strains lacked the pneumococcal-specific target piaA but contained pneumolysin genes with 61–97% sequence identity to the pneumococcal ply gene; suggesting that piaA is pneumococcal-specific, whereas ply may be found within S. mitis strains with varying degrees of similarity with pneumococci.
Figure 2.
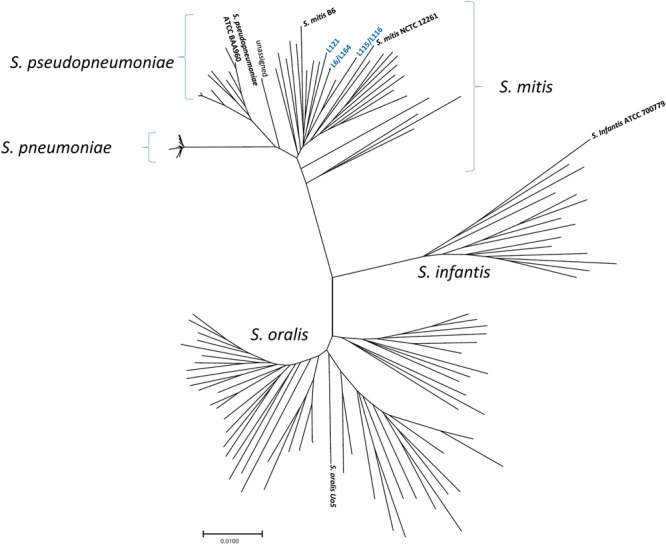
Species assignment of five Streptococcus mitis strains based upon phylogeny of concatenated housekeeping gene segments. Tree showing the positions of well-characterized strains described by Bishop et al.20 and the newly-identify S. mitis strains with pneumococcal cps1 homologues, shown in blue. Evolutionary distances were analyzed using MEGA 722.
Cross-reactivity between S. mitis and serotype 1 S. pneumoniae anti-serum and between S. pneumoniae serotype 1 and anti-S. mitis serotype 1 anti-serum
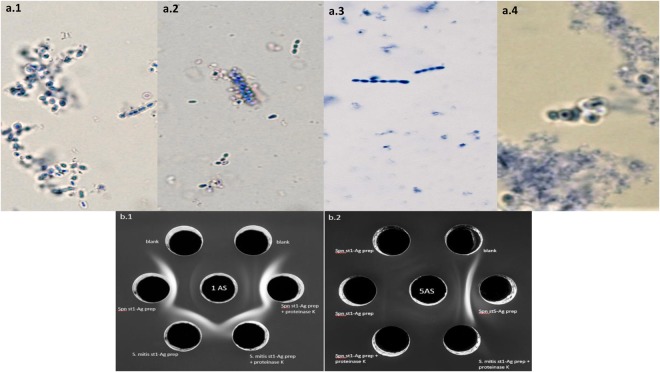
Although subtle, pneumococcal serotype 1 capsule was visualized by Quellung reaction in three of the five S. mitis strains (L006, L121, L164; Table 2, Fig. 3a). Strains L006 and L121 representing two of the three S. mitis clones were tested using double immunodiffusion assays and showed strong cross-reactivity with S. pneumoniae serotype 1 typing anti-serum. Figure 3b shows side-by-side S. mitis serotype 1 antigen and S. pneumoniae serotype 1 antigen that generated symmetrical and fused precipitates in reaction with serotype 1 typing S. pneumoniae antisera. Based on an opsonophagocytosis killing (OPK) assay, capsule expressed by serotype 1 S. mitis and S. pneumoniae strains both triggered anti-serotype 1 antibody mediated opsonophagocytosis, while the S. mitis control strain (wzy1-negative) did not. In addition, opsonophagocytosis of serotype 1 pneumococci was observed with anti-serotype 1 S. mitis serum obtained from rabbits using L006 strain (Fig. 4).
Figure 3.
(a) Quellung reaction using S. pneumoniae serotyping 1 anti-serum for (a.1) S. pneumoniae serotype 1 strain (positive control), (a.2) S. mitis serotype 1 (L121) strain, and (a.3) S. mitis non-serotype 1 (NCTC12261-negative control) strain – no capsule visualization; and Quellung reaction using S. mitis serotyping 1 rabbit anti-serum obtained from L121 strain for (a.4) S. pneumoniae serotype 1 strain. (b) Double immunodiffusion assay testing (b.1) cross-reactive of serotype 1 S. mitis and S. pneumoniae capsule extracts with serotype 1 S. pneumoniae typing anti-serum, and (b.2) control reaction using serotype 1 S. mitis and serotype 1 and non-serotype 1 S. pneumoniae extracts with serotype 5 S. pneumoniae typing anti-serum. Blank – wells with no bacteria; 1AS – serotype 1 S. pneumoniae anti-serum; Spn st1 – S. pneumoniae serotype 1; 5AS – serotype 5 S. pneumoniae anti-serum; Spn st5 – S. pneumoniae serotype 5 (S. mitis serotype 1 L006 strain used in reaction).
Figure 4.
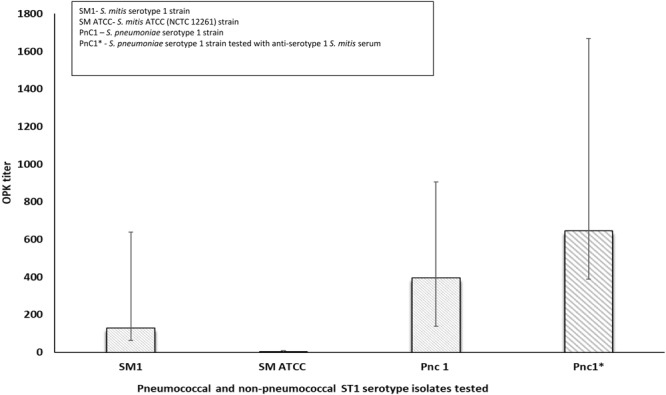
Opsonophagocytosis killing (OPK) testing of S. mitis and serotype 1 S. pneumoniae isolates using anti-serotype 1 pneumococcal and non-pneumococcal serum. Serotype 1 S. pneumoniae = positive control; S. mitis ATCC (NCTC 12261) strain confirmed to be wzy1-negative = negative control. Antibody source for OPK assay: 007 S. pneumoniae human reference serum. Complement: baby rabbit complement; OPK titer: reciprocal of serum dilution with >50% growth compared to serum free complement control. (S. mitis strain L006 and L121 representing two distinct S. mitis clones tested). Error bars represent the lower and upper limit of OPK titer based on the test runs.
Discussion
In our large, 4-site study we found that few older US adults carried pneumococci, as detected either by culture or PCR for lytA on NP and OP specimens. We observed that, in spite of this lack of pneumococci, about 1 in 3 older adults had genes for known pneumococcal polysaccharide serotypes detected by PCR, and the gene for serotype 1 (wzy1) was the type most commonly detected. The finding that genes for PCV13 serotypes were significantly less common among adults who had received PCV13 suggested that the genes detected by PCR were likely linked to organisms that expressed capsules that cross reacted with antibodies generated by PCV13. Indeed, we found that S. mitis identified from 5 wzy1-positive specimens expressed capsules that were nearly identical to pneumococcal serotype 1 capsules by a range of common detection assays, genetic sequencing of the cps locus, and serologic assays.
Over 10% of the adults without pneumococcus detected within their upper respiratory tracts screened positive for the wzy1. This observation, combined with the characterization of the three distinct serotype 1 clones found in our study, suggests that a wide diversity of serotype 1 S. mitis strains may circulate within the adult US population. In our study population of adults aged over 64 years, carriage of wzy1-positive non-pneumococcal streptococci appeared to be more common than overall pneumococcal carriage. S. mitis colonization of the upper respiratory tracts persists from early infancy through adulthood, while S. pneumoniae colonization declines significantly with age27. In contrast to S. pneumoniae, S. mitis is only occasionally associated with disease28. Because the polysaccharide capsule is thought to be an important virulence factor for pneumococci, whether S. mitis that cause disease are typically those expressing a capsule should be explored. Other recent studies have shown that many S. mitis strains express a polysaccharide capsule, and animal models demonstrated that the capsule has a protective function in the clearance of S. mitis23,29,30. In addition, Engen et al.31 have recently identified similar and cross-reactive T helper cells (Th17) responses against commensal S. mitis and pathogenic S. pneumoniae. Exposure to S. mitis can induce T helper memory cells, which may lead to protection against pneumococcal carriage and disease31. The commensal S. mitis strains expressing a serotype 1 capsule that we found in healthy older US adults may induce a T cell response against serotype 1 S. pneumoniae, which in turn could theoretically provide protection against serotype 1 pneumococcal disease among colonized individuals. Our OPK assay results indicate that anti-serotype 1 pneumococcal human antibodies can clear S. mitis expressing serotype 1 capsule and that anti-serotype 1 S. mitis rabbit antibodies can clear serotype 1 pneumococci. If colonization with S. mitis serotype 1 strains induces an antibody response that can clear serotype 1 pneumococci, the notion that immunity induced by these relatively abundant and naturally occurring strains may explain the limited incidence of serotype 1 pneumococcal disease in the United States.
We found three distinct clones among the five S. mitis isolates. All three S. mitis clones shared wzy1 operons that were highly similar to wzy1 of the serotype 1 S. pneumoniae reference strain. We observed more genetic distance between the three S. mitis serotype 1 clones than among the 16 pneumococcal strains of different capsular serotypes, suggesting that S. mitis serotype 1 strain diversity is much higher than that observed between pneumococcal serotype 1 strains. Although some studies have suggested that capsular polysaccharides in S. pneumoniae evolved by importing relevant genes from a range of commensal Streptococcus species23,32, the cps1 locus may have been transferred from pneumococcal strains to S. mitis strains, given the high level of background diversity found among S. mitis with serotype 1 capsule genes, or have been derived from a species that is no longer circulating. Therefore, predicting the directionality of serotype capsule genes transfers between S. pneumoniae and S. mitis is difficult.
Differences in the distribution of pneumococcal serotypes causing disease have been observed among populations4. However, more work is needed to understand if the distribution of capsular polysaccharide serotypes in commensal streptococci affect the pneumococcal serotypes causing disease. Cross-reactivity between encapsulated S. mitis and S. pneumoniae has recently been reported by the Danish group using immunodiffusion assays23. However, serotype 1 S. mitis were not observed among the 66 S. mitis isolates examined, which were mostly from Denmark (N = 42) and non-vaccine serotypes. In a Kenya carriage study, we were also unable to detect wzy1–positive non-pneumococcal Mitis group strains from NP or OP specimens from 158 adults33. In both Denmark and Kenya, serotype 1 is a common serotype causing IPD, while in the United States this serotype has been an uncommon cause of disease in recent decades2,34,35.
Among our study participants, pneumococci were rarely isolated from OP specimens, yet these specimens were much more likely than NP specimens to be positive by PCR for vaccine-serotype polysaccharide capsule genes; often such specimens were lytA-negative, providing evidence that the source of the serotype genes were not pneumococci. The oral cavity contains a great diversity of streptococcal species, and pneumococcal PCR serotyping assays can often identify homologs among mitis group non-pneumococcal strains33. Pneumococcal carriage studies using PCR-based detection of serotype-specific genes directly on OP specimens can overestimate vaccine type pneumococcal carriage rates given the high prevalence of confounding non-pneumococcal species carrying homologs of pneumococcal-serotype specific genes. In addition, the lytA gene for pneumococcal detection is not entirely specific. A recent study from Portugal identified a lytA-positive S. pseudopneumoniae strain in sputum36. In our study, two of 224 non-pneumococcal streptococci colonies screened for lytA were positive (data not shown). The use of PCR-based detection directly on upper respiratory specimens is of questionable value for pneumococcal carriage studies designed to measure vaccine impact based on the high prevalence of serotype 1 and other serotype genes on specimens that did not contain pneumococci. Although OP specimens had higher positivity for wzy1, 7 NP specimens that were lytA-negative were wzy1-positive by real-time PCR. Other studies have also shown problems with real-time PCR for serotyping of NP specimens37.
In conclusion, we found that genes for polysaccharide capsules similar to those expressed by pneumococci, in particular genes for serotype 1, were common among our sample of older US adults. The serotype 1 genes belonged to S. mitis strains that expressed capsules immunologically indistinguishable from pneumococcal capsules. Our findings raise many questions, in particular about the role of commensal streptococci in the development and maintenance of natural immunity to pneumococci. Further work should focus on these questions and whether the presence of capsule-containing non-pneumococcal streptococci affects the serotype distribution and amount of disease caused by pneumococci in different population. These strains could provide a novel approach for enhancing the human microbiome’s ability to protect against serotype 1 pneumococcal disease, which is responsible for 11.7% of all IPD cases in Africa4 and continue to be the main serotype causing pneumococcal meningitis outbreaks in the African meningitis belt countries that have already introduced pneumococcal conjugate vaccine8. Given the genetic similarities between capsular loci, those working with molecular assays to evaluate pneumococcal serotypes in the upper respiratory tract should consider whether non-pneumococcal streptococci strains with capsular genes may affect their results.
Electronic supplementary material
Acknowledgements
The authors want to acknowledge the following personnel for their help with participant recruitment, specimen collection, questionnaire administration and vaccination follow-up: Amy Tunali and Stephanie Thomas from the Georgia Emerging Infections Program and Emory University School of Medicine; Mary Bower, Regina Mosley, Emily Presmanes, Sabrina Williams, and Nina McNair from Emory University; Anna Sharova, Jackie Langdon, and Rosemary Hollick from Johns Hopkins; Diane Kober, Gary Hollick and Christina Felsen from the University of Rochester School of Medicine and Dentistry; Lura McKnight and Gail Hughett from Vanderbilt Medical Center. We also acknowledge the following individuals from the CDC’s Streptococcus and Microbial Pathogenesis and Immune Response Laboratories: Iaci Moura, Shanda Larson and Anne-Kathryn Venero for providing technical support for culture and PCR testing; Sopio Chochua, Theresa Tran, and Hollis Walker for providing the S. mitis genomic sequences, Yuan Li for his interpretation of the potential ancestry of the cps1 loci, and Ellie Kim for providing technical support on the OPK assays. Finally, we are very grateful to Rex Howard and Will Thomas for their expert guidance with antisera preparation. This work was funded by the Emerging Infections Program (EIP) Cooperative Agreement between the four EIP sites and the Centers for Disease Control and Prevention (Grant# CDC-RFA-CK17-1701).
Author Contributions
F.C.L., N.G.R., M.M.F., G.R., M.G.C., B.B., C.G.W. contributed with the design of the study. F.C.L. and J.M. were responsible for training of study staff on data and specimen collection per protocol, and overall oversight and monitoring of study activities. N.G.R., N.M.B., K.T., L.H.H., M.M.F., J.W. implemented and oversaw the study activities at the study sites. F.P. and M.G.C. were responsible for processing all upper respiratory samples and for the isolation of S. mitis strains. R.G. performed the immunodiffusion assays and was responsible for the preparation of anti-Streptococcus mitis sera from rabbits. G.R. conducted and interpreted the opsonophagocytosis killing tests. B.B. was responsible for phylogenetic and whole genome sequence analyses and interpretation, and oversight of laboratory activities related to this study. All authors reviewed epidemiologic and laboratory results and provided input prior to manuscript development. F.C.L., B.B., C.G.W. were responsible for the initial draft of the manuscript. However, all authors critically reviewed and edited the manuscript.
Competing Interests
Dr. Lee Harrison has served on a scientific advisory board for GlaxoSmithKline, and has been a speaker at a Merck scientific symposium. However, none of these commitments had an influence on the design and interpretation of the data. All other authors report no competing financial or non-financial interests related to the work described.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fernanda C. Lessa, Email: flessa@cdc.gov
Bernard Beall, Email: bbeall@cdc.gov.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35921-3.
References
- 1.Feikin DR, Klugman KP. Historical changes in pneumococcal serogroup distribution: implications for the era of pneumococcal conjugate vaccines. Clin Infect Dis. 2002;35:547–55. doi: 10.1086/341896. [DOI] [PubMed] [Google Scholar]
- 2.Moore MR, et al. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect Dis. 2015;3:301–9. doi: 10.1016/S1473-3099(14)71081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byington CL, et al. Temporal trends of invasive disease due to Streptococcus pneumoniae among children in the intermountain west: emergence of nonvaccine serogroups. Clin Infect Dis. 2005;41:21–9. doi: 10.1086/430604. [DOI] [PubMed] [Google Scholar]
- 4.Johnson, H. L. et al. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. Plos Med. 7, 10.1371/journal.pmed.1000348 (2010). [DOI] [PMC free article] [PubMed]
- 5.Scott JA, et al. Serotype distribution and prevalence of resistance to benzylpenicillin in three representative populations of Streptococcus pneumoniae isolates from the coast of Kenya. Clin Infect Dis. 1998;27:1442–50. doi: 10.1086/515013. [DOI] [PubMed] [Google Scholar]
- 6.Leimkugel J, et al. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J Infect Dis. 2005;192:192–9. doi: 10.1086/431151. [DOI] [PubMed] [Google Scholar]
- 7.Yaro S, et al. Epidemiological and molecular characteristics of a highly lethal pneumococcal meningitis epidemic in Burkina Faso. Clin Infect Dis. 2006;43:693–700. doi: 10.1086/506940. [DOI] [PubMed] [Google Scholar]
- 8.Kwambana-Adams BA, et al. An outbreak of pneumococcal meningitis among older children (≥5 years) and adults after the implementation of an infant vaccination programme with the 13-valent pneumococcal conjugate vaccine in Ghana. BMC Infect Dis. 2016;16:575. doi: 10.1186/s12879-016-1914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waight PA, et al. Effect of the 13-valent pneumococcal conjugate vaccine on invasive pneumococcal disease in England and Wales 4 years after its introduction: an observational cohort study. Lancet Infect Dis. 2015;15:535–43. doi: 10.1016/S1473-3099(15)70044-7. [DOI] [PubMed] [Google Scholar]
- 10.Melin M, et al. Serotype-related variation in susceptibility to complement deposition and opsonophagocytosis among clinical isolates of Streptococcus pneumoniae. Infect Immun. 2010;78:5252–61. doi: 10.1128/IAI.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai AP, et al. Decline in pneumococcal nasopharyngeal carriage of vaccine serotypes after the introduction of the 13-valent pneumococcal conjugate vaccine in children in Atlanta, Georgia. Pediatr Infect Dis. 2015;34:1168–74. doi: 10.1097/INF.0000000000000849. [DOI] [PubMed] [Google Scholar]
- 12.Conklin, L. M. et al. High Streptococcus pneumoniae colonization prevalence among HIV-infected Kenyan parents in the year before pneumococcal conjugate vaccine introduction. BMC Infect Dis. 18, 10.1186/s12879-015-1312-2 (2016). [DOI] [PMC free article] [PubMed]
- 13.Centers for Disease Control and Prevention (CDC) Use of 13-valent pneumococcal conjugate vaccine and 23-valent pneumococcal polysaccharide vaccine for adults with immunocompromising conditions: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Morb Mortal Wkly Rep. 2012;61:816–9. [PubMed] [Google Scholar]
- 14.da Gloria Carvalho M, et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J Clin Microbiol. 2010;48:1611–8. doi: 10.1128/JCM.02243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Austrian R. The quellung reaction, a neglected microbiologic technique. Mt Sinai J Med. 1976;43:699–709. [PubMed] [Google Scholar]
- 16.Carvalho MG, et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–6. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pimenta FC, et al. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol. 2013;51:647–52. doi: 10.1128/JCM.02927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metcalf BJ, et al. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect. 2016;22:1002.e1–1002.e8. doi: 10.1016/j.cmi.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Brenner SE, Chothia C, Hubbard TJ. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc Natl Acad Sci USA. 1998;95:6073–8. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop CJ, et al. Assigning strains to bacterial species via the internet. BMC Biol. 2009;26:3. doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skov Sørensen UB, Yao K, Yang Y, Tettelin H, Kilian M. Capsular polysaccharide expression in commensal streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. MBio. 2016;7:e01844–16. doi: 10.1128/mBio.01844-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oudin J. L’analyse immunochemique qualitative methode par diffusion des antigens au sein de l’immunoserum precipitant gelose. Ann Inst Pasteur. 1948;75:109–29. [PubMed] [Google Scholar]
- 25.Ouchterlony, O. Antigen-antibody reactions in gels and the practical applications of this phenomenon in laboratory diagnosis of diphtheria. Med Diss Stockholm (1949).
- 26.Romero-Steiner S, et al. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin Diagn Lab Immunol. 1997;4:415–22. doi: 10.1128/cdli.4.4.415-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granat SM, et al. Epidemiological evidence for serotype-independent acquired immunity to pneumococcal carriage. J Infect Dis. 2009;200:99–106. doi: 10.1086/599364. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell J. Streptococcus mitis: walking the line between commensalism and pathogenesis. Mol Oral Microbiol. 2011;26:89–98. doi: 10.1111/j.2041-1014.2010.00601. [DOI] [PubMed] [Google Scholar]
- 29.Rukke HV, et al. Protective role of the capsule and impact of serotype 4 switching on Streptococcus mitis. Infect Immun. 2014;82:3790–801. doi: 10.1128/IAI.01840-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rukke HV, Hegna IK, Petersen FC. Identification of a functional capsule locus in Streptococcus mitis. Mol Oral Microbiol. 2012;27:95–108. doi: 10.1111/j.2041-1014.2011.00635.x. [DOI] [PubMed] [Google Scholar]
- 31.Engen, S. A. et al. The oral commensal Streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross-reactive with Streptococcus pneumoniae. Plos One. 9, 10.1371/journal.pone.0104306 (2014). [DOI] [PMC free article] [PubMed]
- 32.Kilian, M., Riley, D. R., Jensen, A., Brüggemann, H. & Tettelin, H. Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. MBio. 22, 10.1128/mBio.01490-14 (2014). [DOI] [PMC free article] [PubMed]
- 33.Carvalho, M. G. et al. Non-pneumococcal mitis-group streptococci confound detection of pneumococcal capsular serotype-specific loci in upper respiratory tract. PeerJ. 1, 10.7717/peerj.97 (2013). [DOI] [PMC free article] [PubMed]
- 34.Mudhune S, Wamae M. Network Surveillance for Pneumococcal Disease in the East African Region. Report on invasive disease and meningitis due to Haemophilus influenzae and Streptococcus pneumoniae from the Network for Surveillance of Pneumococcal Disease in the East African Region. Clin Infect Dis. 2019;48(Suppl 2):S147–52. doi: 10.1086/596494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harboe ZB, et al. Impact of 13-valent pneumococcal conjugate vaccination in invasive pneumococcal disease incidence and mortality. Clin Infect Dis. 2014;59:1066–73. doi: 10.1093/cid/ciu524. [DOI] [PubMed] [Google Scholar]
- 36.Simões AS, et al. lytA-based identification methods can misidentify Streptococcus pneumoniae. Diagn Microbiol Infect Dis. doi. 2016;85:141–8. doi: 10.1016/j.diagmicrobio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Wyllie, A.L. et al. Molecular surveillance of nasopharyngeal carriage of Streptococcus pneumoniae in children vaccinated with conjugated polysaccharide pneumococcal vaccines. Sci Rep. 6, 10.1038/srep23809 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.