Abstract
The programmed death 1 receptor (PD-1) and its ligand (PD-L1) are key molecules of immune checkpoint mechanisms in cancer and actually represent one of the main targets of immunotherapy. The predictive and prognostic values of PD-L1 expression alone in cancer patients is currently under debate due to the methodological assessment of PD-L1 expression and its temporal variations. Better detailed studies about the molecular basis of immunotherapy biomarkers are necessary. Here we summarize the current knowledge of PD-L1 gene modifications at genetic and epigenetic levels in different tumors, thus highlighting their reported correlation with cellular processes and potential impact on patient outcomes.
Keywords: CD274/PD-L1, genomic aberrations, polymorphisms, epigenetic modulation
Introduction
Immune checkpoints are regulated by many cellular processes that are in part controlled by PD-1 (programmed death-1) through the interaction with PD-L1 (programmed death-ligand 1) and PD-L2 (programmed death-ligand 2). The aberrant PD-1/PD-L1 binding leads to the activation of a crucial self-tolerance pathway both in immune cells (ICs) and in tumor cells (TCs). Tumors hijack this crucial mechanism to escape immune elimination by deregulating survival and proliferation pathways.1 The translational impact of these findings is rapidly increasing and the stimulation of immune response by using immune checkpoint inhibitors (ICIs) is actually emerging as a dramatic paradigm shift in the treatment of advanced tumors.2 Several biological and clinical studies have shown that the inhibition of the interaction of PD-1 with PD-L1/PD-L2 ligands can overcome the intrinsic resistance to immune surveillance of TCs and may dramatically impact on patients’ outcomes.3,4
However, despite the remarkable success of immunotherapy clinical applications reported in recent years, the efficacy of the therapies appears variable across cancer patients and among different tumor types. The detection by immunohistochemistry (IHC) of PD-L1 protein on TC surface and/or tumor-infiltrating immune cells (TIICs) represents to date the main tool to predict response to Food and Drug Administration (FDA)-approved ICIs.5,6
However, the heterogeneous and time-related staining of PD-L1 within the same tumor and small specimens at different points in treatment may not be fully representative and does not allow absolute discrimination of the real receptorial status.7–10 Mechanisms regulating PD-L1 expression levels are multiple and many relevant questions remain unsolved in this context.
First, a marked efficacy of ICIs was reported in patients with tumors harboring a high tumor mutational burden (TMB) or microsatellite instability, such as melanoma, nonsmall cell lung cancer (NSCLC) and colorectal cancer (CRC).11–13 However, the most recent studies of correlation between TMB and response to immunotherapy suggest that the effects of ICIs could vary concomitant with a dynamic change of tumor DNA mutations.11,14–16 Second, the general evidence that comes from large clinical studies indicates that the predictive value of PD-L1 expression by IHC alone for the use of PD-1/PD-L1 inhibitors should not be exhaustive to stratify patients that could respond and benefit from immunotherapy.
Structural variations of the CD274/PD-L1 gene are emerging as a reasonable and relevant mechanism that can govern the increase of PD-L1 expression. Many fascinating scientific advances have been recently reported in this field. Alternatively, the epigenetic deregulation of PD-L1 is currently emerging in some tumors.
A better elucidation of the mechanisms that directly modulate the expression of the CD274/PD-L1 gene could help to explain and reduce the discrepancies.
CD274/PD-L1 gene structure
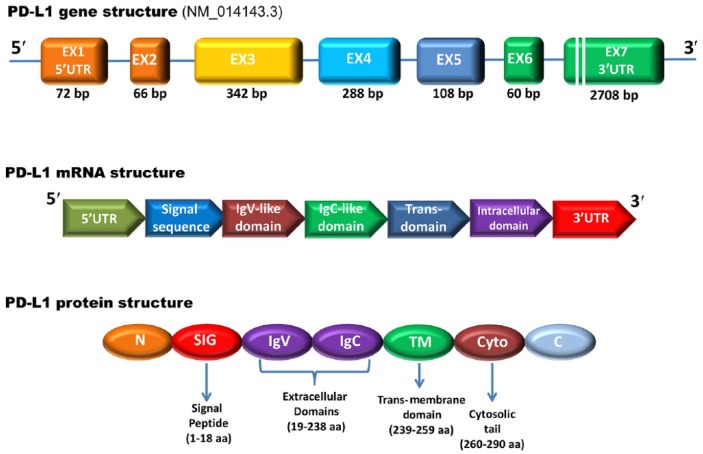
PD-L1, also known as B7 homolog 1 (B7-H1) or cluster of differentiation 274 (CD274), represents the first functionally characterized ligand of the co-inhibitory PD-1. PD-L1 is encoded by the CD274 gene (HGNC accession number: 17635; Ensembl Gene accession: ENSG00000120217), which is located in chromosome 9p24.1 and spans roughly 17.6 kb.17 It is expressed in different tissues, but mainly in activated T and B lymphocyte cells, dendritic cells, monocytes and various types of TCs. The CD274/PD-L1 gene is highly conserved: homologs were found along the vertebrate phylogeny (from Danio rerio to Primates), thereby suggesting its functional importance in many species.18 CD274/PD-L1 promoter has been found to retain CpG methylation sites along the 5′ untranslated region and exon 1, while translation starts from exon 2. Table 1 provides details about the genomic localization of functional elements at the 5′ end of the gene.
Table 1.
Genomic localization of functional elements of CD274/PD-L1 gene.
| Coordinates | Functional element | Transcript accession | Length |
|---|---|---|---|
| 5450410-5450629 | Predicted CpG island | 220 | |
| 5450525-5450596 | Exon 1 (5′ UTR) | NM_014143.3 | 72 |
| 5450503-5450596 | Exon 1 (5′ UTR) | NM_001267706.1 | 94 |
| 5456100-5456165 | Exon 2 | NM_014143.3, NM_001267706.1 | 66 |
| 5456114-5456116 | ATG (Exon 2) | NM_014143.3, NM_001267706.1 | 3 |
| 5457079-5457420 | Exon 3 | NM_014143.3 | 342 |
| 5462834-5463121 | Exon 4 | NM_014143.3 | 288 |
| Exon 3 | NM_001267706.1 | ||
| 5465499-5465606 | Exon 5 | NM_014143.3 | 108 |
| Exon 4 | NM_001267706.1 | ||
| 5466770-5466829 | Exon 6 | NM_014143.3 | 60 |
| Exon 5 | NM_001267706.1 | ||
| 5467860-5467862 | Stop codon (exon 7) |
NM_014143.3, NM_001267706.1 |
3 |
| 5467840-5470547 | Exon 7 + 3′-UTR | NM_014143.3 | 2708 |
| 5467840-5470566 | Exon 6 + 3′-UTR | NM_001267706.1 | 2727 |
Two main alternative transcripts are generated by the CD274/PD-L1 gene: the longest one (3.6 kbp; NCBI accession number: NM_014143.3, Ensembl accession: ENST00000381577.3) encodes for a 290 amino acid protein (NCBI: NP_054862), while the second one (3.3 kbp; NM_001267706.1) encodes for a 176 amino acid isoform (NP_001254635). The longest transcript comprises seven exons, with the coding sequence being approximately 800 bp in length. The encoded PD-L1 protein has a mass of 33 kDa, with two annotated immunoglobulin V-like (encoded by exon 2; amino acid residues: 19–127) and C-like (encoded by exon 3; residues: 133–225) domains, a hydrophobic transmembrane fragment and a cytoplasmic tail of 30 amino acids with a still unclear role in signal transduction (encoded by exons 4–7; residues 240–259, 260–290, respectively).17,21 Due to alternative splicing, the second transcript lacks the third exon, thus generating a shorter PD-L1 isoform with no IgV-like domain.
Similar to other genes that encode transcription factors and cytokines, the CD274/PD-L1 has a long 3′-UTR and a number of cis-acting elements involved in post-transcriptional regulation of mRNA decay, which is a major determinant of mRNA abundance, including an AU-rich element and potential microRNA-binding sites. Structures of the PD-L1 gene, mRNA and its encoded protein are represented in Figure 1.
Figure 1.
Schematic representation of the CD274/PD-L1 gene, mRNA and protein structural domains. The PD-L1 gene comprises seven exons and encodes a putative type I transmembrane protein of 290 amino acids. Exon 1 encodes the 5′ untranslated region (5′-UTR), whereas exon 7 encodes part of the intracellular domain and 3′-UTR of mRNA. The first 18 amino acids contain the signal peptide sequence, removed during protein processing. The PD-L1 protein consists of a large extracellular region that contains IgV-like and IgC-like domains, followed by a hydrophobic transmembrane domain and a cytosolic tail.
The genetic deregulation of CD274/PD-L1 in cancer
PD-L1 expression in cancer can be referred to different molecular mechanisms, some not rigorously genetic (indirect mechanisms) and others mainly genetic and epigenetic (direct mechanisms). In this context, two different representative modes in TCs were described: the innate-immune resistance and adaptive-immune resistance.
In innate-immune resistance, the upregulation of PD-L1 expression is a consequence of constitutive oncogenic signaling within TCs. Multiple mechanisms have been identified so far with regard to the former. The phosphatidylinositol-4,5-bisphosphate 3-kinase/serine/threonine kinase 1/mechanistic Target of Rapamycin (PIK3/AKT/mTOR) signaling represents one of the main pathways to control immune surveillance in several tumors. Phosphatase and tensin homolog (PTEN) loss and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutations induce the activation of the AKT–mTOR pathway and subsequently increase the PD-L1 expression in glioma, breast and prostate cancers.22–24 The same pathway appears to act in a synergistic manner via Janus kinase 2/signal transducer and activator of transcription (JAK/STAT) pathway in NSCLC.25 EGFR and anaplastic lymphoma kinase (ALK) are the master molecular targets of NSCLC targeted therapy. The role of EGFR as a strong independent predictive marker of PD-L1 overexpression was first established in 2013 using an EGFR-murine model of lung cancer and then corroborated in 2014 by analyzing a collection of 164 specimens of surgically resected NSCLCs.26,27 The in vitro inhibition of EGFR activity with erlotinib induces a downregulation of PD-L1 expression, thus corroborating the idea that PD-L1 expression is stimulated by EGFR signaling, enhanced by activating mutations in the EGFR gene.27 Moreover, the induction of PD-L1 expression was demonstrated in NSCLC harboring ALK rearrangements under alectinib treatment.28 The RAS/RAF/MEK/MAPK–ERK pathway was linked to activation of PD-L1 overexpression both in vitro and in vivo in melanoma and NSCLC cells, and pharmacological inhibition of MEK or knockdown of ERK1/2 was shown to reduce PD-L1 expression levels.29–31 More recently, CD274/PD-L1 copy number variations, point mutations and 3′-UTR disruptions have been highlighted as genetic mechanisms of PD-L1 deregulation.32–35
In the adaptive-immune resistance, PD-L1 expression is induced on TCs as a consequence of local inflammatory signals.36 Briefly, when tumor antigen-specific T cells recognize their related antigen expressed by cancer cells, signaling through the T cell receptor (TCR) leads to the expression of activation-induced regulatory receptors, such as PD-1, and to the production of cytokines that are aimed at amplifying the immune response and attracting other ICs, such as macrophages.37 Nevertheless, cytokines lead to the expression of PD-L1 on TCs and inflammatory cells. The interaction of PD-L1 with PD-1 receptor induces the formation of PD-1/TCR inhibitory microclusters, which recruit Src homology 2 domain-containing tyrosine phosphatase 2 (SHP2) molecules. SHP2 molecules induce the dephosphorylation of multiple members of the TCR signaling pathway and turn off T cell activation.37,38
Amplification/copy number gain of CD274/PD-L1
The main genetic mechanism underlying the aberrant PD-L1 expression described is the acquisition of DNA copy number alterations (CNAs) affecting the CD274/PD-L1 locus. CD274/PD-L1 CNAs, which affected focal regions, chromosome 9p24.1 or the whole chromosome 9, were identified across 22 major cancer types and were observed to correlate with PD-L1 mRNA expression changes in several tumor types of different datasets from The Cancer Genome Atlas (TCGA). Deletions of CD274/PD-L1 were generally more frequent than gains (31% versus 12%) and appear most prevalent in melanoma and NSCLC (>50%). Copy number gains (CNGs) most frequently occurred in sarcomas (8%), ovarian (10.7%), head and neck (8.6%), bladder (8.3%), cervical and endocervical tumors (7.1%) and CRCs (14%). Both CD274/PD-L1 amplifications and CD274/PD-L1 deletions were found to be associated with high mutation load.39,40 These findings obtained from TGCA cohorts were also confirmed in independent cohorts of untreated high-grade soft-tissue sarcomas (STS) and lung tumors.41 CD274/PD-L1 CNGs in NSCLC were first described by Goldmann and colleagues using fluorescent in situ hybridization (FISH) analysis in different disease stage specimens. Experiments disclosed CD274/PD-L1 CNGs in 5–9% of NSCLCs, with a perfect correspondence with IHC positivity and a higher than average score of PD-L1 IHC in the amplified cases.42 CD274/PD-L1 CNGs were most commonly observed in smokers and, strikingly, amplifications were found to appear exclusively in the EGFR mutations and ALK rearrangements, both of which were reported to be negatively associated with anti PD-1 (nivolumab, pembrolizumab) and PD-L1 (durvalumab, atezolizumab) immunotherapies.38,43 In NSCLCs, CD274/PD-L1 amplification were found to co-occur with JAK2 (9p24.1) amplification and this co-amplification was associated with PD-L1 expression.44 By contrast, despite the role of PTEN mutations in the cancer-immune context, no association with PTEN CNAs emerged in pulmonary carcinoma specimens. With a lower incidence, genomic rearrangements of CD274/PD-L1 were also described in pulmonary neuroendocrine tumors, though less detailed. A small subset of small cell lung carcinomas (SCLCs, 2%) were reported to bear 9p24 CNGs and focal, high-level amplifications of CD274/PD-L1 with a good correlation with high expression of PD-L1, thus suggesting an alternative mechanism to explain immune evasion in SCLC and sensitivity to ICIs.45 The methodological approach of combined FISH and IHC analyses was successfully used to establish the status of CD274/PD-L1 in squamous cell carcinoma (SqCC) of the cervix and vulva.46 CD274/PD-L1 amplification was observed in 67% of cervical SqCCs and 43% of vulvar SqCCs and was frequently observed in co-occurrence with amplification of the gene encoding for PD-L2 (CD273/PD-L2). Tumors showing co-amplification of CD274/PD-L1 and CD273/PD-L2 genes showed a median of PD-L1 protein expression much higher than tumors with gene disomy. In predominantly HPV-negative SqCC, a CD274/PD-L1 amplification was detected in 19% of cases (a high amplification was observed in 15% and low levels of amplification in 4% of cases).32 Interestingly, the gene amplification was concordant in primary tumors and associated metastases, with a concordance rate with positive PD-L1 immunostaining of 73%.47 On the other hand, by using IHC and in situ mRNA hybridization in breast cancer cell lines and tissues from triple-negative patients, it was showed that PD-L1 strong expression correlates with CD274/PD-L1 gene CNG and suggests a link with reduced mortality in patients.48
CD274/PD-L1 polymorphisms
Very few recent data are available concerning the impact of CD274/PD-L1 polymorphism on PD-L1 expression. The most relevant were two C>G changes recently reported by Tao and colleagues, localized into the UTR of CD274/PD-L1. The first polymorphism, described rs10815225, is located at the promoter region of the CD274/PD-L1 gene into the SP1 consensus sequence.34 SP1 is a zinc finger transcription factor that binds to GC-rich motifs of many promoters and is involved in many cellular processes, including cell differentiation, cell growth, apoptosis and immune responses.49 Abnormal SP1 expression and activation was detected in human gastric cancer and was inversely correlated with patient survival, suggesting that it may represent a potential molecular marker for poor prognosis.50,51 Variations in the expression of SP1 can result in a modification of PD-L1 expression in gastric cancer cells. By in vitro fluorescent assays, Tao and colleagues showed in fact that SP1 bound to rs10815225 mutant allele (G) of the CD274/PD-L1 promoter with more affinity than the wild-type allele, and that the expression level of PD-L1 mRNA in the mutant homozygous cancers was higher than in those heterozygotes for this polymorphism.34 Additionally, rs10815225 was found to be in near-complete linkage disequilibrium with the second functional-relevant polymorphism rs4143815, reported into the CD274/PD-L1 3′-UTR region, and the haplotype blocks of these two polymorphisms were also found to be markedly related to gastric cancer risk. Beside this synergic function, the polymorphic locus rs4143815 seems to contribute to the elevated PD-L1 protein expression in gastric cancer by disrupting the interaction between PD-L1 mRNA and miR-570.52
CD274/PD-L1 3′-UTR disruption
The 3′-UTR portion of many genes codifying for cytokines and transcription factors are generally involved in the decay of mRNAs, thus regulating their cellular levels.53 Using whole-genome and RNA sequencing approaches, Kataoka and colleagues identified in 2016 novel recurrent structural variations (SVs) disrupting the 3′-UTR of the CD274/PD-L1 gene, which may represent an additional important genetic mechanism of immune escape in tumors. The evaluation of TCGA data revealed in fact an extensive recurrence of these SVs in many common cancer types, comprising adult T cell leukemia/lymphoma (27%), diffuse large B cell lymphoma (8%) and stomach adenocarcinoma (2%). It has been shown that the disruption of the CD274/PD-L1 3′-UTR in mice enables immune evasion of TCs with elevated PD-L1 expression in vivo, which is effectively inhibited by PD-1/PD-L1 blockade.
Alterations at 3′-UTR of CD274/PD-L1 should have different biological effects. First, they can induce stabilization and lead to a marked expression of PD-L1 transcripts which are more stable due to the increase of its truncated but functional protein. Second, the long CD274/PD-L1 3′-UTR has a number of AU-rich elements and potential microRNA-binding sites, directly involved in p53-apoptosis escape and intratumoral immunosuppression.54 Finally, it was noticed that the lack of C-terminal portion of PD-L1 could induce variations in the efficiency of different anti-PD-L1 antibodies in protein detection. These considerations could shed some light on the choice of immune checkpoint blockade. The surprisingly high efficacy of anti-PD-1/PD-L1 therapy in Hodgkin lymphoma (HL), in which PD-L1 overexpression is frequently associated with genetic defects in CD274/PD-L1, suggests that the above implication could be relevant for patients with HL and other advanced cancers, particularly those for which no effective therapy is currently available.55
The epigenetic deregulation of CD274/PD-L1 in cancer
CD274/PD-L1 promoter region
A CpG island is predicted to be located within the 5′-UTR of the CD274/PD-L1 gene, according to the University of California, Santa Cruz (UCSC) Genome Browser’s Regulation “CpG Islands” track. Actually, multiple methylation sites within the gene body were found by performing methyl-reduced bisulfite sequencing experiments in GM12878, H1-hESC, K562, HeLa-S3 and HepG2 cell lines.56 CpG segment overlapping the 5′ end of CD274/PD-L1 is reported in Table 1. A total of five relevant CGs within the upstream CpG island located in the CD274/PD-L1 promoter were noted: cg15837913 (Ch 9: 5,449,890), cg02823866 (Ch 9: 5,450,410), cg14305799 (Ch 9: 5,450,550), cg13474877 (Ch 9: 5,450,724) and cg19724470 (Ch 9: 5,450,936) (Figure 2).57,58
Figure 2.
Genomic localization and organization of CD274/PD-L1 promoter region. The transcription start site (TSS), the ATG initiation codon and the seven exons and relative intron are located on the forward strand of chromosome 9 and approximately span the 2.1-kb CpG island (blue bar). The five cg-beads from the Illumina Infinium Human Methylation 450 BeadChip (cg15837913, cg02823866, cg14305799, cg13474877 and cg19724470) are located from exon 1 to intron 1 within the upstream CpG island of the CD274/PD-L1 promoter.
Aberrant promoter methylation of CD274/PD-L1
Current knowledge on the aberrant methylation of the CD274/PD-L1 promoter and its effects on related pathways are summarized in Table 2. The epigenetic control of CD274/PD-L1 via promoter methylation was first hypothesized in vitro in NSCLC cell lines by observing the 5-azacytidine (5-AZA) increased expression signature of immune-related genes at transcript and protein levels. Evidence on cancer tissues come from the TCGA subsets of primary lung NSCLC tumors showing a concordant low expression of AZA induced PD-L1.59 The idea was corroborated in acute myeloid leukemia (AML), where the pharmacological demethylation of the CpG promoter island located into the CD274/PD-L1 and the gene encoded for its receptor CD279/PD-1 was demonstrated to induce a dose-dependent increase in PD-1 mRNA and PD-L1 expression levels.60 Similar investigations and effects were described by Li and colleagues in additional integrative analyses of gene expression and DNA methylation profiling in a collection of several cancer cell lines from breast, colorectal and ovarian.61
Table 2.
Scientific findings related to the regulation of PD-L1 expression by promoter CD274/PD-L1 methylation in human cancers and association with clinical outcomes in patients.
| Target gene | Tissue type | Downstream effects of PD-L1 methylation |
Clinical outcome | References |
|---|---|---|---|---|
|
CD274/
PD-L1 |
Prostate cancer (training cohort from TCGA, n = 498; independent validation cohort, n = 299) | High PD-L1 protein expression and high mRNA of PD-L1 correlates with shorter BCR-free survival (validation cohort) | Better prognosis associated with low methylated subgroup (training and validation cohorts) | Gevensleben et al.57 |
| AML samples from TCGA (n = 197) | PD-L1 mRNA overexpression inversely correlates with low CD274/PD-L1 methylation levels | Poor prognosis associated with CD274/PD-L1 hypomethylation and high methylation levels are inversely associated with the risk of relapse and short OS | Goltz et al.62 | |
| Colorectal cancer samples from the TCGA cohort (n = 383) | Higher methylation levels of CD274/PD-L1 inversely correlates with PD-L1 mRNA levels | Adverse outcome (reduced RFS and OS) related to the increase of CD274/PD-L1 methylation | Goltz et al.63 | |
| NSCLC tissues (n = 384) | Downregulation of PD-L1 mRNA and protein levels related to increased CD274/PD-L1 promoter methylation levels after cancer recurrence with anti-PD-L1 therapy | NA | Zhang et al.64 | |
| HNSCC patient (representative cohort enrolled by TCGA, n = 528; validation cohort, n = 168) | Negative correlation of PD-1 mRNA overexpression with CD274/PD-L1 hypomethylation | CD274/PD-L1 hypomethylation associated with HPV infection and poor HNSCC prognosis | Franzen et al.65 |
AML, acute myeloid leukemia; BCR, biochemical recurrence; HNSCC, head and neck squamous cell carcinoma; HPV, human papilloma virus; NA, not available; NSCLC, nonsmall cell lung cancer; OS, overall survival; RFS, recurrence-free survival; PD-L1, programmed death-ligand 1; TCGA, The Cancer Genome Atlas.
To date, the epigenetic status of CD274/PD-L1 appears to be a multifaceted prognostic factor in cancer patients. Methylation array experiments have revealed a high density of methylation at the CpG island in the promoter region of CD274/PD-L1, located near the transcriptional start site (TSS). A total of five critical cg-beads from the Illumina Infinium Human Methylation 450 BeadChip were identified in this region: cg15837913, cg02823866, cg14305799, cg13474877 and cg19724470. These five CpGs were reported as frequently hypomethylated in prostate cancer tissues of the TCGA dataset matched with their corresponding normal tissues. By contrast, the hypermethylation of PD-L1 promoter was correlated with lower PD-L1 protein expression in prostate cancer patients following radical prostatectomy. Despite the observed biological event, cg19724470 was the only CpG that was proved to inversely correlate with mRNA transcript levels with a significant clinico-pathological and prognostic value and high PD-L1 protein expression seemed to be unrelated to the hypermethylated status of CD274/PD-L1 promoter in the prostate cancer tissues. However, in the same tumor type, the combination between a high PD-L1 promoter methylation and high protein expression was able to predict a shorter recurrence-free survival (RFS) compared to those with low methylation and low expression protein levels.57 The strong association between low methylation status, TP53 mutations and high-risk cytogenetic profile in AML suggested that CD274/PD-L1 hypermethylation might be associated with adverse outcomes as an independent prognostic factor.63 Intriguingly, in colorectal cancer PD-L1 promoter methylation was significantly associated with poor outcome and reduced overall survival (OS) and RFS. Finally, an indirect correlation was observed with MutL Homolog 1 (MLH1) expression, microsatellite instability and BRAF-mutational status, which are strictly associated with PD-L1 mRNA expression.62
A first validation study of TCGA data on CD274/PD-L1 methylation was proposed in NSCLC by Marwitz and colleagues, who analyzed the presence of epigenetic modifications and RNA transcription of PD-1, PD-L1 and cytotoxic T-lymphocyte antigen 4 (CTLA4) using array-based methylation analyses with no effective results. A strong hypomethylation was significantly associated with increased expression of CTLA4 and CD279/PD-1 genes consistent with transcriptome data, while no differences for PD-L1 methylation or mRNA expression were observed.66 However, PD-L1/CD274 promoter methylation may provide a potentially more effective immunotherapeutic strategy in advanced NSCLC patients treated with tyrosine-kinase inhibitor (TKI).27 Taking into account that EGFR activation by EGFR mutations (such as exon 19 deletions, L858R or T790M mutation) was found to be associated with PD-L1 overexpression. Zhang and colleagues suggested that the anti-PD-1 therapy contributes to the tumor microenvironment evolution and subsequently promotes the suppression of PD-L1 expression through CD274/PD-L1 promoter hypermethylation. After cancer recurrence under TKI treatment, PD-L1 expression levels were found to be downregulated in those patients resistant to anti-PD-1 therapy, supporting the idea that PD-L1 in TCs might also be subject to an epigenetic modulation.64,67
The first study that really supports the idea that membranous PD-L1 protein expression generally detected by IHC staining may be alternatively traced back to differential PD-L1 methylation in solid tumors was made on head and neck squamous cell carcinoma (HNSCC).68 CD274/PD-L1 hypomethylation targeted with beads cg15837913 and cg19724470 correlates with transcriptional silencing and HPV infection in HNSCCs and immune cell infiltrates correlated significantly with PD-L1 methylation and mRNA expression, mainly with T cell (CD4+ and CD8+) and dendritic cells. The cg15837913, cg14305799, cg13474877 and cg19724470 methylation showed a significant inverse correlation with infiltrates of dendritic and CD8+ T cells, whereas cg14305799 and cg13474877 methylation correlated with CD4+ T cells.65
Finally, taking into account that PD-L1 is overexpressed in a transient manner during cytokine-driven epithelial–mesenchymal transition (EMT), a strong link between PD-L1 promoter demethylation and the tumor necrosis factor-alpha/transforming growth factor beta 1 (TNF-α/TGF-β1) signaling pathway was shown, which was found to be associated with a loss of DNA methyltransferase 1 (DNMT1) content in lung A549 cell line.69
miRNA targeting PD-L1
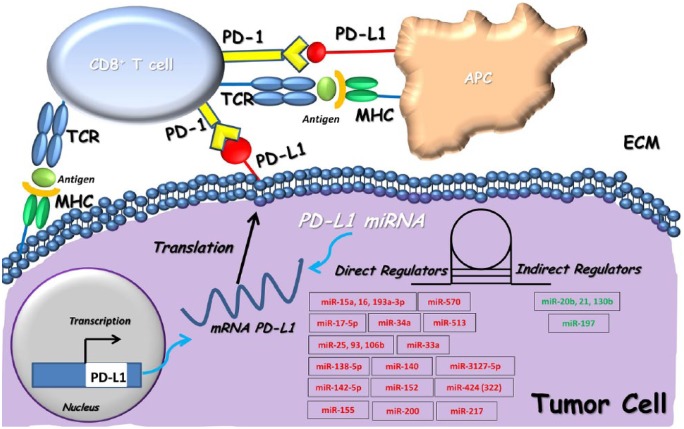
The “dark” unexplored segment of the human genome is represented by the so-called microRNAs (miRNAs): small, endogenous, noncoding RNAs about 20 nucleotides in length.70,71 They act as post-transcriptional regulators of gene expression by binding the 3′-UTR regions of target mRNAs and were recently shown to be directly or indirectly involved in the immune checkpoint modulation and clinical outcome of cancer patients via PD-L1 (Figure 3; Table 3).72 The first evidence of an interplay between miRNA and PD-L1 modulation was presented by Gong and colleagues, who described the role of miR-513 in the inhibition of PD-L1 expression by binding to its 3′-UTR in cholangiocytes.73 Many fascinating results come from this first observation. In malignant pleural mesothelioma (MPM), low expression of the miR-15/16 family was linked to PD-L1-positive samples as a predictive marker of poor prognosis,74,75 miR-17-5p levels in patients with metastatic melanoma were inversely correlated with PD-L1 expression and predict poor clinical prognosis in patients treated with BRAF inhibition.76,77 Several miRNAs predicted to target 3′-UTR of the CD274/PD-L1 gene were identified by using miRNA target prediction tools. A potential immunotherapeutic activity through an inverse correlation with PD-L1 expression was documented for miR-33a in lung adenocarcinoma cells and miR-34a, which appears downregulated in AML and NSCLC. Additionally, in lung cancer cells, low levels of miR34a seem to cooperate in a synergistic manner with P53 levels to suppress PD-L1 activity, thus providing a compelling rationale for why NSCLC patients with high-intensity PD-L1 expression combined with low p53 and miR-34a levels have shorter OS rates and poor clinical outcomes.54,78,79 A role as an independent and poor prognostic factor for clinical outcome of patients with CRC was also suggested for miR-138-5p/PD-L1 interaction. Of note, PD-L1 overexpression reversed the effects of miR-138-5p on cell cycle distribution and blocked the G1/S transition, thereby acting on the downregulation of PD-L1.80 Recent findings also demonstrate enhancement of the anti-tumor immune response by driving PD-L1 expression through miR-140, miR-570, miR-152 and miR-142-5p in NSCLC, gastric adenocarcinoma, hepatocellular carcinoma and pancreatic cells, respectively.81–84 A higher miR-142-5p expression was able to inhibit PD-L1 expression through its link with the 3′-UTR of PD-L1 mRNA and, consequently, led to pancreatic tumor growth inhibition in vivo via stimulation of CD4+ and CD8+ T lymphocyte proliferation.33,81–83 Finally, according to the current state of knowledge about the potential role of interferon-gamma (IFN-γ) and TNF-α, which contribute to the suppression of PD-L1 expression, Yee and colleagues demonstrated by mechanistic investigations in gastric cancer how upregulation of miR-155 co-occurred with significant downregulation of human dermal lymphatic endothelial cells (HDLECs) stimulated upon IFN-γ and TNF-α treatments, thus leading to impaired PD-L1 localization and mRNA levels.85 An interesting link between miR-200 and PD-L1 was described to act as a negative regulator of the EMT process. In NSCLC cells, miR-200 inhibits PD-L1 transcription and drives mesenchymal phenotype changes by creating an immunosuppression microenvironment in the primary NSCLC tissue and promoting widespread metastasis, thus resulting in the suppression of tumor-infiltrating CD8+ T cells.86 Similar evidence has been provided for miR-217/PD-L1 in laryngeal carcinoma cell lines, where the overexpression of miR-217 was able to promote a metastatic repressor activity in EMT and angiogenesis via PD-L1 downregulation.87
Figure 3.
Epigenetic modulation of CD274/PD-L1 by miRNAs. miRNAs mainly downregulate the PD-L1 mRNA expression by linking to the 3′-UTR miRNA of CD274/PD-L1. The representative scheme summarizes two blocks of miRNA regulators that can be distinguished as directly modulating (red miRNAs) and indirectly modulating the PD-1/PD-L1 axis (green miRNAs). APC, adenomatous polyposis coli; ECM, extracellular matrix; MHC, major histocompatibility complex; PD-1, programmed cell death-1; PD-L1, programmed death-ligand 1; TCR, T cell receptor.
Table 3.
miRNAs related to PD-L1 expression in cancer cells and their functional effects.
| Target | MicroRNAs | Cellular context | Cancer models | Functions | Refs |
|---|---|---|---|---|---|
| PD-L1 | hsa-miR-15a hsa-miR-16 hsa-miR-193a-3p |
MPM | MSTO-H211, VMC23 and H28 cells | Directly targeting the 3′-UTR of PD-L1 and leading to its downregulation. | Kao et al.75 |
| hsa-miR-17-5p | MM | A375, SKMEL5 and M14 BRAF V600E-mutated cell lines | Low miR-17-5p and high PD-L1 expression levels are inversely correlated and associated with BRAFi or MEKi sensitivity. | Audrito et al.77 | |
| hsa-miR-20b hsa-miR-21 hsa-miR-130b |
Advanced CRC | CHO cells and human cancer tissues | Indirect regulation of PD-L1 through suppression of PTEN expression. | Zhu et al.88 | |
| hsa-miR-25 hsa-miR-93 hsa-miR-106b |
Primary PDAC | Murine pancreatic cancer models (Ela-KRAS and KPC) and xenografts | Upregulation of miR-25-93-106b cluster results in significant repression of CXCL12 and PD-L1 expression levels in the context of cancer metastasis and immune evasion. | Cioffi et al.89 | |
| hsa-miR-33a | Lung ADC | Tumor tissues | Directly targeting the 3′-UTR of PD-L1/PD-1 and leading to their downregulation. | Boldrini et al.79 | |
| hsa-miR-34a | AML | HL-60 and Kasumi-1 cells | Directly targeting and blocking PD-L1 surface expression with consequent reduction of T cell apoptosis. | Wang et al.78 | |
| NSCLC | H1299 and H460 cells | Inverse correlation with PD-L1 expression mediated by p53 levels. | Cortez et al.54 | ||
| hsa-miR-138-5p | CRC | HCT116, SW620, NCM460 and CCD841CoN cells | Inverse correlation with PD-L1 expression. | Zhao et al.80 | |
| hsa-miR-140 | NSCLC | A549 and NCI-H1650 cells | Inverse correlation with PD-L1 and cyclin E expression levels and inhibition of cell proliferation. | Xie et al.81 | |
| hsa-miR-142-5p | Pancreatic cancer | Panc02 cells | Directly targeting the 3′-UTR of PD-L1 and leading to its downregulation and suppression of mouse tumor growth. | Jia et al.82 | |
| hsa-miR-152 | Gastric cancer | AGS and SGC-7901 cells | Inhibition of PD-L1/PD-1 pathway and increasing T cells proliferation and cytokines production. | Wang et al.83 | |
| hsa-miR-155 | Human dermal lymphatic endothelial cells | HDLECs and HDFs cells | Inhibition of PD-L1 via downregulation of HDLECs stimulated upon IFN-γ and TNF-α treatments. | Yee et al.85 | |
| hsa-miR-197 | NSCLC | A549 and PC14 cells | Inverse correlation with PD-L1 expression mediated by CKS1B/STAT3 pathway in association with Bcl-2, cyclin D1, Survivin and c-Myc expression. | Fujita et al.90 | |
| OSCC | CD3+, CD4+, CD8+, PD-1+, FoxP3+ and CD20+ TILs | Indirect regulation of PD-L1 expression via extrinsic CKS1B/STAT3 pathway. | Ahn et al.91 | ||
| hsa-miR-200 | MLA | CD8+ TIL | Inverse correlation with PD-L1 expression with an effect on tumor development. | Chen et al.86 | |
| hsa-miR-217 | Laryngeal cancer | Hep2 cells | Strong inhibitor effects occurred on PD-L1 through directly repressing its transcription and translation with a concomitant inhibition of cell migration, invasion, proliferation, apoptosis, EMT and angiogenesis. | Miao et al.87 | |
| hsa-miR-424 (322) | Ovarian cancer | OVCAR-3, Skov3 and ID8 cells | Inverse correlation with PD-L1 expression levels and chemoresistance by T-cell immune response activation. | Xu et al.92 | |
| hsa-miR-513 | Biliary epithelial (cholangiocytes) | H69 cells | Directly targeting the 3′-UTR of PD-L1 and leading to its downregulation via IFN-γ. | Gong et al.73 | |
| hsa-miR-570 | Gastric cancer | SGC-7901 cells | Directly targeting the 3′-UTR of PD-L1. Interference with SNP rs4143815 of PD-L1 gene. | Guo et al.84 | |
| hsa-miR-3127-5p | NSCLC | A549, NCI-H1299 cells | Inverse correlation with PD-L1 expression and chemoresistance in the context of cell invasion and proliferation through STAT3 activation. | Tang et al.93 |
3′-UTR, 3′ untranslated region; ADC, adenocarcinoma; AML, acute myeloid leukemia; Bcl-2, B-cell lymphoma 2; BRAF, B-raf proto-oncogene, serine/threonine kinase inhibitor; c-Myc,V-Myc avian myelocytomatosis viral oncogene homolog; CKS1B, cyclin-dependent kinases regulatory subunit 1; CRC, colorectal cancer; EMT, epithelial–mesenchymal transition; CXCL12, C-X-C motif chemokine ligand 12; FOXP3, forkhead box P3; HDFs, dermal fibroblasts; HDLECs, dermal lymphatic endothelial cells; IFN-γ, interferon-γ; IL, interleukin; KPC, LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre mouse model; KRAS, Kirsten rat sarcoma viral oncogene homolog protein; MDSCs, myeloid-derived suppressor cells; MEKi, mitogen-activated protein kinase kinase inhibitor; MLA, mesenchymal lung adenocarcinomas; MM, metastatic melanoma; MPM, malignant pleural mesothelioma; NSCLC, nonsmall cell lung cancer; OSCC, oral squamous cell carcinoma; p53, tumor protein P53; PD-1, programmed cell death protein; PD-L1, programmed death-ligand 1; PDAC, pancreatic ductal adenocarcinoma; PTEN, phosphatase and tensin homolog protein; SNP, single nucleotide polymorphism; STAT3, signal transducer and activator of transcription 3; TILs, tumor-infiltrating lymphocytes; TNF-α, tumor necrosis factor α.
The role of miRNA direct binding to the 3′-UTR region of CD274/PD-L1 in the context of tumor sensitivity to chemo- and immune therapies is emerging.94–96 The interplay between PD-L1 and chemoresistance through the microRNA regulatory cascade was clearly pointed out for miR-424(322). In vitro investigations showed that restoration of miR-424(322) expression reverses chemoresistance, which is accompanied by blockage of the PD-L1 immune checkpoint via CD8+ T cells. In myeloid-derived suppressive cells and regulatory T cells, high levels of miR-424(322) inhibits PD-L1 and CD80 expression and were positively correlated with the progression-free survival of patients.92 A similar role was observed for miR-3127-5p in cisplatin-resistant human lung cancer cells, where the miR-3127-5p/p-STAT3 axis upregulates PD-L1 and induces chemoresistance, whereas its overexpression inhibited cell invasion and proliferation.93
In contrast with the block of miRNAs that directly bind to the 3′-UTR of CD274/PD-L1 and contribute to altering its transcriptional and post-transcriptional regulation, little is known about the indirect modulation of miRNA-mediated signaling networks driving PD-L1 expression. PD-L1 protein expression controlled by miR-20b, miR-21 and miR-130b via the phosphatase and tensin homolog (PTEN) pathway was first hypothesized in CRC and was correlated with metastasis and poor prognosis.88 The loss of PTEN was reported to be significant in NSCLC cells with low levels of miR-197 and inversely correlated with high PD-L1 expression, which was found to be significantly associated with shorter OS in chemoresistant NSCLC patients. The miR-197 was reported to negatively modulate PD-L1 expression via STAT3 in association with oncogene overexpression such as Bcl-2, cyclin D1, Survivin, and c-Myc in NSCLC and OSCC.90,91
Prognostic and predictive value of PD-l1 in the era of immunotherapy
New immunotherapies targeting the PD-1 and PD-L1 to reactivate the suppressed tumor immune system have shown promising results in various cancers. In NSCLC, first nivolumab, an anti-PD-1 monoclonal antibody, demonstrated among patients with previously treated advanced squamous97 and nonsquamous cell lung cancer98 a significantly improved survival benefit and a greater response rate compared with docetaxel. These results were not influenced by PD-L1 expression for squamous cell lung cancer but a correlation with PD-L1 status was shown in the nonsquamous cell lung cancer. In clinical practice there is no need to detect PD-L1 expression to use nivolumab in the second-line setting. Another anti-PD-1 inhibitor, pembrolizumab, showed increased efficacy in terms of response rate and survival when compared with docetaxel in second-line treatment of all-comers NSCLC.99 Pembrolizumab activity seems strictly related to PD-L1 expression and it granted the approval in second-line therapy for patients whose NSCLC expressed PD-L1 ≥1%. In second-line treatment of NSCLC, another immunotherapeutic granted registration without the need for PD-L1 expression, performed on TCs and on TIICs, is atezolizumab, an anti-PD-L1 monoclonal antibody. In a phase III trial, atezolizumab showed statistical and clinical improvements when compared with docetaxel.100 Overall, in second-line treatment of all-comers NSCLC immunotherapy it showed very impressive results when compared with the standard-of-care of docetaxel, with improvements increasing with the expression of PD-L1. In a phase III trial performed in patients affected by all-comers NSCLC with PD-L1 expression ≥50%, first-line pembrolizumab improved all outcomes compared to platinum-based chemotherapy.101 Thus, pembrolizumab was granted approval for first-line treatment of all-comers NSCLC expressing strong positive PD-L1 (≥50%). Despite that PD-L1 expression seems to drive treatment choice, at least in first-line NSCLC therapy, it should be considered as a surrogate for very complicated immune system mechanisms.102 Based on the results available to date, we can define the expression of PD-L1 as a prognostic factor for immunotherapy and a predictive factor for pembrolizumab. However, not all patients with PD-L1-positive tumors responded to these therapies and up to 20% of patients without PD-L1 expression benefit from the therapies.103
In this context, further biomarkers to predict treatment benefits have been explored. Among these, the nonsynonymous mutation burden, the molecular smoking signature and the mismatch-repair deficiency of tumors, all of which would result in neoantigen generation, are in advanced stages of development.102 In particular, TMB has been demonstrated to be another biomarker for patient selection to receive nivolumab plus ipilimumab.104 Moreover, a significant improvement in clinical response to anti-PD-1 has been reported in patients with microsatellite instability-high (MSI-H)/mismatch-repair (MMR)-deficient tumors that accumulate short insertion/deletion mutations, notably in coding microsatellites regions of the genome.105
Future directions
Despite all recent efforts, predicting the response to ICI treatment using a single biomarker remains difficult due to the complex and dynamic interactions between the immune system and tumors. PD-L1 protein expression is not enough to predict response to PD-1/PD-L1 inhibitors, since it cannot identify all cancer patients that may benefit from immune therapies. TMB detected by massive parallel sequencing for each patient in a routine manner seems to be promising and performance and management information obtained from experiments is emerging.106
CNAs of the CD274/PD-L1 gene have received surprisingly little attention until now, even if results are currently highlighting the power of these analyses. Evaluation of CD274/PD-L1 CNGs can be relatively easier to perform even on small biopsy specimens than IHC and can give solid results since it is sustained less dynamically in cancer cells than protein expression. Moreover, CNGs appeared to be helpful in assessing accurate PD-L1 protein expression, TMB and tumor microenvironments, so its predictive significance for therapy response should be prospectively assessed in clinical trials using PD-1/PD-L1 ICIs.
PD-L1 expression appears to be also modulated by epigenetic modifications in epithelial-derived tumors, but a clear value as a predictive/prognostic marker of PD-L1/CD274 promoter methylation has not been reported to date. DNA methylation can be measured accurately in various sample types, so it should be suggested in the future as a robust test to perform in routine diagnostics. Cutoffs and/or ranges need to be first determined to integrate these analyses in clinical trials.
Acknowledgments
The authors thank Dr. Tommaso Mazza for the critical revision of bioinformatics contents of the manuscript, Dr. Paolo Graziano for the revision of the general content of the manuscript and Dr. Andreina Guarnieri for the professional English editing.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Italian Ministry of Health (Ricerca Corrente RC1703LO41, RC1703LO39 to AR) and by the AIRC/MGAF grant 12983 (to LAM) and by the “5x1000” voluntary contributions to IRCCS Casa Sollievo della Sofferenza.
Author contributions: FFP participated in the conception and design of the manuscript and drafted the epigenetic section of the article. TD participated in the conception and design of the manuscript and drafted the genetic section of the article. SA participated in the conception and design of the study and drafted the images for the manuscript. RA participated in and drafted the translational section of the article and critically revised the whole manuscript. CS drafted and revised all the bioinformatics sections of the manuscript. MLA designed the manuscript and critically revised all the sections. All authors approved the final version of the article.
Conflict of interest statement: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ORCID iDs: Federico Pio Fabrizio  https://orcid.org/0000-0002-9122-1348
https://orcid.org/0000-0002-9122-1348
Angelo Sparaneo  https://orcid.org/0000-0002-3211-3072
https://orcid.org/0000-0002-3211-3072
Contributor Information
Federico Pio Fabrizio, Laboratory of Oncology, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy.
Domenico Trombetta, Laboratory of Oncology, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy.
Antonio Rossi, Department of Oncology, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy.
Angelo Sparaneo, Laboratory of Oncology, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy.
Stefano Castellana, Bioinformatic Unit, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy.
Lucia Anna Muscarella, Laboratory of Oncology, Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy.
References
- 1. Coulie Pg, Van den Eynde BJ, van der Bruggen P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014; 14: 135–146. [DOI] [PubMed] [Google Scholar]
- 2. Nishino M, Ramaiya NH, Hatabu H, et al. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol 2017; 14: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018; 359: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vonderheide RH. The immune revolution: a case for priming, not checkpoint. Cancer Cell 2018; 33: 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the “Immunoscore” in the classification of malignant tumours. J Pathol 2014; 232: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsao MS, Kerr KM, Dacic S, et al. IASCL Atlas of PD-L1 testing in lung cancer. 1st ed Aurora, CO: International Associations for the Study of Lung Cancer (IASCL), 2017. [Google Scholar]
- 7. Mclaughlin J, Han G, Schalper KA, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol 2016; 2: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol 2017; 12: 208–222. [DOI] [PubMed] [Google Scholar]
- 9. Gagne A, Enlow W, Pigeon MA, et al. Comprehensive assessment of PD-L1 staining heterogeneity in pulmonary adenocarcinomas using tissue microarrays: impact of the architecture pattern and the number of cores. Am J Surg Pathol 2018; 42: 687–694. [DOI] [PubMed] [Google Scholar]
- 10. Santarpia M, Karachaliou N. Tumor immune microenvironment characterization and response to anti-PD-1 therapy. Cancer Biol Med 2015; 12: 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madore J, Strbenac D, Vilain R, et al. PD-L1 negative status is associated with lower mutation burden, differential expression of immune-related genes, and worse survival in stage III melanoma. Clin Cancer Res 2016; 22: 3915–3923. [DOI] [PubMed] [Google Scholar]
- 14. Hellmann MD, Callahan MK, Awad MM, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 2018; 33: 853–861.e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017; 16: 2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017; 377: 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dong H, Zhu G, Tamada K, et al. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med 1999; 5: 1365–1369. [DOI] [PubMed] [Google Scholar]
- 18. Mitchell AL, Cordell HJ, Soemedi R, et al. Programmed death ligand 1 (PD-L1) gene variants contribute to autoimmune Addison’s disease and Graves’ disease susceptibility. J Clin Endocrinol Metab 2009; 94: 5139–5145. [DOI] [PubMed] [Google Scholar]
- 19. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016; 4: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res 2004; 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J, Jiang CC, Jin L, et al. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol 2016; 27: 409–416. [DOI] [PubMed] [Google Scholar]
- 22. Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med 2007; 13: 84–88. [DOI] [PubMed] [Google Scholar]
- 23. Crane CA, Panner A, Murray JC, et al. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene 2009; 28: 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lastwika KJ, Wilson W, 3rd, Li QK, et al. Control of PD-L1 expression by oncogenic activation of the AKT–mTOR pathway in non-small cell lung cancer. Cancer Res 2016; 76: 227–238. [DOI] [PubMed] [Google Scholar]
- 25. Ikeda S, Okamoto T, Okano S, et al. PD-L1 is upregulated by simultaneous amplification of the PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol 2016; 11: 62–71. [DOI] [PubMed] [Google Scholar]
- 26. Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013; 3: 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014; 25: 1935–1940. [DOI] [PubMed] [Google Scholar]
- 28. Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res 2015; 21: 4014–4021. [DOI] [PubMed] [Google Scholar]
- 29. Jiang X, Zhou J, Giobbie-Hurder A, et al. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res 2013; 19: 598–609. [DOI] [PubMed] [Google Scholar]
- 30. Loi S, Dushyanthen S, Beavis PA, et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin Cancer Res 2016; 22: 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sumimoto H, Takano A, Teramoto K, et al. RAS-mitogen-activated protein kinase signal is required for enhanced PD-L1 expression in human lung cancers. PLoS One 2016; 11: e0166626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 2016; 534: 402–406. [DOI] [PubMed] [Google Scholar]
- 33. Wang W, Sun J, Li F, et al. A frequent somatic mutation in CD274 3′-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum Mutat 2012; 33: 480–484. [DOI] [PubMed] [Google Scholar]
- 34. Tao LH, Zhou XR, Li FC, et al. A polymorphism in the promoter region of PD-L1 serves as a binding-site for SP1 and is associated with PD-L1 overexpression and increased occurrence of gastric cancer. Cancer Immunol Immunother 2017; 66: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inoue Y, Yoshimura K, Mori K, et al. Clinical significance of PD-L1 and PD-L2 copy number gains in non-small-cell lung cancer. Oncotarget 2016; 7: 32113–32128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov 2015; 5: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, et al. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J Exp Med 2012; 209: 1201–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barrett MT, Anderson KS, Lenkiewicz E, et al. Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget 2015; 6: 26483–26493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Budczies J, Bockmayr M, Denkert C, et al. Pan-cancer analysis of copy number changes in programmed death-ligand 1 (PD-L1, CD274): associations with gene expression, mutational load, and survival. Genes Chromosomes Cancer 2016; 55: 626–639. [DOI] [PubMed] [Google Scholar]
- 41. Budczies J, Mechtersheimer G, Denkert C, et al. PD-L1 (CD274) copy number gain, expression, and immune cell infiltration as candidate predictors for response to immune checkpoint inhibitors in soft-tissue sarcoma. Oncoimmunology 2017; 6: e1279777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldmann T, Kugler C, Reinmuth N, et al. PD-L1 copy number gain in nonsmall-cell lung cancer defines a new subset of patients for anti PD-L1 therapy. Ann Oncol 2016; 27: 206–207. [DOI] [PubMed] [Google Scholar]
- 43. Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016; 22: 4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clave S, Pijuan L, Casadevall D, et al. CD274 (PDL1) and JAK2 genomic amplifications in pulmonary squamous-cell and adenocarcinoma patients. Histopathology 2018; 72: 259–269. [DOI] [PubMed] [Google Scholar]
- 45. George J, Saito M, Tsuta K, et al. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin Cancer Res 2017; 23: 1220–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howitt BE, Sun HH, Roemer MG, et al. Genetic basis for PD-L1 expression in squamous cell carcinomas of the cervix and vulva. JAMA Oncol 2016; 2: 518–522. [DOI] [PubMed] [Google Scholar]
- 47. Straub M, Drecoll E, Pfarr N, et al. CD274/PD-L1 gene amplification and PD-L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget 2016; 7: 12024–12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo L, Li W, Zhu X, et al. PD-L1 expression and CD274 gene alteration in triple-negative breast cancer: implication for prognostic biomarker. Springerplus 2016; 5: 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kadonaga JT, Carner KR, Masiarz FR, et al. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 1987; 51: 1079–1090. [DOI] [PubMed] [Google Scholar]
- 50. Wang L, Wei D, Huang S, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res 2003; 9: 6371–6380. [PubMed] [Google Scholar]
- 51. Yao JC, Wang L, Wei D, et al. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res 2004; 10: 4109–4117. [DOI] [PubMed] [Google Scholar]
- 52. Wang W, Li F, Mao Y, et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet 2013; 132: 641–648. [DOI] [PubMed] [Google Scholar]
- 53. Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 2012; 13: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cortez MA, Ivan C, Valdecanas D, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst 2015; 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 2015; 372: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gevensleben H, Holmes EE, Goltz D, et al. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget 2016; 7: 79943–79955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene 2002; 21: 5496–5503. [DOI] [PubMed] [Google Scholar]
- 59. Wrangle J, Wang W, Koch A, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget 2013; 4: 2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang H, Bueso-Ramos C, Dinardo C, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia 2014; 28: 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li H, Chiappinelli KB, Guzzetta AA, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014; 5: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goltz D, Gevensleben H, Dietrich J, et al. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology 2017; 6: e1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goltz D, Gevensleben H, Grunen S, et al. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia 2017; 31: 738–743. [DOI] [PubMed] [Google Scholar]
- 64. Zhang Y, Xiang C, Wang Y, et al. PD-L1 promoter methylation mediates the resistance response to anti-PD-1 therapy in NSCLC patients with EGFR-TKI resistance. Oncotarget 2016; 8: 101535–101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Franzen A, Vogt TJ, Muller T, et al. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget 2017; 9: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marwitz S, Scheufele S, Perner S, et al. Epigenetic modifications of the immune-checkpoint genes CTLA4 and PDCD1 in non-small cell lung cancer results in increased expression. Clin Epigenetics 2017; 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Han JJ, Kim DW, Koh J, et al. Change in PD-L1 expression after acquiring resistance to gefitinib in EGFR-mutant non-small-cell lung cancer. Clin Lung Cancer 2016; 17: 263–270 e262. [DOI] [PubMed] [Google Scholar]
- 68. Lee Y, Shin JH, Longmire M, et al. CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin Cancer Res 2016; 22: 3571–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Asgarova A, Asgarov K, Godet Y, et al. PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology 2018; 7: e1423170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther 2016; 1: 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007; 39: 673–677. [DOI] [PubMed] [Google Scholar]
- 72. Smolle MA, Calin HN, Pichler M, et al. Noncoding RNAs and immune checkpoints: clinical implications as cancer therapeutics. Febs J 2017; 284: 1952–1966. [DOI] [PubMed] [Google Scholar]
- 73. Gong Ay, Zhou R, Hu G, et al. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol 2009; 182: 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Reid G, Pel ME, Kirschner MB, et al. Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann Oncol 2013; 24: 3128–3135. [DOI] [PubMed] [Google Scholar]
- 75. Kao SC, Cheng YY, Williams M, et al. Tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma. J Thorac Oncol 2017; 12: 1421–1433. [DOI] [PubMed] [Google Scholar]
- 76. Massi D, Brusa D, Merelli B, et al. The status of PD-L1 and tumor-infiltrating immune cells predict resistance and poor prognosis in BRAFi-treated melanoma patients harboring mutant BRAFV600. Ann Oncol 2015; 26: 1980–1987. [DOI] [PubMed] [Google Scholar]
- 77. Audrito V, Serra S, Stingi A, et al. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget 2017; 8: 15894–15911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang X, Li J, Dong K, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal 2015; 27: 443–452. [DOI] [PubMed] [Google Scholar]
- 79. Boldrini L, Giordano M, Niccoli C, et al. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell Int 2017; 17: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhao L, Yu H, Yi S, et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016; 7: 45370–45384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xie WB, Liang LH, Wu KG, et al. MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem 2018; 46: 654–663. [DOI] [PubMed] [Google Scholar]
- 82. Jia L, Xi Q, Wang H, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun 2017; 488: 425–431. [DOI] [PubMed] [Google Scholar]
- 83. Wang Y, Wang D, Xie G, et al. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget 2017; 8: 28125–28134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Guo W, Tan W, Liu S, et al. MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumour Biol 2015; 36: 9049–9057. [DOI] [PubMed] [Google Scholar]
- 85. Yee D, Shah KM, Coles MC, et al. MicroRNA-155 induction via TNF-alpha and IFN-gamma suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem 2017; 292: 20683–20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen L, Gibbons D, Goswami S, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 2014; 5: 5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Miao S, Mao X, Zhao S, et al. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget 2017; 8: 62143–62153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhu J, Chen L, Zou L, et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol 2014; 75: 348–353. [DOI] [PubMed] [Google Scholar]
- 89. Cioffi M, Trabulo SM, Vallespinos M, et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget 2017; 8: 21609–21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fujita Y, Yagishita S, Hagiwara K, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther 2015; 23: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ahn H, Yang JM, Kim H, et al. Clinicopathologic implications of the miR-197/PD-L1 axis in oral squamous cell carcinoma. Oncotarget 2017; 8: 66178–66194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xu S, Tao Z, Hai B, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun 2016; 7: 11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tang D, Zhao D, Wu Y, et al. The miR-3127-5p/p-STAT3 axis up-regulates PD-L1 inducing chemoresistance in non-small-cell lung cancer. J Cell Mol Med. Epub ahead of print May 4, 2018. DOI: 10.1111/jcmm.13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lake RA, Robinson BW. Immunotherapy and chemotherapy: a practical partnership. Nat Rev Cancer 2005; 5: 397–405. [DOI] [PubMed] [Google Scholar]
- 95. Galluzzi L, Buque A, Kepp O, et al. Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 2015; 28: 690–714. [DOI] [PubMed] [Google Scholar]
- 96. Black M, Barsoum IB, Truesdell P, et al. Activation of the PD-1/PD-L1 immune checkpoint confers tumor cell chemoresistance associated with increased metastasis. Oncotarget 2016; 7: 10557–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Brahmer J, Reckamp K, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015; 373: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. Nes Engl J Med 2015; 373: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016; 387: 1540–1550. [DOI] [PubMed] [Google Scholar]
- 100. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017; 389: 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016; 375: 1823–1833. [DOI] [PubMed] [Google Scholar]
- 102. Cyriac G, Gandhi L. Emerging biomarkers for immune checkpoint inhibition in lung cancer. Semin Cancer Biol 2018; 52: 269–277. [DOI] [PubMed] [Google Scholar]
- 103. Shen X, Zhao B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: meta-analysis. BMJ 2018; 362: k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with high tumor mutational burden. N Engl J Med 2018; 378: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nebot-Bral L, Coutzac C, Kannouche PL, et al. Why is immunotherapy effective (or not) in patients with MSI/MMRD tumors? Bull Cancer 2018. Epub ahead of print October 17, 2018. DOI: 10.1016/j.bulcan.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 106. Costa J, Reis J, Fernandes M, et al. Assessing tumor mutation load using an NGS-based, routine-friendly target gene panel. Cancer Res. Epub ahead of print July 2018. DOI: 10.1158/1538-7445. [DOI] [Google Scholar]