Abstract
Exosomes mediate cell-cell crosstalk in cancer progression by transferring their molecular cargos, including long noncoding RNAs (lncRNAs). Metastasis‑associated lung adenocarcinoma transcript 1 (MALAT1) is a well-known lncRNA associated with cancer angiogenesis and metastasis. However, the presence of MALAT1 in exosomes and the roles and clinical values of exosomal MALAT1 in epithelial ovarian cancer (EOC) remain unknown. The present study focused on the crosstalk between EOC cells and endothelial cells mediated by exosomal MALAT1 and aimed to explore the roles of exosomes and exosomal MALAT1 in EOC angiogenesis and to reveal the clinical relevance and prognostic predictive value of serum exosomal MALAT1 in EOC. We observed that MALAT1 was increased in both metastatic EOC cells and their secreted exosomes. Exosomal MALAT1 derived from EOC cells was transferred to recipient human umbilical vein endothelial cells (HUVECs) via exosomes. In vitro and in vivo experiments demonstrated that MALAT1 knockdown impaired the exosome-mediated proangiogenic activity of HUVECs through certain key angiogenesis-related genes. Clinically, elevated serum exosomal MALAT1 was highly correlated with an advanced and metastatic phenotype of EOC and was an independent predictive factor for EOC overall survival (OS). Moreover, a prognostic nomogram model we constructed showed a good prediction of the probability of 3-year OS of EOC patients according to the c-index (0.751, 95% confidence interval [CI]=0.691-0.811) and calibration curve. Collectively, our data provide a novel mechanism by which EOC cells transfer MALAT1 via exosomes to recipient HUVECs and influence HUVECs by stimulating angiogenesis-related gene expression, eventually promoting angiogenesis. Additionally, circulating exosomal MALAT1 can serve as a promising serum-based, noninvasive predictive biomarker for EOC prognosis.
Keywords: exosome, lncRNA, intercellular communication, angiogenesis, prognosis
Introduction
Epithelial ovarian cancer (EOC), which has poor five-year survival rate of only 30%, is the deadliest gynaecological cancer 1-3. Widespread metastases account for the poor prognosis and most deaths in EOC patients and pose a major challenge for EOC treatment 2. Therefore, a thorough exploration of the mechanisms underlying EOC metastasis is urgently needed, which may provide potential novel biomarkers for EOC prognosis and novel molecular targets for therapy.
Angiogenesis, a common feature of all tumours, is a prerequisite for tumour metastasis 4,5. Angiogenesis participates in supplying tumours with nutrients and oxygen for continuous tumour growth and metastasis. Additionally, angiogenesis can help malignant cells to leave the primary site for the circulatory system or distant organs, resulting in metastasis 6-11. Therefore, elucidating the mechanisms of EOC angiogenesis is critical for understanding and treating EOC metastasis and progression. To date, numerous proangiogenic molecules, including proteins and noncoding RNAs, have been identified as contributing to EOC angiogenesis 5-11. Metastasis‑associated lung adenocarcinoma transcript 1 (MALAT1), a well-known long noncoding RNA (lncRNA >200 nucleotides), especially caught our attention because it plays powerful roles in promoting cancer angiogenesis and metastases in various cancers 12-20. However, despite the progress of research on the involvement of MALAT1 in cancer, the precise mechanisms, especially how cancer cells transfer MALAT1 to surrounding cells thereby promoting angiogenesis and metastases, remain largely unknown.
Exosomes, small membrane vesicles of 30-150 nm in size that are derived from multiple cells, are of particular interest because they can mediate cell-cell communication and achieve diverse biologic functions through the transfer of their cargo molecules, such as proteins, DNAs, RNAs, microRNAs (miRNAs) and lncRNAs 21-30. Most recently, exosome-mediated transfer of lncRNAs to influence recipient cells is emerging as a novel mechanism for cancer progression. For example, lncRNA-H19 can be taken up by endothelial cells via exosomes to promote adhesion and angiogenesis 31. Exosomal linc-POU3F3 derived from glioma cells can be transferred to endothelial cells to promote angiogenesis 32. Additionally, serum exosomal lncRNAs, such as CRNDE-h, HOTAIR and lncRNA-p21, have emerged as noninvasive cancer diagnostic and prognostic markers 33-35. For MALAT1, although a few recent studies have linked exosomal MALAT1 to cervical cancer, lung cancer and breast cancer 36-38, no published studies have explored the clinical value of exosomal MALAT1 in EOC or clarified its roles in EOC progression, especially in EOC angiogenesis.
The present study demonstrated that EOC cells could transfer MALAT1 via exosomes to recipient HUVECs and influence HUVECs by stimulating angiogenesis-related gene expression, eventually promoting angiogenesis. Additionally, elevated serum exosomal MALAT1 levels were correlated with advanced and metastatic clinical characteristics and poor prognosis of EOC patients. This study is the first to explore the unique crosstalk between EOC cells and endothelial cells mediated by exosomal MALAT1 to promote cancer angiogenesis and the first to reveal the clinical relevance and prognostic value of serum exosomal MALAT1 in EOC.
Materials and methods
Cell lines and cell culture
Two paired EOC cell lines that are metastatic (SKOV3.ip1, HO8910.PM) and their parental (SKOV3, HO8910) sublines 39-42 were gifts from the University of Texas MD Anderson Cancer Center (Houston, TX, USA). Human umbilical vein endothelial cells (HUVECs) used for angiogenesis assays were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). EOC cells and HUVECs were cultured in DMEM medium (Gibco BRL, Gaithersburg, MD, USA) supplemented with 10% foetal bovine serum (FBS; Gibco), 100 units/ml penicillin and 100 mg/ml streptomycin in a humidified 5% CO2 incubator at 37˚C.
Patients and samples
Sixty EOC patients recruited from January 2011 to December 2013 at the Obstetrics and Gynecology Hospital of Fudan University were included based on the availability of blood samples and follow-up data. Staging and grading were evaluated according to the criteria of the International Federation of Gynecologists and Obstetricians (FIGO) and the World Health Organization (WHO). Patients were excluded if they had received preoperative therapies or had more different malignancies. Healthy controls were recruited from outpatients who had undergone a general medical examination. Serum was separated from each participant and stored at -80˚C before use.
The clinical and follow-up data of the EOC patients were collected. Overall survival (OS) was calculated from the date of surgery until the date of death or the end of the follow-up period (December 2017). The Ethics Committee of Obstetrics and Gynecology Hospital of Fudan University approved this study. Informed consent was obtained from all participants.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA extraction from cells and exosomes and qRT-PCR were conducted as previously described 43-44. All assays were performed in triplicate. Statistical analyses of the results were performed using the 2-ΔΔCt relative quantification method. The primer sequences were as follows: MALAT1 forward, 5'-CTTCCCTAGGGGATTTCAGG-3' and reverse, 5'-GCCCACAGGAACAAGTCCTA-3'; GAPDH forward, 5'-GTCAACGGATTTGGTCTGTATT-3' and reverse, 5'-AGTCTTCTGGGTGGCAGTGAT-3'.
Establishment of MALAT1-knockdown stable cell lines
The target sequences of the two MALAT1-shRNAs were 5'-ACGGAAGTAATTCAAGATCAA-3' (MALAT1-sh1) and 5'-TTGCAGAGGCATTTCATCCTT-3' (MALAT1-sh2). Lentiviruses encoding the two shRNAs (MALAT1-knockdown, KD1 and KD2) and the negative control (NC) were constructed by Hanyin Co. (Shanghai, China). After lentiviral infection, stable cell lines were established as follows: SKOV3.ip1-KD1, SKOV3.ip1-KD2, SKOV3.ip1-NC, HO8910.PM-KD1, HO8910.PM-KD2 and HO8910.PM-NC. The efficiency of MALAT1 knockdown was determined using qRT-PCR.
Isolation of exosomes from cell supernatants and patient serum
For isolation of exosomes from cell supernatants, cells were initially cultured in exosome-free medium for 3 days. Then, the exosomes were isolated and purified from the cell supernatants by using the total exosome isolation reagent (from cell culture media) kit #44578259 (Thermo Fisher Scientific, CA, USA) according to the protocol. Exosomes derived from SKOV3.ip1, HO8910.PM, SKOV3 and HO8910 cells were termed Exo/SKOV3.ip1, Exo/HO8910.PM, Exo/SKOV3 and Exo/HO8910. Exosomes derived from SKOV3.ip1-KD cells, SKOV3.ip1-NC cells, HO8910.PM-KD cells and HO8910.PM-NC cells were named Exo/SKOV3.ip1-KD, Exo/SKOV3.ip1-NC, Exo/HO8910.PM-KD and Exo/HO8910.PM-NC. To isolate exosomes from human serum samples, an ExoQuick Exosome Precipitation Solution kit (catalogue no. EXOQ5A-1; System Biosciences, Palo Alto, CA, USA) was used according to the protocol. The exosomes were used immediately or stored at -80˚C.
Transmission electron microscopy (TEM)
Briefly, the exosome suspension was added to an equal volume of 4% paraformaldehyde and applied to a Formvar/carbon film-coated transmission electron microscope grid (Alliance Biosystems, Inc., Osaka, Japan). Subsequently, samples was fixed by incubation with 1% glutaraldehyde, washed with PBS, contrasted with 1% uranyl acetate, embedded in epoxy resin and polymerized. Exosomes were subsequently observed under a Hitachi H-7650 transmission electron microscope (Hitachi, Ltd., Tokyo, Japan).
Nanoparticle tracking analysis (NTA)
Briefly, the exosomes were resuspended in PBS and filtered with a syringe filter (Millipore). Then, the samples were diluted until individual nanoparticles could be tracked. The size distribution of the exosomes was evaluated using a NanoSight NS300 instrument (Malvern Instruments Ltd, Worcestershire, UK).
Western blotting
To identify exosome markers, primary antibodies against CD63 and TSG101 were purchased from Abcam (Cambridge, UK), and primary antibodies against Hsp 70 and Hsp 90 were obtained from Cell Signaling Technology (CST, Beverly, MA, USA). The secondary antibodies were F(ab)2 fragments of donkey anti-mouse immunoglobulin or donkey anti-rabbit immunoglobulin linked to horseradish peroxidase (Jackson ImmunoResearch, Inc., USA). Immunoblotting reagents from an electrochemiluminescence kit were used (Amersham Biosciences, Uppsala, Sweden).
Exosome labelling and tracking
To track exosome internalization, exosomes were fluorescently labelled using the green dye PKH67 (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's protocol. The labelled exosomes were added to HUVECs and incubated for 6 h. The uptake of labelled exosomes by the recipient HUVECs was observed by a Leica TCS SP5 II laser scanning confocal microscope.
Inhibition of exosome release
Exosome release was blocked using GW4869 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), a specific chemical inhibitor of nSMase2. Cells were pre-seeded and cultured for 12 h in complete medium. After incubation, the medium was changed to fresh complete medium with GW4869.
Tube formation assay
Tube formation assay was performed to assess the angiogenic potential. Growth factor-reduced Matrigel (BD Biosciences, San Jose, CA, USA) was placed in 48-well plates. HUVECs were first incubated with serum-free medium for 12 h and then transferred onto the 48-well plates precoated with Matrigel. After incubation for 10 h, tube formation was examined in photographs taken under a microscope. The total tube length was determined by measuring the branches of blood vessels using ImageJ software.
In vivo animal assay
Four-week-old (weighing 20-22 g) female BALB/c athymic nude mice were purchased from Slac Laboratory Animal Co. Ltd. (Shanghai, China) and maintained in a pathogen-free animal facility. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Fudan University. SKOV3.ip1 cells (1 × 106) were injected intraperitoneally into the mice. Then, the mice were injected with PBS, Exo/SKOV3.ip1-NC, Exo/SKOV3.ip1-KD1 or Exo/SKOV3.ip1- KD2 twice each week and were accordingly divided into four groups (n=6 for each condition). After four injections, all the mice were euthanized, and the tumours were collected for weight, implant number and immunohistochemical analyses.
Immunohistochemistry (IHC)
Vascular density was detected using CD31 staining. Xenograft tumours were fixed, embedded in paraffin and sectioned into 4-μm thick slices. After deparaffinization and rehydration, sections were blocked and incubated with a CD31 antibody (Abcam, USA). The quantitative analysis was performed on five randomly selected, independent fields per tumour.
Tumour angiogenesis PCR array
HUVECs co-cultured with PBS or exosomes were collected, and mRNA was isolated using an RNeasy kit according to the manufacturer's protocol (QIAGEN, Valencia, CA). cDNA was generated from 1 ng of total RNA with the Reverse Transcriptase First Strand Kit and analysed via the Angiogenesis PCR Array Kit according to the manufacturer's instructions (SABiosciences, Frederick, MD, USA). Parallel samples were analysed to identify changes in relevant gene expression.
Statistical analysis
Statistical analyses were performed using SPSS for Windows version 16.0 (SPSS, Chicago, IL, USA). Continuous data were analysed using independent t-tests. Categorical data were analysed using chi-square or Fisher's exact tests. OS was calculated using the Kaplan-Meier method and the log-rank test. Multivariate survival analyses were performed using Cox regression models. A predictive model was constructed based on significant variables in the multivariate analysis by R version 3.4.3 (https://www.r-project.org/), and Harrell's concordance index (c-index) was used to assess its predictive efficiency. P values <0.05 were considered significant (P<0.05).
Results
MALAT1 expression is increased in metastatic SKOV3.ip1 and HO8910.PM EOC cells
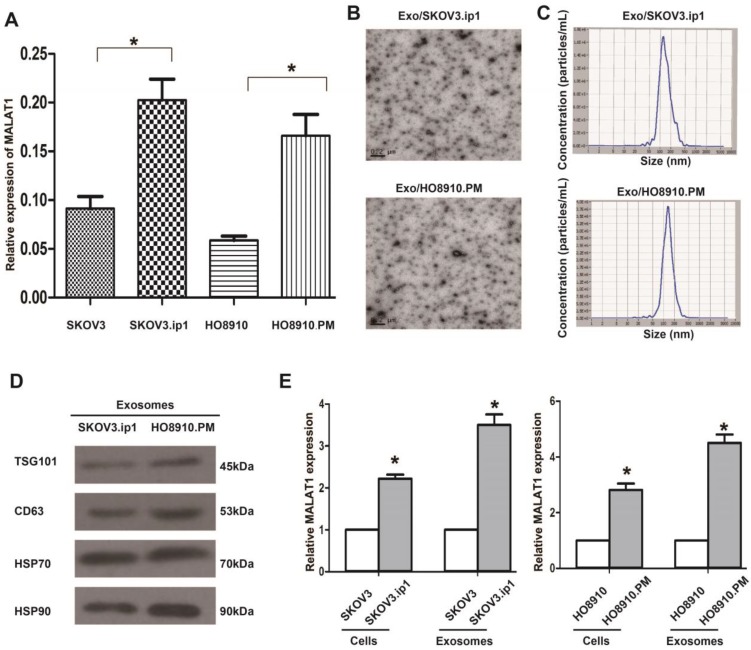
We initially confirmed whether MALAT1 was associated with EOC metastasis. As expected, MALAT1 levels were higher in the metastatic SKOV3.ip1 and HO8910.PM cells than in their parental SKOV3 and HO8910 cells (Fig. 1A), suggesting that MALAT1 contributes to EOC metastasis. Because the metastatic SKOV3.ip1 and HO8910.PM cells exhibited high MALAT1 expression, we focused on them in the following studies.
Figure 1.
Relative MALAT1 levels in EOC cells and their secreted exosomes. (A) qRT-PCR analysis shows that MALAT1 levels in the highly metastatic EOC cells (SKOV3.ip1, HO8910.PM) were higher than those in their parental cells. The data represent the means ± standard deviation (SD) of three independent experiments. The error bars denote the SD. * P<0.05. (B) Transmission electron microscopy images of Exo/SKOV3.ip1 and Exo/HO8910.PM. (C) Nanoparticle tracking analysis to determine the size distribution of Exo/SKOV3.ip1 and Exo/HO8910.PM. The representative size distribution peaks of Exo/SKOV3.ip1 and Exo/HO8910.PM appear at 121 nm and 132 nm, respectively. (D) Western blotting analysis of the exosomal markers CD63, TSG101, Hsp 70 and Hsp 90 in Exo/SKOV3.ip1 and Exo/HO8910.PM. (E) qRT-PCR analysis of MALAT1 expression in EOC cells (SKOV3.ip1, HO8910.PM, SKOV3, and HO8910) and in their secreted exosomes. * P<0.05.
Characterization of exosomes derived from SKOV3.ip1 and HO8910.PM cells
As exosomes are key mediators of cell-cell communication to promote cancer progression, we aimed to explore the exosome-based mechanism of EOC progression. We initially isolated and characterized exosomes derived from SKOV3.ip1 and HO8910.PM cells. TEM analysis showed that Exo/SKOV3.ip1 and Exo/HO8910.PM had similar morphologies (30-150 nm in diameter) and exhibited a round-shaped appearance (Fig. 1B). The NTA results demonstrated that Exo/SKOV3.ip1 and Exo/HO8910.PM showed a similar size distribution, and the peak size range was 80-135 nm (Fig. 1C). Western blot analysis confirmed the presence of four well-known exosomal markers, CD63, TSG101, Hsp 70 and Hsp 90 (Fig. 1D). These results indicated that the exosomes were isolated successfully.
Additionally, we observed that MALAT1 levels were significantly increased not only in metastatic SKOV3.ip1 and HO8910.PM cells but also in their secreted exosomes, and this increase in the exosomes was more evident than that in the cells (Fig. 1E). These results suggested that MALAT1 may act as not only an intracellular lncRNA but also an exosomal lncRNA and implied that both exosomes and exosomal MALAT1 may play potential roles in EOC metastasis.
Exosomes derived from EOC cells promote angiogenesis of HUVECs
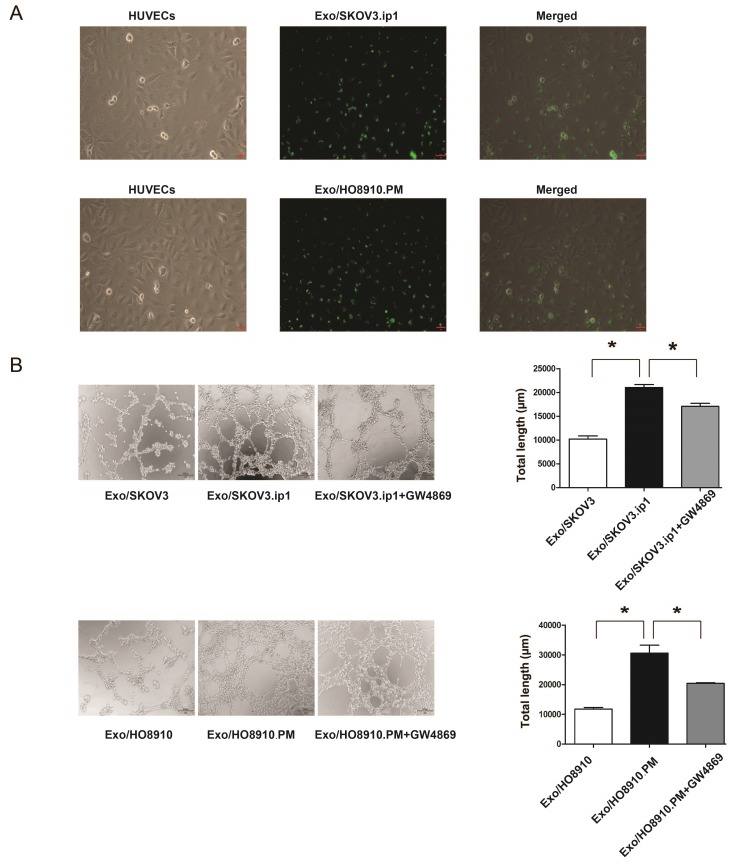
Given that tumour-derived exosomes can be internalized by recipient cells to achieve their function and that angiogenesis is a prerequisite for EOC metastasis, we focused on “tumour cell-endothelial cell” communication and hypothesized that exosomes derived from SKOV3.ip1 and HO8910.PM cells may be internalized by HUVECs and play a role in the angiogenesis of HUVECs. As expected, HUVECs exhibited an uptake of PKH67-labelled Exo/SKOV3.ip1 and Exo/HO8910.PM (Fig. 2A), suggesting that exosomes derived from SKOV3.ip1 and HO8910.PM cells could be effectively internalized by HUVECs.
Figure 2.
Exosomes derived from SKOV3.ip1 and HO8910.PM cells promote angiogenesis of HUVECs. (A) Internalization of exosomes derived from SKOV3.ip1 and HO8910.PM cells by HUVECs. PKH67 (green fluorescent dye)-labelled exosomes derived from SKOV3.ip1 and HO8910.PM cells were taken up by HUVECs. (B) Tube formation of HUVECs. HUVECs were separately co-cultured with Exo/SKOV3, Exo/SKOV3.ip1 and Exo/SKOV3.ip1 + GW4869 and were co-cultured with Exo/HO8910, Exo/HO8910.PM and Exo/HO8910.PM + GW4869. Left: representative photographs illustrating the dynamics of the vascular behaviour of HUVECs. Right: the relative tube lengths in HUVECs. * P<0.05.
Furthermore, tube formation assays revealed that Exo/SKOV3.ip1 and Exo/HO8910.PM increased HUVEC angiogenesis compared with exosomes from their parental cells, while treatment with GW4869 significantly abolished the proangiogenic effect (Fig. 2B). These observations suggested that exosomes derived from SKOV3.ip1 and HO8910.PM cells could promote angiogenesis of HUVECs.
Exosomes derived from EOC cells mediate the transfer of MALAT1 to HUVECs
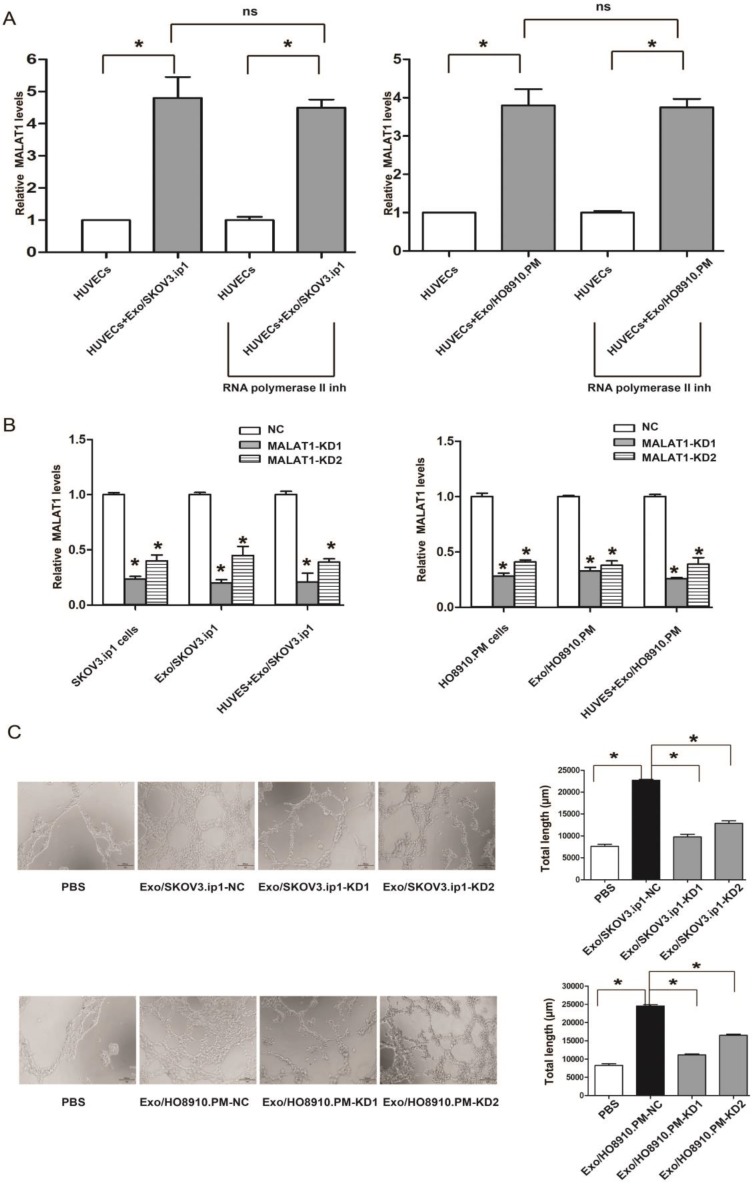
As cargo RNAs contained in exosomes are responsible for exosome function, we hypothesized that MALAT1 contained in exosomes might contribute to such proangiogenic ability. To this end, we first determined whether MALAT1 derived from SKOV3.ip1 and HO8910.PM cells could be transferred to HUVECs via exosomes. We observed that MALAT1 levels were significantly increased in HUVECs treated with Exo/SKOV3.ip1 and Exo/HO8910.PM compared with those in untreated HUVECs, and this increase was not prevented by an RNA polymerase II inhibitor, which was consistent with a previous study 27 (Fig. 3A). Collectively, these findings suggested that MALAT1 could be transferred from SKOV3.ip1 and HO8910.PM cells to recipient HUVECs via exosomes.
Figure 3.
Knockdown of exosomal MALAT1 attenuates the angiogenesis of HUVECs. (A) The transfer of exosomal MALAT1 from SKOV3.ip1 and HO8910.PM cells to HUVECs. MALAT1 levels were increased in HUVECs treated with Exo/SKOV3.ip1 and Exo/HO8910.PM compared to those in untreated HUVECs, and the increase was not prevented by an RNA polymerase II inhibitor. * P<0.05; ns, no significant difference; inh, inhibitor. HUVECs were treated with or without Exo/SKOV3.ip1 and Exo/HO8910.PM in the presence or absence of 5, 6-dichloro-1-β-D-ribofuranosylbenzimidazole (20 μM), a selective inhibitor of transcription elongation by RNA polymerase II, inhibiting mRNA synthesis in eukaryotic cells 49. The RNA isolated from recipient HUVECs were analyzed for MALAT1 levels. (B) Suppression efficiency of MALAT1 knockdown in SKOV3.ip1 and HO8910.PM cells, their secreted exosomes, and the recipient HUVECs treated with Exo/SKOV3.ip1 and Exo/HO8910.PM. (C) Tube formation assays were performed in HUVECs co-cultured with PBS (blank group) and with exosomes derived from SKOV3.ip1 and HO8910.PM cells transfected with MALAT1-NC, MALAT1-KD1 or MALAT1-KD2. Left: representative photographs illustrating the dynamics of the vascular behaviour of HUVECs. Right: the relative tube lengths in HUVECs. * P<0.05.
Exosomal MALAT1 promotes angiogenesis of HUVECs
To further determine whether the proangiogenic ability of Exo/SKOV3.ip1 and Exo/HO8910.PM was dependent on exosomal MALAT1, we first silenced MALAT1 expression and found that both MALAT1-KD1 and MALAT1-KD2 efficiently decreased MALAT1 levels not only in SKOV3.ip1 and HO8910.PM cells but also in their secreted exosomes and the recipient HUVECs (Fig. 3B). Strikingly, the tube formation assay showed that MALAT1 knockdown impaired the proangiogenic ability of Exo/SKOV3.ip1 and Exo/HO8910.PM in HUVECs (Fig. 3C). These results suggested that exosomal MALAT1 was responsible for the proangiogenic ability of Exo/SKOV3.ip1 and Exo/HO8910.PM.
Exosomal MALAT1 promotes tumour angiogenesis, growth and implantation in vivo
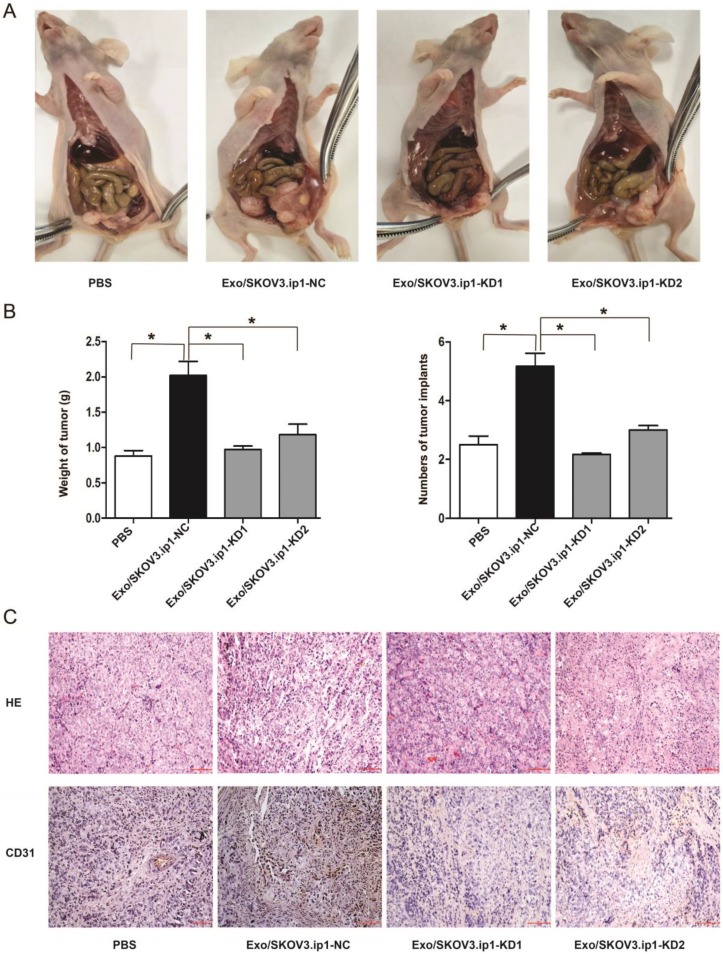
To further estimate the effects of exosomal MALAT1 in vivo, we constructed a mouse model of intraperitoneal implantation as described previously 45-47. After four injections with PBS, Exo/SKOV3.ip1-NC, Exo/SKOV3.ip1-KD1 or Exo/SKOV3.ip1-KD2, we observed significant differences in peritoneal tumor growth and implants between these groups. The mice in Exo/SKOV3.ip1-NC group developed more and larger tumors in the peritoneal region compared to Exo/SKOV3.ip1-KD group and PBS group (Fig. 4A). The average numbers of tumor implants in the different groups were shown in Fig. 4B. These observations indicated that MALAT1 knockdown decreased exosome-mediated tumour growth and implantation in vivo. Additionally, the density of blood vessels (assessed via CD31 staining) was decreased in the Exo/SKOV3.ip1-KD tumours compared with the Exo/SKOV3.ip1-NC tumours (Fig. 4C), confirming that MALAT1 knockdown attenuated the exosome-mediated promotion of tumour angiogenesis in vivo, which corresponds to the in vitro results.
Figure 4.
Exosomal MALAT1 promotes tumour angiogenesis, growth and implantation in vivo. (A) Effects of the exosomes derived from SKOV3.ip1 cells transfected with MALAT1-NC or MALAT1-KD on tumour progression in vivo. Representative images of tumour progression in mice from the 4 groups (the mice were injected with PBS, Exo/SKOV3.ip1-NC, Exo/SKOV3.ip1-KD1 or Exo/SKOV3.ip1-KD2) are shown. (B) The final tumour weight and number of tumour implants were determined after the mice were euthanized. * P<0.05. (C) Cell angiogenesis in vivo was assessed by CD31 immunostaining. Haematoxylin and eosin-stained images and immunohistochemistry analysis of CD31 protein levels in xenograft tumour tissues (bar: 100 µm).
Exosomal MALAT1 promotes angiogenesis through certain angiogenesis-related genes
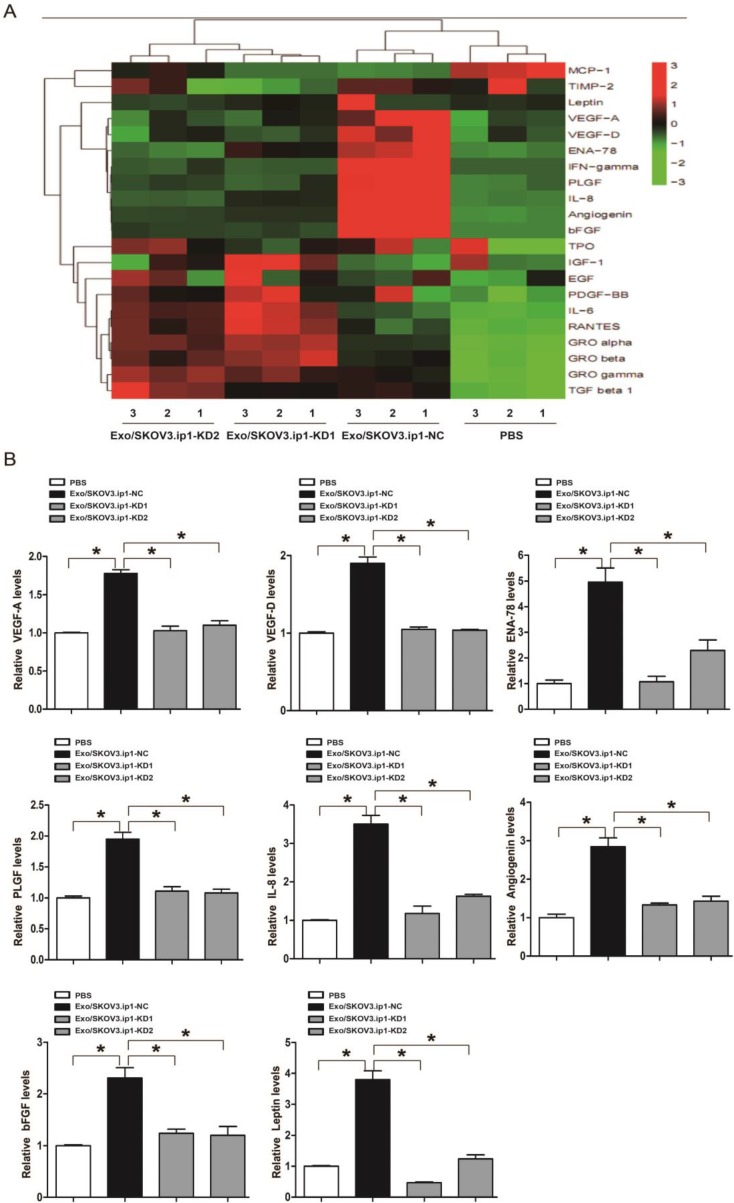
To gain insight into the possible mechanism through which exosomal MALAT1 promoted the angiogenesis of HUVECs, we explored the downstream genes dysregulated by exosomal MALAT1. Angiogenesis-related gene arrays and further qRT-PCR confirmation showed that certain angiogenesis-related genes, including vascular endothelial growth factor (VEGF)-A, VEGF-D, epithelial-neutrophil activating peptide (ENA)-78, placental growth factor (PlGF), interleukin (IL)-8, angiogenin, basic fibroblast growth factor (bFGF) and leptin, were potential downstream mediators of exosomal MALAT1 activity (Fig. 5A and 5B), indicating that exosomal MALAT1 promotes angiogenesis at least in part by regulating VEGF-A, VEGF-D, ENA-78, PlGF, IL-8, angiogenin, bFGF and leptin.
Figure 5.
Array and confirmation analysis of downstream angiogenesis-related genes dysregulated by exosomal MALAT1. (A) The result of hierarchical clustering shows a distinguishable gene expression profile between samples. The rows show 21 differentially expressed angiogenesis-related mRNAs, and the columns show 4 groups. Red represents high expression, whereas green represents low expression. (B) qRT-PCR confirmation of certain angiogenesis-related mRNAs dysregulated by exosomal MALAT1, including VEGF-A, VEGF-D, ENA-78, PlGF, IL-8, angiogenin, bFGF and leptin. * P<0.05.
Serum exosomal MALAT1 is overexpressed and can predict poor prognosis in EOC
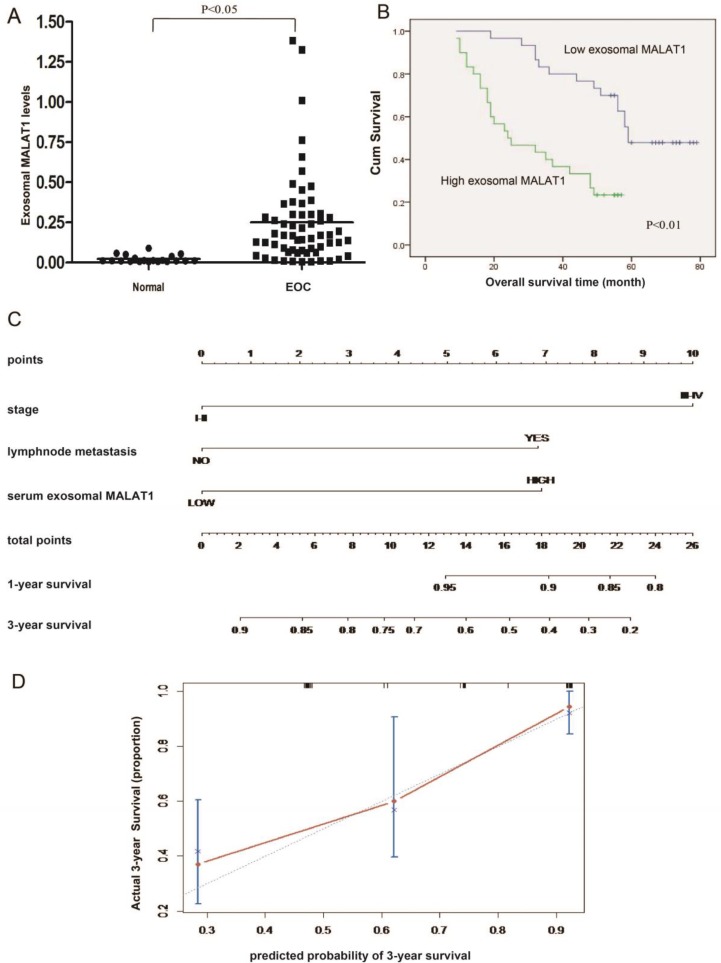
To determine whether exosomal MALAT1 can be detected in the circulation, we tested the expression of serum exosomal MALAT1. The qRT-PCR results showed that serum exosomal MALAT1 levels in EOC patients were significantly higher than those in controls (Fig. 6A). Elevated serum exosomal MALAT1 was correlated with advanced FIGO stage, high histological grade and lymph node metastasis (Table 1). As shown in Fig. 6B, patients with high levels of serum exosomal MALAT1 had shorter OS than those with low levels. In addition, high serum exosomal MALAT1 expression, independent of FIGO stage and lymph node metastasis, was a prognostic factor for OS in EOC patients (Table 2). Moreover, to precisely predict EOC prognosis, a prognostic nomogram model was constructed using the significant factors, including serum exosomal MALAT1 level, FIGO stage and lymph node metastasis (Fig. 6C). The model showed a good prediction of the probability of 3-year OS of EOC patients according to the c-index (0.751, 95% confidence interval [CI]=0.691-0.811) and calibration curve (Fig. 6D). Collectively, these data suggest that serum exosomal MALAT1 is associated with an advanced, metastatic EOC phenotype and poor prognosis and indicate that serum exosomal MALAT1 can serve as a predictive biomarker for EOC prognosis.
Figure 6.
Serum exosomal MALAT1 levels and their association with poor prognosis in ovarian cancer. (A) MALAT1 expression in serum exosomes from EOC patients and normal controls. (B) Kaplan-Meier survival analysis of patients with high and low levels of serum exosomal MALAT1. (C) A predictive nomogram model for the survival of EOC patients according to clinical characteristics, including serum exosomal MALAT1, FIGO stage and lymph node metastasis. (D) Calibration curve for the 3-year survival in EOC patients. The dashed line shows an ideal nomogram, and the solid line refers to the performance of the actual nomogram. The predicted observations matched the actual observations.
Table 1.
The association between serum exosomal MALAT1 expression and clinicopathological variables in EOC patients.
| Variables | Low exosomal MALAT1 expression (n=30) | High exosomal MALAT1 expression (n=30) | P |
|---|---|---|---|
| n(%) | n(%) | ||
| Age (years) | |||
| <50 | 11 (36.7) | 13 (43.3) | 0.598 |
| ≥50 | 19 (63.3) | 17 (56.7) | |
| Histological subtype | |||
| Serous | 22 (73.3) | 26 (86.7) | 0.333 |
| Other | 8 (26.7) | 4 (13.3) | |
| FIGO stage | |||
| I-II | 18 (60.0) | 3 (10.0) | <0.001 |
| III-IV | 12 (40.0) | 27 (90.0) | |
| Histological grade | |||
| G1-G2 | 16 (53.3) | 8 (26.7) | 0.035 |
| G3 | 14 (46.7) | 22 (73.3) | |
| Residual tumour diameter (cm) | |||
| <1 | 26 86.7) | 23 (76.7) | 0.506 |
| ≥1 | 4 (13.3) | 7 (23.3) | |
| Lymph node metastasis | |||
| Absent | 24 (80.0) | 12 (40.0) | 0.002 |
| Present | 6 (20.0) | 18 (60.0) | |
| CA125 level (U/ml) | |||
| <600 | 17 (56.7) | 16 (53.3) | 0.795 |
| ≥600 | 13 (43.3) | 14 (46.7) | |
| Ascites | |||
| <100 | 7 (23.3) | 11 (36.7) | 0.260 |
| ≥100 | 23 (76.7) | 19 (63.3) |
Table 2.
Univariate and multivariate analyses of OS in 60 EOC patients.
| Variables | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| Overall survival | Overall survival | |||||||
| (months) | ||||||||
| Mean±SE | P | β | SE | Wald | P | Exp | 95% CI | |
| Age (years) | ||||||||
| <50 | 55.04±5.23 | 0.201 | — | — | — | — | — | — |
| ≥50 | 44.85±4.14 | — | — | — | — | — | — | |
| Histological subtype | ||||||||
| Serous | 49.55±3.75 | 0.559 | — | — | — | — | — | — |
| Other | 43.75±6.69 | — | — | — | — | — | — | |
| CA125 level (U/ml) | ||||||||
| <600 | 54.37±4.20 | 0.116 | — | — | — | — | — | — |
| ≥600 | 41.45±4.96 | — | — | — | — | — | — | |
| Ascites | ||||||||
| <100 | 42.08±5.48 | 0.292 | — | — | — | — | — | — |
| ≥100 | 50.69±3.95 | — | — | — | — | — | — | |
| FIGO stage | ||||||||
| I-II | 68.36±3.51 | <0.001 | — | — | — | — | — | — |
| III-IV | 34.85±2.91 | 1.286 | 0.507 | 6.443 | 0.011 | 3.620 | 1.341-9.775 | |
| Histological grade | ||||||||
| G1-G2 | 58.30±4.68 | 0.018 | — | — | — | — | — | — |
| G3 | 41.54±4.12 | 0.112 | 0.384 | 0.085 | 0.770 | 1.119 | 0.527-2.375 | |
| Residual tumour | ||||||||
| diameter (cm) | ||||||||
| <1 | 51.61±3.57 | 0.037 | — | — | — | — | — | — |
| ≥1 | 31.73±5.69 | 0.080 | 0.453 | 0.031 | 0.860 | 0.923 | 0. 380-2.244 | |
| exosomal MALAT1 expression | ||||||||
| Low | 61.38±3.56 | — | — | — | — | — | — | |
| High | 31.83±3.25 | <0.001 | 0.891 | 0.396 | 5.070 | 0.024 | 2.437 | 1.122-5.293 |
| Lymph node metastasis |
||||||||
| Absent | 59.61±3.09 | <0.001 | — | — | — | — | — | — |
| Present | 28.33±3.89 | 0.927 | 0.370 | 6.282 | 0.012 | 2.528 | 1.224-5.220 | |
β: Regression coefficient; SE: Standard error; CI: confidence interval.
Discussion
Angiogenesis is a prerequisite for EOC metastasis 4, 5. Mounting evidence indicates that exosomes are critical mediators of communication and information transfer between tumour cells and surrounding cells and that cancer-derived exosomes can play important roles in tumour angiogenesis. For example, exosomes derived from CD90+ cancer cells could influence endothelial cells to achieve a proangiogenic phenotype 31. Exosomes derived from glioma cells have been reported to modulate and promote angiogenesis of recipient human brain microvascular endothelial cells 32. Inspired by these studies, we sought to determine whether exosomes play a role in communication between EOC cells and HUVECs to achieve angiogenesis. As expected, we observed that exosomes derived from metastatic SKOV3.ip1 and HO8910.PM cells were internalized by HUVECs and increased the tube formation of HUVECs, suggesting that exosomes derived from metastatic EOC cells could promote angiogenesis by influencing HUVECs to exhibit proangiogenic activity. Moreover, depletion of exosomes by GW4869 significantly abolished this pro-angiogenesis effect, indicating that such a proangiogenic effect is mainly dependent on the exosomes. Collectively, these data revealed a unique intercellular communication between EOC cells and HUVECs via exosomes and shed new light on EOC angiogenesis from the perspective of exosomes.
Among the cargo components of exosomes, lncRNAs have received increasing interest. Mounting evidence has emphasized that lncRNAs play powerful roles in almost all aspects of tumour progression, such as proliferation, cell cycle, apoptosis, angiogenesis and invasion/metastasis 12-20,43,44. MALAT1, a well-known lncRNA, especially caught our attention because it can promote angiogenesis and metastasis in various cancers 12-20. Although few reports have linked MALAT1 to EOC progression 19,48, the involvement of MALAT1 in EOC angiogenesis and the precise mechanism of how MALAT1 can be transferred to recipient cells to affect angiogenesis is still unknown. In the present study, we observed that MALAT1 was significantly increased not only in metastatic EOC cells but also in their secreted exosomes and the recipient HUVECs, suggesting that MALAT1 can be transferred from EOC cells to recipient HUVECs via exosomes. Subsequent in vitro and in vivo assays showed that MALAT1 knockdown impaired the exosome-mediated proangiogenic ability of HUVECs, indicating that the proangiogenic ability of exosomes was dependent on exosomal MALAT1. Moreover, the array and qRT-PCR results demonstrated that some well-known proangiogenic genes including VEGF-A, VEGF-D, ENA-78, PlGF, IL-8, angiogenin, bFGF and leptin are the potential downstream mediators of exosomal MALAT1 activity. Together, these findings indicated that the metastatic EOC cells promoted angiogenesis partly because they can transfer exosomal MALAT1 to recipient HUVECs and influence HUVECs to exhibit the proangiogenic activity by regulating certain angiogenesis-related genes. Our study proposed a novel mechanism by which EOC cells promote angiogenesis via a previous undescribed regulatory pathway of “exosome-mediated transfer of MALAT1”.
In addition to the important cellular functions of exosomal lncRNAs, current studies have reported that exosomal lncRNAs, such as CRNDE-h, HOTAIR and lncRNA-p21, can be secreted from tumour cells into serum and thus be detected as meaningful and noninvasive prognostic biomarkers 33-35. These studies encouraged us to determine whether exosomal MALAT1 could serve as a biomarker for EOC prognosis. As expected, the present study showed that MALAT1 could be detected in the exosomes of serum collected from EOC patients. The serum exosomal MALAT1 expression in EOC patients was significantly higher than that in controls. High levels of exosomal MALAT1 were associated with an advanced FIGO stage, a high histological grade and lymph node metastasis, suggesting that serum exosomal MALAT1 might be related to advanced and metastatic EOC behaviour. Additionally, based on the multivariate survival analyses and the predictive nomogram model analyses, we concluded that serum exosomal MALAT1 might serve as a predictive biomarker for EOC prognosis.
A major limitation of this study was that we only used a mouse model of intraperitoneal implantation to investigate the effects of exosomal MALAT1 on EOC tumour growth and implantation in vivo. In the future, we aim to construct a mouse model of metastasis by intravenous injection to confirm the roles of exosomal MALAT1 in EOC metastasis.
Conclusion
In conclusion, our data provide a novel mechanism by which EOC cells can transfer MALAT1 via exosomes to recipient HUVECs and, in turn, influence HUVECs by stimulating angiogenesis-related gene expression, eventually promoting angiogenesis. Additionally, elevated serum exosomal MALAT1 was correlated with advanced and metastatic clinical characteristics and could serve as a promising predictor for EOC prognosis. Our study is the first to explore the unique crosstalk between EOC cells and endothelial cells mediated by exosomal MALAT1 to promote cancer angiogenesis and the first to reveal the clinical relevance and prognostic value of serum exosomal MALAT1 in EOC.
Acknowledgments
This project was supported by funding from the National Natural Science Foundation of China (81571404; to Ke-qin Hua), the National Natural Science Foundation for Young Scholars of China (81502240; to Jun-jun Qiu), and the Shanghai Science and Technology Innovation Foundation (16411950500; to Ke-qin Hua).
Author contributions
KQ H and YY L were involved in the study design and in the decision to submit the article for publication. JJ Q was a major contributor in writing the manuscript. XY T analysed and interpreted the patient data. XJ L performed the in vitro experiments. TT Z performed the in vivo experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The clinical study was approved by the Ethics Committee of Obstetrics and Gynecology Hospital of Fudan University, China. Informed consent was obtained from all participants. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Fudan University.
Abbreviations
- lncRNA
long noncoding RNA
- MALAT1
metastasis‑associated lung adenocarcinoma transcript 1
- EOC
epithelial ovarian cancer
- HUVECs
human umbilical vein endothelial cells
- OS
overall survival
- KD
knockdown
- NC
negative control.
References
- 1.Hollis RL, Gourley C. Genetic and molecular changes in ovarian cancer. Cancer Biol Med. 2016;13:236–47. doi: 10.20892/j.issn.2095-3941.2016.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doubeni CA, Doubeni AR, Myers AE. Diagnosis and management of ovarian cancer. Am Fam Phys. 2016;93:937–44. [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J, Kalluri R. Cancer without disease. Nature. 2004;427:787. doi: 10.1038/427787a. [DOI] [PubMed] [Google Scholar]
- 5.Tee AE, Liu B, Song R. et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget. 2016;7:8663–75. doi: 10.18632/oncotarget.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera LB, Bergers G. CANCER. Tumor angiogenesis, from foe to friend. Science. 2015;349:694–5. doi: 10.1126/science.aad0862. [DOI] [PubMed] [Google Scholar]
- 7.Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17:1359–70. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 8.Gordon MS, Mendelson DS, Kato G. Tumor angiogenesis and novel antiangiogenic strategies. Int J Cancer. 2010;126:1777–87. doi: 10.1002/ijc.25026. [DOI] [PubMed] [Google Scholar]
- 9.Xue G, Yan HL, Zhang Y. et al. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2α and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene. 2015;34:1393–406. doi: 10.1038/onc.2014.82. [DOI] [PubMed] [Google Scholar]
- 10.Tate MC, Aghi MK. Biology of angiogenesis and invasion in glioma. Neurotherapeutics. 2009;6:447–57. doi: 10.1016/j.nurt.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Carson-Walter EB, Cooper A. et al. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J NeuroOncol. 2010;99:13–24. doi: 10.1007/s11060-009-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying L, Chen Q, Wang Y. et al. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–94. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 13.Tano K, Mizuno R, Okada T. et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–80. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Liu D, Zhu Y, Pang J. et al. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. 2018;119:1368–80. doi: 10.1002/jcb.26297. [DOI] [PubMed] [Google Scholar]
- 15.Huang JK, Ma L, Song WH. et al. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J Cell Biochem. 2017;118:4821–30. doi: 10.1002/jcb.26153. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Song Y, Liu F. et al. Long non-coding RNA MALAT1 promotes proliferation, angiogenesis, and immunosuppressive properties of mesenchymal stem cells by inducing VEGF and IDO. J Cell Biochem. 2017;118:2780–91. doi: 10.1002/jcb.25927. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Wu Z, Yuan J. et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZC, Tang C, Dong Y. et al. Targeting the long noncoding RNA MALAT1 blocks the pro-angiogenic effects of osteosarcoma and suppresses tumour growth. Int J Biol Sci. 2017;13:1398–408. doi: 10.7150/ijbs.22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Xu X, Lv H. et al. The long noncoding RNA MALAT-1 is highly expressed in ovarian cancer and induces cell growth and migration. PLOS ONE. 2016;11:e0155250. doi: 10.1371/journal.pone.0155250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thum T, Fiedler J. LINCing MALAT1 and angiogenesis. Circ Res. 2014;114:1366–8. doi: 10.1161/CIRCRESAHA.114.303896. [DOI] [PubMed] [Google Scholar]
- 21.Bobrie A, Colombo M, Raposo G. et al. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–68. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 22.Vlassov AV, Magdaleno S, Setterquist R. et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Ge R, Tan E, Sharghi-Namini S. et al. Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers? Cancer Microenviron. 2012;5:323–32. doi: 10.1007/s12307-012-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JE, Tan HS, Datta A. et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–99. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–42. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Luo F, Wang B. et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370:125–35. doi: 10.1016/j.canlet.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Hsu YL, Hung JY, Chang WA. et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–42. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 28.Li XJ, Ren ZJ, Tang JH. et al. Exosomal microRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem. 2017;44:1741–8. doi: 10.1159/000485780. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura K, Sawada K, Kinose Y. et al. Exosomes promote ovarian cancer cell invasion through transfer of CD44 to peritoneal mesothelial cells. Mol Cancer Res. 2017;15:78–92. doi: 10.1158/1541-7786.MCR-16-0191. [DOI] [PubMed] [Google Scholar]
- 30.Xue M, Chen W, Xiang A. et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conigliaro A, Costa V, Lo Dico A. et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang HL, Hu GW, Chen Y. et al. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci. 2017;21:959–72. [PubMed] [Google Scholar]
- 33.Liu T, Zhang X, Gao S. et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–63. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Zhou Y, Lu J. et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. 2014;31:148. doi: 10.1007/s12032-014-0148-8. [DOI] [PubMed] [Google Scholar]
- 35.Işın M, Uysaler E, Özgür E. et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. doi: 10.3389/fgene.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, Zhou H, Lu K. et al. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291–9. doi: 10.2147/OTT.S155134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R, Xia Y, Wang Z. et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–14. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Liu SC, Luo XH. et al. Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J Clin Lab Anal. 2016;30:1116–21. doi: 10.1002/jcla.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu D, Wolf JK, Scanlon M. et al. Enhanced c-erbB-2/neu expression in human ovarian cancer cells correlates with more severe malignancy that can be suppressed by E1A. Cancer Res. 1993;53:891–8. [PubMed] [Google Scholar]
- 40.Bai F, Feng J, Cheng Y. et al. Analysis of gene expression patterns of ovarian cancer cell lines with different metastatic potentials. Int J Gynecol Cancer. 2006;16:202–9. doi: 10.1111/j.1525-1438.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Dong L, Cui H. et al. Up-regulation of mitochondrial antioxidation signals in ovarian cancer cells with aggressive biologic behavior. J Zhejiang Univ Sci B. 2011;12:346–56. doi: 10.1631/jzus.B1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shenhua X, Lijuan Q, Hanzhou N. et al. Establishment of a highly metastatic human ovarian cancer cell line (HO-8910PM) and its characterization. J Exp Clin Cancer Res. 1999;18:233–9. [PubMed] [Google Scholar]
- 43.Qiu JJ, Wang Y, Liu YL. et al. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget. 2016;7:32478–92. doi: 10.18632/oncotarget.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu JJ, Lin YY, Ye LC. et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–8. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 45.Samardzija C, Luwor RB, Volchek M. et al. A critical role of Oct4A in mediating metastasis and disease-free survival in a mouse model of ovarian cancer. Molecular Cancer. 2015;14:152. doi: 10.1186/s12943-015-0417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Y, Hicks MJ, Kaminsky SM. AAV-mediated persistent bevacizumab therapy suppresses tumor growth of ovarian cancer. Gynecol Oncol. 2014;135(2):325–32. doi: 10.1016/j.ygyno.2014.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko SY, Naora H. HOXA9 promotes homotypic and heterotypic cell interactions that facilitate ovarian cancerdissemination via its induction of P-cadherin. Mol Cancer. 2014 Jul 14;13:170. doi: 10.1186/1476-4598-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L, Wang X, Guo Y. Long non-coding RNA MALAT1 is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp Ther Med. 2017;13:3055–60. doi: 10.3892/etm.2017.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poele RHT, Okorokov AL, Joel SP. RNA synthesis block by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) triggers p53-dependentapapoptosis in human colon carcinoma cells. Oncogene. 1999;18:5765–5772. doi: 10.1038/sj.onc.1202961. [DOI] [PubMed] [Google Scholar]