Abstract
Cancer-testis antigen MAGEA3, being restrictedly expressed in testis and various kinds of tumors, has long been considered as an ideal target for immunotherapy. In this study, we report that MAGEA3 interacts with STAT1 and regulates the expression of tyrosine phosphorylated STAT1 (pY-STAT1) in tumor cells. We show that pY-STAT1 is significantly up-regulated when MAGEA3 is silenced by MAGEA3-specific siRNA. RNA sequencing analysis identified 274 STAT1-related genes to be significantly altered in expression level in MAGEA3 knockdown cells. Further analysis of these differentially expressed genes with GO enrichment and KEGG pathway revealed that they are mainly enriched in plasma membrane, extracellular region and MHC class I protein complex, and involved in the interferon signaling pathways, immune response, antigen presentation and cell chemotaxis. The differentially expressed genes associated with chemokines, antigen presentation and vasculogenic mimicry formation were validated by biological experiments. Matrigel matrix-based tube formation assay showed that silencing MAGEA3 in tumor cells impairs tumor vasculogenic mimicry formation. These data indicate that MAGEA3 expression in tumor cells is associated with immune cells infiltration into tumor microenvironment and anti-tumor immune responses, implying that it may play an important role in tumor immune escape. Our findings reveal the potential impact of MAGEA3 on the immunosuppressive tumor microenvironment and will provide promising strategies for improving the efficacy of MAGEA3-targeted immunotherapy.
Keywords: cancer/testis antigen, tumor microenvironment, MAGEA3, STAT1
Introduction
Cancer/testis antigen MAGEA3 is a member of Melanoma Antigen Gene (MAGE) family, which has restricted expression to the testis and is aberrantly expressed in cancer cells. MAGEA3 has been found to be broadly expressed in a variety of malignancies, including melanoma, breast cancer, head and neck cancer, lung cancer, gastric cancer, skin squamous cell carcinoma, colorectal cancer, etc 1. The relatively restricted expression of MAGEA3 and its immunogenicity has made it an ideal target for immunotherapies 1, 2. Recently, emerging data indicated that MAGEA3 expression is associated with hallmarks of aggressive cancer: MAGEA3 expression increased invasive potential in vitro, and orthotopic xenografts of MAGEA3-overexpressing human thyroid carcinoma cells showed increased tumor metastases to the lung 3; MAGEA3 was found to be highly expressed in a cancer stem cell-like side population in bladder cancer which exhibited more robust tumor growth in vivo 4; MAGEA3 expression in cancer cells was associated with poor prognosis, for example, MAGEA3 expression in non-small cell lung cancer was found to be significantly correlated with decreased survival of patients, and MAGEA3 expression in breast cancer is significantly associated with advanced tumor grade, and correlated with worse outcome 5, 6.
Recently, immunotherapy has taken center stage as a novel cancer therapeutic approach. Targeting immune checkpoints such as cytotoxic T lymphocyte antigen 4 (CTLA4), programmed cell death protein 1 (PD1) and its ligand PD-L1 have achieved encouraging results in multiple cancers by inducing a remarkable anti-tumor response 7. However, only a small number of patients have pronounced clinical response with immune checkpoint blockade, and the majority have experienced no clinical benefit when provided the same treatment 8. To date, extensive data have revealed that the tumor microenvironment (TME) is associated with the outcomes of immunotherapy 9. A more recent report indicated that expression of MAGEA members such as MAGEA3, MAGEA6 and MAGEA2 is associated with resistance to blockade of CTLA4 10.
Here we report that MAGEA3 interacts with signal transducer and activator transcription 1 (STAT1), and is involved in remodeling TME by regulating the expression of chemokines, antigen presentation-related genes and formation of vasculogenic mimicry (VM).
Materials and Methods
Cell lines and culture
Human melanoma cell line Hs294T and SK-Mel-28 cells were purchased from ATCC (Manassas, VA, USA) and maintained in DMEM (ATCC) supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA), and human embryonic kidney cell line HEK293T cells were kept by our department and maintained in DMEM (Invitrogen) supplemented with 10% FBS.
Plasmids, siRNA and transfections
The cDNA encoding human MAGEA3 was generated by RT-PCR from Hs294T cells and subcloned into SalI/NotI sites on the pRK-Flag and pRK-HA vectors, and the XhoI/NotI sites on the pCL-Flag vector. The expression vector pCMV-Flag-STAT1 was purchased from Public Protein/Plasmid Library (Nanjing, China). The siRNA sequences for si-MAGEA3#1 (GCGAAUUAGCAAUAACAUACAUGAG), si-MAGEA3#2 (AGAAUGCAAGCGAAAUUAAAUCUGA) and control siRNA were synthesized at RiboBio (Guangzhou, China).
Expression plasmids were transfected with VigoFect (Vigorous Biotechnology, Beijing, China) for HEK293T cells, and MegaTran (OriGene, Rockwell, MD, USA) for SK-Mel-28 cells. siRNAs were transfected with jetPRIME (Polyplus-transfection, Strasbourg, France) for Hs294T cells. All transfections were performed according to the manufacturer's instructions.
Co-immunoprecipitation (Co-IP) and mass spectrometry (MS)
Co-IP was performed as follows: HEK293T cells were transfected with expression vectors pRK-HA-MAGEA3 and pCMV-Flag-STAT1, harvested 48 h following transfection, washed with PBS and lysed with immunoprecipitation (IP)-buffer (20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100 and 1mM EDTA) supplemented with protease inhibitors cocktail (Roche Diagnostics, Indianapolis, IN, USA). Lysates were precipitated with the mouse monoclonal anti-HA antibody (MBL, Nagoya, Japan), the mouse monoclonal anti-Flag antibody (MBL) or normal mouse immunoglobulin IgG (Sigma-Aldrich, St. Louis, MO, USA) at 4˚C overnight, followed by adding protein A-Sepharose (GE Healthcare, Pittsburgh, PA, USA) for additional 4h. For endogenous Co-IP, the lysates from Hs294T cells were precipitated with rabbit monoclonal anti-STAT1 antibody (Cell Signaling Technology, Danvers, MA, USA) or normal rabbit IgG (Sigma-Aldrich). The immunoprecipitates were subsequently washed with IP buffer for 1 h at 4˚C. Complexes were boiled for 5 min in SDS loading buffer and subjected to SDS-PAGE. Western blot analysis was performed with anti-HA, anti-Flag, anti-MAGEA3 (OriGene), or anti-STAT1 antibodies, followed by adding anti-mouse or anti-rabbit antibodies conjugated to horseradish peroxidase (Promega, Madison, WI, USA). Immunoreactive bands were analyzed with chemiluminescence.
Co-immunoprecipitation mass spectrometry (Co-IP/MS) was performed as follows: Flag-MAGEA3-transfected SK-Mel-28 cells were lysed, bound to anti-Flag-Agarose gel (Sigma-Aldrich), eluted with 3×Flag peptide, separated by SDS-PAGE, and stained with silver. Protein bands of interest were excised, in-gel proteolyzed, and identified by MALDI-TOF-MS.
Western blot
Proteins were extracted from transfected Hs294T cells and separated by SDS-PAGE, transferred to a NC membrane (GE Healthcare), blocked and probed with primary specific antibodies, followed by the appropriate secondary antibodies. After washing, the immunoreactive complexes were detected using a chemilumenescence reagent (Biodragon, Beijing, China). Antibodies used in this experiment were as follows: anti-phosphorylated STAT1 (Cell Signaling Technology), anti-CDH5 (Cell Signaling Technology), anti-TFPI2 (Abcam, Cambridge, MA, USA), anti-β-actin (Bioworld Technology, St.Louis, MN, USA).
Immunofluorescence
Hs294T cells were grown directly on glass coverslips for 24 h and then transfected with MAGEA3-specific siRNA, harvested at 36 h following transfection, fixed with 4% formaldehyde in PBS for 15 min at room temperature, and then permeabilized by 100% methanol for 5 min at -20°C. Following 1 h blocking in 5% skimmed bovine serum albumin, the cells were incubated overnight with anti-STAT1 or normal rabbit IgG, and then incubated for 1 h with FITC-conjugated anti-rabbit IgG (ZSGB Bio, Beijing, China). Hoechst33342 (Sigma-Aldrich) staining was performed to visualize the cell nucleus. Images were captured and analyzed using confocal microscope.
RNA sequencing (RNA seq) and bioinformatics analysis
Total RNA was isolated from MAGEA3 silencing or control cells and performed RNA sequencing at the Beijing Genomics Institute (Wuhan, China). Functional enrichment analysis of differentially expressed genes was performed using Gene Ontology (GO) and pathway analysis was performed by Kyoto Encyclopedia of Genes and Genomes (KEGG) mapping based on the Database for Annotation, Visualization, and Integrated Discovery (DAVID) website (http://david.abcc.ncifcrf.gov/).
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA was isolated from MAGEA3 silencing or control cells with Trizol reagent (Invitrogen) and subjected for reverse transcription using Reverse Transcription Kit (Promega) according to the manufacturer's instructions. cDNA was synthesized from 2 μg of total RNA. The resulting complementary DNA was subjected to real-time PCR using SYBR Green qPCR Master Mix (Promega). β-actin was used as an internal standard.
Enzyme-linked immunosorbent assay (ELISA)
Culture supernatants were collected from MAGEA3-silencing Hs294T or control cells, and the quantities of CXCL1, CXCL10 and CXCL11 were detected with commercial available ELISA kits according to the manufacture's procedures. The ELISA kits for CXCL10 and CXCL11 were purchased from Biolegend (San Diego, CA, USA), and CXCL1 from Abcam.
Tube formation assay
Formation of capillary structures by tumor cells was performed as described 11. Briefly, cells were transfected with MAGEA3-specific siRNA, and after 36 h of transfection, the cells were transferred to a 48-well plate containing 0.15 mL matrigel matrix (BD Biosciences, San Jose, CA, USA). After incubation for 12 h, the tubes formed were observed under a microscope and photographed.
Statistical analyses
All experiments were repeated at least twice with consistent results. Quantitative results are presented as mean ± SD. The significance of differences, unless otherwise indicated, was determined by an unpaired two-tailed Student's t-test (with Welch's correction for unequal variances where necessary). All the statistical analysis were performed with SPSS Statistics version 20 (Armonk, NY, USA) and GraphPad Prism 6 (San Diego, CA, USA). Differences were considered statistically significant at p-value<0.05.
Results
Cancer/testis antigen MAGEA3 interacts with STAT1
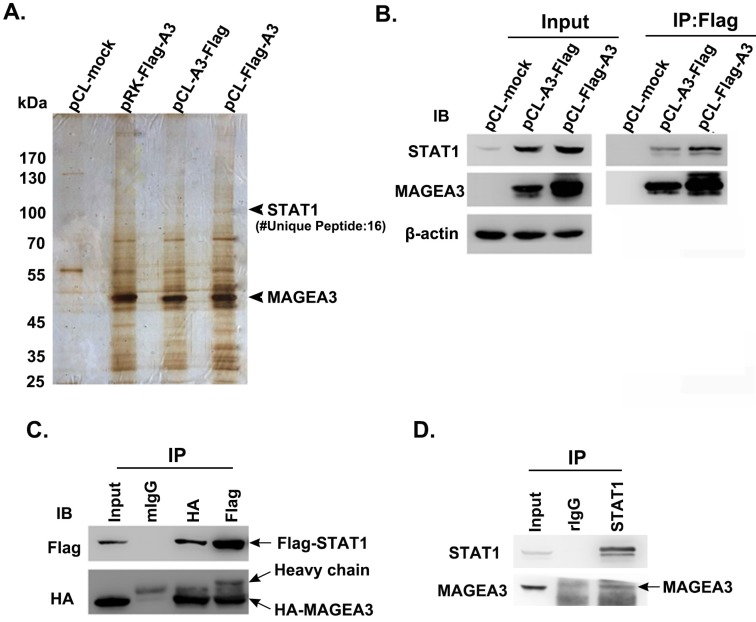
To investigate the biological functions of cancer/testis antigen MAGEA3 in tumor cells, we attempted to screen potential interacting partners of MAGEA3 by Co-IP/MS assay. Considering that the different vectors and the location of tags have impact on the expression levels of transfected genes, three types of expression vectors including pRK-Flag-A3, pCL-Flag-A3 (Flag at N terminal) and pCL-A3-Flag (Flag at C terminal) were transfected into human melanoma SK-Mel-28 cells, respectively. The immunoprecipitated complex with anti-Flag were subject to mass spectrometry analysis, and we identified transcription factor STAT1 (Fig. 1A), which was also validated with anti-STAT1 antibody by western blot (Fig. 1B).
Figure 1.
Cancer/testis antigen MAGEA3 interacts with STAT1. (A) Silver-stained SDS-PAGE gels of anti-Flag immunoprecipitates derived from SK-Mel-28 cells transfected with MAGEA3 expression vectors or control vector. The numbers of polypeptides that yielded unambiguous MS spectra are indicated. (B) SK-Mel-28 cells were transfected with MAGEA3 expression vectors or control vector, whole-cell extracts were immunoprecipitated with anti-Flag, and then STAT1 was detected by western blot. (C) HEK293T cells were co-transfected with Flag-STAT1 and HA-MAGEA3, whole-cell extracts were immunoprecipitated with anti-Flag or anti-HA, followed by immunoblotting (IB) with indicated antibodies. Mouse IgG represents a control antibody used for IP. (D) Whole-cell extracts from Hs294T cells were immunoprecipitated with anti-STAT1, followed by immunoblotting with indicated antibodies. Rabbit IgG represents a control antibody used for IP.
To verify the interaction of MAGEA3 with STAT1, HEK293T cells were co-transfected with expression constructs encoding Flag-STAT1 and HA-MAGEA3, and whole-cell extracts were subjected to Co-IP assay. As shown in Figure 1C, consistent with the mass spectrometry result, exogenously expressed HA-MAGEA3 and Flag-STAT1 were reciprocally precipitated with anti-Flag and anti-HA antibodies. We further investigated the interaction of endogenously expressed MAGEA3 and STAT1 in human melanoma Hs294T cells. As shown in Figure 1D, MAGEA3 was co-precipitated with anti-STAT1 antibody. All these data demonstrated that MAGEA3 is associated with STAT1 within tumor cells.
Silencing MAGEA3 up-regulates pY-STAT1 expression in Hs294T cells
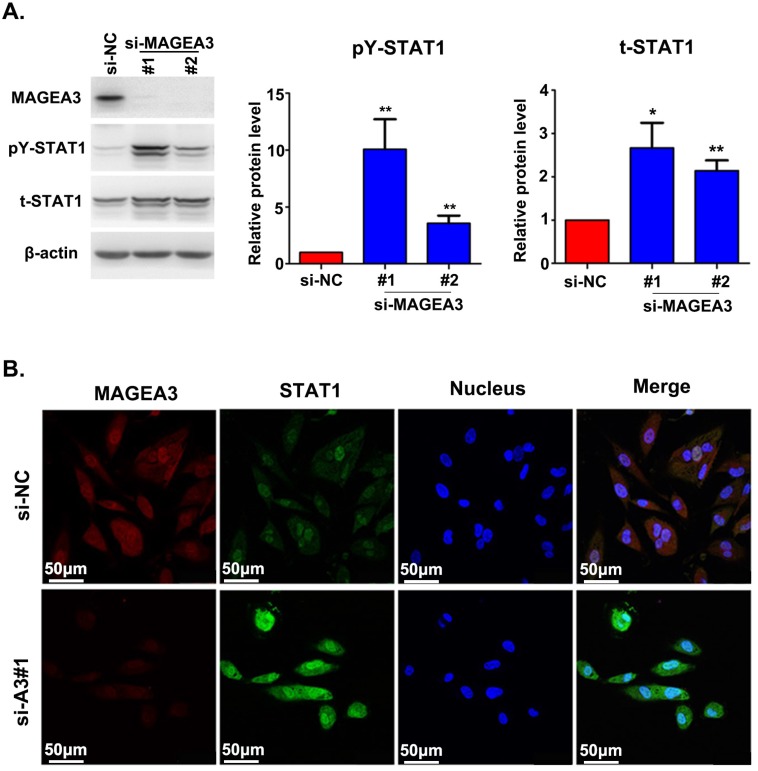
The interaction between MAGEA3 and STAT1 prompted us to investigate the potential impact of MAGEA3 on STAT1 expression. We examined the level of total STAT1 and its active form—STAT1 phosphorylated at tyrosine 701 (pY-STAT1) in the presence or absence of MAGEA3. We observed increased expression of total STAT1 and pY-STAT1 in MAGEA3 knockdown Hs294T cells, and pY-STAT1 was up-regulated more significantly (Fig. 2A). The impact of MAGEA3 on STAT1 expression was further confirmed by immunofluorescence staining. As shown in Figure 2B, knockdown of MAGEA3 with siRNA in Hs294T cells up-regulated the expression of STAT1, especially the active form of STAT1 in the nucleus.
Figure 2.
Silencing MAGEA3 up-regulates pY-STAT1 expression in Hs294T cells. (A) si-MAGEA3 or si-NC (negative control) was transfected into Hs294T cells. Whole-cell extracts were prepared 48 h after transfection and analyzed by immunoblotting with indicated antibodies. Average results from more than two samples are shown in the right panels. (B) si-MAGEA3 or si-NC was transfected into Hs294T cells, followed by immunofluorescence staining with indicated antibodies 48 h after transfection. MAGEA3 (red), STAT1 (green), Hoechst33342 (blue). Scale bar, 50μm. Data are represented as mean ± s.d. *p<0.05; **p<0.01. All p-values were calculated using a two-tailed Student's t-test.
MAGEA3 regulates STAT1-related genes expression
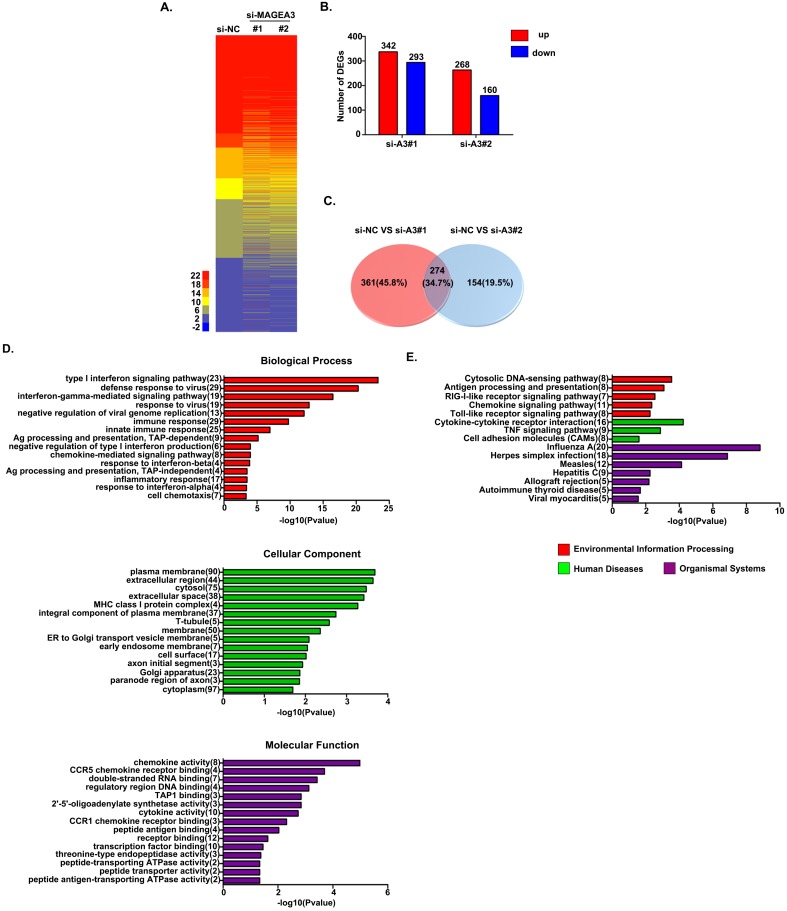
To detect the consequences of MAGEA3 knockdown-induced pY-STAT1 increase in a biological context, we measured mRNA expression in MAGEA3 gene-silenced Hs294T cells (two MAGEA3 specific siRNAs were applied: MAGEA3#1 and MAGEA3#2) by RNA seq analysis. Totally 3083 STAT1-related genes were identified in RNA seq results based on the database (http://software.broadinstitute.org/gsea/msigdb/genesets.jsp), and heatmap of these genes expression in MAGEA3 knockdown and control groups was shown in Figure 3A. With the criteria of at least a 1.5-fold difference and p-value<0.05, 342 genes were found to be up-regulated and 293 down-regulated in MAGEA3#1, 268 up-regulated and 160 down-regulated in MAGEA3#2 compared with control group (Fig. 3B), and of which 274 were common to both MAGEA3#1 and MAGEA3#2 (Fig. 3C).
Figure 3.
MAGEA3 regulates STAT1-related genes expression. (A) Heatmap showed differentially expressed STAT1-related genes. (B) The red and blue bars represented the up-regulated and down-regulated differentially expressed STAT1-related genes in MAGEA3 knockdown Hs294T cells comparing with control cells. (C) Venn diagram showed the common differentially expressed STAT1-related genes between si-MAGEA3#1 and si-MAGEA3#2. (D) GO analysis (across molecular function, biological process and cellular component) was assigned to differentially expressed STAT1-related genes. (E) KEGG pathway analysis was assigned to differentially expressed STAT1-related genes. X axis, negative logarithm (-lg) of the p-value; Y axis, GO or KEGG terms. The top 15 GO or KEGG terms with p-value<0.05 were shown.
The function classification of all 274 differentially expressed genes that were identified in MAGEA3 knockdown Hs294T cells was conducted with GO enrichment analysis. These genes were cataloged according to biological processes (BPs), molecular functions (MFs), and cellular components (CCs) based on GO database, and the top 15 GO terms with p-value <0.05 of each domain were shown in Figure 3D. For BPs, the differentially expressed STAT1- related genes were primarily enriched in type I interferon signaling pathway, immune response, antigen presentation and chemotaxis; for CCs, the differentially expressed genes were distributed mainly in cytoplasm membrane, extracellular region and MHC class I complex; and as for MFs, the differentially expressed genes were mostly enriched in chemokine activity and antigen peptides transporter activity.
The differentially expressed STAT1-related genes were further defined based on KEGG pathway analysis by DAVID tool. The genes differentially expressed in MAGEA3 knockdown Hs294T cells were involved in 27 KEGG pathways, and the top 15 pathways with p-value<0.05 were presented in Figure 3E. Consistent with GO enrichment analysis, differentially expressed genes were prominently enriched in signaling pathways of antigen processing and presentation as well as chemokine.
The above results suggest that the expression of MAGEA3 in tumor cells may be involved in anti-tumor immune responses by regulating antigen presentation and infiltration of immune cells into TME mediated by chemokines.
Knockdown of MAGEA3 in tumor cells increases the expression of chemokines and antigen presentation-related genes
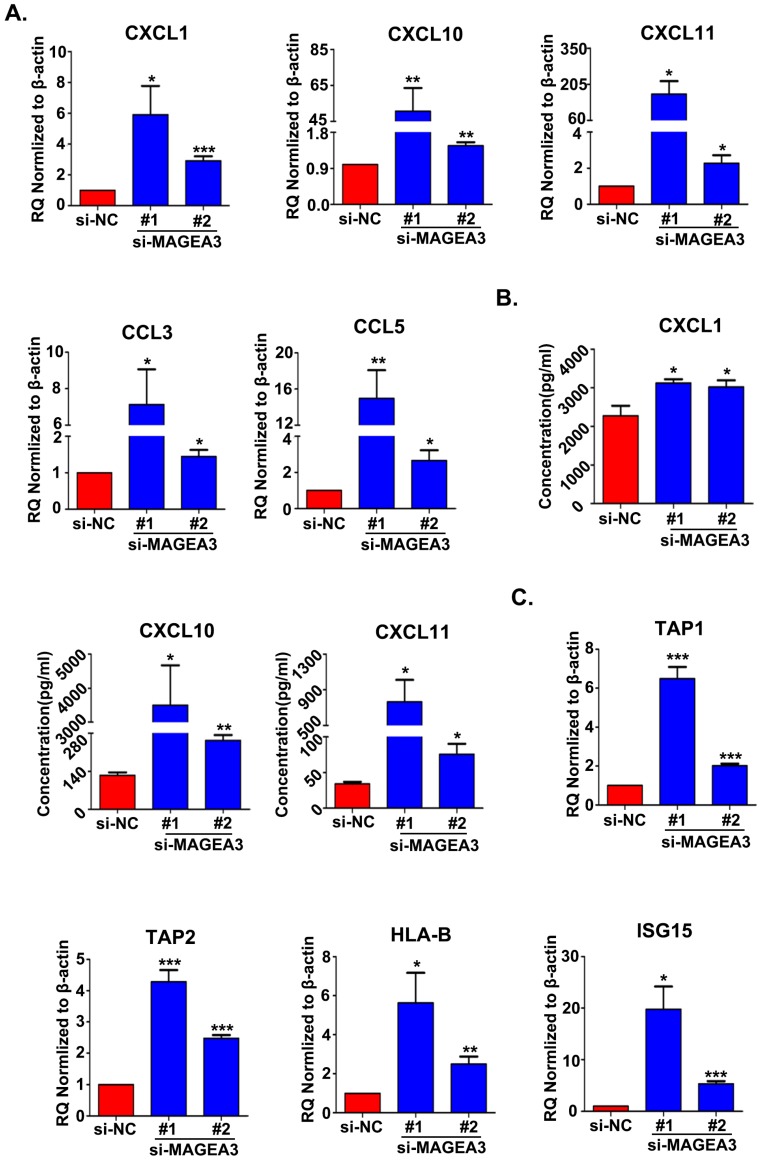
Based on the bioinformatics analysis of the differentially expressed STAT1-related genes, chemokine and antigen processing and presentation signaling pathways appear mostly interesting and relevant, and hence we further validated the genes associated with these two pathways. The expression of chemokines CCL3, CCL5, CXCL1, CXCL10 and CXCL11, which had been reported to bind with the receptors on Th1 cells, CD8+ T cells, NK cells, neutrophils and macrophages and recruit them into TME to regulate immune responses 12, were examined with qRT-PCR in MAGEA3 knockdown Hs294T cells. In accordance with RNA seq results, the expression of CXCL1, CXCL10, CXCL11, CCL3 and CCL5 was up-regulated significantly comparing with control cells (Fig. 4A). Furthermore, the quantities of secreted form of CXCL1, CXCL10 and CXCL11 in the supernatants were determined, and CXCL1, CXCL10 and CXCL11 were found to be increased greatly in MAGEA3 knockdown Hs294T cells compared with control cells (Fig. 4B). These results indicate that the expression of MAGEA3 in tumor cells affects the expression of chemokines and may be involved in regulating infiltration of immune cells into TME.
Figure 4.
Knockdown of MAGEA3 in tumor cells increases the expression of chemokines and antigen presentation-related genes. (A) Chemokines expression was examined in si-MAGEA3-transfected-Hs294T cells and control cells by qRT-PCR. β-actin was used as an endogenous control. (B) Chemokines expression was examined in si-MAGEA3-transfected-Hs294T cells and control cells by ELISA. (C) The expression of antigen presentation-related genes was examined in si-MAGEA3-transfected-Hs294T cells and control cells by qRT-PCR. β-actin was used as an endogenous control. Data are represented as mean ± s.d. *p<0.05; **p<0.01; ***p<0.001. All p-values were calculated using a two-tailed Student's t-test.
Transporter associated with antigen processing (TAP) and MHC class I are essential for antigen presentation and T cell activation 13. Recent report demonstrated that IFN-inducible protein ISG15 could enhance surface expression of MHC class I complex on tumor cells 14. Given the important roles of TAP1, TAP2, HLA-B and ISG15 in antigen processing and presentation as well as their altered expression in RNA seq analysis, we detected the expression level of TAP1, TAP2, HLA-B and ISG15 with qRT-PCR. In line with RNA seq results, the expression of TAP1, TAP2, HLA-B and ISG15 increased largely in MAGEA3 knockdown Hs294T cells comparing with control cells (Fig. 4C), which suggests that the expression of MAGEA3 in tumor cells represses the expression of antigen processing and presentation- related genes and thus may exert a role in inhibiting tumor specific T cells response.
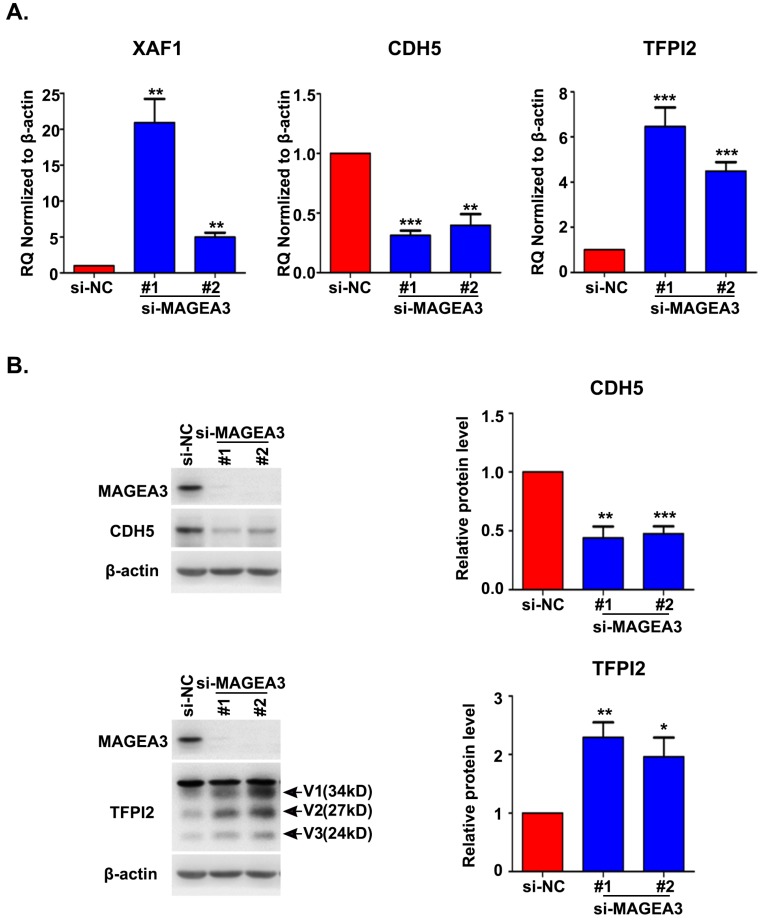
MAGEA3 regulates the expression of vasculogenic mimicry-related genes
Accumulating evidence indicated that VM plays an important role in driving tumor growth and metastasis 15. We noticed the altered expression of vascular endothelial cadherin (CDH5), tissue factor pathway inhibitor 2 (TFPI2) and X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) in RNA seq analysis, which have been reported to be associated with VM formation 16-18. To confirm the results from RNA seq, we analyzed the expression of CDH5, TFPI2 and XAF1 with qRT-PCR and found a significant decrease in CDH5 expression and an increase in XAF1 and TFPI2 expression in MAGEA3 knockdown Hs294T cells (Fig. 5A), consistent with RNA seq results. Further analysis of CDH5 and TFPI2 protein expression by western blot showed that CDH5 was down-regulated and TFPI2 was up-regulated significantly in MAGEA3 knockdown Hs294T cells comparing with control cells (Fig. 5B). These results show that MAGEA3 regulates VM-related genes expression in tumor cells.
Figure 5.
MAGEA3 regulates the expression of vasculogenic mimicry-related genes. (A) The expression of VM-related genes was examined in si-MAGEA3-transfected-Hs294T cells and control cells by qRT-PCR. β-actin was used as an endogenous control. (B) The expression of VM-related genes was examined in si-MAGEA3-transfected-Hs294T cells and control cells by western blot, V1, V2, V3 represent three variants of TFPI2. Average results from more than two samples are shown in the right panels. Data are represented as mean ± s.d. *p<0.05; **p<0.01; ***p<0.001. All p-values were calculated using a two-tailed Student's t-test.
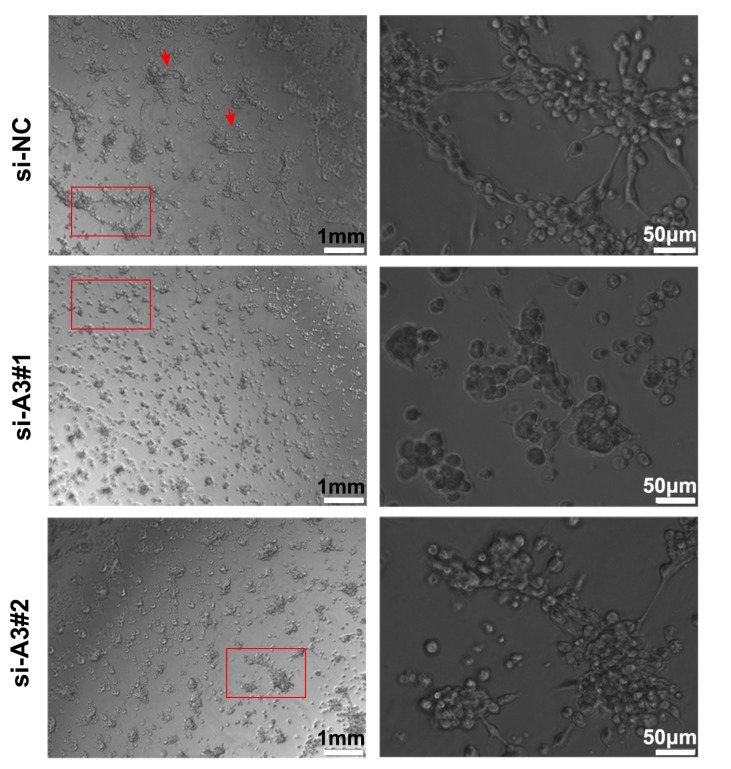
Silencing MAGEA3 impairs formation of tumor vasculogenic mimicry (VM)
To further understand the effects of MAGEA3 on tumor VM formation, we performed tube formation assay. As shown in Figure 6, knockdown of MAGEA3 in Hs294T cells impaired the formation of a complete capillary-like structure compared with control group, which suggests that MAGEA3 expression promotes VM formation mediated by tumor cells.
Figure 6.
Silencing MAGEA3 impairs formation of tumor vasculogenic mimicry (VM). Hs294T cells were transfected with si-MAGEA3 and si-NC, and capillary-like structure formation of the cells was tested on Matrigel and photographed under a light microscope. Right pictures were amplification of the left in red box and red arrows showed capillary-like structures. left scale bar, 1mm; right scale bar, 50μm.
Discussion
Extensive studies revealed that MAGEA3 expression in tumor cells is associated with increased metastases, enrichment in stem cell-like populations and poor clinical prognosis 3, 4, 19. In this study, we found that MAGEA3 interacts with STAT1 and increases the expression level of both total and active form of STAT1. STAT1, a member of signal transducer and activator of transcription (STAT) protein family, is not only crucial for transducing signals from many pathways through the cytoplasm, but also a well-known transcription factor in the nucleus that regulate the expression of a variety of genes 20. STAT1 modulates a variety of cellular processes, such as proliferation and cell death, and more importantly, STAT1 plays a central role in regulating the innate and adaptive arm of immunity 21-24. STAT1 is generally considered as a tumor suppressor, although there are some reports on its tumor promoting functions 23. A correlation of STAT1 expression in tumor cells with good prognosis has been reported in several types of cancers 25, 26. Accumulating evidence indicated that STAT1 acts as a tumor suppressor at multiple levels: tumor-intrinsic growth control and tumor immune environment modulation. It was extensively reported that STAT1 transcriptionally regulates the expression of cell cycle modulators, pro-apoptotic molecules and death-receptors/ligands 23. Moreover, the function of STAT1 in regulation of innate and adaptive immune responses in tumor microenvironment (TME) has been best described, for example, STAT1 regulates the expression of major histocompatibility complex I (MHC class I) molecules on the surface of tumor cells which present antigens to CD8+ T cells for recognition 27; STAT1 promotes polarization of macrophages into M1 type by modulating cytokines expression 28. In addition, it has been reported that many chemokines are target genes of STAT1, such as CCL3, CCL5, CXCL1, CXCL10 and CXCL11, which play important roles in recruiting immune cells into TME 29-32. As different immune cell subsets have distinct effects on tumor progression and therapeutic outcomes, thus STAT1 acts as an important regulator of anti-tumor response in TME.
Our results showed that the expression of active form of STAT1 increased significantly in MAGEA3 knockdown Hs294T cells. To explore the functional consequence of up-regulated pY-STAT1, we performed RNA seq and analyzed the differentially expressed STAT1-related genes in MAGEA3 knockdown Hs294T cells by bioinformatics. Both GO and KEGG pathway analysis unveiled the enrichment of the differentially expressed genes in signaling pathways of chemokine as well as antigen processing and presentation, which are closely associated with anti-tumor immunity in TME 12, 33. We found increased expression of chemokines CXCL1, CXCL10, CXCL11, CCL3 and CCL5 in RNA seq analysis and they were further validated by biological experiments in MAGEA3 knockdown Hs294T cells. It was reported that CXCL10 and CXCL11 can recruit CTLs, Th1 and NK cells into TME by binding to their receptors CXCR3 and develop effective tumor suppression; CXCL1, CCL3 and CCL5 were reported to recruit neutrophils and macrophages to tumor tissues by binding to their corresponding receptors CXCR2, CCR1, CCR3 and CCR5 12. The contributions of CXCL1, CCL3 and CCL5 to anti-tumor immunity in TME are complex, and whether they function as a positively or a negatively regulator in immune responses should depend on the context and occurred on multiple levels 29, 34-37.
Antigen processing and presentation is a crucial step for activating T cells, and inducing MHC class I molecules expression can enhance anti-tumor immune responses and improve effective immunotherapy in a variety of tumors 38. Our results showed that the antigen processing and presentation-related genes TAP1, TAP2, HLA-B and ISG15, which are all target genes of STAT1 39, 40, were up-regulated significantly in MAGEA3 knockdown Hs294T cells. As we know, TAP plays an essential role in antigen presentation of MHC class I molecules by combining with the endogenous peptides and transporting them into ER 13, and ISG15 was recently reported to enhance MHC class I molecules expression on tumor cells 14. The elevated expression of these genes in MAGEA3-silencing cells suggests that MAGEA3 expression suppresses antigen processing and presentation and thus may inhibit anti-tumor immune responses.
Vasculogenic mimicry (VM), a network of channels formed by tumor cells with endothelial cells absent, is usually found in more aggressive tumors. Accumulated evidence shows that tumor cell-mediated VM is associated with tumor invasion, metastasis and poor prognosis 15. CDH5, XAF1 and TFPI2 have been reported to be associated with VM formation, CDH5 and TFPI2 promoting and XAF1 repressing the formation of VM 16-18. Our study indicated that CDH5 was down-regulated, and XAF1 as well as TFPI2 were up-regulated when MAGEA3 was silenced. Furthermore, we found that knockdown of MAGEA3 impairs the VM formation by matrigel-based tube formation assay. These results suggest that the expression of MAGEA3 in tumor cells may facilitate tumor progression by promoting VM formation. It should be pointed out that the altered expression of TFPI2 in our study is in conflict with Wolfram's report that TFPI2 promotes VM formation in melanoma cells C8161 and MUM-2B 17. Further investigation is needed to clarify the role of TFPI2 in VM formation in different cells.
In summary, our study revealed a previously unrecognized regulation of STAT1 signaling pathway by MAGEA3 in tumor cells, and demonstrated that MAGEA3 expression in tumor cells may contribute to tumor immune escape by inhibiting anti-tumor immune responses and accelerating tumor VM formation in TME. Our findings will provide new strategies in MAGEA3-targeted immunotherapy.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (No. 81671637 and 81472645).
References
- 1.Esfandiary A, Ghafouri-Fard S. MAGE-A3: an immunogenic target used in clinical practice. Immunotherapy. 2015;7:683–704. doi: 10.2217/imt.15.29. [DOI] [PubMed] [Google Scholar]
- 2.Straetemans T, van Brakel M, van Steenbergen S, Broertjes M, Drexhage J, Hegmans J. et al. TCR gene transfer: MAGE-C2/HLA-A2 and MAGE-A3/HLA-DP4 epitopes as melanoma-specific immune targets. Clinical & developmental immunology. 2012;2012:586314. doi: 10.1155/2012/586314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer research. 2008;68:8104–12. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 4.Yin B, Zeng Y, Liu G, Wang X, Wang P, Song Y. MAGE-A3 is highly expressed in a cancer stem cell-like side population of bladder cancer cells. International journal of clinical and experimental pathology. 2014;7:2934–41. [PMC free article] [PubMed] [Google Scholar]
- 5.Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S. et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:8055–62. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 6.Ayyoub M, Scarlata CM, Hamaï A, Pignon P, Valmori D. Expression of MAGE-A3_6 in primary breast cancer is associated with hormone receptor negative status, high histologic grade, and poor survival. Journal of immunotherapy. 2014;37:73–6. doi: 10.1097/CJI.0000000000000013. [DOI] [PubMed] [Google Scholar]
- 7.Shi T, Ma Y, Yu L, Jiang J, Shen S, Hou Y, Cancer immunotherapy: A focus on the regulation of immune checkpoints. International journal of molecular sciences; 2018. p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nature medicine. 2018;24:541–50. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MJ, Chang WA, Huang MS, Kuo PL. Tumor microenvironment: a new treatment target for cancer. ISRN biochemistry. 2014;2014:351959. doi: 10.1155/2014/351959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla SA, Bachireddy P, Schilling B, Galonska C, Zhan Q, Bango C. et al. Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell. 2018;173:624–33. doi: 10.1016/j.cell.2018.03.026. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Z, Yu D, Fu S, Zhang G, Pan Y, Bao M. et al. Lycorine hydrochloride selectively inhibits human ovarian cancer cell proliferation and tumor neovascularization with very low toxicity. Toxicology letters. 2013;218:174–85. doi: 10.1016/j.toxlet.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nature reviews Immunology. 2017;17:559–72. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lankat-Buttgereit B, Tampé R. The transporter associated with antigen processing_ function and implications in human diseases. Physiological reviews. 2002;82:187–204. doi: 10.1152/physrev.00025.2001. [DOI] [PubMed] [Google Scholar]
- 14.Desai SD. ISG15: A double edged sword in cancer. Oncoimmunology. 2015;4:e1052935. doi: 10.1080/2162402X.2015.1052935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao L, Liang N, Zhang J, Xie J, Liu F, Xu D. et al. Advanced research on vasculogenic mimicry in cancer. Journal of cellular and molecular medicine. 2015;19:315–26. doi: 10.1111/jcmm.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delgado-Bellido D, Serrano-Saenz S, Fernandez-Cortes M, Oliver FJ. Vasculogenic mimicry signaling revisited: focus on non-vascular VE-cadherin. Molecular cancer. 2017;16:65. doi: 10.1186/s12943-017-0631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruf W, Seftor EA, Petrovan RJ, Weiss RM, Gruman LM, Margaryan NV. et al. Differential role of tissue factor pathway inhibitors 1 and 2 in melanoma vasculogenic mimicry. Cancer research. 2003;63:5381–9. [PubMed] [Google Scholar]
- 18.Wang y, Liu P, Wang X, Mao H. Role of X-linked inhibitor of apoptosis-associated factor-1 in vasculogenic mimicry in ovarian cancer. Molecular medicine reports. 2017;16:325–330. doi: 10.3892/mmr.2017.6597. [DOI] [PubMed] [Google Scholar]
- 19.Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH. et al. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–28. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CK, Rao DT, Gertner R, Gimeno R, Frey AB, Levy DE. Distinct requirements for IFNs and STAT1 in NK cell function. The journal of immunology. 2000;165:3571–7. doi: 10.4049/jimmunol.165.7.3571. [DOI] [PubMed] [Google Scholar]
- 22.Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer immunology immunotherapy. 2011;60:525–35. doi: 10.1007/s00262-010-0961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meissl K, Macho-Maschler S, Muller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20. doi: 10.1016/j.cyto.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Kim HS, Lee MS. STAT1 as a key modulator of cell death. Cellular signalling. 2007;19:454–65. doi: 10.1016/j.cellsig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Fang L, Zhang J, Li G, Ma M, Li C. et al. Blockage of glyoxalase I inhibits colorectal tumorigenesis and tumor growth via upregulation of STAT1, p53, and Bax and downregulation of c-Myc and Bcl-2. International journal of molecular sciences. 2017;18:570. doi: 10.3390/ijms18030570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josahkian JA, Saggioro FP, Vidotto T, Ventura HT, Candido Dos Reis FJ, de Sousa CB. et al. Increased STAT1 expression in high grade serous ovarian cancer is associated with a better outcome. International journal of gynecological cancer. 2018;28:459–465. doi: 10.1097/IGC.0000000000001193. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Wu XP, Zhu XL, Li T, Liu Y. IRG1 increases MHC class I level in macrophages through STAT-TAP1 axis depending on NADPH oxidase mediated reactive oxygen species. International immunopharmacology. 2017;48:76–83. doi: 10.1016/j.intimp.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Liang YB, Tang H, Chen ZB, Zeng LJ, Wu JG, Yang W. et al. Downregulated SOCS1 expression activates the JAK1/STAT1 pathway and promotes polarization of macrophages into M1 type. Molecular medicine reports. 2017;16:6405–11. doi: 10.3892/mmr.2017.7384. [DOI] [PubMed] [Google Scholar]
- 29.Allen F, Rauhe P, Askew D, Tong AA, Nthale J, Eid S. et al. CCL3 enhances antitumor immune priming in the lymph node via IFNgamma with dependency on natural killer cells. Frontiers in immunology. 2017;8:1390. doi: 10.3389/fimmu.2017.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirai K, Shimada T, Yoshida H, Hayakari R, Matsumiya T, Tanji K. et al. Interferon (IFN)-induced protein 35 (IFI35) negatively regulates IFN-beta-phosphorylated STAT1-RIG-I-CXCL10/CCL5 axis in U373MG astrocytoma cells treated with polyinosinic-polycytidylic acid. Brain research. 2017;1658:60–7. doi: 10.1016/j.brainres.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Burke SJ, Lu D, Sparer TE, Masi T, Goff MR, Karlstad MD. et al. NF-kappaB and STAT1 control CXCL1 and CXCL2 gene transcription. American journal of physiology Endocrinology and metabolism. 2014;306:E131–49. doi: 10.1152/ajpendo.00347.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Au KK, Le Page C, Ren R, Meunier L, Clement I, Tyrishkin K. et al. STAT1-associated intratumoural TH1 immunity predicts chemotherapy resistance in high-grade serous ovarian cancer. The journal of pathology Clinical research. 2016;2:259–70. doi: 10.1002/cjp2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology. 2015;4:e1016700. doi: 10.1080/2162402X.2015.1016700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treffers LW, Hiemstra IH, Kuijpers TW, van den Berg TK, Matlung HL. Neutrophils in cancer. Immunological reviews. 2016;273:312–28. doi: 10.1111/imr.12444. [DOI] [PubMed] [Google Scholar]
- 35.Fujimura T, Kambayashi Y, Fujisawa Y, Hidaka T, Aiba S. Tumor-associated macrophages: therapeutic targets for skin cancer. Frontiers in oncology. 2018;8:3. doi: 10.3389/fonc.2018.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao YY, Tsai HC, Chou PY, Wang SW, Chen HT, Lin YM. et al. CCL3 promotes angiogenesis by dysregulation of miR-374b_ VEGF-A axis in human osteosarcoma cells. Oncotarget. 2016;7:4310–25. doi: 10.18632/oncotarget.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaller TH, Batich KA, Suryadevara CM, Desai R, Sampson JH. Chemokines as adjuvants for immunotherapy: implications for immune activation with CCL3. Expert review of clinical immunology. 2017;13:1049–60. doi: 10.1080/1744666X.2017.1384313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, van Hall T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Current opinion in immunology. 2016;39:44–51. doi: 10.1016/j.coi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava RM, Trivedi S, Concha-Benavente F, Hyun-Bae J, Wang L, Seethala RR. et al. STAT1-induced HLA class I upregulation enhances immunogenicity and clinical response to anti-EGFR mAb cetuximab therapy in HNC patients. Cancer immunology research. 2015;3:936–45. doi: 10.1158/2326-6066.CIR-15-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo L, Yoon AR, Yun CO, Chung KC. Covalent ISG15 conjugation to CHIP promotes its ubiquitin E3 ligase activity and inhibits lung cancer cell growth in response to type I interferon. Cell death & disease. 2018;9:97. doi: 10.1038/s41419-017-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]