Abstract
Proper cell proliferation is important for organ homeostasis and normal tissue development. Aberrations in cell proliferation, however, can give rise to degenerative diseases and cancer. Therefore, accurate and simple methods to evaluate cell proliferation are important and necessary to understand the pathways regulating cell proliferation and mechanisms underlying normal development and pathogenesis. The thymidine analog 5-ethynyl-2′-deoxyuridine (EdU), which is incorporated into DNA during active DNA synthesis (e.g., during S phase of the cell cycle), allows easy visualization of proliferating cells. Incorporated EdU can be detected without harsh chemical or enzymatic treatments and is fully compatible with a number of other staining methods, such as immunohistochemistry and in situ hybridization. This protocol describes how to detect proliferating cells using EdU staining in the intestines of Xenopus tadpoles (stages 54–66). Although this method was developed for studying intestinal metamorphosis, it should be applicable to other tissues/organs and other developmental stages as well.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES: Please see the end of this protocol for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
Click-iT Plus EdU Alexa Fluor 594 Imaging Kit (Thermo Fisher Scientific)
Ethanol (100%, 95%, 90%, 80%, and 70%)
Hoechst 33342 (10 mg/mL)
MEMFA fixative <R>
Paraffin wax
Phosphate-buffered saline (PBS, pH 7.2–7.6; 1×, 0.7×) <R>
Phosphate-buffered saline with 0.05% Tween-20 (PBST)
Reagents for situ hybridization, immunostaining, or a TUNEL assay (optional; see Step 7)
Vectashield antifade mounting medium (Vector Laboratories)
Xenopus tropicalis tadpoles and/or frogs (Nieuwkoop and Faber 1965)
Xylene
Equipment
Conical tube (5-mL)
Fluorescence microscope
Hamilton gas-tight syringe (25-μL; gauge: 33; needle length: 12 mm; point style 4; Hamilton Bonaduz)
Microscope slides and coverslips
Microtome
Needle (30-gauge)
Scalpel
Scissors
Slide warmer
Syringe
Water bath
METHOD
Preparation of EdU Solution and EdU Injection
-
1.
Prepare a 10 mg/mL solution of EdU by adding PBS to EdU powder and mixing well in a water bath (55°C-65°C) until the EdU is fully dissolved.
Although the manufacturer recommends dissolving EdU powder in dimethylsulfoxide (DMSO), in our experience most of the tadpoles died after injection of EdU dissolved in DMSO. Thus, we strongly recommend dissolving EdU in PBS. After use, store remaining stock solution at ≤−20°C. -
2.
Place tadpoles in ice water for 5–10 sec until they stop moving. Place tadpoles on a paper towel. With a Hamilton syringe, intraperitoneally inject 1.25, 10, and 10 μL of 10 mg/mL EdU into the abdominal cavity of stage 54, 62, and 66 Xenopus tropicalis, respectively, as shown in Figure 1. Place tadpoles back into the water in which they were reared.
The volumes of injection were determined based on the relative sizes of the animals at different developmental stages.
FIGURE 1.
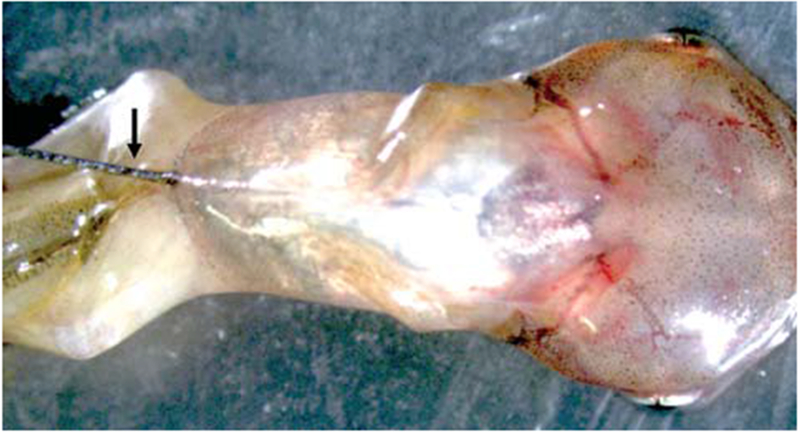
Photographic representation of intraperitoneal injections. The arrow points to the needle (gauge size 33) injecting into a stage 60 tadpole.
Preparation of Intestinal Sections
-
3.
Thirty to 60 min after EdU injection, anesthetize tadpoles by placing them on ice for 5–10 min. Euthanize tadpoles by decapitation with a scalpel or scissors. Isolate the intestine and flush the contents with 0.7× PBS using a 30-gauge needle and syringe.
The flushing helps to reduce auto-fluorescence from the debris, which can make it difficult to visualize the EdU signal. -
4.
Fix the intestine in MEMFA fixative in a 5 mL tube for 2 h at room temperature or overnight at 4°C with shaking. To process the tissue for paraffin embedding, pass the tissue sequentially through graded ethanol solutions for dehydration: 70% ethanol, 80% ethanol, 90% ethanol, 95% ethanol, 100% ethanol, with a 30 min incubation for each ethanol concentration at room temperature. Soak the tissue with xylene three times for 30 min each at room temperature. Melt the paraffin by heating in a heat-proof vessel to 55°C-60°C. Soak the tissue in the paraffin three times for 1 h each. Embed the tissue into paraffin blocks.
-
5.
Cut 5–7 μm thick intestinal sections with a microtome and mount sections on to slides. Dry overnight using a slide warmer (40°C-45°C).
-
6.
Immerse the slides in xylene three times for 5 min each at room temperature, then sequentially in 100% ethanol, 90% ethanol, 80% ethanol, 70% ethanol, and PBST for 5 min each at room temperature.
-
7.
If double labeling is desired, perform in situ hybridization, immunostaining, or a TUNEL assay as described (Okada et al. 2015).
If double labeling will not be performed, proceed to Step 8.See Troubleshooting.
EdU Staining
-
8.
Prepare Click-iT Plus reaction cocktail as per the manufacturer’s instructions.
-
9.
Incubate intestinal sections with 100–200 μL Click-iT Plus reaction cocktail per slide for 30 min at room temperature in the dark.
-
10.
Remove the Click-iT Plus reaction cocktail and wash once with PBST for 5 min at room temperature.
-
11.
Dilute the Hoechst 33342 solution 1:2000 in PBS to obtain a 1× Hoechst 33342 solution. Add 100–200 μL of 1× Hoechst 33342 solution per slide and incubate the sample for 30 min at room temperature in dark.
-
12.
Remove the 1× Hoechst 33342 solution and wash once with PBST for 5 min.
-
13.
Mount sections with coverslips using Vectashield antifade mounting medium.
-
14.
View the slides immediately using a fluorescence microscope or store in the dark.
See Troubleshooting.
TROUBLESHOOTING
Problem (Step 7): When carrying out double labeling with EdU and in situ hybridization, immunostaining, or a TUNEL assay, the signal is weak or absent.
Solution: Consider the following.
Antigen retrieval often interferes with EdU staining. Avoid the antigen retrieval by using a Decloaking Chamber (a pressure cooker which has been designed for heat-induced epitope retrieval) for immunostaining.
Process sections for in situ hybridization, immunostaining, or TUNEL assays first, and then perform EdU staining.
Ensure that the samples are protected from light during incubations.
Problem (Step 14): EdU signal is too low to detect.
Solution: Consider the following.
Ensure that the ingredients in the Click-iT Plus reaction cocktail are mixed in the order listed in the manufacturer’s instructions.
Increase the labeling time. Sacrifice tadpoles 6–12 h after injection for tissues with poor proliferative capacity (see Step 3 above).
Increase the amount of EdU for injection. Test a range of EdU concentrations to determine the optimal concentration (see Step 2 above).
DISCUSSION
Proper spatiotemporal regulation of cell proliferation is critical for the development of multicellular organisms. The measurement of cell proliferation is fundamental to the assessment of the processes underlying development and disease. Historically, measurement of the incorporation of radioactive nucleosides (e.g., 3H-thymidine) was used to assess active DNA synthesis in the S phase of the cell cycle. Additionally, application of the nucleoside analog 5-bromo-2′-deoxyuridine (BrdU) later allowed antibody-based detection of proliferating cells (Gratzner et al. 1976). However, the denaturation step in the BrdU protocol can disrupt DNA integrity, thus affecting nuclear DNA counter-staining, and/or destroy cell morphology and antigen recognition sites. More recently, the thymidine analog EdU was developed and found to be a superior alternative to BrdU (Salic and Mitchison 2008). Incorporated EdU can be detected chemically without DNA denaturation and is fully compatible with a number of other staining methods.
EdU staining allows easy visualization of proliferating cells (Fig. 2). One of the major advantages of EdU staining is that it avoids harsh chemical or enzymatic treatments. Elimination of such treatments results in the preservation of cell morphology and antigen recognition sites, making the approach superior for double-labeling with in situ hybridization, immunohistochemistry, TUNEL assay, or other staining protocols including dye staining (e.g., Methyl green-pyronin Y staining) (Okada et al. 2015). Such double-labeling allows a direct view of the spatiotemporal localization of proliferating cells with other markers such as those for specific cell types or apoptotic cells and should find applications in both basic research and clinical development such as disease diagnosis.
FIGURE 2.
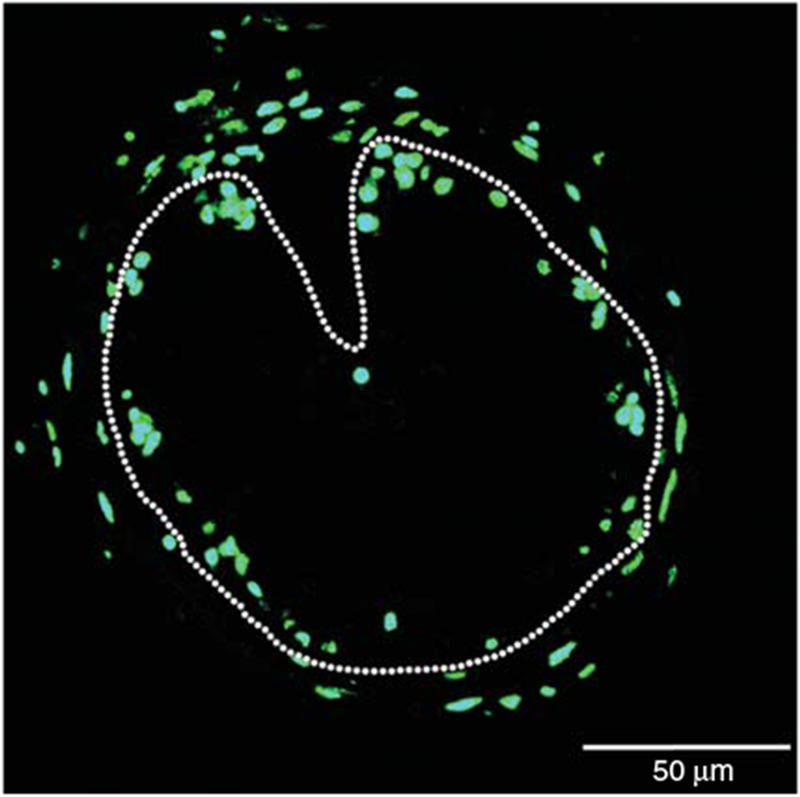
EdU staining of Xenopus intestinal section. A cross-section of the intestine from premetamorphic stage 54 tadpole treated with 10 nm thyroid hormone for 6 d was used for EdU staining. The dotted lines depict the epithelium-mesenchyme boundary. See Okada et al. (2015) for more details.
RECIPES
MEM Salts (10×)
| Reagent | Quantity (for 500 mL) | Final concentration |
|---|---|---|
| MOPS (pH 7.4) | 104.65 g | 1 m |
| EGTA | 3.804 g | 20 mm |
| MgSO4 | 0.602 g | 10 mm |
| H2O | to 500 mL |
This solution can be stored for several months at room temperature, protected from light.
MEMFA Fixative
| MEM salts (10×) <R> | 0.4 mL |
| Formaldehyde (37%) | 0.4 mL |
| H2O | 3.2 mL |
Prepare fresh before use.
Phosphate-Buffered Saline (PBS)
| Reagent | Amount to add (for 1× solution) | Final concentration (1×) | Amount to add (for 10× stock) | Final concentration (10×) |
|---|---|---|---|---|
| NaCl | 8 g | 137 mm | 80 g | 1.37 m |
| KCl | 0.2 g | 2.7 mm | 2 g | 27 mm |
| Na2HPO4 | 1.44 g | 10 mm | 14.4 g | 100 mm |
| KH2PO4 | 0.24 g | 1.8 mm | 2.4 g | 18 mm |
| If necessary, PBS may be supplemented with the following: | ||||
| CaCl2•2H2O | 0.133 g | 1 mm | 1.33 g | 10 mm |
| MgCl2•6H2O | 0.10 g | 0.5 mm | 1.0 g | 5 mm |
PBS can be made as a 1× solution or as a 10× stock. To prepare 1 L of either 1× or 10× PBS, dissolve the reagents listed above in 800 mL of H2O. Adjust the pH to 7.4 (or 7.2, if required) with HCl, and then add H2O to 1 L. Dispense the solution into aliquots and sterilize them by autoclaving for 20 min at 15 psi (1.05 kg/cm2) on liquid cycle or by filter sterilization. Store PBS at room temperature.
ACKNOWLEDGMENTS
Work in our laboratory is supported by the Intramural Research Program of National Institute of Child Health and Human Development, National Institutes of Health. M.O. was supported in part by a Japan Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researchers at the National Institutes of Health.
Footnotes
From the Xenopus collection, edited by Hazel L. Sive.
REFERENCES
- Gratzner HG, Pollack A, Ingram DJ, Leif RC. 1976. Deoxyribonucleic acid replication in single cells and chromosomes by immunologic techniques. J Histochem Cytochem 24: 34–39. [DOI] [PubMed] [Google Scholar]
- Okada M, Wen L, Miller TC, Su D, Shi YB. 2015. Molecular and cytological analyses reveal distinct transformations of intestinal epithelial cells during Xenopus metamorphosis. Cell Biosci 5: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci 105: 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. 1965. Normal table of Xenopus laevis. 1st edn. North Holland Publishing, Amsterdam. [Google Scholar]


