Abstract
Resistance to adjuvant systemic treatment, including taxanes (docetaxel and paclitaxel) is a major clinical problem for breast cancer patients. lncRNAs (long non-coding RNAs) are non-coding transcripts, which have recently emerged as important players in a variety of biological processes, including cancer development and chemotherapy resistance. However, the contribution of lncRNAs to docetaxel resistance in breast cancer and the relationship between lncRNAs and taxane-resistance genes are still unclear. Here, we performed comprehensive RNA sequencing and analyses on two docetaxel-resistant breast cancer cell lines (MCF7-RES and MDA-RES) and their docetaxel-sensitive parental cell lines. We identified protein coding genes and pathways that may contribute to docetaxel resistance. More importantly, we identified lncRNAs that were consistently up-regulated or down-regulated in both the MCF7-RES and MDA-RES cells. The co-expression network and location analyses pinpointed four overexpressed lncRNAs located within or near the ABCB1 (ATP-binding cassette subfamily B member 1) locus, which might up-regulate the expression of ABCB1. We also identified the lncRNA EPB41L4A-AS2 (EPB41L4A Antisense RNA 2) as a potential biomarker for docetaxel sensitivity. These findings have improved our understanding of the mechanisms underlying docetaxel resistance in breast cancer and have provided potential biomarkers to predict the response to docetaxel in breast cancer patients.
Introduction
Breast cancer is the most frequently diagnosed cancer among females and the leading cause of cancer death among women worldwide1. Multiple treatment modalities are used in breast cancer, such as surgery, radiation therapy, chemotherapy and target therapy. Taxanes, including docetaxel and paclitaxel, are among the most commonly used chemotherapeutic agents to treat breast cancer2. However, patients receiving a taxane treatment may become resistant to taxanes, thus resulting in recurrence and metastatic disease.
To reduce patient suffering from disease recurrence caused by taxane resistance, researchers focus on the molecular mechanisms and predictive biomarkers of taxane resistance at multiple levels. Several genes are associated with taxane resistance. The differential expression of the ABCB1 gene is one of the most studied putative biomarkers in taxane-resistant cancers3,4. Pgp (permeability glycoprotein), encoded by the ABCB1 gene, has been reported to act as an ATP-dependent efflux pump and reduce taxane concentration by expelling the drug5. Other genes, such as ABCG2 (ATP binding cassette subfamily G member 2)6 ABCB4 (ATP binding cassette subfamily B member 4)7 are also involved in the process of taxane resistance. However, the underlying mechanism of taxane resistance in breast cancer is still not fully elucidated, and the regulators of the taxane-resistant genes remain unknown. As a result, there are presently no predictive biomarkers for taxanes in clinical use.
Long non-coding RNAs (lncRNAs) are defined as RNAs longer than 200 nucleotides, with little potential in protein coding. Many lncRNAs are described to influence mRNA generation and expression8. Recent studies have shown that lncRNAs are also implicated in chemotherapy resistance. The lncRNA HOTAIR (HOX Transcript Antisense RNA) is reported to contribute to cisplatin resistance in human lung adenocarcinoma cells via down-regulating p21WAF1/CIP1 expression9. Another study found that the lncRNA MRUL (multidrug resistance related and up-regulated lncRNA) promotes ABCB1 expression in multidrug-resistant gastric cancer10. More recently, data have shown that two lncRNAs, ROR (regulator of reprogramming) and CCAT1 (colon cancer-associated transcript-1), regulate docetaxel resistance in lung adenocarcinoma11,12. Furthermore, from a transcriptome microarray study, the lncRNAs HIF1A-AS2 (HIF1A Antisense RNA 2) and AK124454 were shown to promote cell proliferation and invasion in TNBC (triple-negative breast cancers) cells and contribute to paclitaxel resistance13. However, the contribution of lncRNAs to docetaxel resistance in breast cancer is still unclear.
In this study, we carried out whole transcriptome sequencing in two cell lines, MCF-7 and MDA-MB-231, and in their docetaxel-resistant sublines, MCF7-RES and MDA-RES. We identified significantly differentially expressed (SDE) mRNAs and lncRNAs between the parental and resistant sublines, and we also uncovered the potential relationship between the SDE mRNAs and lncRNAs. Compared with previous studies, we discovered several novel genes in addition to ABCB1 which might contribute to the taxane-resistant phenotype of breast cancers. More important, we identified a group of lncRNAs that might potentially regulate taxane sensitivity by controlling the expression of chemotherapy-resistant genes.
Results
Sequencing results and quality control
By performing Illumina-based RNA-Seq sequencing, a total of 1,825,984,984 raw reads were produced from the 12 RNA samples (3 independent samples from each cell line (Table 1)). After quality control, 1,750,124,272 clean reads (157.5 Gb) were obtained.
Table 1.
Summary of data yield and quality control.
| Sample | MCF7_12_P | MCF7_14_P | MCF7_15_P | MCF7_13_T | MCF7_14_T | MCF7_15_T | MDA_13_P | MDA_14_P | MDA_16_P | MDA_14_T | MDA_15_T | MDA_16_T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of raw reads | 155,368,324 | 150,086,362 | 154,603,830 | 151,890,702 | 146,565,380 | 150,531,888 | 154,601,446 | 150,960,982 | 155,560,554 | 150,067,038 | 154,602,638 | 151,145,840 |
| Raw data size(Gb) | 13.98 | 13.51 | 13.91 | 13.67 | 13.19 | 13.55 | 13.91 | 13.59 | 14.00 | 13.51 | 13.91 | 13.60 |
| Number of clean reads | 147,254,586 | 145,503,292 | 148,444,124 | 145,691,082 | 141,393,020 | 143,013,044 | 149,175,500 | 146,265,718 | 146,883,552 | 144,846,612 | 149,085,610 | 142,568,132 |
| Clean data size(Gb) | 13.25 | 13.10 | 13.36 | 13.11 | 12.73 | 12.87 | 13.43 | 13.16 | 13.22 | 13.04 | 13.42 | 12.83 |
| Clean data/Raw data(%) | 94.78 | 96.95 | 96.02 | 95.92 | 96.47 | 95.01 | 96.49 | 96.89 | 94.42 | 96.52 | 96.43 | 94.32 |
| Q30 reads(%) | 88.66 | 92.75 | 89.63 | 89.73 | 92.72 | 88.81 | 90.36 | 92.57 | 89.23 | 92.96 | 90.54 | 88.38 |
| GC(%) | 49.87 | 47.26 | 49.83 | 49.46 | 47.33 | 49.89 | 47.54 | 46.84 | 47.42 | 46.58 | 48.24 | 49.61 |
| Reads mapped to genome | 126,241,357 | 129,177,823 | 134,000,511 | 133,059,665 | 125,613,559 | 122,233,249 | 133,556,825 | 129,562,173 | 126,202,348 | 128,884,515 | 132,119,668 | 118,246,009 |
| Mapping rate(%) | 85.73 | 88.78 | 90.27 | 91.33 | 88.84 | 85.47 | 89.53 | 88.58 | 85.92 | 88.98 | 88.62 | 82.94 |
For the sample names, P represents the parental cells, T represents the docetaxel-resistant cells, number represents the passage.
The proportion of clean reads among the raw reads of the 12 samples ranged from 94.32% to 96.95%. The proportion of clean reads with a Phred quality value greater than 30 among the samples ranged from 88.38% to 92.96%. The GC (guanine and cytosine nucleotides) content of the clean reads of the 12 samples ranged from 47.26% to 49.89%. In total, 82.94% to 91.33% of the clean reads were aligned against the human reference genome (Table 1).
Overview of mRNA expression and the significantly differentially expressed mRNAs between the parental and docetaxel-resistant cell groups
A total of 13,714 mRNAs were detected. More than 97.81% of the mRNAs was detected simultaneously among 3 passages of the same cell line (Sup Fig. 1A). The FPKM (fragments per kilobase million) values of the 13,714 mRNAs from 12 samples were centralized and normalized for the PCA (Principal component analysis). The results show that PC1 (Principal component 1) accounted for 52.28% of the total variance and could separate the MDA cells from the MCF7 cells; PC2 (Principal component 2) accounted for 13.91% of the total variance and separated the parental and docetaxel-resistant MDA cells. However, parental and docetaxel-resistant MCF7 cells clustered together (Sup Fig. 1B).
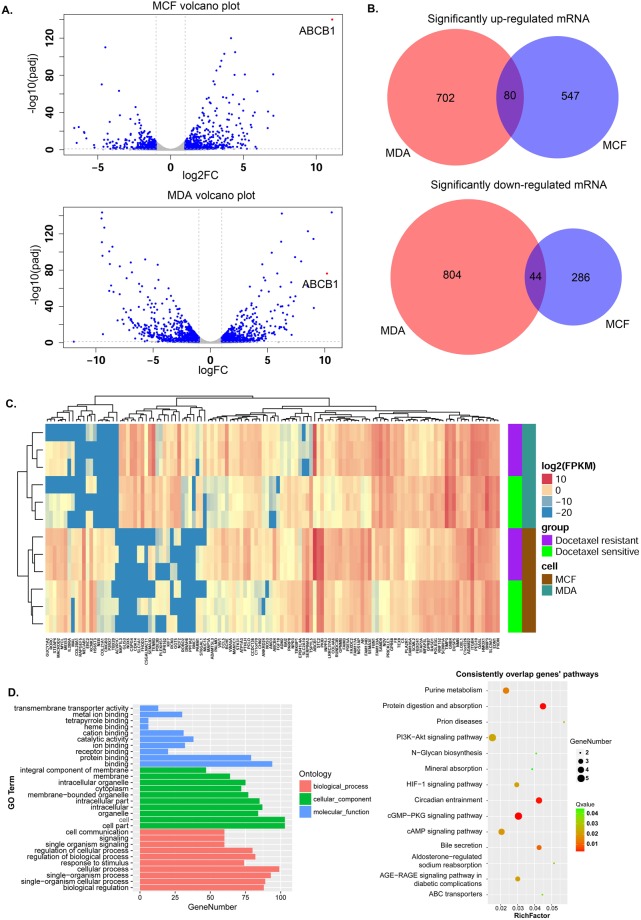
Differentially expressed mRNAs between the parental and docetaxel-resistant cell groups were calculated by DESeq in the two cell lines. The criteria of |Log2FC| > 1 (Log2 (fold change)), FDR < 0.1 (false discovery rate) was used to select the SDE mRNAs. We identified 1630 and 957 SDE mRNAs in the MDA-RES and MCF7-RES groups by comparing them to their parental groups, respectively, as shown in the volcano figure (Fig. 1A).
Figure 1.
Overview of the mRNA expression and significantly differentially expressed mRNAs between the parental and docetaxel-resistant cell groups. (A) Volcano figure showing the significantly differentially expressed genes in the MDA-RES and MCF7-RES cells in comparison to their parental cells. The blue dots represent mRNAs with |Log2FC| > 1, FDR < 0.1. (B) Venn diagrams of the significantly up-regulated or down-regulated mRNAs in both of the cells. (C) Complete-linkage clustering with the 124 consistent SDE mRNAs in the 12 samples. D. KEGG pathway enrichment and GO analysis for the consistent SDE genes. Left panel: GO functional classification of the consistent SDE genes. Red, green, and blue represent the three classes of the GO term. The top 10 enriched GO terms are shown in each class. Right panel: Scatter plot for the KEGG enrichment of the consistent SDE genes. RichFactor is the ratio of the consistent SDE genes annotated in a pathway to all the genes in this pathway. The Q value is the corrected p-value. The top 14 pathways with a Q value < 0.05 are shown in the figure.
In the MDA-RES cells, 782 mRNAs were significantly up-regulated, and 848 mRNAs were significantly down-regulated. In the MCF7-RES cells, 627 mRNAs were significantly up-regulated, and 330 mRNAs were significantly down-regulated.
The most studied MDR (multidrug resistance) related gene, ABCB1 was dramatically up-regulated in both the MDA-RES and MCF7-RES cells (Fig. 1A), which was consistent with a previous report4. The top ten significantly up- or down-regulated genes in the MDA-RES or MCF7-RES cells are shown in Sup Table 1.
By analyzing the list of significantly up-regulated and down-regulated mRNAs in the MDA-RES and MCF7-RES cells, we found 80 mRNAs that were consistently up-regulated and 44 mRNAs that were consistently down-regulated in both of the cells (Fig. 1B, Sup Table 2).
Using these 124 consistent SDE mRNAs, the 12 samples were clustered into two groups (MDA-MB-231 and MCF-7) (Fig. 1C), which indicated that the mRNA diversity between the MCF-7 cells and MDA-MB-231 cells was greater than that between the docetaxel-resistant and parental cells.
GO (Gene ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses of the significantly differentially expressed genes
A KEGG pathway analysis was performed in each cell line. In the MCF7-RES cells, the three most enriched KEGG pathways associated with the SDE mRNAs were the metabolic pathways, axon guidance, and neuroactive ligand-receptor interaction. However, other pathways, such as the PI3K-Akt signaling pathway, Wnt signaling pathway, ECM-receptor interaction and ABC transporters, were also significantly enriched (Sup Fig. 2).
In the MDA-RES cells, the PI3K-Akt signaling pathway, cytokine-cytokine receptor interaction, and pathways in cancer were the top three pathways, followed by the metabolic pathway, AGE-RAGE signaling pathway, and the cell adhesion molecules (CAMs) pathway. Furthermore, ABC transporters, the MAPK signaling pathway and ECM-receptor interaction were also significantly enriched (Sup Fig. 3).
We also performed the KEGG pathway analysis on the 124 consistent SDE mRNAs shared by the MCF7 and MDA cells. As shown in Fig. 1D, 14 signaling pathways were significantly enriched (Q value < 0.05). The most significantly enriched pathway was the cGMP-PKG signaling pathway. However, the ABC transporters, PI3K-Akt signaling pathway, AGE-RAGE signaling pathway in diabetic complications, and HIF-1 signaling pathway were also significantly enriched.
A GO analysis for the consistent SDE mRNAs was also performed to identify the function of the coding transcripts. With regard to biological processes, the most enriched GO term associated with the consistent SDE mRNAs was cellular process. With regard to the cellular component, the most enriched GO terms were cell and cell part. Moreover, the most enriched GO terms associated with the consistent SDE mRNAs for molecular function were binding and protein binding (Fig. 1D).
Overview of lncRNA expression and the significantly differentially expressed lncRNAs between the parental and docetaxel-resistant cell groups
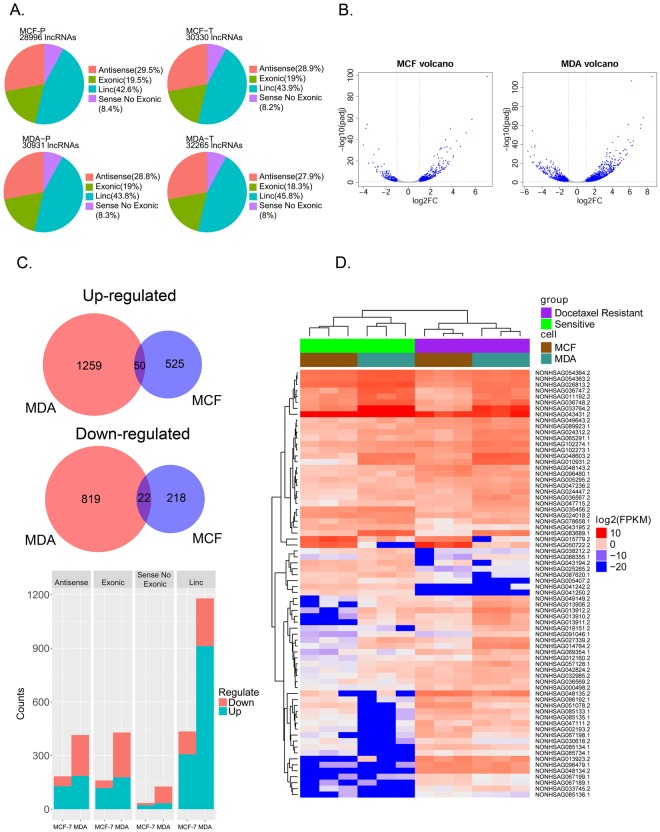
NONCODE 2016 was used to annotate the lncRNAs. The lncRNAs were classified into 4 subgroups as antisense, exonic, linc (long intergenic non-coding), and sense no exonic lncRNAs, according to their location and their relationship with nearby genes. The classification of the lncRNAs in the NONCODE database was provided online (http://www.noncode.org/datadownload/NONCODE2016_human_cc.tgz). We found that lincRNAs and antisense lncRNAs were the two most abundant subtypes in all the cell lines (Fig. 2A).
Figure 2.
Overview of the lncRNA expression and significantly differentially expressed lncRNAs between the parental and docetaxel-resistant cell groups. (A) The subgroups and number of lncRNAs detected in the docetaxel-resistant and parental cell lines (P represents the parental cell line; T represents the docetaxel-resistant cell line). (B) Volcano figure showing the significantly differentially expressed lncRNAs in the MDA-RES and MCF7-RES cells compared to their parental cells. The blue dots represent lncRNAs with |Log2FC| > 1, FDR < 0.1. (C) Upper panel: Venn diagrams of consistently up-regulated or down-regulated lncRNAs in both of the MDA-RES and MCF7-RES cells. Lower panel: the bar chat shows the subgroups and numbers of significantly up-regulated or down-regulated lncRNAs. (D) Complete-linkage clustering with the 72 consistent SDE lncRNAs in the 12 samples.
A total of 39,363 lncRNAs were identified in the 12 RNA samples. More than 81.55% of the lncRNAs were detected simultaneously among three independent passages of the same cell line (Sup Fig. 3A). The differentially expressed lncRNAs between the parental and docetaxel-resistant cell groups were calculated by DESeq. We identified 2,150 and 815 SDE lncRNAs (|Log2FC| > 1, FDR < 0.1) in MDA-RES and MCF7-RES groups (Fig. 2B,C). The SDE lncRNAs identified in the MDA-RES cells included 415 antisense lncRNAs, 1179 intergenic lncRNAs, 429 exonic lncRNAs and 127 sense no exonic lncRNAs. While in the MCF7-RES cells, the SDE lncRNAs included 184 antisense lncRNAs, 435 intergenic lncRNAs, 161 exonic lncRNAs, and 35 sense no exonic lncRNAs (Fig. 2C).
By analyzing the list of SDE lncRNAs in the MCF7-RES and MDA-RES cells, we found 50 lncRNAs that were consistently up-regulated, while 22 lncRNAs were consistently down-regulated in both the MCF7-RES and MDA-RES cell lines (Fig. 2C, Sup Table 3).
By performing complete-linkage clustering with these consistent SDE lncRNAs, the 12 samples were clustered into two groups (parental and resistant) perfectly (Fig. 2D).
lncRNA-mRNA co-expression network construction and docetaxel-resistant module detection
It is well documented that lncRNAs function as regulators of target mRNAs8,14. Additionally, lncRNAs and their target mRNAs usually present similar expression patterns15–17. To predict the target mRNAs of the lncRNAs, we used the WGCNA (weighted gene co-expression network analysis)18 R software package to detect similar expression patterns between the SDE lncRNAs and mRNAs.
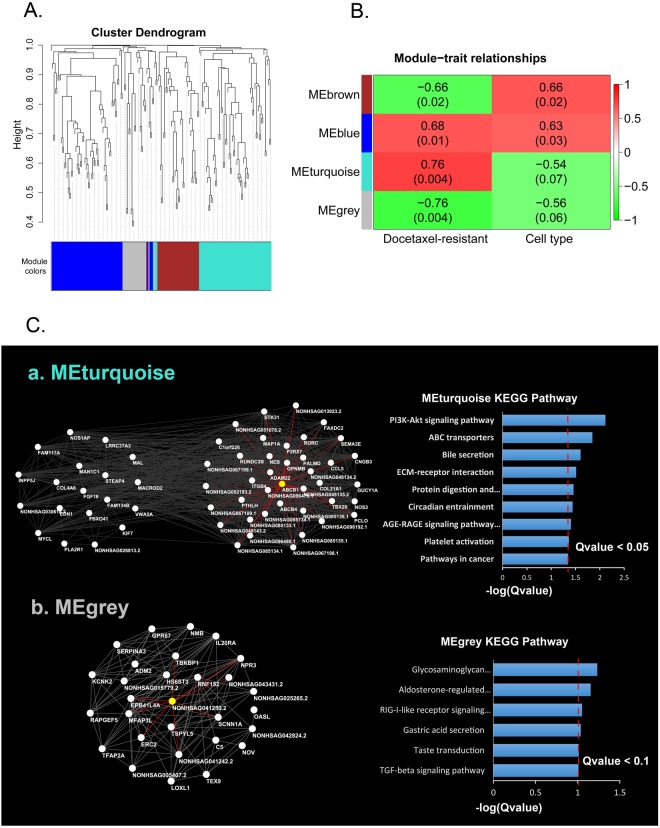
All the SDE genes (72 lncRNAs and 124 mRNAs) were subjected to the WGCNA analysis. We identified 4 groups of co-expressed genes, termed ‘modules’ (Fig. 3A). Each module was labeled with a unique color underneath the cluster tree19.
Figure 3.
lncRNA-mRNA co-expression network construction and docetaxel-resistant module detection using the consistent SDE mRNAs and lncRNAs. (A) A total of 72 SDE lncRNAs and 124 SDE mRNAs were subjected to the WGCNA analysis. Using the hierarchical clustering method, 4 groups of co-expressed genes, termed ‘modules’, were identified. (B) The 4 modules were related to the docetaxel-resistant status. The correlation coefficients representing the module-trait relationships are indicated in the center of the modules, and the corresponding P-values are in the brackets. (C) The hub genes in the Turquoise and Gray modules were visualized by Cytoscape. Pathway enrichment was performed with the hub genes in the two modules. a. The connections of the 59 hub genes in the Turquoise module. The white nodes represent the hub genes, and the red lines indicate the connections to ABCB1. b. The connections of the 29 hub genes in the Gray module. The white nodes represent the hub genes, and the red lines indicate the connections to NONHSAG041250.2 (EPB41L4A-AS2).
Inside a given module, the expression profiles of the transcripts can be summarized by the ‘module eigengene’, which is the weighted average of the module gene expression profiles19. To identify the module associate with docetaxel-resistant status, we regressed each of the 4 module eigengenes on docetaxel-resistant status. We found that the Turquoise and Gray modules were most significantly associated with docetaxel-resistant status (Fig. 3B). Of these 2 associated modules, the Turquoise was positively correlated with docetaxel resistance, meaning that the genes in this module are predominantly up-regulated in docetaxel-resistant cells. In contrast, the Gray module was negatively correlated with a docetaxel-resistant status, meaning that most genes in this module were down-regulated in the docetaxel-resistant cells.
To identify the genes that are central and highly connected in the docetaxel-resistance-associated modules, we selected Hub genes with |GS| > 0.4 (gene significance) and |MM| > 0.8 (module membership) in the two modules and visualized these genes by Cytoscape (Fig. 3C).
In the Turquoise module, we identified 59 hub genes, including 40 protein coding genes and 19 lncRNAs (Sup Table 4). Several well-known chemotherapy-resistance-associated genes including ABCB1 were found in the protein coding genes list. We found that 39 genes, including 22 protein coding and 17 non-coding, were linked to ABCB1 (Table 2). Among the 22 coding genes linked to ABCB1, we found that many of the genes are implicated in drug or chemotherapy resistance, such as ABCB4, ADAM22 (disintegrin and metalloproteinase domain-containing protein 22), GPNMB (glycoprotein nmb), COL21A1 (collagen type XXI alpha 1 chain), and ITGB4 (integrin subunit beta 4) (Table 2, marked with asterisks). Strikingly, we found a list of lncRNAs closely linked to ABCB1 in the Turquoise module (Table 2), indicating that these lncRNAs may function as regulators of ABCB1.
Table 2.
Nodes links to ABCB1 in the Turquoise module.
| Node links to ABCB1 | Region | Log2 (MCF-RES/MCF-7) | MCF padj | Log2 (MDA-RES/MDA-MB-231) | MDA padj |
|---|---|---|---|---|---|
| ABCB4* | chr7:87031361-87105019 | 4.391873563 | 3.0392E-81 | 4.798895486 | 1.78397E-10 |
| ADAM22* | chr7:87563566-87832204 | 3.031204518 | 5.24574E-71 | 2.042465751 | 8.48915E-17 |
| C1orf226 | chr1:162348696-162356608 | 1.30617006 | 2.08367E-08 | 1.218567139 | 0.002493763 |
| CCL5 | chr17:34198495-34207377 | 4.281358914 | 2.28917E-14 | 3.425308925 | 0.016192984 |
| CNGB3 | chr8:87586024-87755903 | 2.927154653 | 8.53446E-07 | 4.3137896 | 4.17421E-05 |
| COL21A1* | chr6:55921388-56112378 | 4.89902163 | 1.49237E-23 | 3.061434362 | 0.004363835 |
| FAXDC2 | chr5:154198052-154230213 | 1.236440283 | 7.16651E-08 | 2.057091728 | 0.002687024 |
| GPNMB* | chr7:23286316-23314729 | 3.634939682 | 3.41382E-06 | 3.979523007 | 5.07143E-21 |
| GUCY1A2 | chr11:106557910-106889171 | 3.923885806 | 3.04573E-08 | 2.419615258 | 0.008199061 |
| ITGB4* | chr17:73717516-73753899 | 1.264990464 | 3.44083E-05 | 1.28963119 | 1.4676E-10 |
| MAP1A | chr15:43809806-43823818 | 1.820284794 | 0.005866863 | 2.930015235 | 4.37426E-12 |
| NEB | chr2:152341853-152591001 | 5.071449042 | 1.95162E-15 | 1.272976565 | 0.074560694 |
| NOS3 | chr7:150688144-150711687 | 4.229375395 | 6.48733E-15 | 1.677234009 | 0.009459785 |
| P2RX7 | chr12:121570631-121624354 | 3.174388573 | 3.15828E-08 | 3.742594891 | 0.024211279 |
| PALMD | chr1:100111431-100160097 | 3.418917876 | 1.04212E-12 | 3.476080265 | 0.000185888 |
| PCLO | chr7:82383321-82792197 | 5.256273053 | 6.619E-18 | 1.320153124 | 0.000404159 |
| PTHLH | chr12:28111017-28124916 | 5.660286658 | 2.89989E-37 | 1.102001097 | 0.022862275 |
| RORC | chr1:151778547-151804348 | 2.438996823 | 2.19322E-10 | 2.525862773 | 0.011217021 |
| RUNDC3B | chr7:87257729-87461613 | 3.889364599 | 2.65512E-25 | 2.527876138 | 1.00736E-12 |
| SEMA3E | chr7:82993222-83278479 | 3.4028601 | 3.31146E-90 | 3.774037375 | 3.23296E-38 |
| STK31 | chr7:23749838-23872132 | 4.004572338 | 6.89337E-33 | 1.460119768 | 0.003048511 |
| TBX20 | chr7:35242042-35293711 | 3.537902823 | 0.000168179 | 3.453322279 | 0.025040493 |
| NONHSAG002193.2 | chr1:100111502-100152712 | 2.306183482 | 0.00000108 | 1.765770312 | 0.035443541 |
| NONHSAG013923.2 | chr13:89793303-89815434 | 2.858465834 | 4.82E-10 | 2.473788666 | 0.000298561 |
| NONHSAG048134.2 | chr7:87150777-87151726 | 4.661655972 | 4.16E-34 | 4.849823297 | 1.08E-20 |
| NONHSAG048135.2 | chr7:87229434-87342570 | 3.06347186 | 6.79E-13 | 3.485170506 | 3.16E-09 |
| NONHSAG048143.2 | chr7:87829109-87832202 | 3.266694083 | 3.73E-33 | 1.013242299 | 0.016081777 |
| NONHSAG051078.2 | chr8:115582746-115686229 | 3.00855 | 2.46E-23 | 1.929201108 | 0.008762611 |
| NONHSAG067189.1 | chr13:89594992-89616155 | 5.165615 | 7.06E-47 | 3.947107328 | 1.03E-13 |
| NONHSAG067198.1 | chr13:91322334-91371571 | 3.65213 | 2.43E-20 | 2.165973989 | 0.003256962 |
| NONHSAG067199.1 | chr13:91465148-91472109 | 4.027848 | 2.53E-24 | 2.187822303 | 0.000869085 |
| NONHSAG085133.1 | chr3:165592410-165780111 | 2.821382 | 1.38E-20 | 5.039315468 | 1.02E-23 |
| NONHSAG085134.1 | chr3:165609026-165630375 | 2.314128 | 1.26E-09 | 3.197328113 | 0.000000284 |
| NONHSAG085135.1 | chr3:165633442-165636497 | 2.229836 | 1.85E-09 | 3.518443205 | 9.92E-10 |
| NONHSAG085136.1 | chr3:165651727-165888351 | 1.995099 | 0.000138828 | 1.567863816 | 0.09203305 |
| NONHSAG085734.1 | chr3:126760621-126763555 | 1.264793 | 0.05765945 | 1.590391302 | 0.083859182 |
| NONHSAG096192.1 | chr7:23296654-23301401 | 2.382385 | 1.7E-09 | 3.160102464 | 2.15E-08 |
| NONHSAG096479.1 | chr7:87075330-87214789 | 7.059226 | 1.77E-99 | 7.209915426 | 7.23E-58 |
| NONHSAG096480.1 | chr7:87445537-87459755 | 2.385153266 | 9.27E-10 | 1.169846587 | 0.062398465 |
*Asterisk indicates the gene which has been implicated in chemotherapy resistant; padj refer to adjusted P value.
In the Gray module, we identified 29 hub genes, including 22 protein coding genes and 7 lncRNAs (Sup Table 5). We found that some of the protein coding genes are reported to be associated with chemotherapy sensitivity. The over-expression of TSPYL5 (testis-specific Y-encoded-like protein 5) and TFAP2A (transcription factor AP-2 alpha) are reported to increase the cell sensitivity to chemotherapy in different cancers20–23. Moreover, in the Gray module, we also identified several lncRNAs that were consistently down-regulated in the docetaxel-resistant MCF7-RES and MDA-RES cells. Notably, an antisense lncRNA, EPB41L4A-AS2 (NONHSAG041250.2), which is reported to inhibit tumor proliferation and is associated with favorable prognoses in breast cancer24, was identified as a hub gene in this module.
Location and functional analysis of the important lncRNAs and mRNAs in the resistance- associated modules
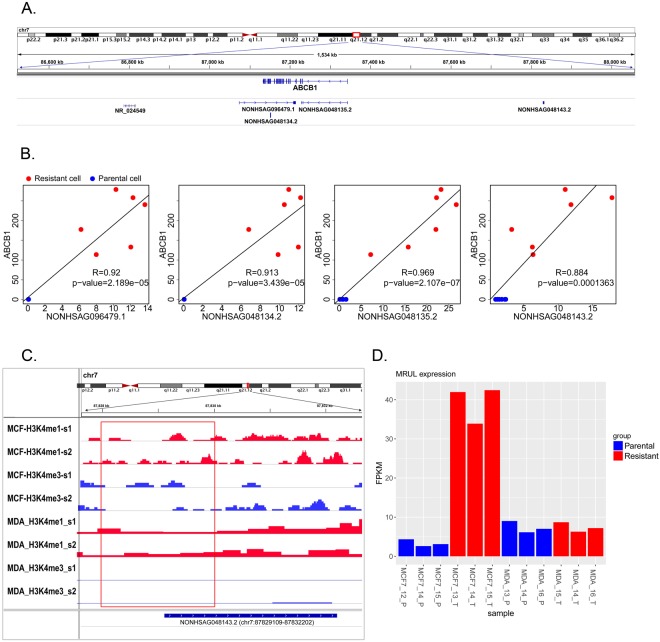
To uncover the relationship of the lncRNAs and the most studied MDR-related gene, ABCB1, we analyzed the genomic locations of the lncRNAs that were linked with ABCB1 in the Turquoise module. Our results showed that several lncRNAs were localized at the regions overlapping or near the ABCB1 gene locus. The locations of lncRNAs NONHSAG048134.2, NONHSAG048135.2 and NONHSAG096479.1 overlapped with ABCB1. NONHSAG048135.2 was transcribed in the same direction and was localized to the exons of the ABCB1 gene, indicating that it was an exonic lncRNA, whereas NONHSAG048134.2 and NONHSAG096479.1 were located in the antisense strand to the ABCB1 gene locus, indicating that these two lncRNAs were antisense lncRNAs for the gene ABCB1 (Fig. 4A). Another lncRNA, NONHSAG048143.2, was located 480 kb upstream of ABCB1 (Fig. 4A).
Figure 4.
Location and functional analysis of important lncRNAs closely linked to ABCB1 mRNAs in the Turquoise module. (A) The location of MRUL, ABCB1 and the four lncRNAs near the ABCB1 locus. (B) Correlation between the expression of the 4 lncRNAs and the ABCB1 gene. (C) The H3K4me1/H3K4me3 status at the TSS region (indicated by the red box) of the lncRNA NONHSAG048143.2. (D) The MRUL expression (FPKM) in the docetaxel-resistant cells (red bars) and parental cells (blue bars).
The correlation analysis of the expression data of these four lncRNAs and ABCB1 mRNA revealed a strong positive correlation between the expression of these lncRNAs and the ABCB1 gene (Fig. 4B), suggesting that there was regulatory relationship between these lncRNAs and the ABCB1 gene.
We downloaded the ChIP-seq (chromatin immunoprecipitation sequencing) datasets for MCF-7 and MDA-MB-231 breast cancer cells from the GEO (Gene expression omnibus) and ENCODE (encyclopedia of DNA elements) databases. By calculating the H3K4me1/H3K4me3 ratios at each TSS (transcriptional start site) of the lncRNAs near ABCB1, we found that the lncRNA NONHSAG048143.2, upstream of ABCB1, was associated with an enhancer-like chromatin state (H3K4me1/H3K4me3 ratio > 1.2) in both the MCF-7 and MDA-MB-231 cells (Fig. 4C). This indicated that NONHSAG048143.2 might function as an eRNA (enhancer lncRNA)25 in the regulation of the ABCB1 gene.
We also identified a previously reported lncRNA, NR_024549 (termed MRUL)10, which located 400 kb downstream of ABCB1. This lncRNA was found to be down-regulated in MCF7-RES cells (Fig. 4D).
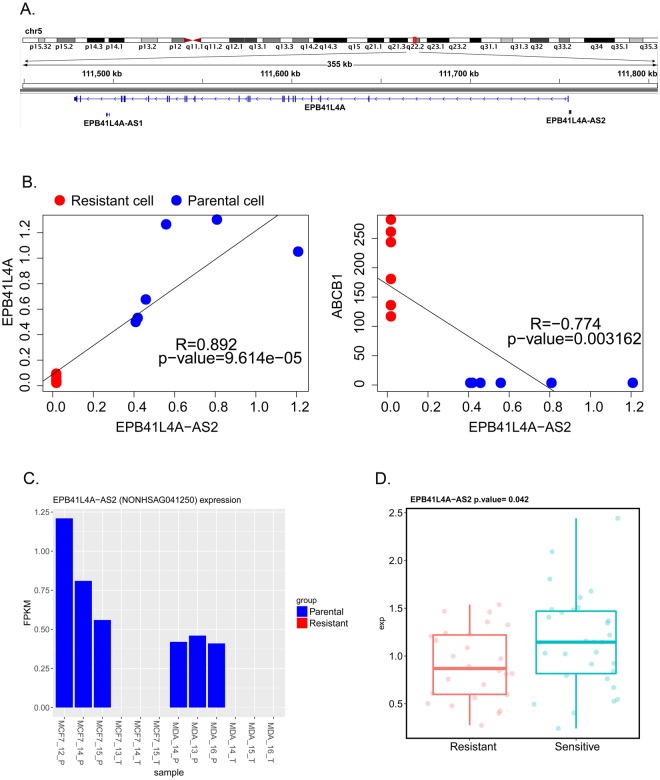
The other lncRNA, EPB41L4A-AS2, which had been reported to be a potential tumor suppressor and prognostic biomarker was located head to head with the EPB41L4A gene in the antisense orientation (Fig. 5A). The correlation analysis on the expression data of this lncRNA and EPB41L4A and ABCB1 mRNAs revealed strong positive correlation between the expression of EPB41L4A-AS2 and EPB41L4A (Pearson correlation test, R = 0.892, P = 9.61 × 10−5, Fig. 5B) but strong negative correlation between the expression of EPB41L4A-AS2 and ABCB1 genes (Pearson correlation test, R = 0.774, P = 0.003, Fig. 5B).
Figure 5.
Location and functional analysis of the lncRNA EPB41L4A-AS2. (A) The location of the EPB41L4A-AS2 and EPB41L4A genes. (B) Correlation between the expression of EPB41L4A-AS2 and EPB41L4A and ABCB1. (C) The expression of EPB41L4A-AS2 (FPKM) in the docetaxel-resistant cells (red bars) and parental cells (blue bars). (D) The expression of EPB41L4A-AS2 in clinical breast cancers tissues (GSE21997) before neoadjuvant chemotherapy (doxorubicin and docetaxel).
To validate the relationship between EPB41L4A-AS2 expression and chemotherapy resistance, we downloaded the primary microarray data GSE21997, which profile the RNA expression in primary breast cancers before neoadjuvant chemotherapy (doxorubicin and docetaxel)26, from the GEO repository database. And the residual breast cancer burden (RCB) index was adopted to classified the breast cancer patients as previous report27. Patients of RCB-III class were considered to be therapeutic resistant. By analyzing the microarray data, we found that the expression of EPB41L4A-AS2 was significantly lower (Student’s t-test, P = 0.042, Fig. 5D) in the resistant group than the non-resistant group.
Real-time qPCR validation of important SDE lncRNAs and mRNAs
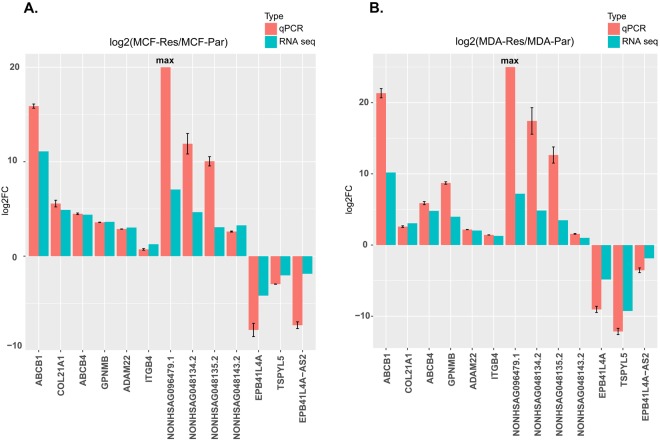
The qPCR results showed that the expression of the mRNAs, including ABCB1, ABCB4, ADAM22, COL21A1, GPNMB, and ITGB4 and the expressions of the lncRNAs, including NONHSAG096479.1, NONHSAG048134.2, NONHSAG048135.2, and NONHSAG048143.2 were up-regulated in docetaxel resistant MCF7-RES and MDA-RES cell lines compared to their parental cells lines. On the contrary, the expressions of the mRNAs EPB41L4A and TSPYL5 and the expressions of the lncRNAs EPB41L4A-AS2 were down-regulated in docetaxel resistant MCF7-RES and MDA-RES cell lines compared to their parental cells lines (Fig. 6). The deregulated genes were identically regulated in both the MCF (Fig. 6A) and MDA (Fig. 6B) groups. Importantly, the qPCR analyses validated the results obtained by RNA-seq data.
Figure 6.
Real-time qPCR validation of important SDE lncRNAs and mRNAs. Validation of the important SDE lncRNAs and mRNAs in the MCF cells (A) and MDA cells (B). The heights of the red columns represent the logarithmically transformed mean fold-changes between the resistant and parental cells detected by QPCR. Data from three duplications are presented as mean ± standard error. The blue columns represent the logarithmically transformed fold-changes between the resistant and parental groups detected by RNA-seq. ‘Max’ represents an infinitely large number due to the absence of NONHSAG096479.1 in the parental cells.
Discussion
Taxanes are a class of anti-microtubule agents that have been demonstrated to be more efficient than anthracycline-based regimens and are listed as the first-line regimens for breast cancer28,29. However, patients receiving taxane treatment usually develop resistance to taxanes, thus resulting in recurrence and metastasis30.
To uncover the mechanisms underlying taxane resistance and find novel potential predictive biomarkers for clinical use, many studies have been carried out in both clinical cancer samples and resistant cell model systems4,31–38. Through this research, a number of genes were identified to be associated with taxane resistance, and the differential expression of the ABCB1 gene was extensively investigated and identified as one of the most credible biomarkers in chemotherapy-resistant cancers3,4,33,34,36,37. However, these previous studies were mainly based on microarray hybridization technologies, and therefore, limited concordant differentially expressed genes are shared between the different studies3,4,33,34,36,37, and the common denominator associated with taxane resistance remains largely unknown. Moreover, the underlying mechanism of taxane resistance in breast cancer and the regulators of taxane-resistant genes still needs to be uncovered.
lncRNAs regulate gene expression at the transcriptional and epigenetic levels39. Recent studies have shown that lncRNAs are also implicated in tumor chemotherapy resistance9–13. However, the contribution of lncRNAs to docetaxel resistance in breast cancer is still unclear.
Here, we used RNA-Seq technology (Illumina platform), which does not rely on pre-designed complement probes and shows a higher sensitivity in the detection of genes with very low expression and a higher accuracy in the detection of extremely abundant genes compared with microarray technology40, to gain a more comprehensive and global profile of mRNAs and lncRNAs in docetaxel-resistant breast cancer cells (MCF7-RES and MDA- RES).
We detected an average of 26,329 lncRNAs and 13,010 mRNAs per sample. Importantly, the lncRNAs and mRNAs showed a high consistency between different passages from the same cell subline (Sup Figs 1A, 4A), which was in concordance with the reports on the high reproductivity of Illumina RNA-Seq40 and demonstrated the reliability of these data.
We identified 1630 and 957 SDE coding genes in the MDA-RES and MCF7-RES groups compared to their parental cells, respectively. In comparison to the previous microarray analysis research in the same cell lines4, we identified more consistently up-regulated or down-regulated genes shared by the MCF7-RES and MDA-RES cells (124 vs 16), which are the potential common denominators associated with taxane resistance. The KEGG pathway analyses were performed on the SDE genes in each cell line, which identified the common pathways shared by the two cell lines and found that the most enriched pathways were different between the two cell lines (Sup Figs 2, 3). The 124 consistent SDE genes were also significantly enriched in 14 signaling pathways (Q-value < 0.05). The most significantly enriched pathway was the cGMP-PKG signaling pathway. However, the ABC transporters, the PI3K-Akt signaling pathway, and the AGE-RAGE signaling pathway in diabetic complications signaling pathway were also significantly enriched. Cyclic GMP (cGMP) is the intracellular second messenger that mediates the action of nitric oxide (NO) and natriuretic peptides (NPs), regulating a broad array of physiological processes. The role of this pathway in chemotherapy is unclear. However, it is implicated in the regulation of apoptosis in cancer cells41 and has been reported to mediate the inhibition of Pgp efflux function by YC-142. The PI3K-Akt signaling pathway was the most significantly enriched pathway in the MDA-MB-231 cells (Sup Fig. 3) and was also significantly enriched in the MCF-7 cells. Although it has not been reported in the previous taxane-resistance studies in breast cancer3,4,33,34,36,37, we found that it was linked to docetaxel resistance in prostate cancer43,44. These results indicated that in addition to the classical pathways, such as the ABC transporters and ECM-receptor interaction pathways, other signaling pathways may also play important roles in taxane resistance in breast cancer.
In addition to the mRNA analysis, the more novel and important work here was the profiling of the lncRNAs in the parental and docetaxel-resistant breast cancer cells. We identified 72 consistently SDE lncRNAs (50 up-regulated and 22 down-regulated) in both of the docetaxel-resistant MCF-7 and MDA-MB-231 cell lines. By performing complete-linkage clustering with these consistently SDE lncRNAs, the 12 samples (3 samples for each of the 4 cell lines) perfectly clustered into two groups: the parental and resistant groups (Fig. 2D). A comparison of the clustering results with a similar number of SDE mRNAs (Fig. 1C) suggested that lncRNAs might be more specific biomarkers for distinguishing docetaxel-resistant cells from the non-resistant cells than mRNAs. Furthermore, this highlighted the potential application of lncRNAs as predictive biomarkers and for the prediction of response to chemotherapy.
lncRNAs have been described to exert their biological functions by regulating the generation and expression of target mRNAs8. Co-expression network analyses have been widely used to predict the target mRNAs of lncRNAs by detecting their similar expression patterns15,16, and thus, the WGCNA R software package was used in our study to decipher the relationship between the SDE mRNAs and the SDE lncRNAs. By using a hierarchical clustering method, we identified 4 groups of co-expressed lncRNAs and mRNAs, termed ‘modules’. Among these, the modules “Turquoise” and “Gray” were most significantly associated with a docetaxel-resistant status.
In the Turquoise module, which was positively correlated with docetaxel resistance, ABCB1 was identified as one of the central and highly connected genes. We found that 39 genes, including 22 coding and 17 non-coding, were linked to ABCB1 in this module. In the 22 coding genes linked to ABCB1, we found that many of the genes were implicated in drug or chemotherapy resistance. For instance, ABCB4, another ATP-binding cassette gene, which was reported to be involved in taxane resistance7, also clustered in this module. ADAM22 was also closely linked to ABCB1. This gene has been reported to function as an estrogen receptor-independent mediator of endocrine-resistant breast cancer45. However, it has not previously been linked to docetaxel resistance. Similarly, the gene GPNMB was also linked to ABCB1 in the Turquoise module and was up-regulated in all the docetaxel-resistant cells. GPNMB has been shown to induce aggressive cellular phenotypes in breast cancer and has been identified as a potential therapeutic target for patients with basal-like breast cancer (BLBC) and TNBC46,47. The concordant expression of ABCB1 and other potential drug-resistant genes in docetaxel-resistant breast cancer cells indicates that besides ABCB1, there are also other genes that may play an important role in the generation of docetaxel resistance, and these genes may contribute to docetaxel resistance in a cooperative way with ABCB1. Pathway enrichment with the genes in this module highlighted the important roles of the PI3K-Akt and ABC transporter signaling pathways.
Moreover, a striking finding here is that many lncRNAs were also co-expressed with ABCB1 and other potential drug-resistant genes in this module. We identified a list of lncRNAs closely linked to ABCB1 in the Turquoise module (Table 2), indicating that some of these lncRNAs may function as regulators of ABCB1 By analyzing the genomic location of the lncRNAs linked with ABCB1, we found that several lncRNAs were located at the regions overlapping or near the ABCB1 gene locus. The lncRNA NONHSAG048135.2 was identified as an exonic lncRNA of ABCB1, whereas NONHSAG048134.2 (termed ABCB1-AS1) and NONHSAG096479.1 (termed ABCB1-AS2) were identified as antisense lncRNAs for ABCB1 Another lncRNA, NONHSAG048143.2, was identified as an intergenic lincRNA located 480 kb upstream of ABCB1.
lncRNAs are implicated in a wide-spectrum of biological processes at different molecular levels, including epigenetic modifications, transcription, and post-transcriptional processing48, and the functions of lncRNAs appears to always to be associated with the location of the lncRNA and the nearby mRNA49,50. Antisense lncRNAs have been shown to bind to the complementary pre-mRNAs, regulate RNA editing and splicing and, finally, affect the expression levels of mRNAs and proteins51,52. For instance, lncRNA ZEB2 NAT (zinc finger e-box binding homeobox 2 natural antisense transcript) was reported to bind to the ZEB2 pre-mRNA and obstruct the splicing of the Zeb2 5’-UTR, thus up-regulating the expression of the Zeb2 protein52. Antisense lncRNAs have also been shown to increase the stability of mRNAs by forming a lncRNA-mRNA duplex and, thereby, up-regulate gene expression53,54. Although, the exact functions of the two novel antisense lncRNAs ABCB1-AS1 and ABCB1-AS2 are still unclear, and further research needs to be performed, our correlation analysis revealed a strong positive correlation between the expressions of these antisense lncRNAs and ABCB1 mRNA (Fig. 4B) and raised the possibility that these lncRNAs may up-regulate the expression of the ABCB1 gene via RNA splicing or stabilization mechanisms.
In addition to the lncRNAs located within the locus of the mRNAs, accumulating evidence suggests that lncRNAs located in the regulatory regions are also critical to the regulation of gene expression39,55. Many lncRNA transcripts from enhancer regions have been shown to facilitate the transcription of protein-coding genes56. For instance, CCAT1-L, which is transcribed from a super-enhancer region 500 kb upstream of the MYC (Proto-Oncogene C-Myc) gene, promotes chromatin interactions between MYC and its upstream regulatory elements and, thereby, up-regulates MYC transcription57. Recently, a study of lncRNAs in doxorubicin- and vincristine-resistant gastric cancer cells also identified an lncRNA, MRUL, which is located 400 kb downstream of ABCB1 and promotes the expression of ABCB110. To our knowledge, this is the only lncRNA reported to promote ABCB1 expression in chemotherapy-resistant cancer cells. Since this lncRNA has not been recorded in the NONCODE 2016 database, we analyzed the differential expression of the transcripts of MRUL between the parental MCF-7 and MDA-MB-231 cells and their docetaxel-resistant sublines. We found that MRUL was dramatically up-regulated in the docetaxel-resistant MCF-7 subline compared to the parental MCF-7 cells. However, this up-regulation was not apparent in the docetaxel-resistant MDA-MB-231 subline (Fig. 4D). This finding suggested that MRUL may also contribute to docetaxel resistance in ER (estrogen receptor) positive breast cancer but maybe not in the TNBC type.
Most intriguingly, in our study, we also identified the lncRNA NONHSAG048143.2 located 480 kb upstream of ABCB1. The correlation analysis revealed a strong positive correlation between the expression of this lncRNA and ABCB1 mRNA (Fig. 4B), suggesting that this lncRNA may be another enhancer lncRNA for ABCB1. The histone H3K4me1/H3K4me3 ratio has been used to characterize enhancer elements across the genome25,58,59. Therefore, we downloaded the ChIP-seq datasets of the MCF-7 and MDA-MB-231 breast cancer cells to calculate the H3K4me1/H3K4me3 ratios at the TSS of the lncRNA NONHSAG048143.2. Our result revealed that lncRNA NONHSAG048143.2 was located in a region presenting an enhancer-like chromatin state (H3K4me1/H3K4me3 ratio > 1.2) in both the MCF-7 and MDA-MB-231 cells (Fig. 4C). This result strongly suggested that this lncRNA might function as an enhancer lncRNA in the up-regulation of the ABCB1 gene in docetaxel-resistant breast cancer cells.
In the Gray module, which was negatively correlated with the docetaxel-resistant status, we found that the gene TSPYL5 was dramatically and consistently down-regulated in the docetaxel-resistant MCF-7 and MDA-MB-231 cells. This gene has been involved in the modulation of cell growth and the cellular response to gamma radiation probably via the regulation of the Akt signaling pathway60. The overexpression of TSPYL5 has also been linked to chemotherapy sensitivity in both lung and prostate cancer cells20,61. The dramatic down-regulation of TSPYL5 in the docetaxel-resistant breast cancer cells may be involved in the loss of docetaxel sensitivity. Interestingly, we found that the lncRNA EPB41L4A-AS2 was expressed in all the parental breast cancer cell lines but was absent in all the docetaxel-resistant descendants (Fig. 5C). According to a recent study, this lncRNA may inhibit tumor cell proliferation and is associated with favorable prognoses in breast cancer24. However, the role of EPB41L4A-AS2 in chemotherapy-resistant cancer has not been deciphered yet. Here, we report, for the first time, the depletion of EPB41L4A-AS2 in docetaxel-resistant breast cancer cells. Our correlation analysis also showed that the decreased level of EPB41L4A-AS2 in the docetaxel-resistant breast cancer cells was significantly associated with an increased level of ABCB1 mRNA (Fig. 5B). These results strongly indicate that the depletion of EPB41L4A-AS2 contributes to the docetaxel-resistant phenotype in breast cancer cells. In support of this notion, our analysis in clinical breast cancers tissues (GSE21997) before neoadjuvant chemotherapy (doxorubicin or docetaxel) also showed that the expression of EPB41L4A-AS2 was significantly lower in the patients resistant to neo-adjuvant chemotherapy than in non-resistant patients (Fig. 5D).
In conclusion, we performed a comprehensive Illumina-based RNA sequencing and analysis on two docetaxel-resistant breast cancer cell lines and their parental breast cancer cell lines. In addition to the most studied docetaxel-resistant gene, ABCB1, we also identified other SDE protein-coding genes and pathways, which might contribute to the generation of docetaxel resistance. These findings improved our understanding of the mechanism underlying docetaxel resistance in breast cancer and suggest that a number of lncRNAs are promising candidates as predictive biomarkers for the docetaxel response. To our knowledge, this is the first study to report on a global profile of mRNAs and lncRNAs in docetaxel-resistant breast cancer. We identified groups of lncRNAs, which were consistently up-regulated or down-regulated in both the docetaxel-resistant MCF-7 and MDA-MB-231 cell lines and constructed a co-expression network to decipher the potential regulatory relationship between the SDE lncRNAs and mRNAs. Our results highlighted four overexpressed lncRNAs that overlapped or were near the ABCB1 locus, which might up-regulate the expression of ABCB1 via different mechanisms. We also identified lncRNA EPB41L4A-AS2 as a potential biomarker for docetaxel sensitivity in clinical breast cancer samples. Although further basic biological and clinical research still needs to be performed to explore the exact biological function of these newly identified lncRNAs, our study provided potential biomarkers to predict the response to docetaxel in breast cancer patients and might help improve the strategy for the treatment of advanced breast cancer.
Methods
Cell culture and treatment
The two human breast cancer cell lines MDA-MB-231 and MCF-7 and their docetaxel-resistant sublines MDA-RES and MCF7-RES were obtained from Hansen et al.’s study4.
The MCF-7 cells were cultured in DMEM (Gibco, Thermo Fisher Scientific, USA) plus 1% non-essential amino acids (Gibco, Thermo Fisher Scientific, USA) and 5% fetal calf serum (FCS) (Gibco, Thermo Fisher Scientific, USA). The MDA-MB-231 cells were cultured in DMEM plus 10% FCS. The culture medium for the MDA-RES and MCF7-RES cells was the same as their parental cells except for the supplementation of 65 or 150 nM of docetaxel (Sanofi Aventis, France), respectively. All the cell lines were grown in 5% carbon dioxide at 37 °C.
Total RNA purification and rRNA depletion transcriptome Sequencing
For the Illumina-based RNA-Seq, the total RNA from 3 passages of MCF-7, MCF7-RES, MDA-MB-231 and MDA-RES, was purified with RNAiso Plus Kit (TAKARA, Japan). After the RNA purification and DNase I digestion, rRNAs were removed from the total RNA with RiboMinus Eukaryote Kit (Qiagen, USA). The remained RNAs were fragmented using Ambion Fragmentation Solution (Thermo Fisher Scientific, USA). Then the mRNA fragments were used to synthesize cDNAs, followed by end repairing and adenine connection. Sequencing adaptors were ligated the fragments. Suitable fragments were selected according to the agarose gel electrophoresis results for PCR amplification. The StepOnePlus System (Applied Biosystems, Thermo Fisher Scientific, USA) and the Agilent 2100 Bioanaylzer (Agilent Technologies, USA) were used to quantify and qualify the sample libraries in the QC steps. Finally, all the libraries were sequenced by the HiSeq. 2000 sequencer (Illumina, USA).
lncRNA and mRNA expression profiling
Firstly, adaptor sequences were removed from the raw reads. And low quality reads, which contained more than 50% of low quality bases (base quality < 10) and 5% of undefined nucleotides [N] on the read, were also removed. To avoid the reads mapped to the remaining rRNAs disturbing the next analysis, we aligned the reads to the human ribosomal RNA sequences (rRNA) and filtered out the perfect mapping reads. After the above two-step filtering, the remaining reads were considered clean reads and were used in next expression profile. In the mRNA analysis, the clean reads were mapped to the human reference genome (hg19) by TopHat262. Then, the genes annotated in RefSeq were quantified by Cufflinks tools63. For lncRNA, the clean reads were mapped to the noncoding RNA transcriptome (NONCODE 201664) by STAR65 and quantified by RSEM66. The parameters used in the analysis tools referred to the ENCODE RNA-seq data processing pipelines (https://www.encodeproject.org/pipelines/). Finally, we obtained all the mRNA and lncRNA expression tables represented in both the FPKM and read-count format.
Identification of significantly differentially expressed lncRNAs and mRNAs
The differentially expressed lncRNAs and mRNAs between the parental and docetaxel-resistant cells groups were calculated by DESeq.67 in the two cell lines, respectively. To ensure that the denominator was not zero when performing the division operation, we added 1 to every element in the read-count matrix. The P-value was calculated based on a negative binomial distribution model, and adjusted by Benjamini-Hochberg mothed. The criteria of |Log2FC| > 1, FDR < 0.1 was used to select the significantly differentially expressed lncRNAs and mRNAs.
GO and KEGG pathway enrichment
Functional annotation enrichment analyses for GO and the KEGG pathway were performed using the KOBAS server68. P-value is calculated by hypergeometric test. The Q-value is the P-value corrected by Benjamini-Hochberg mothed. The GO terms and pathways with a Q value less than 0.05 were considered significantly enriched.
Co-expression network analysis
The WGCNA R package18 was used to construct the co-expression network between the significantly differentially expressed lncRNAs and mRNAs.
By hierarchical clustering of the expression data, several modules, which are clusters of highly interconnected genes, were identified in the co-expression network. Then, we related the drug-resistant feature to the module eigengene to identify the drug-resistant modules (Pearson correlation test, P-value < 0.05). In the drug-resistant modules, hub genes were selected by |MM| > 0.8 and |GS| > 0.4, and they were highly connected with other genes. The connections among the hub genes were visualized as an unsigned network by Cytoscape69.
Classification of lncRNAs near ABCB1 with the downloaded ChIP-seq (H3K4me1/H3K4me3) data
We downloaded the ChIP-seq datasets for the MCF-7 and MDA-MB-231 breast cancer cells from the GEO and ENCODE website. The NCBI (National Center for Biotechnology Information) accession number for the MDA-MB-231 cell ChIPseq data is GSE49651. The ENCODE accessions for the MCF-7 cell datasets are ENCSR493NBY and ENCSR985MIB. Then, we calculated the H3K4me1/H3K4me3 ratio at each TSS (mean over interval, 1 kb up and down TSS).
Validation of the SDE mRNAs and lncRNAs
The PCR primers for the eight selected mRNAs (including ABCB1, ABCB4, ADAM22, COL21A1, GPNMB, ITGB4, and EPB41L4A) and the five lncRNAs (including EPB41L4A-AS2, NONHSAG096479.1, NONHSAG048134.2, NONHSAG048135.2, and NONHSAG048143.2) were designed using Primer 5.0 (Sup Table 6). The PCR primers for GAPDH was also designed to serve as the endogenous control. The length of the PCR products ranged from 100 bp to 400 bp (Sup Table 6). Total RNAs from each cell line was extracted using RNAiso Plus (TAKARA, Japan), and examined Qubit fluorometer (Invitrogen, Thermo Fisher Scientific, USA). QPCR was performed in triplicate on an StepOnePlus instrument (Applied Biosystems, Thermo Fisher Scientific, USA) using the SYBR Premix Ex Taq II (TAKARA, Japan) reagent. ROX dye II was applied for baseline correction. The relative expressions of the mRNA and lncRNAs were calculated with the 2−ΔΔCt method.
Electronic supplementary material
Acknowledgements
This work was funded by the National Natural Science Foundation of China (81502412), the Scientific Research Projects of Shenzhen Health System (201501015), and the Guangzhou science and technology program key projects (201604020005). The authors would like to thank the contributors of the microarray dataset GSE21997 and the ChIP-seq datasets of GSE49651, ENCSR493NBY and ENCSR985MIB.
Author Contributions
N.B., P.H. and J.S. conceived the study and analyses. Y.Y., S.L., and Z.D. performed the sequencing. P.H., F.L. and L.L. performed the bioinformatics analysis. P.H. wrote the manuscript. J.S., N.B. and S.W. revised the manuscript. N.B., S.W. and Q.G. supervised and supported this project.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peide Huang, Fengyu Li and Lin Li contributed equally.
Contributor Information
Song Wu, Email: doctor_wusong@126.com.
Nils Brünner, Email: nbr@sund.ku.dk.
Jan Stenvang, Email: stenvang@sund.ku.dk.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36231-4.
References
- 1.Torre, L. A., Islami, F., Siegel, R. L., Ward, E. M. & Jemal, A. (AACR, 2017). [DOI] [PubMed]
- 2.King KM, et al. Optimal use of taxanes in metastatic breast cancer. Current oncology. 2009;16:8–20. doi: 10.3747/co.v16i3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HJ, et al. ABCB1 polymorphism as prognostic factor in breast cancer patients treated with docetaxel and doxorubicin neoadjuvant chemotherapy. Cancer science. 2015;106:86–93. doi: 10.1111/cas.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen SN, et al. Acquisition of docetaxel resistance in breast cancer cells reveals upregulation of ABCB1 expression as a key mediator of resistance accompanied by discrete upregulation of other specific genes and pathways. Tumor Biology. 2015;36:4327–4338. doi: 10.1007/s13277-015-3072-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-j, et al. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 6.Leonessa F, Clarke R. ATP binding cassette transporters and drug resistance in breast cancer. Endocrine-related cancer. 2003;10:43–73. doi: 10.1677/erc.0.0100043. [DOI] [PubMed] [Google Scholar]
- 7.Hansen SN, et al. The stepwise evolution of the exome during acquisition of docetaxel resistance in breast cancer cells. BMC genomics. 2016;17:442. doi: 10.1186/s12864-016-2749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature reviews Molecular cell biology. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21 WAF1/CIP1 expression. PloS one. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Molecular and cellular biology. 2014;34:3182–3193. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y, et al. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget. 2017;8:33144. doi: 10.18632/oncotarget.16562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, et al. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016;7:62474. doi: 10.18632/oncotarget.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang YZ, et al. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer research. 2016;76:2105–2114. doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 14.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Q, et al. Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic acids research. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, et al. Analysis of long noncoding RNA and mRNA using RNA sequencing during the differentiation of intramuscular preadipocytes in chicken. PloS one. 2017;12:e0172389. doi: 10.1371/journal.pone.0172389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, et al. Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma. Oncogenesis. 2017;6:e297. doi: 10.1038/oncsis.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong S, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PloS one. 2012;7:e39498. doi: 10.1371/journal.pone.0039498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakshmanan, I. et al. MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing p53. Clinical Cancer Research (2017). [DOI] [PMC free article] [PubMed]
- 21.Nordentoft I, et al. Increased expression of transcription factor TFAP2α correlates with chemosensitivity in advanced bladder cancer. BMC cancer. 2011;11:135. doi: 10.1186/1471-2407-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonckheere N, et al. Tumour growth and resistance to gemcitabine of pancreatic cancer cells are decreased by AP-2α overexpression. British journal of cancer. 2009;101:637–644. doi: 10.1038/sj.bjc.6605190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan E-H, et al. A multicentre phase II gene expression profiling study of putative relationships between tumour biomarkers and clinical response with erlotinib in non-small-cell lung cancer. Annals of oncology. 2010;21:217–222. doi: 10.1093/annonc/mdp520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu S, et al. The long non-coding RNA EPB41L4A-AS2 inhibits tumor proliferation and is associated with favorable prognoses in breast cancer and other solid tumors. Oncotarget. 2016;7:20704. doi: 10.18632/oncotarget.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilott, N. E. et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nature communications5 (2014). [DOI] [PMC free article] [PubMed]
- 26.Romero, A. et al. Glutathione S-transferase P1 c. 313 A>G polymorphism could be useful in the prediction of doxorubicin response in breast cancer patients. Annals of oncology, mdr483 (2011). [DOI] [PubMed]
- 27.Symmans WF, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. Journal of Clinical Oncology. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 28.Nabholtz J-M, et al. Docetaxel and doxorubicin compared with doxorubicin and cyclophosphamide as first-line chemotherapy for metastatic breast cancer: results of a randomized, multicenter, phase III trial. Journal of Clinical Oncology. 2003;21:968–975. doi: 10.1200/JCO.2003.04.040. [DOI] [PubMed] [Google Scholar]
- 29.Nabholtz, J., Paterson, A. & Dirix, L. In Proc Am Soc Clin Oncol
- 30.Brooks T, et al. Taxane-based reversal agents modulate drug resistance mediated by P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Molecular cancer therapeutics. 2003;2:1195–1205. [PubMed] [Google Scholar]
- 31.Chang JC, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. The Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 32.Martín M, et al. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast cancer research and treatment. 2011;128:127. doi: 10.1007/s10549-011-1461-y. [DOI] [PubMed] [Google Scholar]
- 33.İşeri ÖD, Kars MD, Gündüz U. Two different docetaxel resistant MCF-7 sublines exhibited different gene expression pattern. Molecular biology reports. 2012;39:3505–3516. doi: 10.1007/s11033-011-1123-5. [DOI] [PubMed] [Google Scholar]
- 34.Kars MD, Işeri ÖD, Gündüz U. A microarray based expression profiling of paclitaxel and vincristine resistant MCF-7 cells. European journal of pharmacology. 2011;657:4–9. doi: 10.1016/j.ejphar.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Işeri ÖD, Kars MD, Arpaci F, Gündüz U. Gene expression analysis of drug-resistant MCF-7 cells: implications for relation to extracellular matrix proteins. Cancer chemotherapy and pharmacology. 2010;65:447–455. doi: 10.1007/s00280-009-1048-z. [DOI] [PubMed] [Google Scholar]
- 36.Kars MD, Işeri ÖD, Gündüz U. Drug resistant breast cancer cells overexpress ETS1 gene. Biomedicine & Pharmacotherapy. 2010;64:458–462. doi: 10.1016/j.biopha.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Villeneuve DJ, et al. cDNA microarray analysis of isogenic paclitaxel-and doxorubicin-resistant breast tumor cell lines reveals distinct drug-specific genetic signatures of resistance. Breast cancer research and treatment. 2006;96:17–39. doi: 10.1007/s10549-005-9026-6. [DOI] [PubMed] [Google Scholar]
- 38.Duan Z, Lamendola DE, Duan Y, Yusuf RZ, Seiden MV. Description of paclitaxel resistance-associated genes in ovarian and breast cancer cell lines. Cancer chemotherapy and pharmacology. 2005;55:277–285. doi: 10.1007/s00280-004-0878-y. [DOI] [PubMed] [Google Scholar]
- 39.Xiong X-d, et al. Long non-coding RNAs: An emerging powerhouse in the battle between life and death of tumor cells. Drug Resistance Updates. 2016;26:28–42. doi: 10.1016/j.drup.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PloS one. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren Y, Zheng J, Yao X, Weng G, Wu L. Essential role of the cGMP/PKG signaling pathway in regulating the proliferation and survival of human renal carcinoma cells. International journal of molecular medicine. 2014;34:1430–1438. doi: 10.3892/ijmm.2014.1925. [DOI] [PubMed] [Google Scholar]
- 42.Hung C-C, Liou H-H. YC-1, a novel potential anticancer agent, inhibit multidrug-resistant protein via cGMP-dependent pathway. Investigational new drugs. 2011;29:1337–1346. doi: 10.1007/s10637-010-9496-1. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, Li H, Chen Q. INPP4B reverses docetaxel resistance and epithelial-to-mesenchymal transition via the PI3K/Akt signaling pathway in prostate cancer. Biochemical and Biophysical Research Communications. 2016;477:467–472. doi: 10.1016/j.bbrc.2016.06.073. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Zhu G, Getzenberg RH, Veltri RW. The Upregulation of PI3K/Akt and MAP Kinase Pathways is Associated with Resistance of Microtubule‐Targeting Drugs in Prostate Cancer. Journal of cellular biochemistry. 2015;116:1341–1349. doi: 10.1002/jcb.25091. [DOI] [PubMed] [Google Scholar]
- 45.McCartan D, et al. Global characterization of the SRC-1 transcriptome identifies ADAM22 as an ER-independent mediator of endocrine-resistant breast cancer. Cancer research. 2012;72:220–229. doi: 10.1158/0008-5472.CAN-11-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maric G, Rose A, Annis MG, Siegel PM. Glycoprotein non-metastatic b (GPNMB): A metastatic mediator and emerging therapeutic target in cancer. Onco Targets Ther. 2013;6:839–852. doi: 10.2147/OTT.S44906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yardley, D. et al. (AACR, 2012).
- 48.Li X, Wu Z, Fu X, Han W. Long noncoding RNAs: insights from biological features and functions to diseases. Medicinal research reviews. 2013;33:517–553. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 49.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 50.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salameh A, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proceedings of the National Academy of Sciences. 2015;112:8403–8408. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes & development. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang B, et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927–4936. doi: 10.1038/onc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nature medicine. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natoli G, Andrau J-C. Noncoding transcription at enhancers: general principles and functional models. Annual review of genetics. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 57.Xiang J-F, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marques AC, et al. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome biology. 2013;14:R131. doi: 10.1186/gb-2013-14-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim EJ, et al. TSPYL5 is involved in cell growth and the resistance to radiation in A549 cells via the regulation of p21 WAF1/Cip1 and PTEN/AKT pathway. Biochemical and biophysical research communications. 2010;392:448–453. doi: 10.1016/j.bbrc.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 61.Kumar SR, Bryan JN, Esebua M, Amos-Landgraf J, May TJ. Testis specific Y-like 5: gene expression, methylation and implications for drug sensitivity in prostate carcinoma. BMC cancer. 2017;17:158. doi: 10.1186/s12885-017-3134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic acids research. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic acids research. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.