The extraordinary ability of human immunodeficiency virus type 1 (HIV-1) to evade host immunity represents a major obstacle to the development of a protective vaccine. Thus, elucidating the mechanisms whereby HIV-1 protects its external envelope (Env), which is the sole target of virus-neutralizing antibodies, is an essential step toward vaccine design. We identified a key structural element that maintains the HIV-1 Env trimer in a closed, antibody-resistant conformation. A major role is played by two conserved tyrosines at the apex of the Env spike, whose mutation causes a global opening of the trimer structure, exposing multiple concealed targets for neutralizing antibodies. We also found that HIV-infected individuals produce very large amounts of antibodies that neutralize the open Env form; however, the bulk of these antibodies are unable to penetrate the tight defensive shield of the native virus. This work may help to devise new strategies to overcome the viral defensive mechanisms and facilitate the development of an effective HIV-1 vaccine.
KEYWORDS: human immunodeficiency virus, immune evasion, neutralizing antibodies, viral envelope
ABSTRACT
The human immunodeficiency virus type 1 (HIV-1) envelope (Env) trimer evades antibody recognition by adopting a closed prefusion conformation. Here, we show that two conserved tyrosines (Y173, Y177) within the second variable (V2) loop of the gp120 Env glycoprotein are key regulators of the closed, antibody-protected state of the trimer by establishing intramolecular interaction with the base of the third variable (V3) loop. Mutation of Y177 and/or Y173 to phenylalanine or alanine dramatically altered the susceptibility of diverse HIV-1 strains to neutralization, increasing sensitivity to weakly and nonneutralizing antibodies directed against diverse Env regions, consistent with the adoption of an open trimer configuration. Conversely, potent broadly neutralizing antibodies (bNAbs) against different supersites of HIV-1 vulnerability exhibited reduced potency against V2 loop tyrosine mutants, consistent with their preferential targeting of the closed trimer. Mutation of V3 loop residues predicted to interact with the V2 loop tyrosines yielded a similar neutralization phenotype. Sera from chronically HIV-1-infected patients contained very high titers of antibodies capable of neutralizing V2 loop tyrosine mutants but not wild-type viruses, indicating that the bulk of antibodies produced in infected hosts are unable to penetrate the protective shield of the closed trimer. These results identify the tyrosine-mediated V2-V3 loop complex at the trimer apex as a key structural constraint that facilitates HIV-1 evasion from the bulk of host antibodies.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) has evolved an extraordinary assortment of immune-evasion mechanisms, including antigenic variation, extensive N-linked glycosylation, and conformational camouflage of its envelope (Env) glycoproteins, gp120 and gp41, all of which contribute to the ability of this virus to establish lifelong persistent infection in the face of an initially competent immune system (1–5). Over the past few years, major progress has been made in our knowledge of the molecular anatomy of HIV-1 Env, offering new opportunities to elucidate the structural basis of HIV-1 immune evasion and, thereby, to devise effective strategies to overcome it. Key advancements have been made with the design and crystallization of soluble cleaved gp140 trimers stabilized in a near-native configuration (SOSIP.664) (6–17), as well as with the refinement of cryoelectron microscopy (cryoEM) imaging of both soluble and native membrane-bound Env spikes (18–22). High-resolution structures have illustrated the tightly welded assemblage of the HIV-1 Env trimer, which shields highly conserved neutralization epitopes, most notably the binding sites for the primary viral receptor, CD4, and the coreceptors CCR5 and/or CXCR4, from the reach of host antibodies.
The concealed configuration of most conserved neutralization epitopes represents a major obstacle to the development of a protective HIV-1 vaccine. Nevertheless, the HIV-1 vaccine field has undergone a dramatic acceleration in recent years with the precise characterization of several antigenic supersites of HIV-1 vulnerability following the cloning and crystallization of an increasing number of potent broadly neutralizing antibodies (bNAbs) from infected individuals. These sites include the CD4-binding site (CD4-BS), glycan-dependent regions at the first and second variable (V1-V2) loop apex and V3 loop base, the gp120/gp41 interface (bridging) region, which encompasses the fusion peptide, and the membrane-proximal external region (MPER) of gp41 (5, 23, 24). The CD4 supersite has recently been expanded with the identification of a second, quaternary CD4-BS, which is also a contact site for selected bNAbs (25). Although measurable titers of bNAbs develop only in ∼30 to 50% of HIV-1-infected individuals and only after a considerable duration of active viremia, their identification has provided a proof of concept that the human immune system is capable of mounting a protective humoral response; therefore, an HIV-1 vaccine may be feasible. Indeed, passive infusion of bNAbs was shown to protect nonhuman primates from infection with chimeric simian-human immunodeficiency virus (SHIV) bearing a native HIV-1 Env protein (24).
Elucidating the structural elements involved in the shielding of highly conserved Env epitopes from antibody recognition is essential for the development of effective strategies to counter the mechanisms of HIV-1 immune evasion and design a protective vaccine. We recently identified two conserved tyrosine (Y) residues within the V2 loop of gp120 (Y173, Y177) that can be posttranslationally modified by O-sulfation and appear to play an important role in modulating HIV-1 sensitivity to antibody-mediated neutralization (26). We also reported that these tyrosines are structural and functional mimics of two sulfotyrosines present within the N-terminal domain of CCR5 which are critical for interaction with the CCR5 coreceptor and HIV-1 infection (27). Indeed, these V2 loop tyrosines establish intramolecular interaction with elements of the coreceptor-binding site at the base of the V3 loop, resulting in the occlusion of this site in unliganded native Env (28). The present study was designed to further dissect the functional role of the V2 loop tyrosine-mediated V3 loop interaction as a regulator of the open/closed state of the Env trimer, which determines the sensitivity of the virus to antibody-mediated neutralization. Using V2 loop tyrosine mutants that facilitate the adoption of an open trimer configuration, we provide evidence that most bNAbs, including those that bind with high affinity to monomeric gp120, preferentially interact with the closed trimer configuration, which is a necessary condition for neutralization of native virions. Furthermore, we show that the bulk of host-produced anti-Env antibodies potently neutralize the open form of the trimer but lack neutralizing capacity against wild-type (WT) viruses, as they are unable to penetrate the tight protective shield of the closed, native structure of HIV-1 Env.
RESULTS
Critical role of V2 loop tyrosines in modulating HIV-1 neutralization sensitivity.
We previously demonstrated that two tyrosines in the V2 loop of gp120, namely, Y173 and Y177, can be posttranslationally sulfated, resulting in a decreased sensitivity to weakly/nonneutralizing antibodies and soluble CD4 (sCD4) while at the same time increasing the sensitivity to trimer-preferring bNAbs (26). In high-resolution structures of the HIV-1 Env trimer, Y173 and Y177, both of which are highly conserved in most HIV-1 clades, appear to participate in intramolecular interactions between the V2 and V3 loops, establishing bonds with specific V3 loop residues (Fig. 1A and B) (12, 13, 17, 20). To validate this model, we mutagenized the two tyrosines, individually or in combination, in a clade B, tier 1B HIV-1 Env protein (BaL) by replacing them with either phenylalanine (F) or alanine (A). Mutation to F prevents sulfation because it lacks the hydrophilic hydroxyl group in the ortho position of the phenyl ring; however, it maintains the bulky aromatic side chain. In contrast, mutation to A removes both the hydroxyl group and the phenyl ring. Infectious pseudoviruses bearing the mutated Envs were produced and tested for neutralization sensitivity. The mutants exhibited a neutralization phenotype similar to that induced by tyrosine sulfation inhibition (26), with increased sensitivity to sCD4 and to weakly neutralizing monoclonal antibodies (MAbs) against the tip of the V3 loop (447-52D) and the coreceptor-binding site (412d), while at the same time showing decreased sensitivity to trimer-preferring bNAbs against the V1-V2 loop glycan supersite at the trimer apex (PG9, PG16); no significant changes were observed with MAb 2G12 used as a control (Fig. 1C). Thus, mutagenesis of the V2 loop tyrosines appears to induce the Env trimer to adopt an open configuration, more accessible to sCD4 and poorly neutralizing antibodies but less efficiently recognized by trimer-preferring bNAbs.
FIG 1.
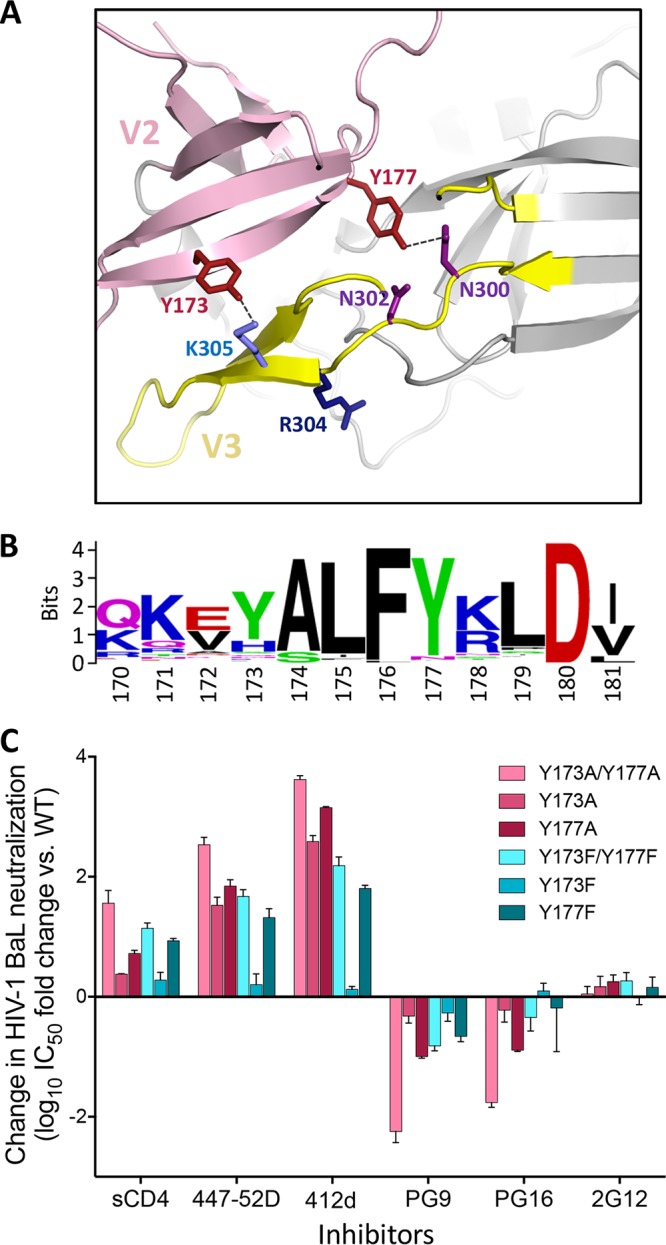
Critical role of two tyrosine residues in the gp120 V2 loop in modulating HIV-1 neutralization sensitivity. (A) Intramolecular interaction between the gp120 V2 loop (pink) and V3 loop (yellow) visualized in a high-resolution structure of the HIV-1 envelope trimer shown in cartoon representation (PDB accession number 4VTP). Specific residues are highlighted in stick representation. The V2 loop tyrosines Y173 and Y177 (brick red) are in close proximity to V3 loop residues K305 and N300, respectively. (B) Conservation of the C-terminal portion of the V2 loop among group M HIV-1 strains. Sequence alignment of a segment of the V2 loop region of gp120 (amino acids 170 to 181) from all the available group M HIV-1/SIVcpz isolates (Los Alamos HIV database). The height of each stack indicates the degree of conservation for each residue; the relative height of each letter within individual stacks represents the frequency of the indicated amino acid at that site. (C) Mutagenesis of the V2 loop tyrosines Y173 and/or Y177 to phenylalanine (F) or alanine (A) dramatically alters the HIV-1 neutralization profile. Pseudoviruses bearing the wild-type (WT) or mutated HIV-1 BaL Env sequence (tier 1B) were tested for their sensitivity to neutralization by a panel of inhibitors (sCD4 and monoclonal antibodies). The neutralization tests were performed using the TZM-bl assay. The data represent the mean (±SEM) from three independent experiments.
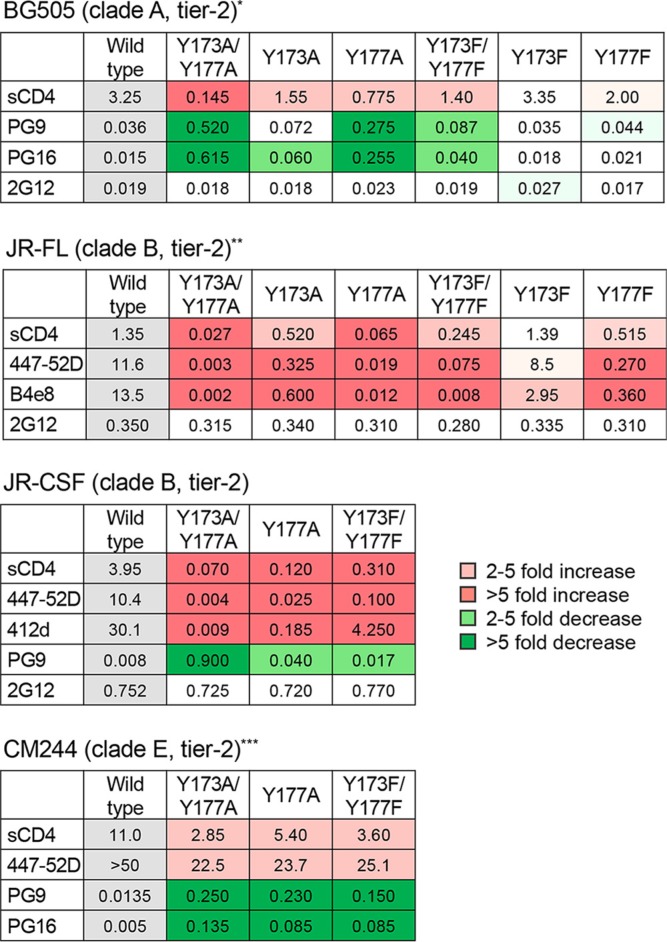
A consistent hierarchy of neutralization sensitivity was seen among the mutants, with a greater impact of Y177 over Y173 single mutations, of double over single mutations, and of A over F substitutions (Fig. 1C). The last result demonstrates the importance of the phenyl ring of Y/F as an anchoring structure that bolsters the V2-V3 loop interaction. Similar results were obtained by mutagenesis of tier 2 HIV-1 Envs from different clades, namely, BG505 (clade A), JR-FL and JR-CSF (clade B), and CM244 (clade E) (Table 1), confirming that the observed changes in neutralization phenotype were not unique to a specific HIV-1 strain, clade, or neutralization tier. The broad changes observed in the neutralization profiles of the mutants rule out a direct role of the V2 loop tyrosines in individual epitopes recognized by the tested antibodies, which target distinct and distant regions of the Env trimer. Thus, these data corroborate the role of the V2 loop tyrosines in regulating the global neutralization sensitivity of HIV-1 through stabilization of the intramolecular interaction between the V2 and V3 loops.
TABLE 1.
Effect of V2 loop tyrosine mutations on the neutralization of different HIV-1 strains and cladesa
Half-maximal neutralization titers (IC50s) against different HIV-1 wild-type and mutated pseudoviruses were determined using the TZM-bl assay. Color codes denote various degrees of neutralization increase (different shades of red) or decrease (different shades of green). *, MAb 447-52D was not tested on BG505 because this Env protein has a GPGQ motif at the tip of the V3 loop that abrogates reactivity with 447-52D. **, MAbs PG9 or PG16 was not tested on JR-FL because this Env has an acidic residue (E) at position 168 that abrogates reactivity with these bNAbs. ***, MAb 2G12 was not tested on CM244 because it does not react with this Env.
Increased sensitivity of V2 loop tyrosine mutants to a wide panel of weakly/nonneutralizing antibodies.
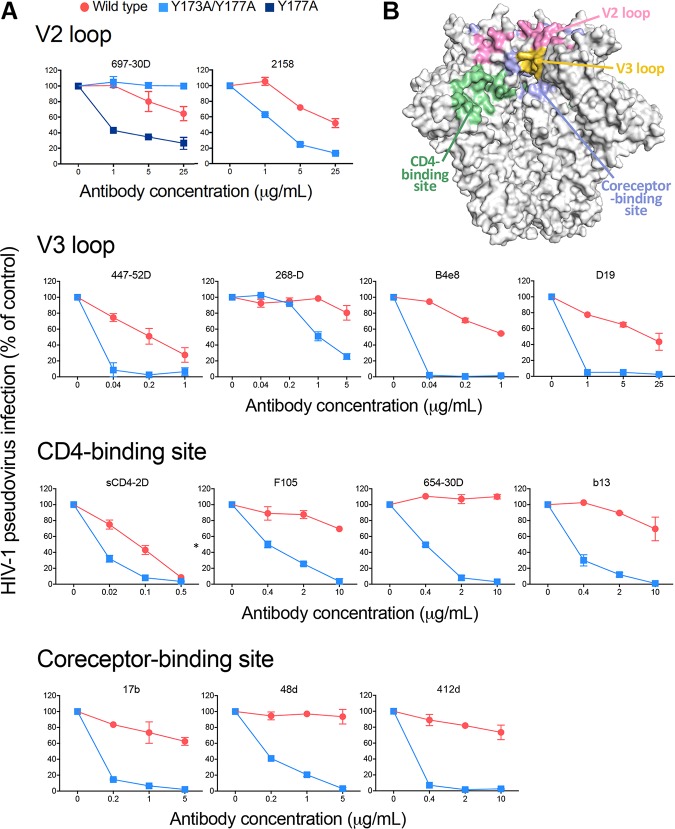
To confirm the role of the V2 loop tyrosines in maintaining the HIV-1 trimer in a closed, antibody-protected configuration, we tested the sensitivity of the double-alanine mutant HIV-1 BaL Y173A Y177A to neutralization by a larger panel of weakly or nonneutralizing antibodies directed against diverse Env antigenic regions, including non-glycan-dependent V2 loop epitopes, the tip of the V3 loop, the CD4-BS, and the coreceptor-binding site. The double-alanine mutation markedly increased sensitivity to all the weakly neutralizing MAbs tested, as well as to sCD4, and even rendered the virus sensitive to nonneutralizing antibodies against the CD4-BS, such as F105, 654-30D, and b13 (Fig. 2). It is noteworthy that for the anti-V2 loop MAb 697-30D, tyrosine 173 is part of the binding epitope (29), and therefore mutation of this residue completely abrogated binding and neutralization by this antibody. Thus, 697-30D was also tested on the Y177A single mutant, which showed a markedly increased sensitivity compared to that of the WT (Fig. 2). Together, these results demonstrate that removal of the V2 loop tyrosine-mediated constraint allows the Env trimer to transition toward a more open configuration that exposes cryptic epitopes in diverse antigenic regions of gp120.
FIG 2.
V2 loop tyrosine mutation increases neutralization sensitivity to a wide array of weakly/nonneutralizing antibodies. (A) Neutralization sensitivities of the double-alanine mutant HIV-1 BaL Y173A Y177A compared to those of the wild-type virus. Broad arrays of weakly and nonneutralizing antibodies against diverse antigenic regions (V2 loop, V3 loop, CD4-binding site, and coreceptor-binding site) are shown. The Y177A single mutant was tested only for MAb 697-30D because residue Y173 is part of the epitope recognized by this antibody, and thus, the Y173 mutation abrogates its binding. The neutralization tests were performed using the TZM-bl assay. (B) Surface representation of a high-resolution structure of the HIV-1 Env trimer (PDB accession number 4VTP) with the different antigenic regions recognized by these antibodies highlighted in different colors, as indicated.
Preferential recognition of the closed HIV-1 Env trimer by the majority of potent bNAbs.
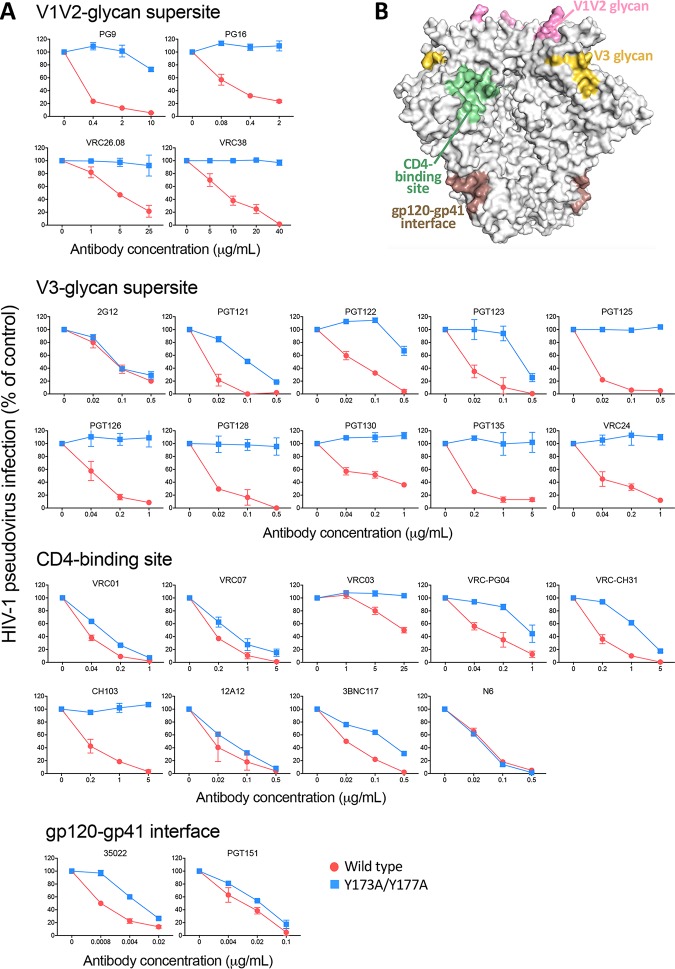
While selected bNAbs, such as V1-V2 loop glycan-specific PG9 and PG16, were initially reported as trimer-preferring or trimer-specific antibodies (30), many potent bNAbs, such those against the CD4-BS or the V3 loop base glycan, efficiently recognize both trimeric and monomeric gp120. To investigate the efficiency of bNAb interaction with the open trimer, we tested the sensitivity of the double-alanine HIV-1 BaL mutant (Y173A Y177A) to bNAbs directed against different supersites of HIV-1 vulnerability, i.e., glycan-dependent epitopes at the V1-V2 loop apex and V3 loop base, the CD4-BS, the gp120/gp41 interface, and the MPER of gp41. Strikingly, most of the bNAbs against these different supersites showed a marked reduction in neutralizing potency against the double mutant, compared to that against WT Env (Fig. 3). The effect was more extreme for bNAbs to the V1-V2 loop glycan and V3 loop glycan supersites (30–32), most of which displayed little, if any, neutralizing activity against the double-alanine mutant, while others, including VRC-PG04, VRC-CH31, and 3BNC117 (anti-CD4-BS) and 35O22 (anti-gp120/gp41 interface), showed a lesser effect, and a few, such as the anti-CD4-BS VRC01, VRC07, and 12A12, showed only a limited potency reduction (Fig. 3). Notable exceptions were N6, one of the most potent anti-CD4-BS bNAbs thus far identified, the unconventional outer domain glycan-dependent antibody 2G12, and the anti-gp120/gp41 interface antibody PGT151, all of which were equally potent against WT and mutated Env (Fig. 3). Among bNAbs against the MPER of gp41, two (2F5 and 4E10) were more effective against the double-alanine mutant, in line with previous results obtained with more-open trimers (33), while 10E8, the most potent anti-MPER bNAb, was equally effective against both Env forms (Fig. S1). The fact that two of the most potent bNAbs in their respective categories showed similar activities against the WT and V2 loop-mutated Envs may reflect a unique mode or angle of interaction of these antibodies, irrespective of the conformational state of the trimer. Together, these data demonstrated that most anti-gp120 bNAbs preferentially interact with the closed HIV-1 Env trimer, corroborating the concept that efficient recognition of the bona fide native Env spike is critical in order to achieve neutralization.
FIG 3.
V2 loop tyrosine mutation decreases neutralization sensitivity to potent bNAbs against diverse antigenic supersites. (A) Neutralization sensitivities of the double-alanine mutant HIV-1 BaL Y173A Y177A compared to those of the wild-type virus. A broad array of broadly neutralizing antibodies (bNAbs) were tested and are shown grouped according to the Env antigenic region targeted by the MAbs (V1-V2 loop glycan supersite, V3 loop glycan supersite, CD4-binding site, gp120-gp41 interface [indicated at the top of each group of graphs]). The neutralization tests were performed using the TZM-bl assay. (B) Surface representation of a high-resolution structure of the HIV-1 Env trimer (PDB accession number 4VTP), with the different antigenic regions recognized by these antibodies highlighted in different colors, as indicated.
Effect of V2 loop tyrosine mutation on neutralization by bNAbs against the gp41 MPER. Neutralization sensitivity of the double-alanine mutant HIV-1 BaL Y173A Y177A compared to that of the wild-type virus. The neutralization tests were performed using the TZM-bl assay. Download FIG S1, PDF file, 0.1 MB (62.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Identification of V2 loop tyrosine-interactive residues in the V3 loop.
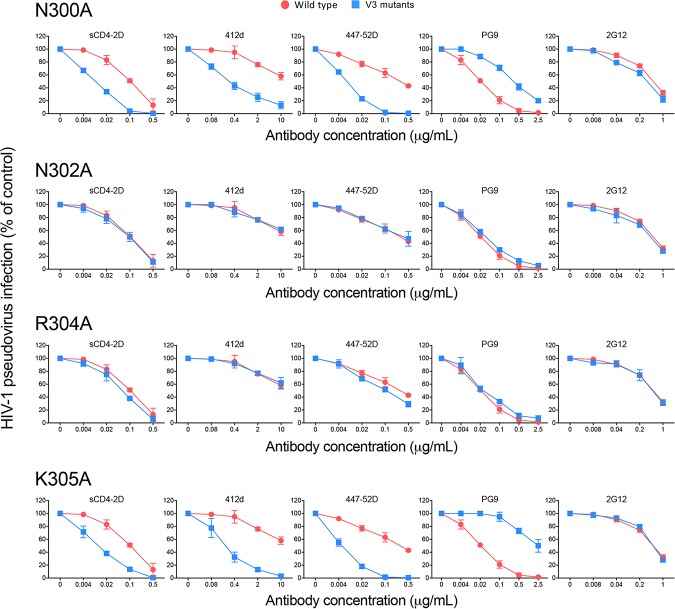
Next, we sought to identify residues that interact with the V2 loop tyrosines on the V3 loop side. Analysis of high-resolution structures of different SOSIP trimers pointed to residues in the ascending limb at the base of V3 loop as potential candidates for direct interaction with the V2 loop tyrosines (Fig. 1A). In particular, in some structures, the side chain of N300 is favorably positioned to form a hydrogen bond with the hydroxyl group of Y177, while the positively charged side chain of K305 may establish a π interaction with the phenyl ring of Y173 (12, 13, 17, 20). Thus, both N300 and K305 were mutated to alanine in HIV-1 BaL, and pseudoviruses bearing the mutated Env were produced; two contiguous residues with similar characteristics, N302 and R304, were also mutated to alanine as controls. Both the N300A and the K305A mutants exhibited a neutralization profile similar to that of V2 loop tyrosine mutants, with increased sensitivity to sCD4 and weakly neutralizing antibodies 412d and 447-52D, while conversely showing decreased sensitivity to the trimer-preferring bNAb PG9; in contrast, mutation of N302 or R304 did not significantly alter the neutralization profile (Fig. 4). Similar data were obtained with the tier 2 Env JR-CSF (data not shown). These results provided complementary evidence to validate our model of intramolecular V2-V3 loop interaction and identify N300 and K305 as critical residues within the V3 loop involved in V2 loop interaction.
FIG 4.
Identification of V3 loop residues that stabilize the Env trimer via interaction with the V2 loop tyrosines. Neutralization sensitivities of single V3 loop mutants (N300A, N302A, R304A, and K305A mutants) of HIV-1 BaL compared to those of the wild-type virus. A selected panel of inhibitors were tested to assess the neutralization profile of V3 loop mutants, including sCD4, weakly neutralizing antibodies (412d, 447-52D), and broadly neutralizing antibodies (PG9, 2G12). The neutralization tests were performed using the TZM-bl assay.
Increased sensitivity of V2 loop tyrosine mutants to serum antibodies from HIV-1-infected patients.
Having established that mutation of the V2 loop tyrosines can change the global HIV-1 Env configuration resulting in a more open trimer state, we evaluated the sensitivity of V2 loop tyrosine mutants to neutralization by antibodies present in the sera of chronically HIV-1-infected individuals. Initially, we tested purified immunoglobulin G (IgG) from a pool of HIV-1-infected patient sera (HIV-Ig) and a representative patient serum (N25) against different V2 loop tyrosine mutants of HIV-1 BaL. A dramatic increase in neutralization potency against various V2 loop tyrosine mutants was seen with both the pooled IgG and the patient serum, with a distinct hierarchy that closely reflected the results obtained with human MAbs (Fig. 1B). Thus, the double-alanine mutant showed the greatest gain in neutralization sensitivity, followed by the Y177A single mutant, while the double-phenylalanine mutant and the Y173A and Y177F single mutants showed an intermediate sensitivity; in contrast, the Y173F single mutant had the same sensitivity as the WT (Fig. 5A).
FIG 5.
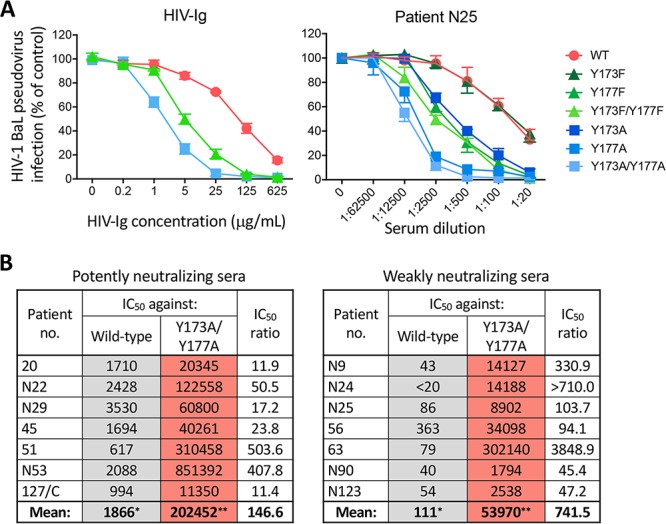
Sera from HIV-1-infected patients contain high titers of neutralizing antibodies against V2 loop tyrosine mutants. (A) Neutralization sensitivities of HIV-1 BaL (tier 1b) V2 loop tyrosine mutants to pooled sera from chronically HIV-1-infected individuals (HIV-Ig) or a representative HIV-1-seropositive patient serum (N25). (B) Half-maximal neutralization titers (IC50s) against HIV-1 BaL wild-type virus versus the Y173A Y177A double-alanine mutant in a panel of sera with different neutralization potencies (potent neutralizers, left panel; weak neutralizers, right panel). The fold increases in neutralization (IC50 ratio) between the wild-type and the Y173A Y177A mutant are shown in the right column. The asterisks indicate the data columns that were compared by unpaired t test: *, neutralization titers against WT virus; **, neutralization titers against Y173A Y177A mutated virus. The neutralization tests were performed using the TZM-bl assay.
Next, we tested the efficacy of a panel of patient sera with different neutralization potencies (strong versus weak neutralizers), which had been included in a previous study of serum neutralization breadth and potency in subjects infected with clade B HIV-1 (34). A striking discrepancy was observed between neutralization of WT BaL pseudovirus and that of the double-alanine mutant (Fig. 5B). While the WT was neutralized with the expected potency (50% inhibitory concentration [IC50] ranges, 1:617 to 1:3,530 for strong neutralizers [mean, 1:1,866] and <1:20 to 1:363 for weak neutralizers [mean, 1:111] [P = 0.00039]), the Y173A Y177A mutant was extremely sensitive to neutralization by all the patient sera tested, with IC50 titers ranging between 1:11,350 and 1:851,392 (mean, 1:202,452) for strong neutralizers and between 1:1,794 and 1:302,140 (mean, 1:53,970) for weak neutralizers (P was not significant) (Fig. 5B). The fold increase in neutralization (IC50 ratio) between the WT and the Y173A Y177A mutant ranged between 11 and 504 (mean, 146.6) for potently neutralizing sera and between 45 and 3,849 (mean, 741.5) for weakly neutralizing sera (Fig. 5B). Thus, although weakly neutralizing sera were less effective than potently neutralizing sera against both the WT and the Y173A Y177A mutant, the neutralizing titers against the open mutant in the two groups were within the same range, despite the markedly lower titers against the WT among weakly neutralizing sera, suggesting that infected individuals develop high titers of antibodies that recognize open trimer configurations regardless of their neutralization capacity against the shielded WT trimer.
To extend these observations to a tier 2 Env protein, we tested the ability of selected patient sera (4 potently and 2 weakly neutralizing sera) to neutralize V2 loop tyrosine mutants from JR-CSF, a tier 2 clade B virus. The double-alanine JR-CSF mutant showed an overall increase in sensitivity to neutralization by all the sera tested, while there was a less marked gain against the double-phenylalanine mutant (Fig. 6). Thus, despite the more protected configuration of tier 2 Envs, these results confirmed the importance of the V2 loop tyrosines as global regulators of the open/closed state of the trimer even in a tier 2 strain. Together, these data provide evidence that HIV-1-infected patients mount robust antiviral antibody responses, although the bulk of such antibodies are unable to penetrate the tight defensive shield of the closed native trimer since they are directed against concealed epitopes that are only transiently exposed and accessible predominantly on monomeric gp120.
FIG 6.
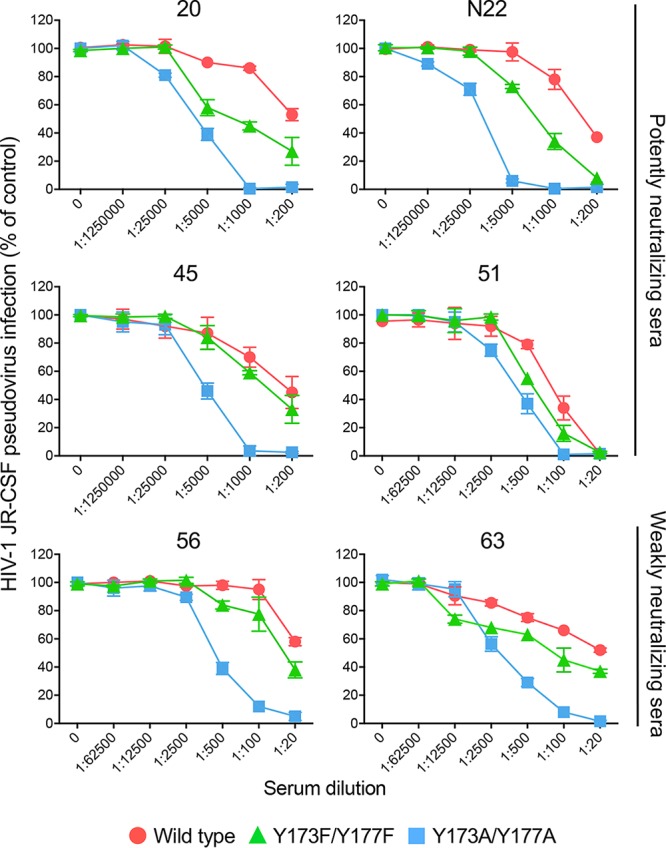
Sera from HIV-1-infected patients contain high titers of neutralizing antibodies against V2 loop tyrosine mutants. Neutralization sensitivity of HIV-1 JR-CSF (tier 2) V2 loop tyrosine mutants to potently neutralizing (top 4 panels) or weakly neutralizing (bottom two panels) patient sera. The neutralization tests were performed using the TZM-bl assay.
DISCUSSION
The HIV-1 Env trimer eludes recognition by the host immune system through a variety of mechanisms. One of the most effective such mechanism is the adoption of a tightly bundled trimer configuration that conceals many of the epitopes targeted by neutralizing antibodies. This configuration, which corresponds to the prevalent state of native, membrane-bound prefusion Env spikes, has been referred to as state 1 (35). Elucidating the structural constraints that hold the trimer in this closed, antibody-protected configuration may provide critical information to overcome the virus defensive strategies and aid in the development of an effective vaccine. In this work, we identified the intramolecular interaction between the V2 and V3 loops as a key regulator of the open/closed state of the Env trimer, with global effects on distinct and distant neutralization target sites, and we provide evidence that this constraint is critically dependent on two tyrosine residues within the V2 loop. We previously reported that these V2 loop tyrosines can be posttranslationally sulfated, particularly in viral progenies produced by primary CD4+ T cells, and that this modification further increases trimer stability and antibody resistance (26). However, the levels of tyrosine sulfation in continuous cell lines, such as HEK 293T, which was used in the present study, are constitutively low, indicating that the V2 loop tyrosines can stabilize the V2-V3 loop interaction regardless of their sulfation level.
We found that mutagenesis of the V2 loop tyrosines to phenylalanine or alanine had dramatic effects on the neutralization profile of different HIV-1 strains by inducing global changes in the Env trimer, leading to a more open, antibody-accessible configuration. Thus, we observed dichotomous effects on HIV-1 neutralization, with increased susceptibility to sCD4 and weakly or nonneutralizing antibodies, while at the same time the susceptibility to most bNAbs was either reduced or absent. This dichotomy likely reflects the release of V2-V3 loop intramolecular constraints that contribute to maintaining the prefusion trimer in high-energy, metastable state 1. Notably, some of our V2 loop tyrosine mutants showed reduced infectious activity, despite their increased CD4-binding affinity, a phenomenon that was more marked upon mutation to alanine and correlated with the degree of phenotypic changes; this loss in replication fitness may be related, at least in part, to reduced trimer association, as we observed increased levels of gp120 shedding by the double-alanine mutants (P. Zhang, C. Guzzo, and P. Lusso, unpublished data). Only a few of the bNAbs tested in this study (i.e., 2G12, N6, PGT151, and 10E8) exhibited similar neutralization potencies against WT and V2 loop tyrosine mutants, presumably due to their ability to target unique epitopes that maintain the same structure and accessibility regardless of the conformational state of the trimer. Divergent effects on weak versus potent neutralizing antibodies were recently reported by Herschhorn et al., who mutated a single hydrophobic residue at the V2 loop C-terminal end, leucine 193 (L193), which is not involved in contact with the V3 loop, suggesting that V3 loop-independent interactions also play a role in maintaining the closed trimer conformation (36). Instead, Zolla-Pazner and colleagues documented the role of hydrophobic intraprotomer interactions, along with specific gp120 glycans, in stabilizing the packing of the V3 loop onto the V1-V2 loops at the trimer apex; however, the impact of their mutations on the global trimer configuration was more limited than that observed by V2 loop tyrosine mutagenesis (37, 38), further corroborating the importance of the V2 loop tyrosines as global regulators of the open/closed state of the trimer.
Our results provide evidence for a dominant role of Y177 in consolidating the intramolecular interaction between the V2 loop and V3 loop, as previously suggested by in silico modeling, nuclear magnetic resonance (NMR) spectroscopy, and peptide-mediated inhibition studies (28), while neighboring Y173 seems to play an accessory role. It is noteworthy that Y177 is conserved in ∼98% of group M HIV-1 isolates, while Y173 is conserved in ∼67%. Also, replacement of Y173 with phenylalanine did not significantly modify the neutralization profile, suggesting that the presence of a phenyl ring at this position is sufficient for consolidating the V2-V3 loop interaction. This finding is consistent with the nature of the predicted chemical bond between Y173 and the V3 loop K305, which appears to be a π interaction between the positively charged side chain of K305 and the phenyl ring of Y173, thus making it independent of the hydroxyl group at the phenyl ring apex. Mutagenesis on the V3 loop side confirmed the critical role of K305 in maintaining the trimer in a closed configuration, as well as of N300, a residue that appears to directly interact with the side chain of Y177 (12, 13, 17, 20). However, additional residues participate in the formation of the Y177-binding pocket at the V3 loop base (26, 28) as well as in the establishment of strong hydrophobic interactions along the V2-V3 loop interface. Furthermore, there is significant strain-to-strain variation in the fine topography of the V2-V3 loop interaction (9–20).
The finding that mutations of the V2 loop tyrosines had global effects on the configuration and stability of the entire HIV-1 Env trimer, as shown by their profound impact on distant antigenic sites, argues against a direct role of epitope alteration as a major mechanism for the disrupted neutralization profile of the mutants. Only in the case of antibody 697-30D was the direct epitope involvement of tyrosine Y173 obvious, which prompted us to use the Y177 single mutant, instead of the double mutant, to verify the global effects on the trimer. With regard to the anti-V2 loop glycan antibodies, Walker et al. reported the involvement of Y173 in the binding epitope for bNAbs PG9 and PG16, as suggested by a marked reduction in binding upon alanine substitution of Y173, but not of Y177, in strain JR-CSF (30). Although we did not produce the Y173A single mutant of JR-CSF, we observed a limited effect of the single Y177A versus the double Y173A Y177A mutation in this Env, suggesting that Y173 not only may be a critical component of the epitope to which the antibody binds but also may play the dominant role in stabilizing the V2-V3 loop interaction in JR-CSF. In other HIV-1 strains, however, we consistently observed a more pronounced effect of the Y177 mutation than that of Y173, in agreement with our previous results (28).
One of the most striking observations emerging from the present study was the specificity for the closed trimer that we documented for the vast majority of potent bNAbs. Indeed, we found that not only anti-V2 loop glycan bNAbs, such as PG9 and PG16, but also bNAbs against the V3 loop glycan and the CD4-BS have a strong bias toward the recognition of the closed trimer, in some cases with a nearly complete loss of neutralizing activity against viruses with a more open configuration. This finding has a logical basis because bNAbs represent rare breeds of antibodies selected for their ability to bind pure Env spikes in order to effect neutralization (39), and therefore they must efficiently recognize the closed native Env conformation. While some bNAbs still bind monomeric gp120, their preferential recognition of the trimer reveals the footprints of a long and grueling affinity maturation and selection process, which reaches completion only for a small proportion of antibodies and only in a fraction of infected individuals. These considerations help to explain why immunization with monomeric gp120 or with open or partially unprotected trimers has consistently failed to induce bNAb responses (23, 24, 40). Likewise, the lack of detectable bNAbs in the majority of chronically infected individuals attests to the inherent challenges that the human immune system has to overcome in order to recognize highly conserved epitopes in the native HIV-1 trimer and to manufacture/expand the rare antibodies that are able to reach such difficult targets. In this regard, we found that chronically HIV-1-infected individuals produce large amounts of anti-Env antibodies capable of neutralizing the more open Env forms. This finding extended to subjects with poor neutralizing capacity against the respective WT virus. Similar results were reported upon mutation of L193, another V2 loop residue that appears to contribute to trimer stabilization (36). The abundance of antibodies that efficiently neutralize V2 loop tyrosine mutants authenticates the remarkable effort made by the immune system to control HIV-1 infection, which collides with the defensive shield of the virus. However, some of these poorly neutralizing antibodies might mediate alternative effector functions, such as antibody-dependent cellular cytotoxicity (ADCC) (41), which has been suggested as a potential protective mechanism in the RV144 human trial, the only HIV-1 vaccine so far to have shown partial efficacy (42).
In summary, the present work provides new insights into the molecular constraints that maintain the HIV-1 Env trimer in its closed, metastable state, allowing it to explore only transiently more-open conformations competent for receptor and coreceptor binding without compromising its defensive shield against antibody neutralization. Identifying strategies to disentangle the inherent antibody resistance of the HIV-1 Env trimer may open new avenues for the prevention and treatment of HIV/AIDS.
MATERIALS AND METHODS
HIV-1 gp160 mutagenesis.
The gp160 genes from different HIV-1 strains were mutagenized by site-directed mutagenesis using QuikChange II site-directed mutagenesis kit (Agilent Technologies).
Antibodies and proteins.
Human antibodies were obtained through the NIH AIDS Reagent Program or produced and purified by the core facility of the Vaccine Research Center (VRC). Two-domain sCD4 was obtained through the NIH AIDS Reagent Program.
Patient sera.
Serum was obtained from chronically HIV-1-infected patients attending the NIAID AIDS Clinic for routine medical visits and laboratory testing. All patients gave written informed consent for donating blood products for research use, and the study protocol (https://clinicaltrials.gov/ct2/show/NCT00039689?term=02-I-0202&rank=1) received approval by the NIH Institutional Review Board. All samples were coded, and no patient identifier was accessible to the laboratory personnel involved in this study. Both male and female patients were enrolled without discrimination.
Pseudovirus preparation, infectivity, and neutralization assays.
HIV-1 pseudoparticles expressing WT or mutated gp160 were produced in HEK 293T cells (female human embryonic kidney cells, obtained from the ATCC), by cotransfecting Env-expressing plasmids with a backbone plasmid, pSG3Δenv, expressing a full-length HIV-1 clone with a defective Env gene. To produce the pseudoviruses, 2 μg of each Env-expressing plasmid and 4 μg of the backbone plasmid were mixed in Opti-MEM medium (Gibco), and 24 μl of Mirus293 Transfection Reagent was added at a 1:4 DNA-to-reagent ratio, followed by a 15-min incubation at room temperature. A DNA-Mirus293 complex was then added to the cells and incubated overnight at 37°C. After replacing the culture medium with 2 ml fresh 10% fetal bovine serum (FBS)-Dulbecco’s modified Eagle’s medium (DMEM), the cells were incubated for 48 h, and the supernatants containing pseudoviruses were harvested by centrifugation. For infection, TZM-bl cells (female epithelial carcinoma cells, HeLa derivative, obtained from the AIDS Reagent Program) were seeded into 96-well flat-bottom plates at 10,000 cells/well in 100 μl 10% FBS-DMEM. After 30 min at 37°C, pseudovirus stocks were added to the cells, yielding a total volume of 200 μl/well. Reporter gene (luciferase) expression was detected 2 days later after removal of 150 μl medium and addition of 40 μl per well of luciferase assay reagent (Promega). The cell lysates were transferred to Lumino plates and measured with a luminometer (PerkinElmer). Relative light units (RLU) were recorded, and the final values were normalized against the values obtained with the WT Env, set at 100%. All the samples were tested in duplicate or triplicate wells. Calculation of half-maximal inhibitory concentrations (IC50) and two-tailed t tests were performed using GraphPad Prism 7 (Prism). All graphs were also plotted using Prism 7.
Sequence conservation analysis.
Sequence alignment of a segment of the V2 loop region of gp120 (amino acids 170 to 181) from the all the available group M HIV-1/SIV from chimpanzee (SIVcpz) isolates (including both subtypes A to K and recombinant forms) was obtained from the Los Alamos HIV database (http://www.hiv.lanl.gov). The sequence logo was created using WebLogo (http://weblogo.berkeley.edu).
Statistical analysis.
Calculation of IC50s and statistical tests were performed using the GraphPad Prism 7 (Prism) software. Unless differently indicated, data from in vitro experiments are presented as means ± standard errors of the means (SEM) from duplicate or triplicate samples. Statistically significant differences were determined by Mann-Whitney U test or paired or unpaired t test. Statistical details can be found directly in the figures or in the corresponding figure legends.
Figures. All graphs were plotted using Graphpad Prism 7, and structural figures were constructed and are displayed using PyMOL.
Data availability.
All data are available in the figures and tables of the paper. Primary data are also available upon request.
ACKNOWLEDGMENTS
We thank Susan Zolla-Pazner and James E. Robinson for the gift of anti-Env monoclonal antibodies, Anthony S. Fauci for critical reading of the manuscript, the patients and nurses of the NIAID AIDS Clinic, and the AIDS Reagent Program for providing the reagents indicated in Materials and Methods.
This research was supported by the Intramural Programs of the Division of Intramural Research, NIAID, NIH, and of the Vaccine Research Center, NIAID, NIH. The funding agency had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We declare that we have no conflict of interest.
Footnotes
Citation Guzzo C, Zhang P, Liu Q, Kwon AL, Uddin F, Wells AI, Schmeisser H, Cimbro R, Huang J, Doria-Rose N, Schmidt SD, Dolan MA, Connors M, Mascola JR, Lusso P. 2018. Structural constraints at the trimer apex stabilize the HIV-1 envelope in a closed, antibody-protected conformation. mBio 9:e00955-18. https://doi.org/10.1128/mBio.00955-18.
Contributor Information
Salim Abdool Karim, University of KwaZulu-Natal.
Barton Haynes, Duke University Medical Center.
Marzena Pazgier, Uniformed Services University of the Health Sciences.
REFERENCES
- 1.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Cimbro R, Lusso P, Berger EA. 2011. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci U S A 108:20148–20153. doi: 10.1073/pnas.1104840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonsignori M, Liao HX, Gao F, Williams WB, Alam SM, Montefiori DC, Haynes BF. 2017. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 275:145–160. doi: 10.1111/imr.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schön A, Van Galen J, Chuang G-Y, Gorman J, Harned A, Pancera M, Stewart-Jones GBE, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schön A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, Yang Y, Doria-Rose NA, Georgiev IS, Gorman J, Joyce MG, O'Dell S, Zhou T, McDermott AB, Mascola JR, Kwong PD. 2017. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J Virol 91:e02268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pancera M, Lai YT, Bylund T, Druz A, Narpala S, O'Dell S, Schon A, Bailer RT, Chuang GY, Geng H, Louder MK, Rawi R, Soumana DI, Finzi A, Herschhorn A, Madani N, Sodroski J, Freire E, Langley DR, Mascola JR, McDermott AB, Kwong PD. 2017. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol 13:1115–1122. doi: 10.1038/nchembio.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Gorman J, Geng H, Liu Q, Lin Y, Tsybovsky Y, Go EP, Dey B, Andine T, Kwon A, Patel M, Gururani D, Uddin F, Guzzo C, Cimbro R, Miao H, McKee K, Chuang GY, Martin L, Sironi F, Malnati MS, Desaire H, Berger EA, Mascola JR, Dolan MA, Kwong PD, Lusso P. 2018. Interdomain stabilization impairs CD4 binding and improves immunogenicity of the HIV-1 envelope trimer. Cell Host Microbe 23:832–844.e6. doi: 10.1016/j.chom.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu G, Liu J, Taylor KA, Roux KH. 2011. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 Loop. J Virol 85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HL, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mascola JR, Haynes BF. 2013. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev 254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, Chuang GY, Druz A, Zhou T, Rice WJ, Wigge C, Carragher B, Potter CS, Kwong PD, Lusso P. 2017. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat Struct Mol Biol 24:370–378. doi: 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cimbro R, Gallant TR, Dolan MA, Guzzo C, Zhang P, Lin Y, Miao H, Van Ryk D, Arthos J, Gorshkova I, Brown PH, Hurt DE, Lusso P. 2014. Tyrosine sulfation in the second variable loop (V2) of HIV-1 gp120 stabilizes V2-V3 interaction and modulates neutralization sensitivity. Proc Natl Acad Sci U S A 111:3152–3157. doi: 10.1073/pnas.1314718111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676. doi: 10.1016/S0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 28.Cimbro R, Peterson FC, Liu Q, Guzzo C, Zhang P, Miao H, Van Ryk D, Ambroggio X, Hurt DE, De Gioia L, Volkman BF, Dolan MA, Lusso P. 2016. Tyrosine-sulfated V2 peptides inhibit HIV-1 infection via coreceptor mimicry. EBioMedicine 10:45–54. doi: 10.1016/j.ebiom.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol 68:8312–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cale EM, Gorman J, Radakovich NA, Crooks ET, Osawa K, Tong T, Li J, Nagarajan R, Ozorowski G, Ambrozak DR, Asokan M, Bailer RT, Bennici AK, Chen X, Doria-Rose NA, Druz A, Feng Y, Joyce MG, Louder MK, O’Dell S, Oliver C, Pancera M, Connors M, Hope TJ, Kepler TB, Wyatt RT, Ward AB, Georgiev IS, Kwong PD, Mascola JR, Binley JM. 2017. Virus-like particles identify an HIV V1V2 apex-binding neutralizing antibody that lacks a protruding loop. Immunity 46:777–791.e10. doi: 10.1016/j.immuni.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A 105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol 84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2016. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. doi: 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powell RLR, Totrov M, Itri V, Liu X, Fox A, Zolla-Pazner S. 2017. Plasticity and epitope exposure of the HIV-1 envelope trimer. J Virol 91:e00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong T, Crooks ET, Osawa K, Binley JM. 2012. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol 86:3574–3587. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haynes BF. 2015. New approaches to HIV vaccine development. Curr Opin Immunol 35:39–47. doi: 10.1016/j.coi.2015.05.007. doi: 10.1016/j.coi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis GK, Finzi A, DeVico AL, Pazgier M. 2015. Conformational masking and receptor-dependent unmasking of highly conserved Env epitopes recognized by non-neutralizing antibodies that mediate potent ADCC against HIV-1. Viruses 7:5115–5132. doi: 10.3390/v7092856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of V2 loop tyrosine mutation on neutralization by bNAbs against the gp41 MPER. Neutralization sensitivity of the double-alanine mutant HIV-1 BaL Y173A Y177A compared to that of the wild-type virus. The neutralization tests were performed using the TZM-bl assay. Download FIG S1, PDF file, 0.1 MB (62.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
All data are available in the figures and tables of the paper. Primary data are also available upon request.