Abstract
Background
Bud sport mutants of apple (Malus domestica Borkh.) trees with a highly blushed colouring pattern are mainly caused by the accumulation of anthocyanins in the fruit skin. Hormones are important factors modulating anthocyanin accumulation. However, a good understanding of the interplay between hormones and anthocyanin synthesis in apples, especially in mutants at the molecular level, remains elusive. Here, physiological and comparative transcriptome approaches were used to reveal the molecular basis of color pigmentation in the skin of ‘Red Delicious’ (G0) and its mutants, including ‘Starking Red’ (G1), ‘Starkrimson’ (G2), ‘Campbell Redchief’ (G3) and ‘Vallee spur’ (G4).
Results
Pigmentation in the skin gradually proliferated from G0 to G4. The anthocyanin content was higher in the mutants than in ‘Red Delicious’. The activation of early phenylpropanoid biosynthesis genes, including ASP3, PAL, 4CL, PER, CHS, CYP98A and F3’H, was more responsible for anthocyanin accumulation in mutants at the color break stage. In addition, IAA and ABA had a positive regulatory effect on the synthesis of anthocyanins, while GA had the reverse effect. The down-regulation of AACT1, HMGS, HMGR, MVK, MVD2, IDI1 and FPPS2 involved in terpenoid biosynthesis influences anthocyanin accumulation by positively regulating transcripts of AUX1 and SAUR that contribute to the synthesis of IAA, GID2 to GA, PP2C and SnRK2 to ABA. Furthermore, MYB and bHLH members, which are highly correlated (r=0.882–0.980) with anthocyanin content, modulated anthocyanin accumulation by regulating the transcription of structural genes, including CHS and F3’H, involved in the flavonoid biosynthesis pathway.
Conclusions
The present comprehensive transcriptome analyses contribute to the understanding of the the relationship between hormones and anthocyanin synthesis as well as the molecular mechanism involved in apple skin pigmentation.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1595-8) contains supplementary material, which is available to authorized users.
Keywords: Apple, Bud sport, Fruit skin, Pigmentation, RNA-seq, Hormone
Background
Somatic mutations which are also referred to as 'bud sports’ usually occur in woody plant species [1, 2]. and the genetic background of these mutants is nearly identical to that of their parents [3, 4]. However, epigenetic changes causing fruit colour alteration in apple have been identified (Malus domestica Brokh.) [2, 5]. Fruit skin colour is a key appearance and nutrition quality attribute of apple fruit [6]. Anthocyanins are among the secondary metabolites that contribute to the colours of fruits [7]. Pigmentation in the skin of apple fruit varies among cultivars and is influenced by environmental factors, including temperature [8] and the level of sunlight irradiation [9, 10]. Furthermore, hormones are likely to be important factors that modulate light-dependent anthocyanin accumulation [11–13]. In summary, exploring the molecular mechanisms of hormones and anthocyanin synthesis in apple fruit and its bud sport mutants is crucial to research on pigment accumulation and plant somatic mutation.
Previous studies have shown that auxin (IAA), cytokinin (CTK), gibberellins (GA), jasmonate acid (JA), abscisic acid (ABA) and ethylene (ETH) interact in controlling anthocyanin biosynthesis [11, 13–16]. In addition, the identification and functional characterization of MYB and bHLH transcriptional factors revealed that they play a role in structural gene transcription. These structural genes include chalcone isomerase (CHI), chalcone synthase (CHS), flavonol synthase (FLS), leucoanthocyanidin reductase (LAR), flavonoid 3’-hydroxylase (F3’H) and anthocyanidin reductase (ANR), which are involved in the anthocyanin biosynthesis pathway [17–21]. MYB proteins are characterized by two imperfect repeats of the DNA-binding motifs R2 and R3 [22], and bHLH proteins are characterized by the basic helix-loop-helix domain, which is responsible for sequence-specific DNA binding [23].
The publication of the apple reference genome [24] and the development of new tools for transcriptomics have facilitated recent advances in the genome-wide analysis of dynamic gene expression during fruit development [2, 25]. The strategies of hormone and anthocyanin synthesis are often applied without a full understanding of the effect at the molecular level, with the exception of a few studies that have correlated biochemical and physiological outcomes with transcriptomic changes [11–13]. Apple cultivar ‘Red Delicious’ (M. domestica) is the most frequently captured sport apple variety that is usually selected on the phenotypic basis of spur type and intense red fruit colour. The cultivar’s four continuous generation mutants, namely, ‘Starking Red’, ‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee Spur’, have been screened. Therefore, a comparative transcriptome analysis combined with physiological and biochemical characteristics were employed to investigate the relationship between hormone and anthocyanin synthesis on the accumulation of apple skin pigment.
Materials and methods
Plant material
‘Red Delicious’ is the most frequently captured sport apple variety, featuring four continuous generation mutants. ‘Starking Red’ is a bud sport from ‘Red Delicious’ and a typical representative of the first generation. The second generation is ‘Starkrimson’, which is a bud sport from the first-generation ‘Starking Red’. The fourth generation, ‘Vallee Spur’, is a bud sport of the third generation, ‘Campbell Redchief’, which is bud sport of ‘Starkrimson’. Mature apple fruit of these five cultivars range from having red vertical stripes to being completely red (Fig. 1a).
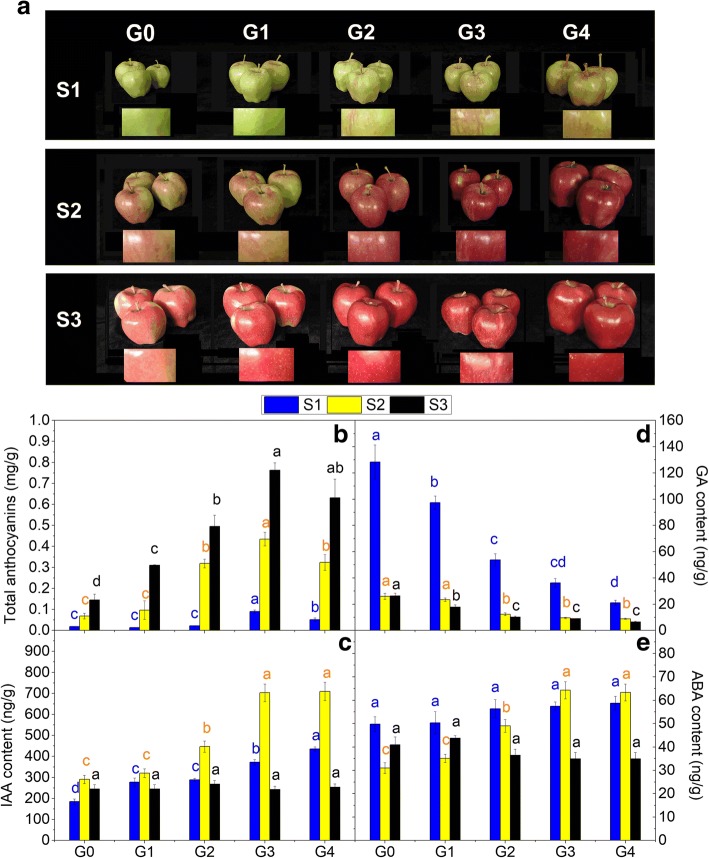
Fig. 1.
a Close-up views of ‘Red Delicious’ and its four generation mutants (‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee spur’), named G0 to G4, at three developmental stages (S1–S3) used for anthocyanin quantification, transcriptome profiling and qRT-PCR. b The changes in total anthocyanin concentrations in the fruit skin of the five strains at S1 to S3. Changes in endogenous hormone levels, including IAA (c), GA (d) and ABA (e), in the fruit skin of the five strains at S1 to S3. Values are means ± SE. Different lower case letters indicate significant differences among the five strains (P=0.05)
Fruit samples of ‘Red Delicious’ and its four continuous generation mutants (‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee spur’) were named G0 to G4 and collected in 2017 from 12-year-old trees grown in apple demonstration gardens at Tianshui, China. Briefly, 20−30 fruits from each of the five strains were sampled at three developmental stages, i.e., 5 August (S1), 25 August (S2), and 14 September (S3) (Fig. 1a). Before color break, at color break and fruit maturity stages are designated by S1, S2 and S3, respectively. The entire skin samples were removed with 1 mm of the cortical tissue and collected from 6 fruits per replicate with three independent biological replicates. All samples were collected at the same time of day (9–10 AM), immediately frozen in liquid nitrogen and stored at −80 °C for further analysis of anthocyanin contents, endogenous hormone contents and gene expression profiles (qRT-PCR). In addition, samples from S2 were used for RNA-seq analysis.
Anthocyanin quantification
Lyophilized apple skin samples were finely ground, and 500 mg of powdered samples was homogenized in 10 mL of methanol with 1% HCl. The homogenate was transferred to a calibration test tube with a constant volume of 20 mL and kept for 20 min at 4 °C with shaking under dark conditions. Then, the samples were filtered through a 0.2 μm polyethersulfone filter (Krackeler Scientific, Inc., Albany, NY, USA) and analysed using a TU-1900 double beam UV-visible spectrophotometer (Beijing Purkinje General Instrument Co. LTD). Anthocyanin levels were calculated by dividing the absorbance by the coefficient of regression (0.0214) acquired by standard scale measurements.
Determination of IAA, GA and ABA concentrations
A total of 1.0 g of each lyophilized apple skin samples were used for phytohormone extractions [26]. The apparatus used for high-performance liquid chromatography (HPLC) was the LC-20AD system (Shimadzu, Kyoto, Japan) equipped with a Zorbax Eclipse Plus C18 column (4.6 mm × 250 mm × 5.0 μm, Agilent, Palo Alto, CA, USA) and an SPD-20A UV detector. The determination method was performed with different concentrations of IAA, GA, and ABA, standard samples, which were used to construct a standard curve [27].
RNA extraction
Total RNA was extracted from approximately 200 mg of lyophilized apple skin samples ground in liquid nitrogen using the RNase-Free DNase Set (Qiagen, Valencia, CA, USA) and then cleaned with the RNeasy Mini Kit (Qiagen). RNA quality and quantity were determined using a Pultton P200 Micro Volume Spectrophotometer (Pultton Technology Limited).
RNA-seq library preparation and construction
The 5 triplicate samples (5 varieties at S2) yielded 15 nondirectional cDNA libraries with a total of 68.18 million reads (Table 1), which were prepared from 3.0 μg of total RNA using the NEBNext, UltraTM RNA Library Prep Kit (NEB, USA). The process of library construction was described in detail by Mao et al. (2017) [26, 27], and the 15 libraries were sequenced on an Illumina HiSeq 2000 platform.
Table 1.
Summary of RNA-Seq data and mapping metrics
| Variety | Replicate | Total reads | Clean reads | Mapped reads | Average reads | Average mapped reads | Quality (%) |
|---|---|---|---|---|---|---|---|
| G0 | 1 | 41,914,250 | 20,957,125 | 35,506,898 (84.71%) | 43,351,070 | 36,583,507 | 84.39 |
| 2 | 44,423,646 | 22,211,823 | 37,674,222 (84.81%) | ||||
| 3 | 43,715,314 | 21,857,657 | 36,569,401 (83.65%) | ||||
| G1 | 1 | 42,443,636 | 21,221,818 | 35,442,650 (83.51%) | 42,675,702 | 35,772,242 | 83.82 |
| 2 | 44,392,916 | 22,196,458 | 37,298,311 (84.02%) | ||||
| 3 | 41,190,554 | 20,595,277 | 34,575,765 (83.94%) | ||||
| G2 | 1 | 40,378,914 | 20,189,457 | 34,083,517 (84.41%) | 40,665,685.33 | 34,093,629 | 83.84 |
| 2 | 40,932,238 | 20,466,119 | 34,090,846 (83.29%) | ||||
| 3 | 40,685,904 | 20,342,952 | 34,106,524 (83.83%) | ||||
| G3 | 1 | 53,099,790 | 26,549,895 | 44,666,920 (84.12%) | 51,522,379.33 | 43,153,440.33 | 83.76 |
| 2 | 51,901,662 | 25,950,831 | 43,334,048 (83.49%) | ||||
| 3 | 49,565,686 | 24,782,843 | 41,459,353 (83.65%) | ||||
| G4 | 1 | 46,540,170 | 23,270,085 | 38,908,154 (83.60%) | 49,058,894 | 41,193,441 | 83.97 |
| 2 | 47,105,492 | 23,552,746 | 39,399,685 (83.64%) | ||||
| 3 | 53,531,020 | 26,765,510 | 45,272,484 (84.57%) |
RNA-seq data analysis
The files of raw reads were cleaned by removing adapter sequences and the clean reads were aligned onto the apple reference genome (https://iris.angers.inra.fr/gddh13//) [24]. An average of 83.95% of reads were mapped for each sample (Table 1). All usable reads were then calculated and normalized by an absolute value of log2 (fold change) with Fragments Per Kilobase of transcript per million mapped reads (FPKM) ≥ 1.0 was used as a threshold to determine significant differentially expressed genes (DEGs) [28]. A Gene Ontology (GO) database (http://www.geneontology.org) was used to assign apple genes to GO categories [29]. Besides, a Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/pathway.html) was used for KEGG pathway analyses. GO terms coupled with KS<0.01 was considered significantly enriched by DEGs (Additional file 1: Table S1). KEGG pathways with a Q-value ≤ 0.05 were considered significantly enriched (Table 2).
Table 2.
Significantly enriched pathways of DEGs in ‘Red Delicious’ and its four generation mutants
| Pathway ID | Pathway | Number of DEGs | Enrichment factor | Q-value (<0.05) |
|---|---|---|---|---|
| G0 versus G1 | ||||
| ko04626 | Plant-pathogen interaction | 14 | 3.97 | 0.000516177001036455 |
| ko04075 | Plant hormone signal transduction | 16 | 3.46 | 0.000621209033772607 |
| ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 4 | 9.64 | 0.0409888977775863 |
| G0 versus G2 | ||||
| ko00941 | Flavonoid biosynthesis | 16 | 8.82 | 1.55373225396715e-09 |
| ko04626 | Plant-pathogen interaction | 25 | 3.08 | 3.9124668653967e-05 |
| ko04712 | Circadian rhythm - plant | 9 | 4.96 | 0.00618212628781434 |
| ko04075 | Plant hormone signal transduction | 24 | 2.26 | 0.0120396678678723 |
| ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 6 | 6.28 | 0.027494105974142 |
| G0 versus G3 | ||||
| ko04075 | Plant hormone signal transduction | 53 | 2.60 | 7.99624033653856e-09 |
| ko00941 | Flavonoid biosynthesis | 15 | 4.31 | 0.000139485985032328 |
| ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 10 | 5.46 | 0.000908007889189655 |
| ko04626 | Plant-pathogen interaction | 34 | 2.18 | 0.00135034354389485 |
| ko04712 | Circadian rhythm - plant | 12 | 3.45 | 0.0158793441067531 |
| ko00900 | Terpenoid backbone biosynthesis | 12 | 3.16 | 0.0371242613055638 |
| G0 versus G4 | ||||
| ko00196 | Photosynthesis - antenna proteins | 11 | 6.29 | 6.37521150037568e-05 |
| ko04712 | Circadian rhythm - plant | 15 | 3.84 | 0.000640829801345832 |
| ko00941 | Flavonoid biosynthesis | 15 | 3.84 | 0.000640829801345832 |
| ko00900 | Terpenoid backbone biosynthesis | 15 | 3.51 | 0.00196937336544101 |
| ko04075 | Plant hormone signal transduction | 44 | 1.92 | 0.00212458752282485 |
| G1 versus G2 | ||||
| ko00944 | Flavone and flavonol biosynthesis | 2 | 173.44 | 0.00100183440108559 |
| ko00941 | Flavonoid biosynthesis | 4 | 18.26 | 0.00125854608191001 |
| G2 versus G3 | ||||
| ko04075 | Plant hormone signal transduction | 22 | 3.04 | 0.000165774159370358 |
| G3 versus G4 | ||||
| ko04075 | Plant hormone signal transduction | 16 | 4.21 | 4.46200952750608e-05 |
Common expression pattern clustering analysis of DEGs
The different expression patterns of DEGs among the five strains were analysed using the R language, Cluster package, Biobase package, and Q-value package. The DEGs with a common expression trend were divided into a data set, which was expressed as a model map. The distance measure used was Euclidean distance, and the clustering method was K-means clustering or hierarchical clustering.
Correlation analysis
A correlation matrix was prepared using SPSS statistical software and Pearson’s correlation coefficient as the statistical metric. The analysis was performed using the anthocyanin content at S2 and the FPKM average of each candidate DEG. Correlation values were converted to distance coefficients to define the height scale of the dendrogram.
Quantitative reverse-transcription PCR (qRT-PCR) validation of RNA-Seq data
DNase-treated RNA (2 μg) was reverse transcribed in a reaction volume of 20 μl using PrimerScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China). Gene-specific primers were designed using Primer Express software (Applied Biosystems) (Additional file 2: Table S2). qRT-PCR assays were performed with SYBR Green PCR Master Mix (Takara, Dalian, China). The qRT-PCR reaction solution (20 μL total volume) was composed of 1.0 μL of each primer, 2 μL of cDNA, 10.0 μL 2×SYBR Green Master Mix, and 6.0 μL RNase-free water. Three biological and three technical replicates for each reaction were analysed on a LightCycler® 96 SW 1.1 instrument (Roche). The amplification program was described in detail by Mao et al. (2017) [27]. All relative expression was normalized by comparing with G0 expression and analysed using the comparative 2-ΔΔCT method [30] against the reference gene MdGADPH.
Statistical analysis
Data regarding the anthocyanin content and the relative expression level of specific genes were analysed by ANOVA. SPSS 21.0 software was used for statistical analysis of treatment means, which were separated by Duncan’s multiple range test at P < 0.05. For correlation analysis, the Pearson correlation coefficient (r) was calculated, and a two-tailed test was carried out.
Results
Fruit skin pigmentation increased with bud sport generation and maturation
Visual inspection of apple skin colour during development revealed that the G2, G3 and G4 strains began colouring at S1, with the most visibly intense red colouring occurring in G3 and G4 (Fig. 1a). Pigmentation of these five strains progressively advanced to much higher levels from S1 to the subsequent stage S3, resulting in red fruit at maturity. In addition, red coloration gradually proliferated from G0 to G4 during the three stages.
Consistent with visual inspection, the analysis of apple skin anthocyanin contents showed that the levels of total anthocyanins in the five strains were initially low and sharply increased with maturation (from S1 to S3) (Fig. 1b). Relative to that at G0, the total anthocyanin level in fruit skin was ~0.78-, ~1.20-, ~5.02- and ~2.85-fold higher in G1, G2, G3 and G4 at stage S1, respectively. At S2, the level in fruit skin was ~1.41-, ~4.65-, ~6.33- and ~4.72-fold higher in G1, G2, G3 and G4, respectively. Finally, at S3, the level in fruit skin was ~2.14-, ~3.41-, ~5.25- and ~4.35-fold higher in G1, G2, G3 and G4, respectively. Briefly, anthocyanin content was lowest in G0 and highest in G3, followed by the contents in G4, G2, and G1. It was concluded that the more intense red colouring pattern in the apple skin of bud sport mutants was mainly caused by the accumulation of anthocyanins. In addition, a more blushed colouring pattern was observed with an increase in the number of bud sport generations. However, the third-generation mutant G3 showed greater blushing than did the fourth-generation mutant G4 at S1 and S2, and there was no significant difference at S3.
The contents of IAA and ABA in apple skin increased with bud sport generation at color break stage, while the content of GA decreased
Hormone levels were also analyzed at three time points (S1, S2, and S3). The overall trend of IAA concentrations in apple skin first increased and then decreased from S1 to S3 and peaked at S2 (Fig. 1c). However, the GA content decreased from S1 to S3 (Fig. 1d). ABA concentrations of G0, G1 and G2 peaked at S1, whereas those of G3 and G4 peaked at S2 (Fig. 1e). Importantly, the IAA and ABA contents of G3 and G4 in apple skin at color break stage S2 were considerably higher than those of G2, while G0 and G1 showed considerably lower levels than did G2. Nevertheless, GA concentrations, which displayed a trend opposite that of IAA and ABA, decreased with bud sport generation, that is, from G0 to G4.
Transcriptomic profiling of the fruit skin from ‘Red Delicious’ and its four continuous generation mutants
Triplicate sampling of the fruit skin from ‘Red Delicious’ and its four continuous generation mutants at S2 yielded 15 RNA samples for RNA sequencing (RNA-seq) analysis, and the mapping rate of 20,189,457-26,765,510 clean reads onto the apple reference genome (https://iris.angers.inra.fr/gddh13//) (Daccord et al. 2017) ranged from 83.29% to 84.81% (Table 1). The average number of mapped reads ranged from 34,093,629 in G2 to 43,153,440.33 in G3. An FDR < 0.01 and fold changes ≥ 2 were used as screening criteria for DEGs; in addition, an FPKM (fragments per kilobase of mapped reads) ≥ 1.0 in at least one of the 5 triplicate samples was considered to be expressed. The mean normalized expression value (FPKM) per transcript of the three biological replicates was calculated for each sample using the geometric normalization method. The resulting dataset comprising 33,192 transcripts was used for subsequent analysis (Additional file 3: Dataset S1).
Differentially expressed genes (DEGs) in ‘Red Delicious’ versus its mutants gradually increased with bud sport generation at color break stage
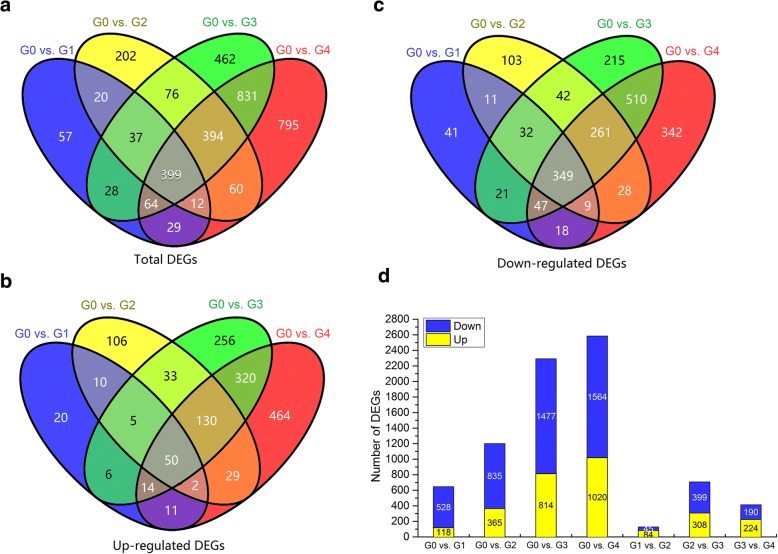
To identify DEGs in ‘Red Delicious’ and its four generation mutants, seven pairwise transcriptome comparisons (i.e., G0 vs. G1, G0 vs. G2, G0 vs. G3, G0 vs. G4, G1 vs. G2, G2 vs. G3, and G3 vs. G4) were performed at S2 (Fig. 2, Additional file 4: Figure S1 and Additional file 5: Table S3). The total number of DEGs was 3,466, including 1,456 up-regulated DEGs and 2029 down-regulated DEGs (Fig. 2 and Additional file 6: Dataset S2). Among them, the number of both up-regulated and down-regulated DEGs increased considerably from the first generation mutant G1 to the fourth generation mutant G4 versus the number observed in G0, and the smallest number was observed in G1 versus that in G2. Moreover, we found that more genes were down-regulated than up-regulated in G1, G2, G3 and G4 relative to G0.
Fig. 2.
Summary of the number of differentially expressed genes (DEGs) identified by RNA-seq analysis in the fruit skin tissues of ‘Red Delicious’ and its four generation mutants (‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee spur’) at S2, named G0 to G4. The number of total DEGs (a), up-regulated DEGs (b), and down-regulated DEGs (c) are presented by Venn diagrams (FDR < 0.01 and fold change ≥ 2). d, Number of total up-regulated and down-regulated DEGs. The histogram represents the number of commonly down-regulated (blue) and up-regulated (yellow) DEGs
Comparative transcriptome enrichment analysis identified key processes responsible for anthocyanin accumulation in ‘Red Delicious’ and its mutants
To understand the major functional categories represented by the DEGs, GO functional enrichment analysis was carried out using all reference genes as background. The results of GO functional enrichment analysis are displayed in three main categories: Biological process, cellular component and molecular function (Additional file 1: Table S1). The biological process category was significantly enriched into three terms, namely, defence response to fungus, 1-aminocyclopropane-1-carboxylate biosynthetic process and negative regulation of growth, were observed in G0 vs. G1, G0 vs. G2, G0 vs. G3 and G0 vs. G4; among these terms, 1-aminocyclopropane-1-carboxylate biosynthetic process and negative regulation of growth were also enriched in G1 vs. G2, G2 vs. G3 and G3 vs. G4. Furthermore, in addition these two GO terms, three other significantly enriched GO terms, namely, chitin catabolic process, regulation of leaf development and DNA conformation change, were shared in G0 vs. G1, G1 vs. G2, G2 vs. G3 and G3 vs. G4. In the cellular component category, GO term of elongator holoenzyme complex was enriched in G0 vs. G1, G0 vs. G2, G0 vs. G3 and G0 vs. G4, whereas, elongator holoenzyme complex, mitochondrial intermembrane space and U12-type spliceosomal complex were shared in G0 vs. G1, G1 vs. G2, G2 vs. G3 and G3 vs. G4. In the molecular function category, nine terms were observed in the seven abovementioned comparison groups: ADP binding, L-iditol 2-dehydrogenase activity, acyl-CoA hydrolase activity, 3-beta-hydroxy-delta5-steroid dehydrogenase activity, O-methyltransferase activity, 1-aminocyclopropane-1-carboxylate synthase activity, naringenin-chalcone synthase activity, catechol oxidase activity and trans-cinnamate 4-monooxygenase activity. Moreover, chitinase activity, sulfur compound binding and caffeate O-methyltransferase activity were only shared in G0 vs. G1, G1 vs. G2, G2 vs. G3 and G3 vs. G4.
To further systematically understand the molecular interactions among the DEGs, we performed KEGG analysis using KOBAS software. The significantly enriched KEGG pathway term sesquiterpenoid and triterpenoid biosynthesis was shared in G0 vs. G1, G0 vs. G2 and G0 vs. G3, but not in G0 vs. G4 (Table 2). However, the sesquiterpenoid and triterpenoid biosynthesis pathway was derived from terpenoid backbone biosynthesis, which occurred both in G0 vs. G3 and G0 vs. G4. Furthermore, the number of DEGs belonging to the triterpenoid biosynthesis pathway gradually increased from G0 vs. G1 to G0 vs. G4. In addition, the flavonoid biosynthesis pathway was enriched in G0 vs. G2, G0 vs. G3 and G0 vs. G4. Moreover, the plant hormone signal transduction pathway was enriched in G0 vs. G1, G0 vs. G2, G0 vs. G3, G0 vs. G4, G2 vs. G3 and G3 vs. G4 but not in G1 vs. G2. Thus, the four abovementioned candidate pathways were considered to be heavily involved in anthocyanin accumulation.
Functional classification of DEGs in ‘Red Delicious’ and its four continuous generation mutants
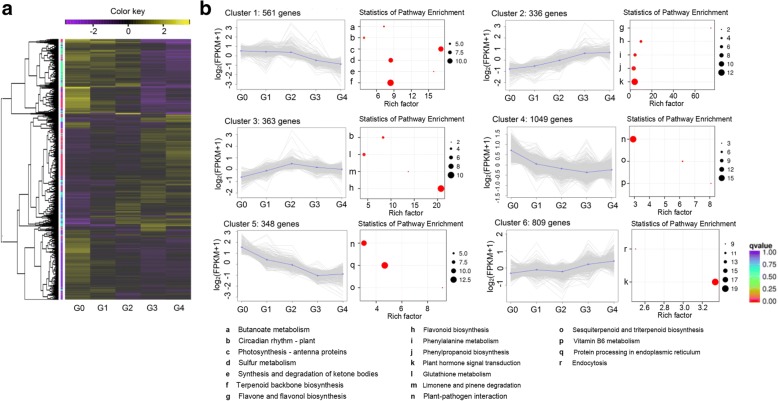
To further identify the major functions of DEGs and establish the skin pigment transcriptome, clustering analysis was applied to the 3,466 DEGs. These genes were grouped into six expression patterns (Fig. 3 and Additional file 7: Dataset S3). Cluster 1 contained 561 DEGs whose expression peaked at G0/G1/G2. Cluster 2 contained 336 DEGs whose expression peaked at G3/G4. Cluster 3 contained 363 DEGs whose expression peaked at G2. Furthermore, 1049 and 348 DEGs whose expression peaked at G0 were included in clusters 4 and 5, respectively. Cluster 6 contained 809 DEGs whose expression peaked at G4. KEGG analysis was also carried out for DEGs belonging to each pattern with a P-value ≤ 0.01. The expression pattern of cluster 2 was positively consistent with total anthocyanin content (Fig. 1b), whereas clusters 4 and 5 were negatively aligned. DEGs in cluster 2 were significantly enriched in plant hormone signal transduction, flavonoid biosynthesis, flavone and flavonol biosynthesis, phenylalanine metabolism, and phenylpropanoid biosynthesis. Among those, some of the final metabolites of flavonoid biosynthesis, flavone and flavonol biosynthesis, phenylalanine metabolism, and phenylpropanoid biosynthesis are anthocyanins (Massonnet et al. 2017). Remarkably, the pathway that was co-enriched by clusters 4 and 5 was sesquiterpenoid and triterpenoid biosynthesis, which was hypothesized to be negatively related to the accumulation of anthocyanins. In addition, the pathway of terpenoid backbone biosynthesis was also enriched in cluster 1, and flavonoid biosynthesis and phenylalanine metabolism were enriched in cluster 3. Overall, the results of the functional classification analysis of the common expression patterns of DEGs combined with KEGG agreed with the results of the aforementioned comparative transcriptome enrichment analysis. Therefore, pathways including flavonoid/phenylpropanoid biosynthesis, terpenoid biosynthesis and plant hormone signal transduction were selected for subsequent analysis.
Fig. 3.
Gene expression profiles and KEGG category distribution of the DEGs in the six common expression clusters composing the fruit skin transcriptome at S2. Clusters were derived by coupled clustering analysis of the 3,466 commonly modulated genes. a Heat map of the overall common expression pattern. b Each line represents the log2-transformed average of the mean FPKM values for an individual transcript. Significantly overrepresented KEGG categories are represented by red dots. KEGG category enrichment was computed using the R language, Cluster package, Biobase package, and Q-value package (P ≤ 0.01)
Key candidate DEGs responsible for anthocyanin accumulation in ‘Red Delicious’ and its mutants
Genes involved in phenylpropanoid/flavonoid biosynthesis pathway
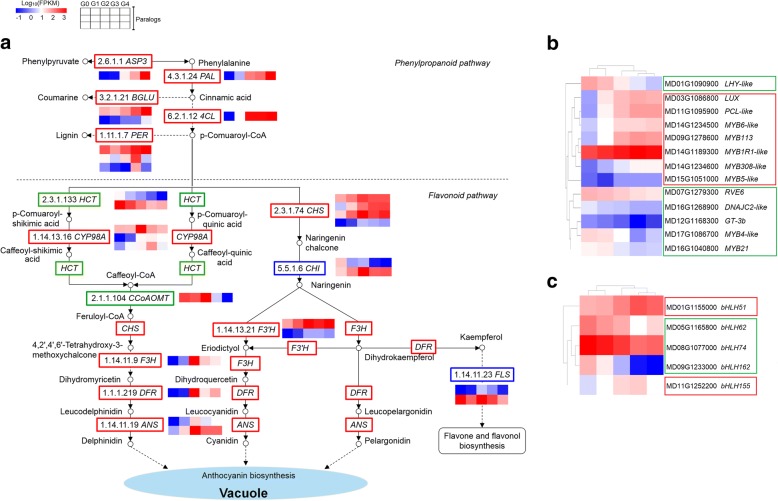
Variety-specific trends in the expression of phenylpropanoid/flavonoid biosynthesis pathway genes at S2 were investigated by preparing heat maps (Fig. 4a). We focused on the 28 DEGs involved in this pathway, including 4 from cluster 1, 13 from cluster 2, 10 from cluster 3, and 1 from cluster 5 (Additional file 8: Table S4). Differences in the expression pattern of these genes from cluster 2 were found among ‘Red Delicious’ and its four continuous generation mutants, closely mirroring the differences in total anthocyanin concentration at S2. For example, all DEGs involved in the phenylpropanoid biosynthesis pathway, including one aspartate aminotransferase cytoplasmic (ASP3), one phenylalanine ammonia-lyase (PAL), two beta-glucosidase (BGLU), one 4-coumarate-CoA ligase (4CL) and three peroxidases (PER), were from cluster 2, demonstrating a gradually increasing expression pattern from G0 to G4 (Fig. 4a and Additional file 9: Dataset S4). Among the DEGs, ASP3 participates in the synthesis of phenylalanine and phenylpyruvate, which are precursors of anthocyanin synthesis. BGLU and PER are involved in coumarine and lignin biosynthesis, respectively. PAL and 4CL are phenylpropanoid genes. In addition, down-regulated genes, including two quinate hydroxycinnamoyl transferase (HCT) and one caffeoyl-CoA O-methyltransferase (CCoAOMT), and up-regulated genes, including three cytochrome P450 98A2-like (CYP98A), four CHS, one avanone-3-hydroxylase (F3H), one dihydroflavonol reductase (DFR), and one anthocyanidin synthase (ANS), are involved in the synthesis of delphinidin from the phenylpropanoid biosynthesis pathway. Furthermore, the four CHS genes described above, one down-regulated and one up-regulated CHI gene, two F3’H genes, and the aforementioned F3H, DFR and ANS are involved in the synthesis of cyanidin. F3’H and other genes involved in pelargonidin synthesis are the same as those involved in cyanidin synthesis. These findings confirm that delphinidin, cyanidin and pelargonidin are the three main substances responsible for the synthesis of anthocyanins via the phenylpropanoid biosynthesis pathway in apple. Moreover, two down-regulated FLS genes are involved in flavone and flavonol biosynthesis.
Fig. 4.
a Differential expression of genes involved in phenylpropanoid/flavonoid biosynthesis pathway hormone signal transduction pathway in ‘Red Delicious’ and its four generation mutants (‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee spur’), named G0 to G4. Differential expression of transcription factors that encode MYB-like (b) and helix-loop-helix (c) DNA-binding domains. Heat maps depict the normalized gene expression values, which represent the means ± SD of three biological replicates. Expression values of five libraries are presented as FPKM normalized log10-transformed counts
Genes involved in MYB-like and helix-loop-helix DNA-binding domain transcriptional factors
Correlative analysis was carried out between anthocyanin content at S2 and the expression levels of transcription factors that encode MYB-like and helix-loop-helix DNA-binding domains in DEGs (Additional file 10: Dataset S5 and Additional file 11: Dataset S6). As a result, 13 MYB and 5 bHLH transcription factors were screened. Among them, the expression of MYB members, including LUX, MYB113, PCL-like, MYB1R1-like, MYB6-like, MYB308-like and MYB5-like, bHLH members, including bHLH51 and bHLH155, presented a notable positive correlation with anthocyanin content at S2 (P<0.05) (Fig. 4b, c). MYB members LHY-like, RVE6, GT-3b, MYB21, DNAJC2-like and MYB4-like and bHLH members bHLH62, bHLH74 and bHLH162 showed a remarkably negative correlation with anthocyanin content at color break stage S2 (P<0.05).
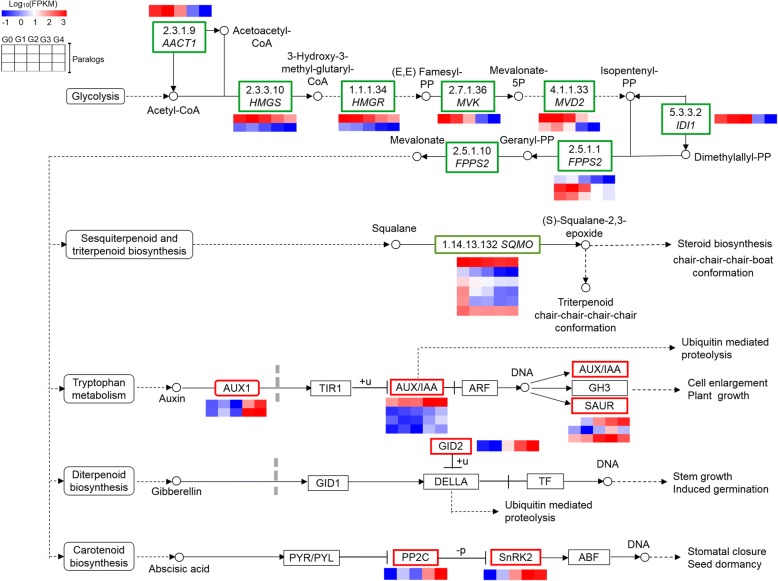
Genes involved in terpenoid biosynthesis and plant hormone signal transduction
KEGG analysis showed that terpenoid backbone biosynthesis was over-presented in cluster 1 and that sesquiterpenoid and triterpenoid biosynthesis branching from terpenoid backbone biosynthesis was over-presented in clusters 4 and 5 (Fig. 3), indicating that this pathway may play important roles in anthocyanin accumulation in apple. Moreover, the plant hormone signal transduction pathway, which is modulated by terpenoid biosynthesis, was enriched in cluster 2. A total of 18 DEGs in the terpenoid biosynthesis pathway (Additional file 12: Dataset S7 and Additional file 13: Table S5) and 12 DEGs in the plant hormone signal transduction pathway (Additional file 14: Dataset S8 and Additional file 15: Table S6) were also investigated by preparing heat maps (Fig. 5). The expression of DEGs involved in terpenoid backbone biosynthesis, including one acetyl-CoA acetyltransferase, cytosolic 1 (AACT1), two hydroxymethylglutaryl-CoA synthase-like (HMGS), two 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1-like (HMGR), one mevalonate kinase-like (MVK), two diphosphomevalonate decarboxylase MVD2-like (MVD2), one isopentenyl-diphosphate Delta-isomerase I (IDI1), three farnesyl pyrophosphate synthase 2-like (FPPS2) and six squalene monooxygenase-like (SQMO), were gradually down-regulated from G0 to G4. Nevertheless, two auxin transporter-like 1 (AUX1), four auxin-responsive protein AUX/IAA and three auxin-responsive protein SAUR involved in tryptophan metabolism of auxin biosynthesis; one F-box protein GID2-like (GID2) involved in diterpenoid biosynthesis of GA biosynthesis; and one protein phosphatase 2C (PP2C) and one sucrose non-fermenting-1-related protein kinase 2 (SnRK2) involved in carotenoid biosynthesis of ABA biosynthesis were up-regulated from G0 to G4.
Fig. 5.
Differential expression of genes involved in sesquiterpenoid and triterpenoid/terpenoid biosynthesis coupled with hormone signal transduction pathway in ‘Red Delicious’ and its four generation mutants (‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee spur’), named G0 to G4. Heat maps depict the normalized gene expression values, which represent the means ± SD of three biological replicates. Expression values of five libraries are presented as FPKM normalized log10-transformed counts
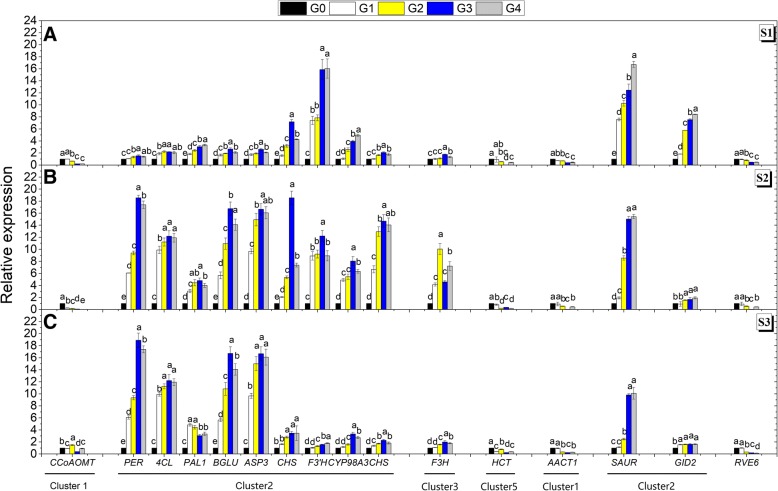
To further evaluate the validity of our results, 16 representative DEGs used previously as shown in Figs. 4 and 5 were selected for expression level examination by qRT-PCR (Additional file 2: Table S2). The overall trend of relative expression levels at three stages was consistent with that of deep sequencing at S2 (Fig. 6), suggesting that the candidate genes involved in pathways such as phenylpropanoid/flavonoid biosynthesis, terpenoid biosynthesis and plant hormone signal transduction, appended with MYB and bHLH transcriptional factors, were directly correlated with anthocyanin accumulation.
Fig. 6.
Relationships between total anthocyanin contents and transcript levels of the sixteen representative genes from Figs. 4 and 5. For each accession, the expression was determined in three developmental stages (S1 to S3) of fruit skin tissues. For the qRT-PCR assay, the mean was calculated from three biological replicates, each with three technical replicates (n=9). These replicates were then normalized relative to the expression of MdGADPH. The x-axis in each chart is the same and represents different Malus accessions, as indicated by names in the bottom panel, which are arranged in different clusters. The left y-axis represents relative expression levels determined by qRT-PCR
Discussion
The activation of early phenylpropanoid biosynthesis genes was more responsible for anthocyanin accumulation in apple skin of bud sport mutants
In plants, phenylpropanoid biosynthesis gives rise to a large number of secondary metabolites, including hydroxycinnamic acids, monolignols/lignin, coumarins, benzoic acids, stilbenes, anthocyanins and flavonoids, serving different functions in plant development, reproduction, defence, and protection against biotic/abiotic stresses [31–33]. Differences in the expression pattern of genes involved in phenylpropanoid/flavonoid biosynthesis result in diverse anthocyanin profiles [2, 25]. Our survey provided a comprehensive profile of the phenylpropanoid/flavonoid biosynthesis pathway in ‘Red Delicious’ and its four continuous generation mutants. The results showed that all of the early phenylpropanoid biosynthesis pathway genes, including ASP3, PAL, 4CL, BGLU and PER, were aggregated in cluster 2 (Fig. 4a and Additional file 8: Table S4), which matched the anthocyanin content (Fig. 1a, b). Other genes in cluster 2 containing CHS, CYP98A and F3’H are involved in the middle steps of the phenylpropanoid pathway, that is, the early steps of the flavonoid biosynthesis pathway. Nevertheless, genes encoding CHS, CYP98A, CHI, F3H, DFR, FLS and ANS were involved in the middle and late steps of the phenylpropanoid biosynthesis pathway and gathered in clusters 3, which was contrary to the findings in previous reports where the late biosynthetic genes usually correlated positively with the anthocyanin content [34–37]. Remarkably, anthocyanin concentrations of glucose-treated Paeonia suffruticosa ‘Luoyang Hong’ cut flowers were higher than those of the sucrose-treated flowers and increased significantly from the pre-opening stage to the full opening stage, whereas the expression of PsCHI1, PsF3H1 and PsDFR1 was differentially induced by sucrose and glucose at different development stages [38]. This report is in line with our results which shows that the expression levels of F3H which was selected for qRT-PCR verification from cluster 3 were consistent with the total anthocyanin contents at the corresponding stages, S1 and S3 (Fig. 6). In addition, members of CHS were also enriched in cluster 1, 2 and 3. Similarly, transcript levels of BrCHS1, -4, -5, F3H, DFR, and ANS were high in pigmented epidermis of light-exposed swollen hypocotyls from Brassica rapa, while those of BrCHS2, -3, and -6 were almost undetectable [39]. MdUFGT2 which was involved in the late step of the phenylpropanoid biosynthesis pathway was up-regulated only in non-red apple cultivars, while MdUFGT4 was up-regulated only in red skin cultivar [40]. In this case, different gene family members encoding structural genes were expressed at different levels and these need to be further investigate in future. To sum up, the activation of early phenylpropanoid biosynthesis pathway genes was demonstrated to be most responsible for pigment accumulation in the apple skin of bud sport mutants at the color break stage. In addition, ASP3, BGLU and PER were confirmed to be involved in the synthesis of phenylpyruvate, coumarine and lignin, respectively (Fig. 4a). Interestingly, 44 stilbene synthase (STS) genes involved in stilbene biosynthesis were characterized to influence anthocyanin accumulation during grapevine (Vitis vinifera) maturation as reported by Massonnet et al. (2017) [25]. Nevertheless, these genes do not exist among our 3,466 DEGs, possibly because they are variety-specific in nature [41].
MYB and bHLH modulated anthocyanin accumulation in apple skin by regulating the transcription of genes involved in the phenylpropanoid/flavonoid pathway
MYB and bHLH autonomously mediated the transcription of genes involved in the middle steps of the phenylpropanoid pathway, that is, the early steps of the flavonoid biosynthetic pathway (CHS, CHI, F3H, F3’H and FLS), which leads to the production of colourless dihydroflavonol compounds [20, 42–45]. The heterologous expression of OjMYB1 in Arabidopsis could enhance anthocyanin content and up-regulate the expression levels of structural gene-related anthocyanin biosynthesis [46]. The red radish (Raphanus sativus L.) bHLH transcription factor RsTT8 acts as a positive regulator of anthocyanin biosynthesis [47]. Nevertheless, AtMYB4, AmMYB308, FaMYB1, ZmMYB31, ZmMYB42, PhMYB4, VvMYBC2-L1 and PtrMYB57 have been demonstrated to repress phenylpropanoid synthesis, likely via repression of synthesis genes [48–52]. We corroborated that MYB members, including LUX, MYB113, PCL-like, MYB1R1-like, MYB6-like, MYB308-like and MYB5-like, and bHLH members, including bHLH51 and bHLH155, showed a notable positive correlation with anthocyanin content (Additional file 10: Dataset S5, Additional file 11: Dataset S6 and Fig. 4b, c) and were considered to promote anthocyanin synthesis by mediating the transcription of structural genes, CHS and F3’H, which are involved in the flavonoid pathway. Other MYB members, including LHY-like, RVE6, GT-3b, MYB21, DNAJC2-like and MYB4-like, and bHLH members, including bHLH62, bHLH74 and bHLH162 showed a remarkably negative correlation with anthocyanin content and were demonstrated to repress anthocyanin synthesis. In addition, HD-Zip I transcription factor MdHB1 was involved in the regulation of anthocyanin accumulation [53]. When MdHB1 is silenced, MdMYB10, MdbHLH3, and MdTTG1 are released to activate the expression of MdDFR and MdUFGT and anthocyanin biosynthesis, resulting in red flesh in apple cv. ‘Granny Smith’ [54]. The expression of F3’5’H, DFR and ANS is strongly inhibited by the increase in the expression of MYBL1, which is a novel R3 MYB transcription factor classified as an MYB transcriptional repressor [55]. However, a full understanding of the mechanism by which structural genes involved in anthocyanin synthesis are specifically mediated by MYB and bHLH remains elusive clear and requires further investigation.
Terpenoid biosynthesis modulated anthocyanin accumulation by positively regulating plant hormone signal transduction in apple skin of bud sport mutants
Hormones are important factors inducing anthocyanin accumulation [56–58]. Carvalho et al. (2010) [12] provided evidence that anthocyanin accumulation was promoted by exogenous ABA and CTK and inhibited by GA in tomato hypocotyls. Co-treatment of IAA and CTK significantly enhanced the cytokinin-induced increase in anthocyanin levels, but an auxin concentration that was too high strongly inhibited anthocyanin synthesis even in the presence of cytokinin in callus cultures of red-fleshed apple (M. sieversii f.niedzwetzkyana), as shown by Ji et al. (2014) [16]. Our results showed that sesquiterpenoid and triterpenoid biosynthesis along with plant hormone transduction, including tryptophan metabolism for IAA, diterpenoid biosynthesis for GA and carotenoid biosynthesis for ABA, branches from the general terpenoid backbone synthesis pathway and shares the same precursors as glycolysis (Fig. 5). The sesquiterpenoid and triterpenoid biosynthesis pathways comprised six SQMO genes in clusters 4 and 5, which were negatively correlated with anthocyanin content. The genes AACT1, HMGS, HMGR, MVK, MVD2, IDI1 and FPPS2, involved in the early steps of terpenoid backbone synthesis, were in cluster 1, which also reflected the differential accumulation of anthocyanin to some extent. Moreover, AUX1, AUX/IAA and SAUR, associated with tryptophan metabolism for IAA; GID2, associated with diterpenoid biosynthesis for GA; and PP2C and SnRK2, associated with carotenoid biosynthesis for ABA, were in cluster 2, which positively reflected the anthocyanin content. Likewise, the contents of IAA and ABA increased, while GA decreased with maturation and pigment accumulation from G0 to the fourth-generation mutant G4 (Fig. 1c, e). Therefore, the down-regulation of genes involved in terpenoid biosynthesis positively induced the expression of AUX1, AUX/IAA, SAUR, GID2, PP2C and SnRK2, resulting in increased synthesis of IAA and ABA and decreased synthesis of GA, thus modulating anthocyanin accumulation. Loreti et al. (2010) [13] suggested the existence of crosstalk between the sucrose and hormone signalling pathways in the regulation of the anthocyanin biosynthetic pathway. Similarly, exogenous ABA promoted fruit ripening by increasing anthocyanin content in sweet cherry (Prunus avium L.) cv. Sato Nishiki, and the expression of PaPP2C3, PaPP2C5 and PaPP2C6 was significantly induced by exogenous ABA [58]. In general, there may be some form of crosstalk between the activation of phenylpropanoid biosynthesis and plant hormone signal transduction in pigment accumulation of apple bud sport mutants.
Conclusions
We have investigated the fruit skin transcriptome of ‘Red Delicious’ and its four continuous generation mutants (‘Starking Red’, ‘Starkrimson’, ‘Campbell Redchief’ and ‘Vallee spur’) and identified specific processes that lead to the accumulation of anthocyanin. The results indicate that apple skin pigmentation and anthocyanin content were increased in the mutants due to bud sport. Terpenoid biosynthesis influences anthocyanin accumulation by positively regulating the synthesis of IAA and ABA and negatively regulating the synthesis of GA. MYB and bHLH modulate anthocyanin accumulation in apple skin by regulating the transcription of genes CHS and F3’H. ASP3, CYP98A and CCoAOMT are novel anthocyanin-associated genes of apple first reported the present study. This novel set of genes provides not only new insights into anthocyanin biosynthesis but also important clues for more dedicated studies to broaden our knowledge of the anthocyanin pathway in apple.
Additional files
Table S1. Gene ontology (GO) enrichment analyses for DEGs in ‘Red Delicious’ and its four generation mutants (XLS 6282 kb)
Table S2. Sequence of primers used for qRT-PCR analysis. (XLS 7198 kb)
Dataset S1. The 5-sample data set. Mean of the normalized expression value per transcript (FPKM fragments per kilobase of mapped reads) of the 33,191 transcripts at S2. (XLS 116 kb)
Figure S1. Summary of the number of differentially expressed genes (DEGs) identified by RNA-seq analysis in the fruit skin tissues of ‘Red Delicious’ and its four generation mutants (XLS 17 kb)
Table S3. Statistical table of the number of annotated differentially expressed genes (DEGs) (XLS 2153 kb)
Dataset S2. Differentially expressed genes identified as commonly up-regulated or down-regulated in each pairwise comparison of ‘Red Delicious’ and its four generation mutants. (XLS 36 kb)
Dataset S3. Gene composition of the six clusters identified using gene expression clustering analysis. (XLS 13 kb)
Table S4. List of the 28 genes in phenylpropanoid biosynthesis and flavonoid biosynthesis pathways (ko00940 and ko00941) identified in ‘Red Delicious’ and its four generation mutants, their descriptions, loci, expression patterns and functional annotations. (XLS 12 kb)
Dataset S4. Gene composition and mean FPKM value of the phenylpropanoid/flavonoid biosynthesis pathway. (TIF 392 kb)
Dataset S5. Correlation analysis of anthocyanin content in S2 and MYB-like DNA-binding domain transcriptional factors. (DOC 359 kb)
Dataset S6. Correlation analysis of anthocyanin content in S2 and helix-loop-helix DNA-binding domain transcriptional factors (DOC 29 kb)
Dataset S7. Gene composition and mean FPKM value of the terpenoid biosynthesis pathway. The GO cellular component is reported. (DOC 34 kb)
Table S5. List of the 18 genes in terpenoid backbone biosynthesis (ko00900) and steroid biosynthesis/sesquiterpenoid and triterpenoid biosynthesis pathways (ko00100/ko00909) identified in ‘Red Delicious’ and its four generation mutants, their descriptions, loci, expression patterns and functional annotations. (DOC 62 kb)
Dataset S8. Gene composition and mean FPKM value of the plant hormone signal transduction. (DOC 49 kb)
Table S6. List of the 12 genes in the plant hormone transduction pathway (ko04075) identified in ‘Red Delicious’ and its four generation mutants, their descriptions, locus, expression patterns and functional annotations. (DOC 35 kb)
Acknowledgments
None.
Funding
This work was financially supported by the Discipline Construction Fund Project of Gansu Agricultural University (GSAU-XKJS-2018-230), the Fostering Foundation for the Excellent Ph.D. Dissertation of Gansu Agricultural University (2017002), and the Science and Technology Major Project of Gansu Province (18ZD2NA006).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Abbreviations
- 4CL
4-coumarate-CoA ligase
- AACT1
Acetyltransferase, cytosolic 1
- ANR
Anthocyanidin reductase
- ANS
Anthocyanidin synthase
- ASP3
Aspartate aminotransferase cytoplasmic
- AUX1
Auxin; transporter-like 1
- BGLU
Beta-glucosidase
- CCoAOMT
Caffeoyl-CoA O-methyltransferase
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- CYP98A
Cytochrome P450 98A2-like
- DEGs
Differentially expressed genes
- DFR
Dihydroflavonol reductase
- F3’H
Flavonoid 3’-hydroxylase
- FDR
False discovery rate
- FLS
Flavonol synthase
- FPPS2
Farnesyl pyrophosphate synthase 2-like
- GO
Gene ontology
- HCT
Quinate hydroxycinnamoyl transferase
- HMGR
3-hydroxy-3-methylglutaryl-coenzyme A reductase 1-like
- HMGS
Hydroxymethylglutaryl-CoA synthase-like
- HPLC
High-performance liquid chromatography
- IDI1
Isopentenyl-diphosphate Delta-isomerase I
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LAR
Leucoanthocyanidin reductase
- MVD2
Diphosphomevalonate decarboxylase MVD2-like
- MVK
Mevalonate kinase-like
- PAL
Phe ammonia lyase
- PER
Peroxidases
- PP2C
Protein phosphatase 2C
- SnRK2
Sucrose non-fermenting-1-related protein kinase 2
- SQMO
Squalene monooxygenase-like
Authors’ contributions
BHC designed the research. WFL carried out the experiments with the help of JM, SJY, ZGG, ZHM, CWZ and MYC. WFL collected the experimental data and drafted the manuscript. MMD reviewed the manuscript and part of the data analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experiments did not utilize transgenic technology or involve protected species. ‘Red Delicious’ and its four generation bud sport mutants used in this study were obtained from an orchard in Tianshui, which was a demonstration base of Gansu Agricultural University, China. It does not require specific permits.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wen-Fang Li, Email: 1304784689@qq.com.
Juan Mao, Email: maojuan-81@163.com.
Shi-Jin Yang, Email: 1947919171@qq.com.
Zhi-Gang Guo, Email: 3148123508@qq.com.
Zong-Huan Ma, Email: mazohu@163.com.
Mohammed Mujitaba Dawuda, Email: mmdawuda@yahoo.com.
Cun-Wu Zuo, Email: zuocunwu2006@126.com.
Ming-Yu Chu, Email: chu.my@foxmail.com.
Bai-Hong Chen, Email: bhch@gsau.edu.cn.
References
- 1.Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annu Rev Ecol Evol S. 2006;37:187–214. doi: 10.1146/annurev.ecolsys.37.091305.110215. [DOI] [Google Scholar]
- 2.El-sharkawy I, Dong L, Xu K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J Exp Bot. 2015;66(22):7359–7376. doi: 10.1093/jxb/erv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nwafor CC, Gribaudo I, Schneider A, Wehrens R, Grando MS, Costantini L. Transcriptome analysis during berry development provides insights into co-regulated and altered gene expression between a seeded wine grape variety and its seedless somatic variant. BMC Genomics. 2014;15:1030. doi: 10.1186/1471-2164-15-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto D, Petersen R, Brauksiepe B, Braun P, Schmidt ER. The columnar mutation (“Co gene”) of apple (Malus×domestica) is associated with an integration of a Gypsy-like retrotransposon. Mol Breeding. 2014;33:863–880. doi: 10.1007/s11032-013-0001-3. [DOI] [Google Scholar]
- 5.Xu Y, Feng S, Jiao Q, Liu C, Zhang W, Chen W, Chen X. Comparison of MdMYB1 sequences and expression of anthocyanin biosynthetic and regulatory genes between Malus domestica Borkh. cultivar ‘Ralls’ and its blushed sport. Euphytica. 2012;185:157–170. doi: 10.1007/s10681-011-0494-y. [DOI] [Google Scholar]
- 6.Williams RS, Benkeblia N. Biochemical and physiological changes of star apple fruit (Chrysophyllum cainito), during different “on plant” maturation and ripening stages. Sci Hortic. 2018;236:36–42. doi: 10.1016/j.scienta.2018.03.007. [DOI] [Google Scholar]
- 7.Meng R, Zhang J, An L, Zhang B, Jiang X, Yang Y, Zhao Z. Expression profiling of several gene families involved in anthocyanin biosynthesis in apple ( Malus domestica, Borkh.) skin during fruit development. J Plant Growth Regul. 2016;35(2):449–464. doi: 10.1007/s00344-015-9552-3. [DOI] [Google Scholar]
- 8.Arrizabalaga M, Morales F, Oyarzun M, Delrot S, Gomès E, Irigoyen JJ, Hilbert G, Pascual I. Tempranillo clones differ in the response of berry sugar and anthocyanin accumulation to elevated temperature. Plant Sci. 2018;267:74. doi: 10.1016/j.plantsci.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol. 2008;165(8):886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Honda C, Moriya S. Anthocyanin biosynthesis in apple fruit. Horticult J. 2018;87(3):305–14.
- 11.Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 2004;167(2):247–252. doi: 10.1016/j.plantsci.2004.03.021. [DOI] [Google Scholar]
- 12.Carvalho RF, Vera Q, Lep P. Hormonal modulation of photomorphogenesis-controlled anthocyanin accumulation in tomato (Solanum lycopersicum L. cv Micro-Tom) hypocotyls: physiological and genetic studies. Plant Sci. 2010;178(3):258–264. doi: 10.1016/j.plantsci.2010.01.013. [DOI] [Google Scholar]
- 13.Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179(4):1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- 14.El-Kereamy A, Chervin C, Roustan JP, Cheyhnier V, Souquet JM, Moutounet M, Raynal J, Ford C, Latché A, Pech JC, Bouzayen M. Exogenous ethylene stimulates the long term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol Plantarum. 2003;119:175–282. doi: 10.1034/j.1399-3054.2003.00165.x. [DOI] [Google Scholar]
- 15.Liu Z, Shi MZ, Xie DY. Regulation of anthocyanin biosynthesis in Arabidopsis thaliana red pap1-D cells metabolically programmed by auxins. Planta. 2014;239(4):765. doi: 10.1007/s00425-013-2011-0. [DOI] [PubMed] [Google Scholar]
- 16.Ji XH, Wang YT, Zhang R, Wu SJ, An MM, Li M, Wang CZ, Chen XL, Zhang YM, Chen XS. Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii, f. niedzwetzkyana) Plant Cell Tiss Org. 2015;120(1):325–337. doi: 10.1007/s11240-014-0609-y. [DOI] [Google Scholar]
- 17.Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008;147:2041–2053. doi: 10.1104/pp.108.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 19.Telias A, Linwang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EE, Bradeen JM. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011;11(1):93. doi: 10.1186/1471-2229-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 21.An XH, Tian Y, Chen KQ, Wang XF, Hao YJ. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol. 2012;169:710–717. doi: 10.1016/j.jplph.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Massari ME, Murre C. Helix-loop-helix proteins:regulators of transcription in eucaryotic organisms. J Mol Cell Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49(7):1099. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- 25.Massonnet M, Fasoli M, Tornielli GB, Altieri M, Sandri M, Zuccolotto P, Paci P, Gardiman M, Zenoni S, Pezzott M. Ripening transcriptomic program in red and white grapevine varieties correlates with berry skin anthocyanin accumulation. Plant Physiol. 2017;174(4):2376–2396. doi: 10.1104/pp.17.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao J, Li W, Mi B, Dawuda MM, Calderón-Urrea A, Ma Z, Zhang Y, Chen B. Different exogenous sugars affect the hormone signal pathway and sugar metabolism in “Red Globe” (Vitis vinifera L.) plantlets grown in vitro as shown by transcriptomic analysis. Planta. 2017;246(8):1–16. doi: 10.1007/s00425-017-2712-x. [DOI] [PubMed] [Google Scholar]
- 27.Mao J, Li W, Mi B, Ma Z, Dawuda MM, Zuo C, Zhang Y, Jiang X, Chen B. Transcriptome analysis revealed glucose application affects plant hormone signal transduction pathway in “Red Globe” grape plantlets. Plant Growth Regul. 2017;84(7):1–12. [Google Scholar]
- 28.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 29.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. Agrigo: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Web Server issue):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Butelli E, De Stefano R, Schoonbeek H, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol. 2013;23:1094–1100. doi: 10.1016/j.cub.2013.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh RS, Kesari R, Kumar U, Jha VK, Kumar A, Kumar T, Pal AK, Singh PK. Candidate genes of flavonoid biosynthesis in Selaginella bryopteris (L.) baker identified by RNA-seq. Funct Integr Genomic. 2018;18:505–517. doi: 10.1007/s10142-018-0603-2. [DOI] [PubMed] [Google Scholar]
- 34.Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem. 2002;40(11):955–962. doi: 10.1016/S0981-9428(02)01454-7. [DOI] [Google Scholar]
- 35.Ban Y, Kondo S, Ubi BE, Honda C, Bessho H, Moriguchi T. UDP-sugar biosynthetic pathway: contribution to cyanidin 3-galactoside biosynthesis in apple skin. Planta. 2009;230(5):871–881. doi: 10.1007/s00425-009-0993-4. [DOI] [PubMed] [Google Scholar]
- 36.Aza-González C, Herrera-Isidrón L, Núñez-Palenius HG, Martínez De La Vega O, Ochoa-Alejo N. Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development. Biologia Plantarum. 2013;57(1):49–55. doi: 10.1007/s10535-012-0265-1. [DOI] [Google Scholar]
- 37.Rahim MA, Busatto N, Trainotti L. Regulation of anthocyanin biosynthesis in peach fruits. Planta. 2014;240(5):913–929. doi: 10.1007/s00425-014-2078-2. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Fu J, Wang Y, Gao S, Du D, Wu F, Guo J, Li D. Glucose supply improves petal coloration and anthocyanin biosynthesis in Paeonia suffruticosa ‘Luoyang Hong’ cut flowers. Postharvest Biol Technol. 2015;101:73–81. doi: 10.1016/j.postharvbio.2014.11.009. [DOI] [Google Scholar]
- 39.Zhou B, Wang Y, Zhan Y, Li Y, Kawabata S. Chalcone synthase family genes have redundant roles in anthocyanin biosynthesis and in response to blue/UV-A light in turnip (Brassica rapa; Brassicaceae) Am J Botany. 2013;100(12):2458–2467. doi: 10.3732/ajb.1300305. [DOI] [PubMed] [Google Scholar]
- 40.Shi X, Qu D, Zhao Z, Gao Z, Liu Y, Yang H, Shi X, Zhao Z. Anthocyanin accumulation and related gene family expression in theskin of dark-grown red and non-red apples (Malus domestica Borkh.)in response to sunlight. Scientia Horticulturae. 2015;189:66–73. doi: 10.1016/j.scienta.2015.03.046. [DOI] [Google Scholar]
- 41.Zenoni S, Fasoli M, Guzzo F, Dal Santo S, Amato A, Anesi A, Commisso M, Herderich M, Ceoldo S, Avesani L, Pezzotti M, Tornielli GB. Disclosing the molecular basis of the postharvest life of berry in different grapevine genotypes. Plant Physiol. 2016;172:1821–1843. doi: 10.1104/pp.16.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 43.Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu DG, Sun CH, Ma QJ, You CX, Cheng L, Hao YJ. MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 2016;170(3):1315–1330. doi: 10.1104/pp.15.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An JP, Liu X, Li HH, You CX, Wang XF, Hao YJ. Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant Cell Physiol. 2017;58(11):1953–1962. doi: 10.1093/pcp/pcx129. [DOI] [PubMed] [Google Scholar]
- 46.Feng K, Xu ZS, Que F, Liu JX, Wang F, Xiong AS. An R2R3-MYB transcription factor, OjMYB1, functions in anthocyanin biosynthesis in Oenanthe javanica. Planta. 2017;247(4):1–15. doi: 10.1007/s00425-017-2783-8. [DOI] [PubMed] [Google Scholar]
- 47.Lim SH, Kim DH, Kim JK, Lee JY, Ha SH. A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front Plant Sci. 2017;8:1917. doi: 10.3389/fpls.2017.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 50.Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG. PhMYB4 fine-tunes the floral volatile signature of Petunia×hybrida through PhC4H. J Exp Bot. 2011;62:1133–1143. doi: 10.1093/jxb/erq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang YF, Vialet S, Guiraud JL, Torregrosa L, Bertrand Y, Cheynier V, This P, Terrier N. A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 2014;201(3):795–809. doi: 10.1111/nph.12557. [DOI] [PubMed] [Google Scholar]
- 52.Wan S, Li C, Ma X, Luo K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017;36(8):1–14. doi: 10.1007/s00299-017-2151-y. [DOI] [PubMed] [Google Scholar]
- 53.Lü P, Zhang C, Liu J, Liu X, Jiang G, Jiang X, Khan MA, Wang L, Hong B, Gao J. RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J. 2014;78(4):578–590. doi: 10.1111/tpj.12494. [DOI] [PubMed] [Google Scholar]
- 54.Jiang Y, Liu C, Yan D, Wen X, Liu Y, Wang H, Dai J, Zhang Y, Liu Y, Zhou B, Ren H. MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith. J Exp Bot. 2017;68(5):1055. doi: 10.1093/jxb/erx029. [DOI] [PubMed] [Google Scholar]
- 55.Gates DJ, Olson B, Clemente TE. Smith SD. A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytol. 2018;217(3):1–11. doi: 10.1111/nph.14830. [DOI] [PubMed] [Google Scholar]
- 56.Jiang C, Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145(4):1460–1470. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shan X. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009;60(13):3849–3860. doi: 10.1093/jxb/erp223. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Chen P, Sun L, Li Q, Dai S, Sun Y, Wenbin K, Zhang Y, Liang B, Leng P. Transcriptional regulation of PaPYLs, PaPP2Cs and PaSnRK2s during sweet cherry fruit development and in response to abscisic acid and auxin at onset of fruit ripening. Plant Growth Regul. 2015;75(2):455–464. doi: 10.1007/s10725-014-0006-x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Gene ontology (GO) enrichment analyses for DEGs in ‘Red Delicious’ and its four generation mutants (XLS 6282 kb)
Table S2. Sequence of primers used for qRT-PCR analysis. (XLS 7198 kb)
Dataset S1. The 5-sample data set. Mean of the normalized expression value per transcript (FPKM fragments per kilobase of mapped reads) of the 33,191 transcripts at S2. (XLS 116 kb)
Figure S1. Summary of the number of differentially expressed genes (DEGs) identified by RNA-seq analysis in the fruit skin tissues of ‘Red Delicious’ and its four generation mutants (XLS 17 kb)
Table S3. Statistical table of the number of annotated differentially expressed genes (DEGs) (XLS 2153 kb)
Dataset S2. Differentially expressed genes identified as commonly up-regulated or down-regulated in each pairwise comparison of ‘Red Delicious’ and its four generation mutants. (XLS 36 kb)
Dataset S3. Gene composition of the six clusters identified using gene expression clustering analysis. (XLS 13 kb)
Table S4. List of the 28 genes in phenylpropanoid biosynthesis and flavonoid biosynthesis pathways (ko00940 and ko00941) identified in ‘Red Delicious’ and its four generation mutants, their descriptions, loci, expression patterns and functional annotations. (XLS 12 kb)
Dataset S4. Gene composition and mean FPKM value of the phenylpropanoid/flavonoid biosynthesis pathway. (TIF 392 kb)
Dataset S5. Correlation analysis of anthocyanin content in S2 and MYB-like DNA-binding domain transcriptional factors. (DOC 359 kb)
Dataset S6. Correlation analysis of anthocyanin content in S2 and helix-loop-helix DNA-binding domain transcriptional factors (DOC 29 kb)
Dataset S7. Gene composition and mean FPKM value of the terpenoid biosynthesis pathway. The GO cellular component is reported. (DOC 34 kb)
Table S5. List of the 18 genes in terpenoid backbone biosynthesis (ko00900) and steroid biosynthesis/sesquiterpenoid and triterpenoid biosynthesis pathways (ko00100/ko00909) identified in ‘Red Delicious’ and its four generation mutants, their descriptions, loci, expression patterns and functional annotations. (DOC 62 kb)
Dataset S8. Gene composition and mean FPKM value of the plant hormone signal transduction. (DOC 49 kb)
Table S6. List of the 12 genes in the plant hormone transduction pathway (ko04075) identified in ‘Red Delicious’ and its four generation mutants, their descriptions, locus, expression patterns and functional annotations. (DOC 35 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.