Abstract
We investigated multivoxel proton magnetic resonance spectroscopy (1H-MRS) biometrics for preoperative differentiation and prognosis of patients with brain metastases (MET), low-grade glioma (LGG) and high-grade glioma (HGG). In total, 33 patients (HGG, 14; LGG, 9; and 10 MET) were included. 1H-MRS imaging (MRSI) data were assessed and neurochemical profiles for metabolites N-acetyl aspartate (NAA) + NAAG(NAA), Cr + PCr(total creatine, tCr), Glu + Gln(Glx), lactate (Lac), myo-inositol(Ins), GPC + PCho(total choline, tCho), and total lipids, and macromolecule (tMM) signals were estimated. Metabolites were reported as absolute concentrations or ratios to tCho or tCr levels. Voxels of interest in an MRSI matrix were labeled according to tissue. Logistic regression, receiver operating characteristic, and Kaplan–Meier survival analysis was performed. Across HGG, LGG, and MET, average Ins/tCho was shown to be prognostic for overall survival (OS): low values (≤1.29) in affected hemisphere predicting worse OS than high values (>1.29), (log rank < 0.007). Lip/tCho and Ins/tCho combined showed 100% sensitivity and specificity for both HGG/LGG (P < .001) and LGG/MET (P < .001) measured in nonenhancing/contrast-enhancing lesional tissue. Combining tCr/tCho in perilesional edema with tCho/tCr and NAA/tCho from ipsilateral normal- appearing tissue yielded 100% sensitivity and 81.8% specificity (P < .002) for HGG/MET. Best single biomarker: Ins/tCho for HGG/LGG and total lipid/tCho for LGG/MET showed 100% sensitivity and 75% and 100% specificity, respectively. HGG/MET; NAA/tCho showed 75% sensitivity and 84.6% specificity. Multivoxel 1H-MRSI provides prognostic information for OS for HGG/LGG/MET and a multibiometric approach for differentiation may equal or outperform single biometrics.
Keywords: MRS, magnetic resonance spectroscopy, brain tumor, brain, metastasis, sensitivity, specificity, glioma, machine learning algorithm
Introduction
The radiological differentiation between primary and secondary brain lesions is often difficult to establish (1). Especially difficult is the separation of ring or focal solid enhancing primary brain tumors from brain metastases (MET) (1). Radiological differentiation between high-grade glioma (HGG) and low-grade glioma (LGG) tumors will in most cases be based on their imaging characteristics. Additional information such as clinical symptoms provided by the referring physician is sometimes helpful for the neuroradiologist in the final radiological decision-making and reporting. However, major radiological problems exist because of similar clinical presentation as imaging characteristics do not always exclude the possibility of HGG presenting as LGG (2). Furthermore, it has been shown that the amount of residual tumor left after resection of solid tumor negatively affects the prognosis (3). This benefit in prognosis from complete resection is an established fact in glioblastomas but near complete resection in LGG and even grade 3 HGG may result in further malignant degeneration (4, 5). It has to be noted that ∼10% of glioblastoma multiforme and 30% of anaplastic astrocytoma show no contrast enhancement (CE), while LGGs occasionally enhance (6). Important for the management and also for treatment planning is that overall survival (OS) significantly differs between high-grade tumors like glioblastomas (World Health Organisation [WHO] grade 4), and low-grade tumors (LGG) (WHO grade 2) (2). The differentiation between a small solid or ring-enhancing glioblastoma and a single metastasis is often more difficult (7). The imaging method of choice for evaluation of primary or secondary brain lesions is magnetic resonance imaging (MRI) using conventional magnetic resonance (MR) sequences, as well as more advanced imaging techniques like perfusion, diffusion, and magnetic resonance spectroscopy (MRS) (8).
There is a need for improvement in diagnostic accuracy by means of aiding the radiologist's subjective assessment of conventional MR images through optimizing methods and further using quantitative methods to benefit patient management for improving prognosis estimations and treatment response assessment. Diagnosing by using light microscopical assessment of phenotype along with MRI characteristics such as amount of edema, necrosis, and ring enhancement, all being somewhat hampered by a component of subjectivity, has been found to be biased owing to the subjective nature of conventional diagnostics and heterogeneity in tumor manifestations. The new 2016 CNS (central nervous system) WHO classification infers molecular along with histological pattern assessment for diagnosis (9), which leaves less space for subjective interpretation (10). Multivoxel MRS offers a great degree of objectivity through the quantification of metabolites of patients suffering from primary or secondary brain neoplasms (11).
Various previous studies have shown different metabolic patterns for histologically different gliomas and MET (7, 12, 13). The majority of previous studies focused on differentiating low- from high-grade astrocytoma while using 1.5 T field strengths and single-voxel spectroscopy (11, 14). Recent MRS studies show moderate success in differentiating HGG from LGG by using metabolic ratios such as choline/creatine (Cho/Cr), Cho/N-acetyl aspartate (Cho/NAA) and NAA/Cr with metabolic ratios reported superior to absolute concentrations (12). Notably, the majority of recent studies only used a single metabolite ratio to differentiate LGG from HGG (12). Furthermore, there were no studies comparing tumoral tissue and healthy-appearing tissue for differentiation of LGG from HGG (12).
This study arises from the need to increase diagnostic accuracy for malignant tumors owing to their poor prognosis and rapid infiltrative nature. The hypothesis is that by using a machine learning algorithm and combining significantly different metabolic data, from healthy-appearing regions and tumor regions in LGG/HGG/MET, extraction of more accurate information can be accomplished to increase diagnostic accuracy in the clinical setting. The aim of this study was to overcome these obstacles by investigating multivoxel 1H-MRS biometrics for preoperative differentiation and prognosis of patients with MET, LGG, and HGG.
Materials and Methods
Presurgical MR spectroscopy examinations were retrospectively obtained in 33 patients (HGG, 14; LGG, 9; MET, 10). Subjects consisted of 11 females and 22 males with a mean age at diagnosis; 57 years for MET (range, 30–77 years), 64 years for HGG (range 48–77 y), and 47 years for LGG (range, 27–66). Deviations from total subjects for different metabolites occurred owing to nonobtainable spectra of adequate quality with LCModel because of either technical issues such as initial placement of MRS grid or low signal-to-noise (S/N) ratio, baseline distortions, and broad line width. Discrepancies in number of subjects analyzed also occurred because of individual characteristics of the target tissue. This is exemplified by the presence and absence of edema in 5 and 4 of the patients with LGG, respectively. Histopathological diagnosis was attained after either resection or biopsy. Demographics are presented in Table 1.
Table 1.
Demographics, Sex, Histopathology, Tumor Location, Type of Surgery, OS
| Patient/Sex/Age of Diagnosis (Years)/Type of Disease | Histopathology | Location of Tumor | Type of Surgery | Overall Survival (Months) |
|---|---|---|---|---|
| 1/M/48/HGG | Oligoastrocytoma Grade 3 | Temporal sin | Resection | >18 |
| 2/M/66/HGG | GBM | Frontal dx | Resection | 22 |
| 3/M/63/HGG | GBM | Frontal dx | Resection | 16 |
| 4/M/62/HGG | Oligoastrocytoma Grade 3 | Frontal dx | Resection | >20.5 |
| 5/M/60/HGG | GBM | Temporal sin | Resection | >3 |
| 6/F/68/HGG | GBM | Frontal sin | Resection | >8 |
| 7/M/74/HGG | GBM | Parietal sin | Needle Biopsy | 9 |
| 8/F/60/HGG | GBM | Parietooccipital dx | Resection | >27 |
| 9/F/59/HGG | GBM | Temporal-occipital sin | Resection | >3 |
| 10/M/66/HGG | GBM | Temporal sin | Open Biopsy | 2 |
| 11/M/60/HGG | GBM | Parietal-Temporal dx | Biopsy via Craniotomy | >20 |
| 12/M/77/HGG | GBM | Temporal-occipital sin | Needle Biopsy | >7 |
| 13/M/71/HGG | GBM | Frontal sin | Extirpation | >9 |
| 14/M/60/HGG | Anaplastic Oligoastrocytoma Grade 3 | Parietal sin | Biopsy via Craniotomy | 15 |
| 15/F/34/LGG | Oligoastrocytoma Grade 2 | Frontal sin | Resection | >39 |
| 16/M/27/LGG | Diffuse astrocytoma Grade 2 | Frontal dx | Extirpation | >31 |
| 17/F/20/LGG | Oligodendroglioma Grade 2 | Frontal sin, Temporal sin | Macroscopic Radical Surgery | >21 |
| 18/M/56/LGG | Astrocytoma Grade 2 | Temporal sin | Open Biopsy | >31 |
| 19/M/66/LGG | Diffuse astrocytoma Grade 2 | Parietoocciptal sin | Needle Biopsy | 12 |
| 20/M/51/LGG | Diffuse astrocytoma Grade 2 | Temporal sin | Extirpation | >26 |
| 21/M/52/LGG | Astrocytoma grade 2 | Temporal sin | Needle Biopsy | >8 |
| 22/M/66/LGG | Diffuse Astrocytoma Grade 2 | Temporal sin, Parietal sin | Open Biopsy | >13 |
| 23/M/49/LGG | Oligodendroglioma Grade 2 | Frontal dx | Resection | >6 |
| 24/M/77/MET | Metastasis-Malignant Melanoma | Frontal sin | Resection | >6 |
| 25/F/54/MET | Metastasis- Primary Breast cancer | Cerebellum dx | Resection | >2 |
| 26/M/49/MET | Metastasis-primary Lung cancer | Cerebellum sin | Extirpation | 8 |
| 27/M/45/MET | Metastasis-malignant Melanoma | Parietal sin | Extirpation | 4 |
| 28/F/73/MET | Metastasis-breast Adenocarcinoma | Cerebellum sin | Extirpation | 16 |
| 29/M/59/MET | Metastasis-malignant Melanoma | Frontal dx | Extirpation | 9 |
| 30/F/73/MET | Metastasis-lung Adenocarcinoma | Cerebellum dx | Extirpation | >31 |
| 31/F/56/MET | Metastasis-lung Adenocarcinoma | Temporal sin | Extirpation | 6 |
| 32/F/30/MET | Metastasis-breast Adenocarcinoma | Parietal sin | Extirpation | >26 |
| 33/F/53/MET | Metastasis-colorectal Adenocarcinoma | Frontal sin | Extirpation | >1 |
The retrospective nature of this study does not allow for grouping according to the new classification (9), as not all subjects have undergone adequate genetic analysis of their lesions. Therefore, grades 1–2 glioma will be henceforth labelled as LGG and grades 3–4 as HGG. MET have been selected randomly with regards to their origin and histopathological diagnosis.
Study Protocol
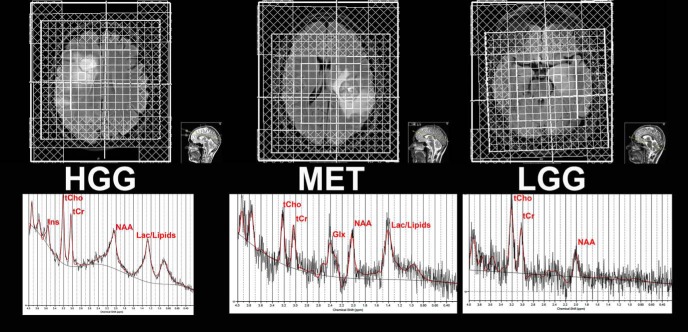
Proton MRS imaging (1H-MRSI) data were acquired in a 3 T Magnetom Skyra scanner (Siemens, Germany) with a 20-channels head coil (Siemens, Germany). Localization of the volume of interest (VOI) in the brain was performed with axial T1WI magnetization prepared rapid acquisition gradient echo (MPRAGE) (repetition time [TR] = 1900 milliseconds, echo time [TE] = 2.5 milliseconds) and axial fluid-attenuated inversion recovery MR images (TR = 9000 milliseconds, TI = 2500 milliseconds, TE = 81 milliseconds) obtained after the injection of gadolinium contrast agent. Magnetic field inhomogeneities were compensated by using the 3D gradient shimming protocol to obtain well-resolved metabolite peaks. Water suppression and outer volume suppression protocols were performed. 1H-MRSI scans were acquired using the Point-RESolved Spectroscopy sequence (PRESS, TR = 2000 milliseconds, TE = 144 milliseconds) with a voxel size of 10 × 10 × 15 mm acquired in a VOI of 4 × 4 × 1 to 8 × 8 × 1 depending on size and or location of tumor (Figure 1). A total acquisition time of 6.12 minutes was used in the 1H-MRSI measurements for each patient.
Figure 1.
Volume of interest grid for HGG, LGG and MET with spectra from one voxel exemplified.
Postprocessing
The in vivo neurochemical profile was analyzed using the LCModel software (LCModel version 6.3-1L, Steven Provencher, Ontario, Canada) (15, 16). Quantities of 6 metabolites N-acetyl aspartate + N-acetyl aspartyl glutamate (NAA), creatine + phosphocreatine (total creatine, tCr), glutamate + glutamine (Glx), lactate (Lac), myo-inositol (Ins), glycerophosphocholine + phosphocholine (total choline, tCho) and the total lipids (tLip), and total macromolecule (tMM) signals were estimated for each MR spectrum. Concentrations were reported as ratios to tCho or tCr levels. Each voxel in the MRSI VOI was labelled on the basis of the tissue type as solid tumor tissue; noncontrast-enhanced tumor (nCE) in all LGG; and partially contrast-enhanced tumor tissue (CE) in all HGG and MET and also cystic/necrotic tumor tissue, perilesional edema, ipsilateral normal-appearing white matter (iNAWM), or contralateral NAWM (cNAWM); the average of the metabolite ratios for each tissue type was reported. Data quality was evaluated by visual inspection and data were excluded on the basis of presence of artifacts, heavy baseline distortions, or poor S/N ratio in the spectra. Owing to the low amount of VOI with nCE in HGG and MET for in this cohort, nCE in HGG, MET, and LGG was pooled together with CE.
Statistical Analysis
Statistical analysis was performed with SPSS® v. 23.0 (IBM Corp., New York, NY; formerly SPSS Inc., Chicago, IL). Shapiro–Wilk tests with normality plots were performed. Kruskal–Wallis H was chosen for the 3 groups—LGG, HGG and MET. Bonferroni adjustment yielded a P-value of < .017 as significance-threshold á priori post hoc testing (17). Mann–Whitney U was performed for pairwise comparison of groups and tissue analyzed: 100% tumor, 100% CE, 100% edema, with regard to tumor compared with iNAWM and tumor compared with cNAWM. The machine learning algorithm logistic regression was used for construction of a model that is able to incorporate significant metabolites/metabolite ratios. Receiver operating characteristic (ROC) analysis with omnibus model, Nagelkerke R2, area under the curve (AUC), and sensitivity and specificity were obtained for significant metabolite concentrations and metabolite ratios (18). Kaplan–Meier survival analysis was performed for LGG, HGG, and MET, based on the reviewed OS status in the period from initial MRI for diagnosis to patient death during the period from 03072012 to 01012018, as well as metabolite concentrations and metabolic ratios with a P-value set to <.05. Spectra obtained within cerebrospinal fluid or where voxels contained ≥25% cerebrospinal fluid were discarded from analysis as most metabolites are deemed of low concentration and therefore negligible (19).
Results
In total, 33 patients were included in this study (HGG, 14; LGG, 9; MET, 10) (Table 2). No exclusions were made, but spectra were obtained for all but a few subjects across all metabolite concentrations or ratios (Table 3). The analysis exclusion criteria included S/N ratio <5 and the presence of significant spectra baseline distortions.
Table 2.
ROC Analysis Performed on Biometrics for LGG, HGG, and MET With Sensitivity/Specificity/Cutoff Value/AUC/P-value
| Group and Biometric | Sensitivity (%) | Specificity (%) | Cutoff Value | AUC (95% CI) | P-Value < |
|---|---|---|---|---|---|
| HGG/LGG | |||||
| tLip/tChoa | 83.3 | 100 | 2.35 | 0.905 (0.746-1.00) | .004 |
| Ins/tChoa | 100 | 75 | 1.61 | 0.905 (0.767-1.00) | .004 |
| Combined Biometricsa | 100 | 100 | 0.499b | 1.00 (1.00-1.00) | .001 |
| HGG/MET | |||||
| tCr/tChoc | 77.8 | 76.7 | 2.65 | 0.824 (0.647-1.00) | .013 |
| tCho/tCrd | 75 | 84.6 | 0.37 | 0.788 (0.567-1.00) | .030 |
| NAA/tChod | 75 | 84.6 | 0.80 | 0.817 (0.634-1.00) | .017 |
| Combined Biometricsc,d | 100 | 81.8 | 0.311b | 0.935 (0.825-1.00) | .002 |
| LGG/MET | |||||
| tLip/tChoe | 100 | 100 | 3.59 | 1.00 (1.00-1.00) | .001 |
| Ins/tChoe | 85.7 | 100 | 1.69 | 0.984 (0.935-1.00) | .001 |
| Combined Biometricse | 100 | 100 | 0.50b | 1.00 (1.00-1.00) | .001 |
Significance level set at P-value <.05:
aComparing non-enhancing hypo intense volume on T1-MPRAGE (100% anticipated tumor tissue) for LGG with contrast enhancing tissue for HGG;
bProbability cut-off value generated by regression model;
cComparing edematous tissue between HGG and MET;
dComparing ipsilateral normal appearing white matter in HGG and MET;
eComparing NE hypointense volume on T1-MPRAGE (100% anticipated tumor tissue) for LGG with CE tissue for MET.
Table 3.
Kruskal–Wallis H Test, Comparison Across 3 Groups; LGG, HGG, and MET and Listed Metabolites
| Kruskal–Wallis Test For–Tissue Type | EDEMA HGG (n = 12), LGG (n = 5), MET (n = 9) | Contrast EnhancementHGG (n = 12), LGG (n = 7), MET (n = 10) | Cyst/Necrosis HGG (n = 9), MET (n = 4) | Ipsilateral NAWM HGG (n = 13), LGG (n = 8), MET (n = 9) | Contralateral NAWM HGG (n = 13), LGG (n = 8), MET (n = 8) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite Concentration or Ratio with P- and Chi-Square Value | Chi-Square | P-Value < | Chi-Square | P-Value < | Chi-Square | P-Value < | Chi-Square | P-Value < | Chi-Square | P-Value < |
| tLip/tCho | 0.012 | .994 | 12.984 | .002 | 0.857 | .355 | 3.820 | .148 | 0.482 | .786i |
| tLip/tCr | 1.858 | .395 | 4.038 | .133 | 0.024 | .877 | 2.118 | .347 | 1.368 | .505i |
| tMM/tCho | 2.791 | .248 | 2.157 | .340 | 5.357 | .021 | 0.013 | .994 | 1.451 | .484 |
| tMM/tCr | 1.072 | .585a | 0.955 | .620 | 3.429 | .064 | 1.248 | .536 | 0.432 | .806 |
| Ins/tCho | 5.384 | .068b | 13.941 | .001d | 1.444 | .229f | 4.243 | .120 | 1.306 | .520i |
| Ins/tCr | 1.263 | .532b | 1.218 | .544d | 0.419 | .518f | 1.963 | .375 | 2.211 | .331i |
| Ins conc | 2.244 | .326b | 0.862 | .65d | 0.009 | .926f | 2.243 | .326 | 7.005 | .030i |
| Lac/tCho | 4.769 | .092c | 3.645 | .162e | 0.857 | .355 | 1.952 | .377g | 3.262 | .196i |
| Lac/tCr | 4.311 | .116c | 1.087 | .581e | 0.024 | .877 | 2.322 | .313g | 4.924 | .085i |
| Lac conc | 2.807 | .246c | 3.701 | .157e | 0.024 | .877 | 2.058 | .357g | 0.025 | .987 |
| tCho/tCr | 0.265 | .876 | 2.389 | .303 | 0.381 | .537 | 8.204 | .017 | 1.868 | .393 |
| tCho conc | 0.244 | .885 | 0.733 | .693 | 4.024 | .045 | 4.702 | .095 | 0.771 | .680 |
| NAA/tCho | 1.666 | .435 | 0.387 | .824 | 3.429 | .064 | 6.701 | .035h | 1.756 | .416 |
| NAA/tCr | 4.602 | .100 | 0.406 | .816d | 0.214 | .643 | 0.973 | .615 | 0.049 | .976 |
| NAA conc | 0.969 | .616 | 0.066 | .967 | 0.095 | .758 | 1.664 | .435 | 4.928 | .085 |
| tCr/tCho | 6.824 | .033 | 6.439 | .040 | 0.024 | .877 | 1.997 | .368h | 0.890 | .641 |
| tCr conc | 0.566 | .754 | 0.371 | .831 | 0.381 | .537 | 1.495 | .474 | 0.422 | .810 |
| Glx/tCho | 7.421 | .024 | 3.455 | .178 | 1.167 | .280 | 1.310 | .520h | 0.536 | .765 |
| Glx/tCr | 3.904 | .142 | 2.695 | .260 | 1.524 | .217 | 5.237 | .073 | 0.503 | .778 |
| Glx conc | 0.777 | .678 | 1.348 | .510 | 0.857 | .355 | 3.103 | .212 | 0.092 | .955 |
Significance before Bonferroni adjustment set at P-value < .05:
aHGG: n = 11; LGG: n = 5; MET: n = 9;
bHGG: n = 11; LGG: n = 5; MET: n = 8;
cHGG: n = 12; LGG: n = 4; MET: n = 9;
dHGG: n = 12; LGG: n = 7; MET: n = 9;
eHGG: n = 11; LGG: n = 7; MET: n = 9;
fHGG: n = 9; MET: n = 3;
gHGG: n = 12; LGG: n = 8; MET: n = 9;
hHGG: n = 13; LGG: n = 8; MET: n = 8;
iHGG: n = 12; LGG: n = 8; MET: n = 8.
In the nCE/CE tissue (HGG, 12; LGG, 7), tLip/tCho and Ins/tCho were significantly different: P < .004 and < 0.004, respectively (Tables 2–4). In addition, significance was evident for tCho/tCr in iNAWM (HGG, 13; MET, 9) with P-value < .015 for HGG and MET along with NAA/tCho with P-value < .017 (Tables 2–4). For cNAWM (HGG, 13; LGG, 8; MET, 8), no significant differences were observed with P-values > .05. Similarly, for nCE/CE tissue, Mann–Whitney U test showed P-value < .004 for both tLip/tCho and Ins/tCho for HGG and LGG. Both these metabolite ratios were found to be significant for LGG and MET with P-values < .001 (Table 2). For iNAWM, tCho/tCr and NAA/tCho were significantly different according to MW-U with P < .0015 and P < .017, respectively.
Table 4.
Mann–Whitney U test Pairwise Across Groups and Listed Metabolites
| Mann–Whitney U Test for LGG, HGG, and MET Across Tissue Types and Listed Metabolites | Mann–Whitney U Values with P-Value | tLip/tCho | Ins/tCho | tCho/tCr | NAA/tCho | tCr/tCho |
|---|---|---|---|---|---|---|
| LGG vs. HGG for EDEMA | Mann–Whitney U | 29.000 | 7.000 | 25.000 | 25.000 | 24.000 |
| HGG n = 12, LGG n = 5, | P-value < | .916 | .020a | .598 | .598 | .527 |
| HGG vs. MET for EDEMA | Mann–Whitney U | 53.000 | 39.000 | 49.000 | 46.000 | 19.000 |
| HGG n = 12, MET n = 9 | P-value < | .943 | .680b | .722 | .570 | .013 |
| LGG vs. MET for EDEMA | Mann–Whitney U | 22.000 | 8.000 | 21.000 | 11.000 | 10.000 |
| LGG n = 5, MET n = 9 | P-value < | .947 | .079c | .841 | .125 | .096 |
| HGG vs. LGG for Contrast-Enhancement | Mann–Whitney U | 8.000 | 8.000 | 25.000 | 41.000 | 14.000 |
| HGG n = 12, LGG n = 7 | P-value < | .004 | .004 | .151 | .933 | .018 |
| HGG vs. MET for Contrast-Enhancement | Mann–Whitney U | 48.000 | 29.000 | 47.000 | 51.500 | 38.000 |
| HGG n = 12, MET n = 10 | P-value < | .429 | .076d | .391 | .575 | .147 |
| LGG vs. MET for Contrast-Enhancement | Mann–Whitney U | 0.000 | 1.000 | 26.000 | 30.000 | 21.000 |
| LGG n = 7, MET n = 10 | P-value < | .001 | .001e | .380 | .626 | .172 |
| HGG vs. MET for Cystic/Necrosis | Mann–Whitney U | 12.000 | 7.000 | 14.000 | 6.000 | 17.000 |
| HGG n = 9, MET n = 4 | P-value < | .355 | .229f | .537 | .064 | .877 |
| HGG vs MET for Ipsilateral NAWM | Mann–Whitney U | 32.000 | 29.000 | 22.000 | 19.000 | 34.000 |
| HGG n = 13, MET n = 9 | P-value < | .077 | .049 | .015 | .017g | .192g |
| HGG vs. LGG for Ipsilateral NAWM | Mann–Whitney U | 45.000 | 37.000 | 36.500 | 50.000 | 41.000 |
| HGG n = 13, LGG n = 8 | P-value < | .612 | .277 | .261 | .885 | .426 |
| LGG vs. MET for Ipsilateral NAWM | Mann–Whitney U | 20.000 | 26.000 | 12.000 | 12.000 | 25.000 |
| LGG n = 8, MET n = 9 | P-value < | .124 | .336 | .021 | .036h | .462h |
| HGG vs. MET for Contralateral NAWM | Mann–Whitney U | 45.000 | 36.000 | 51.000 | 36.000 | 41.000 |
| HGG n = 13, MET n = 8 | P-value < | .817i | .355i | .942 | .247 | .426 |
| HGG vs. LGG for Contralateral NAWM | Mann–Whitney U | 41.000 | 45.000 | 33.000 | 50.000 | 42.000 |
| HGG n = 13, LGG n = 8 | P-value < | 0.589j | .817j | .169 | .885 | .469 |
| LGG vs. MET for Contralateral NAWM | Mann–Whitney U | 26.000 | 22.000 | 23.000 | 21.000 | 29.000 |
| LGG n = 8, MET n = 8 | P-value < | .529 | .294 | .344 | .248 | .753 |
Significance level of <.017 after Bonferroni–Sidàk correction :
aHGG n = 11, LGG n = 5;
bHGG n = 11, MET n = 9;
cLGG n = 5, MET n = 8;
dHGG n = 12, MET n = 9;
eLGG n = 7, MET n = 9;
fHGG n = 9, MET n = 3;
gHGG n = 13, MET n = 8;
hLGG n = 8, MET n = 8;
iHGG n = 12, MET n = 8;
jHGG n = 12, LGG n = 8.
For edematous tissue (HGG, 10; LGG, 4; MET, 8) Kruskal–Wallis H for LGG/HGG/MET showed significant differences for tCr/tCho and tGlx/tCho with P-value < .033 and < .024, respectively, but it failed to show significant difference with P-values >.017. Pairwise Kruskal–Wallis-H along with Sidák–Bonferroni correction with subsequent Mann–Whitney U test excluded significant differences in the remaining absolute concentrations and ratios of metabolites to either tCr or tCho (NAA, tCr, tGlx, tLac, Ins, tCho, tLipids, and tMM) with P > .05 or P > .017 after correction according to Sidàk–Bonferroni correction.
Ins/tCho and tLip/tCho for HGG/LGG and LGG/MET
When comparing noncontrast/contrast-enhancing tissue in HGG and LGG regarding tLip/tCho and Ins/tCho; mean tLip/tCho was significantly higher (10.28) in HGG vs. LGG (1.22). In contrast, mean Ins/tCho was found to be higher in LGG (4.67) vs HGG (1.43). ROC-analysis for separate metabolites showed 83.3% and 100% sensitivity as well as 100% and 75% specificity for tLip/tCho and Ins/tCho respectively for HGG/LGG with cut-offs 2.35 and 1.61 respectively.
Similarly, MET and LGG tLip/tCho was significantly higher in MET (14.42) vs LGG (1.22). Ins/tCho was found to be higher in LGG (4.67) vs MET (0.92). ROC-analysis showed 100% and 85.7% sensitivity as well as 100% and 100% specificity for tLip/tCho and Ins/tCho respectively, for LGG/MET with cut-offs 3.59 and 1.69.
Cr/Cho, Cho/Cr and NAA/Cho for HGG and MET
In iNAWM, tCho/tCr was found to be significantly higher in MET (0.39) than in HGG (0.34; p-value < 0.015). NAA/tCho was found to be higher in MET (2.43) vs HGG (1.14; P-value < 0.017). tCr/tCho in edema was observed to be higher in MET (2.95) vs (2.43) in HGG. ROC-analysis for the single metabolic ratios yielded 77.8%, 75.0%, 75.0% sensitivity as well as 76.7%, 84.6% and 84.6% specificity for tCr/tCho, tCho/tCr and NAA/tCho with cut-offs 2.65, 0.37 and 0.80, respectively.
Combined ROC-Analysis
Cr/Cho, Cho/Cr and NAA/Cho MET vs HGG.
The omnibus model showed a Nagelkerke R2 of 0.79 and P-value < .001 (11 HGG; 7 MET). The combination tCr/tCho in edema with tCho/tCr and NAA/tCho from iNAWM showed 100% sensitivity and 81.8% specificity (P < .002) and AUC = 0.935 when comparing HGG to MET.
Ins/tCho and tLip/tCho LGG vs HGG and MET vs LGG.
The omnibus models showed a Nagelkerke R2 of 1.00 and P < .001 for HGG/LGG (12 HGG; 7 LGG) and a Nagelkerke R2 = 1.00; P < .001 LGG/MET (7 LGG; 9 MET). The combination of tLip/tCho and Ins/tCho, measured in nonenhancing/contrast-enhancing lesional tissue, demonstrated 100% sensitivity/specificity for both HGG/LGG (P < .001) and LGG/MET (P < .001) and probabilistic cut-off values 0.499 and 0.50, respectively. No significant differences between HGG and MET were found.
Overall Survival
There were significant differences between the HGG, LGG and MET with regards to OS; Mantel-Cox, Generalized Wilcoxon, Tarone–Ware P-value < 0.014; 0.009; and 0.009, respectively. The median OS for HGG (n = 18), LGG (n = 10), and MET (n = 15) were 17, 29 and 8 months, respectively. Mean OS ranged from 14-23, 35-58 and 7-33 months for HGG, LGG and MET.
Across HGG/LGG/MET (n = 33); the average Ins/tCho of all voxels in the affected hemisphere was shown to be prognostic for OS: with low values (≤1.29, n = 17) in affected hemisphere predicting worse OS than high values (>1.29n = 16), (Log Rank < 0.007) (Figure 2).
Figure 2.
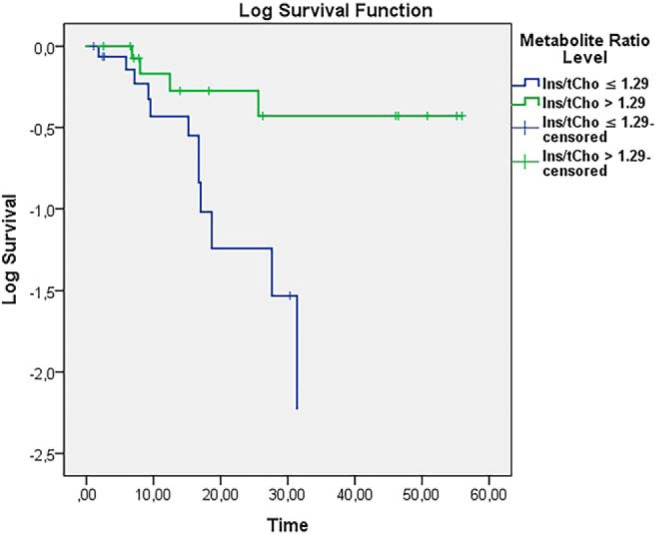
Log survival Ins/tCho mean of all voxels in affected hemisphere for LGG/MET/HGG based on a cutoff at 1.29 for Ins/tCho (N = 33 in total; n = 16 for Ins/tCho > 1.29; n = 17 for Ins/tCho ≤ 1.29).
Discussion
Differentiation of HGG from MET and HGG from LGG solely using conventional MRI are of particular concern to clinicians (20). Furthermore, one systematic review and two meta-analyses also pointed out the difficulty in differentiating HGG and MET presenting as solitary lesions with spectroscopy, not just with conventional MRI (7, 11, 13). At best, the differentiation was deemed to be of moderate success for HGG and MET by utilization of 1H-MRS (7).
Although several studies obtained 100% specificity with Cho/Cr ratio and Cho/NAA ratio, they did not reach 100% sensitivity in discrimination between HGG and MET (7). Furthermore, these studies (7) exhibited poorer CI 95% in comparison with the present study. Another key point was the inconsistency in reported AUC-values for peritumoral ratios of Cho/Cr and Cho/NAA (7). The best AUC-values obtained with Cho/NAA and Cho/Cr were 0.9504 and 0.8959 with sensitivity 85% and specificity 0.93 for Cho/NAA and 86% and 86% for Cho/Cr respectively (7).
Our combined approach with tCr/tCho in iNAWM, tCho/tCr in edematous tissue and NAA/tCho in iNAWM yielded 100% sensitivity and 81.8% specificity with an AUC of 0.935 (Table 2) for MET and HGG. Our study not only confirms already established diagnostic value in the peritumoral regions for differentiation between MET and HGG (7), but also uniquely, and equally important, shows the impact of utilizing metabolic information from tumor to ipsilateral and normal appearing tissue on conventional MRI. Furthermore, in the present cohort, the combination of metabolic information in peritumoral regions as well as normal appearing tissue ipsilateral to tumor give sufficient diagnostic information for 100% sensitivity (Table 2).
MRS has been shown to provide information about metabolic tissue composition, tumor metabolism (glucose), proliferation/membrane turnover (tCho), homeostasis of energy (tCr) and glioneural integrity (NAA) (12). Moreover, A systematic review has found that differentiation of glioblastomas from MET is poor, regardless of long or short echo time 1H-MRS (11).
This present study shows equal or improved accuracy, that is, sensitivity/specificity and AUC, for distinction between LGG, HGG and MET by utilization of metabolic information obtained with multivoxel 1H-MRS from edematous tissue and tumor tissue, as well as from ipsi- and contralateral NAWM (Table 2, Figures 3–5).
Figure 3.
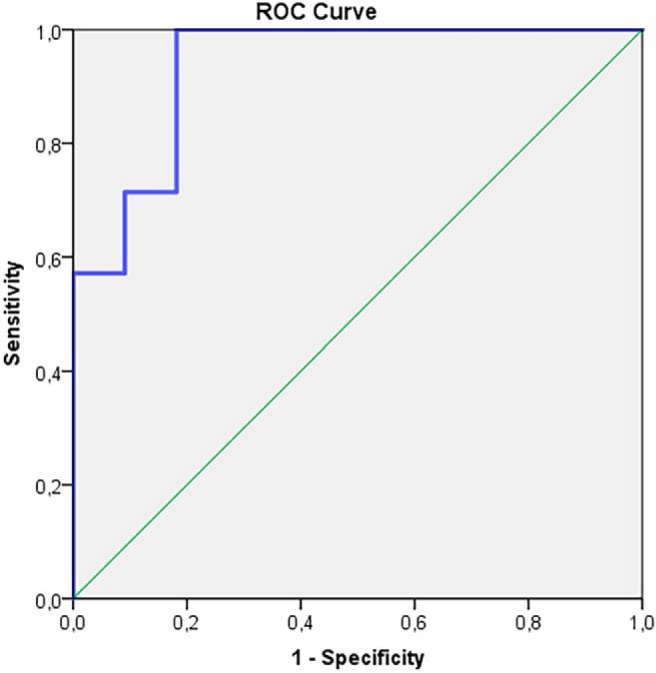
Receiver operating characteristic (ROC)-curve for high-grade glioma (HGG) and metastases (MET) tCr/tCho in edema, tCho/tCr, and NAA/tCho in iNAWM. AUC = 0.935; 95% CI 0.825-1.000; P-value < 0.002.
Figure 5.
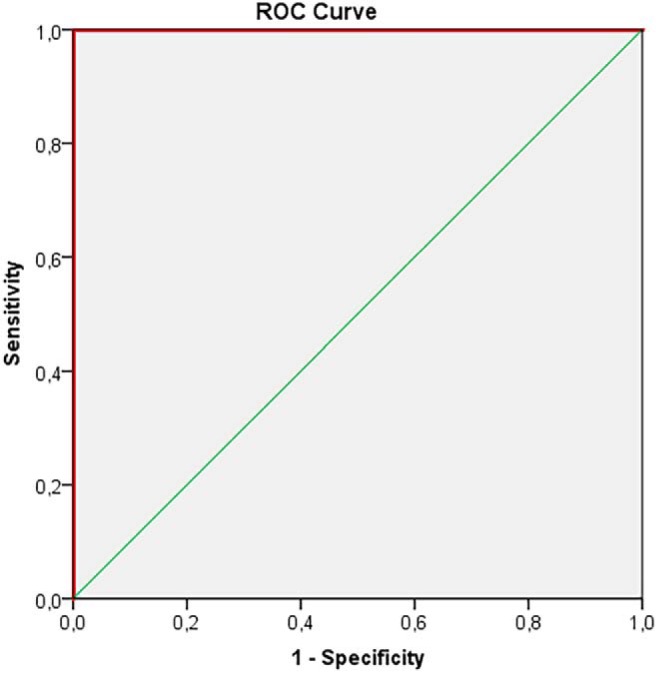
ROC-curve for MET and LGG in tumor tissue (nCE, CE) for tLip/tCho, Ins/tCho. AUC = 1.00; 95% CI 1.00–1.00; P-value < 0.001 (9 MET, 7 LGG).
Figure 4.
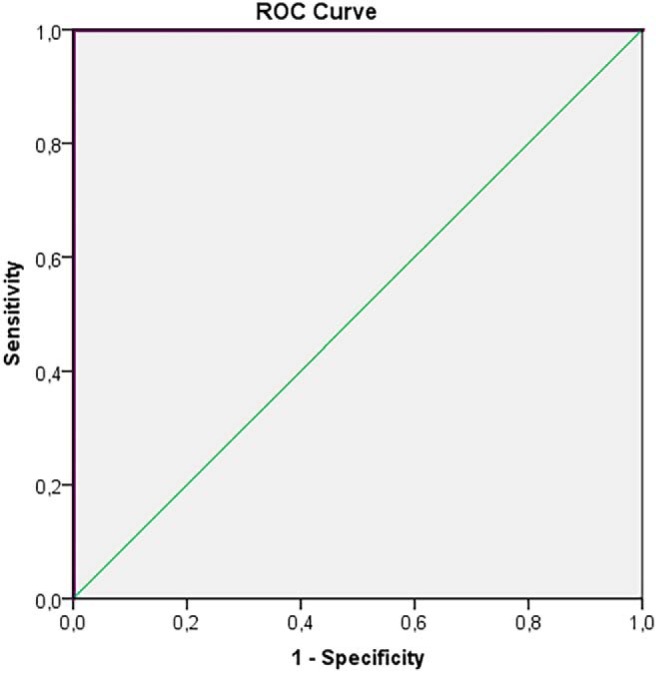
ROC-curve for HGG and low-grade glioma (LGG) in tumor tissue (nCE, CE) for tLip/tCho, Ins/tCho. AUC = 1.00; 95% CI 1.00-1.00; P-value < 0.001.
Different metabolic marker concentration deviations can be attributed to specific mechanisms on a cellular and genetic level. NAA, derived from the neurotransmitter aspartate, reflects neuronal viability. Cr levels indicate the cellular energy state, while Cho, being a component in cell membranes through phospholipids, becomes dysregulated and accumulated by oncogenic signaling, that is, malignant transformation. Cho is also a source of the methyl group and can, through epigenetics, methylate the O6-methylguanine-DNA methyltransferase in glioblastoma. Furthermore, the increased Cho concentrations often seen in grades 2 and 3 glioma are correlated with cellular density (21) and Ki-76 proliferation index (22); similarly glioblastoma exhibit high levels of Cho (23). Increase in Cho/Cr is associated with higher grades of glioma (24).
Previous reports show that diagnostic differentiation capacity between LGG and HGG by utilization of tCho/tCr can reach 100% sensitivity and specificity, but this high level of discrimination is evident in only 1 of 4 previous studies (12, 25–28). The tCho/tCr ratio with single voxel 1H-MRS is reported as significantly higher in HGG than in LGG (12), although this is not the case in the present cohort for LGG and HGG, as tCho/tCr is significantly higher in MET than in HGG. Furthermore, our study shows 100% sensitivity and specificity by stratification of regions and utilization of several metabolite ratios: Ins/tCho and tLip/tCho for LGG and HGG, as well as MET and LGG. Equal sensitivity and specificity with higher AUC in our study with more plausible 95% CI than in 26 of 30 studies included in review and meta-analysis on LGG and HGG by Wang et al. (12) arguably set out the combined approach with logistic regression from the singular approach with only 1 metabolite or ratio. Only 4 studies obtained similar results to ours in terms of sensitivity and specificity albeit with lower 95% CI than our study, probably owing to the chosen method of forest plots, with 1 study (26) reporting 95% CI between 0.16 and 1.00 (12, 25–28). A more complicated task for differentiation is MET from LGG and HGG (11). Our results of 100%/81.8% sensitivity/specificity and AUC = 0.935; 95% CI 0.825–1.000 for tCr/tCho, tCho/tCr, and NAA/tCho MET vs. HGG are in concordance with previous evidence, and although NAA in the mentioned studies is normalized to Cr, it clearly shows that the combined approach yields higher sensitivity and specificity and AUC (AUC = 0.84 and 80% sensitivity and 80% specificity) (11, 29).
Where conventional contrast-enhanced MRI may not reveal the true extent of tumor, MRS may provide insight if a tissue is metabolically deranged and hence raise suspicion of infiltration by neoplastic cells. An increase in tCho, tCr, Ins concentration may be because of a range of causes; gliosis, decreased intra- or extracellular H2O content with subsequent density rise, elevation in osmolarity in the brain, deviations in volume of the cortex, and lastly, increased synthesis or release of metabolites (19). Another aspect is that of clonal heterogeneity that may be reflected by variations in levels of tCho and tCr in tumors and is as such an established characteristic of HGG.
The spreading of tumor within brain parenchyma has been observed to occur by the formation of clusters of minor tumor cell colonies spreading away in all directions from the primary tumor (30). With regards to malignant spread of gliomas, brain parenchyma with low tCho/tCr ratios have been observed to manifest reduced cluster size and reduced frequencies of tumor clusters in brain parenchyma, when compared to tissue with high tCho/tCr ratios that were observed to have tumor clusters with more rapid growth in size and with higher frequencies of these clusters (30). Furthermore, different regions of gliomas seem to be prone to different frequencies of tumor-clusters being formed. Additionally, there is evidence that MRS-guided resection yields a higher survival probably because of either better margins to tumor or eradication of more of the stem cell-like cancer cells in tissue around the primary lesion (30).
In our study, the mean tCho/tCr ratio across the entire MRS grid in the affected hemisphere shows predictive capacity. A tCho/tCr ratio of ≥1.29 is associated with a higher mortality risk across the three groups. While a tCho/tCr ratio <1.29 has better survival prognosis. These findings may suggest a more aggressive spread of disease with cancer stem cell–like cells being formed more rapidly and in higher frequency across the affected hemisphere.
Limitations
The present study is performed in a fairly small cohort. However the number of patients is more than sufficient for the selected statistical tests. Bootstrapping may also be chosen for evaluation of the specific statistical model, as it is a better estimation tool of overfitting. The predictive probability value is clustered to 0.311–0.50 (Table 2), suggesting an underfitted model, which in differential diagnostic efforts is more plausible than having closer to 1.0, that is, perfect fit, as underfitting may yield better separation of LGG, HGG, and MET from each other.
Although the model is underfitted and may produce less optimistic results than an overfitted model, findings need to be validated in a larger cohort before external validation is performed. Partial volume effect owing to tumors expanding in a nonuniform manner and occasionally toward bony areas may also have affected spectral quality owing to suboptimal grid placement.
Conclusion
This study shows that 1H-MRS can be used to successfully differentiate between brain lesions such as LGG, HGG, and MET and it adds to a growing body of evidence deploying machine learning algorithms for tumor diagnostics. Stratification of tissue-type and combination of several metabolite ratios in a logistic regression model is evidently superior to a single metabolite ratio and nonstratification of tissue type. The best single marker for differentiation of tumor type was tLip/tCho, which showed a 100% sensitivity and specificity. tLip/tCho and Ins/tCho combined showed 100% sensitivity and specificity for both HGG/LGG (P < .001) and LGG/MET (P < .001) measured in tumor tissue.
Acknowledgments
The study was supported by F2014/354 (ALF (Swedish Regional Research Funds), CAN 2016/365 (The Swedish Cancer Society) and P01CA085878 (National Institute of Health, USA).
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare
Footnotes
- 1H-MRS
- Proton magnetic resonance spectroscopy
- MET
- metastases
- LGG
- low-grade glioma
- HGG
- high-grade glioma
- tCr
- total creatine
- MRSI
- MRS imaging
- tCho
- total choline
- OS
- overall survival
- Ins
- myo-inositol
- CE
- contrast enhancement
- MRI
- magnetic resonance imaging
- MR
- magnetic resonance
- MRS
- magnetic resonance spectroscopy
- S/N
- signal-to-noise
- TR
- repetition time
- TE
- echo time
- iNAWM
- ipsilateral normal-appearing white matter
- cNAWM
- contralateral NAWM
- VOI
- volume of interest
- nCE
- noncontrast-enhanced tumor
- ROC
- receiver operating characteristic
- AUC
- area under the curve
References
- 1. Schwartz KM, Erickson BJ, Lucchinetti C. Pattern of T2 hypointensity associated with ring-enhancing brain lesions can help to differentiate pathology. Neuroradiology. 2006;48:143–149. [DOI] [PubMed] [Google Scholar]
- 2. Diwanji TP, Engelman A, Snider JW, Mohindra P. Epidemiology, diagnosis, and optimal management of glioma in adolescents and young adults. Adolesc Health Med Ther. 2017;8:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blomstergren A, Rydelius A, Abul-Kasim K, Lätt J, Sundgren PC, Bengzon J. Evaluation of reproducibility in MRI quantitative volumetric assessment and its role in the prediction of overall survival and progression-free survival in glioblastoma. Acta Radiol. 2018. 10.1177/0284185118786060 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 4. Stummer W, Reulen H-J, Meinel T, Pichlmeier U, Schumacher W, Tonn J-C, Rohde V, Oppel F, Turowski B, Woiciechowsky C, Franz K, Pietsch T, ALA-Glioma Study Group. Extent of resection and survival in glioblastoma multiforme: Identification of and adjustment for bias. Neurosurgery. 2008;62:564–576. [DOI] [PubMed] [Google Scholar]
- 5. Chaichana KL, McGirt MJ, Laterra J, Olivi A, Quiñones-Hinojosa A. Recurrence and malignant degeneration after resection of adult hemispheric low-grade gliomas. J Neurosurg. 2010;112:10–17. [DOI] [PubMed] [Google Scholar]
- 6. Scott JN, Brasher PMA, Sevick RJ, Rewcastle NB, Forsyth PA. How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology. 2002;59:947–949. [DOI] [PubMed] [Google Scholar]
- 7. Wang Q, Zhang J, Xu W, Chen X, Zhang J, Xu B. Role of magnetic resonance spectroscopy to differentiate high-grade gliomas from metastases. Tumor Biol. 2017;39:101042831771003. [DOI] [PubMed] [Google Scholar]
- 8. Durmo F, Lätt J, Rydelius A, Engelholm S, Kinhult S, Askaner K, Englund E, Bengzon J, Nilsson M, Björkman-Burtscher IM, Chenevert T, Knutsson L, Sundgren PC. Brain tumor characterization using multibiometric evaluation of MRI. Tomography. 2018;4:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44:139–150. [DOI] [PubMed] [Google Scholar]
- 10. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. [DOI] [PubMed] [Google Scholar]
- 11. Hollingworth W, Medina LS, Lenkinski RE, Shibata DK, Bernal B, Zurakowski D, Comstock B, Jarvik JG. A systematic literature review of magnetic resonance spectroscopy for the characterization of brain tumors. AJNR Am J Neuroradiol. 2006;27:1404–1411. [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q, Zhang H, Zhang J, Wu C, Zhu W, Li F, Chen X, Xu B. The diagnostic performance of magnetic resonance spectroscopy in differentiating high-from low-grade gliomas: A systematic review and meta-analysis. Eur Radiol. 2016;26:2670–2684. [DOI] [PubMed] [Google Scholar]
- 13. Usinskiene J, Ulyte A, Bjørnerud A, Venius J, Katsaros VK, Rynkeviciene R, Letautiene S, Norkus D, Suziedelis K, Rocka S, Usinskas A, Aleknavicius E. Optimal differentiation of high- and low-grade glioma and metastasis: a meta-analysis of perfusion, diffusion, and spectroscopy metrics. Neuroradiology. 2016;58:339–350. [DOI] [PubMed] [Google Scholar]
- 14. Burtscher IM, Ståhlberg F, Holtås S. Proton (1H) MR spectroscopy for routine diagnostic evaluation of brain lesions. Acta Radiol. 1997;38:953–960. [DOI] [PubMed] [Google Scholar]
- 15. Provencher SW. Automatic quantitation of localized in vivo1H spectra with LCModel. NMR Biomed. 2001;14:260–264. [DOI] [PubMed] [Google Scholar]
- 16. Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. [DOI] [PubMed] [Google Scholar]
- 17. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat Scand J Stat. 1979;623566:65–70. [Google Scholar]
- 18. Halligan S, Altman DG, Mallett S. Disadvantages of using the area under the receiver operating characteristic curve to assess imaging tests: a discussion and proposal for an alternative approach. Eur Radiol. 2015;25:932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang L, Ernst T, Poland RE, Jenden DJ. In vivo proton magnetic resonance spectroscopy of the normal aging human brain. Life Sci. 1996;59:2049–2056. [DOI] [PubMed] [Google Scholar]
- 20. Caivano R, Lotumolo A, Rabasco P, Zandolino A, D'antuono F, Villonio A, Lancellotti MI, Macarini L, Cammarota A. 3 Tesla magnetic resonance spectroscopy: cerebral gliomas vs. metastatic brain tumors. Our experience and review of the literature. Int J Neurosci. 2013;123:537–543. [DOI] [PubMed] [Google Scholar]
- 21. Gupta RK, Cloughesy TF, Sinha U, Garakian J, Lazareff J, Rubino G, Becker DP, Vinters HV, Alger JR. Relationships between choline magnetic resonance spectroscopy, apparent diffusion coefficient and quantitative histopathology in human glioma. J Neurooncol. 2000;50:215–226. [DOI] [PubMed] [Google Scholar]
- 22. McKnight TR, Smith KJ, Chu PW, Chiu KS, Cloyd CP, Chang SM, Phillips JJ, Berger MS. Choline metabolism, proliferation, and angiogenesis in nonenhancing grades 2 and 3 astrocytoma. J Magn Reson Imaging. 2011;33:808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He T, Qiu T, Wang X, Gui H, Wang X, Hu Q, Xia H, Qi G, Wu J, Ma H. Multivoxel magnetic resonance spectroscopy identifies enriched foci of cancer stem-like cells in high-grade gliomas. Onco Targets Ther. 2017;10:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeun SS, Kim MC, Kim BS, Lee JM, Chung ST, OH CH, Lee SY, Choe BY. Assessment of malignancy in gliomas by 3T 1H MR spectroscopy. Clin Imaging. 2005;29:10–15. [DOI] [PubMed] [Google Scholar]
- 26. Chen CY, Lirng JF, Chan WP, Fang CL. Proton magnetic resonance spectroscopy-guided biopsy for cerebral glial tumors. J Formos Med Assoc. 2004;103:448–458. [PubMed] [Google Scholar]
- 27. Shokry A. MRS of brain tumors: Diagrammatic representations and diagnostic approach. Egypt J Radiol Nucl Med. 2012;43:603–612. [Google Scholar]
- 28. Stadlbauer A, Gruber S, Nimsky C, Fahlbusch R, Hammen T, Buslei R, Tomandl B, Moser E, Ganslandt O. Preoperative grading of gliomas by using metabolite quantification with high-spatial-resolution proton MR spectroscopic imaging. Radiology. 2006;238:958–969. [DOI] [PubMed] [Google Scholar]
- 29. Opstad KS, Wright AJ, Bell BA, Griffiths JR, Howe FA. Correlations between in vivo 1H MRS and ex vivo 1H HRMAS metabolite measurements in adult human gliomas. J Magn Reson Imaging. 2010;31:289–297. [DOI] [PubMed] [Google Scholar]
- 30. Li Z, Wang H, Eyler CE, Hjelmeland AB, Rich JN. Turning cancer stem cells inside out: an exploration of glioma stem cell signaling pathways. J Biol Chem. 2009;284:16705–16709. [DOI] [PMC free article] [PubMed] [Google Scholar]