Abstract
Even as medical data sets become more publicly accessible, most are restricted to specific medical conditions. Thus, data collection for machine learning approaches remains challenging, and synthetic data augmentation, such as generative adversarial networks (GAN), may overcome this hurdle. In the present quality control study, deep convolutional GAN (DCGAN)–based human brain magnetic resonance (MR) images were validated by blinded radiologists. In total, 96 T1-weighted brain images from 30 healthy individuals and 33 patients with cerebrovascular accident were included. A training data set was generated from the T1-weighted images and DCGAN was applied to generate additional artificial brain images. The likelihood that images were DCGAN-created versus acquired was evaluated by 5 radiologists (2 neuroradiologists [NRs], vs 3 non-neuroradiologists [NNRs]) in a binary fashion to identify real vs created images. Images were selected randomly from the data set (variation of created images, 40%–60%). None of the investigated images was rated as unknown. Of the created images, the NRs rated 45% and 71% as real magnetic resonance imaging images (NNRs, 24%, 40%, and 44%). In contradistinction, 44% and 70% of the real images were rated as generated images by NRs (NNRs, 10%, 17%, and 27%). The accuracy for the NRs was 0.55 and 0.30 (NNRs, 0.83, 0.72, and 0.64). DCGAN-created brain MR images are similar enough to acquired MR images so as to be indistinguishable in some cases. Such an artificial intelligence algorithm may contribute to synthetic data augmentation for “data-hungry” technologies, such as supervised machine learning approaches, in various clinical applications.
Keywords: AI, artificial intelligence, magnetic resonance imaging, MRI, DCGAN, GAN, stroke, machine learning
Introduction
In recent years, the use of artificial intelligence (AI) has attracted interest for medical imaging tasks. However, small data sets are major obstacles, in particular for supervised machine learning and for rare conditions for which only a small number of cases may exist even in large databases (1–3). Even as medical data sets become more publicly accessible, most of those data sets are restricted to specific medical conditions, and data collection for machine learning approaches remains challenging (2).
To overcome this hurdle, some efforts have turned to the augmentation of existing data. In this regard, several methods for data augmentation have been suggested, but minor alterations such as overfitting in learning processes or geometric transformations have not met the urgent need to provide data sets on a larger basis (4, 5). However, considerable progress has been made by the introduction of synthetic data augmentation to enlarge training sets. By generating synthetic data, novel created images can be added to existing data sets. Such an approach may provide a larger number of images to enhance the variety within a data set and, ultimately, to improve machine learning algorithms (2).
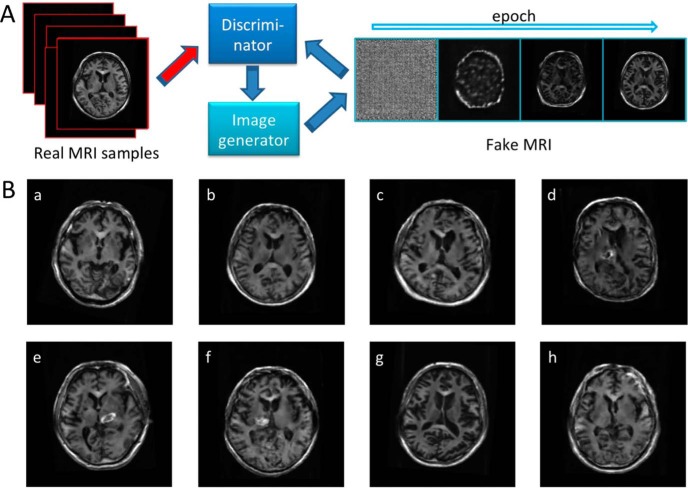
Generative adversarial networks (GANs) may contribute toward meeting the need for synthetic data augmentation. In principle, GANs are based on an adversarial process where one network is creating artificial images, while the other network continuously learns to differentiate between real and generated images (Figure 1A) (6, 7). Several studies have applied the concept of GAN to medical imaging, for example, by producing new retinal images from a data set of pairs of retinal vessel trees (8). Apart from ophthalmologic applications, GANs have also found applicability in the field of molecular oncology imaging to test the detection rate of malignant liver lesions by using computed tomography and generated positron emission tomography (PET) images (9). However, although current applications within radiology aim to assist in diagnosis, the quality of GAN-generated data has not yet been validated by human perception in this context. Thus, in the present feasibility study, we aimed to test the capability of a GAN to create brain magnetic resonance (MR) images and to perform quality assessment of GAN-generated artificial images by a visual assessment conducted by blinded radiologists.
Figure 1.
Work flow chart (A): generative adversarial networks (GANs) are based on an adversarial process where one network is creating artificial images, while the other network continuously learns to differentiate between real and generated images. Interactive quiz (B): mixed data set of real and created brain magnetic resonance (MR) images as provided to both human observers. Artificial magnetic resonance (MR) images have been created using deep convolutional generative adversarial networks (DCGAN). Created images are b, c, d, f, and h (B).
Methodology
Patient Population and Imaging Acquisition
This retrospective study was approved by the local medical ethics committee (National Cardiovascular and Cerebral Research Center, Suita, Japan) and conducted in strict accordance with the World Medical Association Declaration of Helsinki and Ethical Principles for Medical Research Involving Human Subjects. All subjects signed written informed consent. In total, 96 T1-weighted (T1W) brain images of 30 healthy volunteers and 33 patients with a history of cerebrovascular accident (women, 26; mean age, 69 ± 10) were enrolled, while the latter group underwent MR scans both at acute and chronic phases (9 ± 6 and 101 ± 13 days after disease onset, respectively). MR images were acquired by 3-T whole-body scanner (Signa Excite HD V12M4; GE Healthcare, Milwaukee, WI) with an 8-channel phased-array brain coil and a spoiled gradient-recalled sequence (repetition time = 8.6 milliseconds, echo time = 1.8 milliseconds, flip angle = 8°) in the sagittal planes, reconstructed matrix size, 256 × 256 × 124; voxel size, 0.94 × 0.94 × 1.5 mm3.
Training Data Set
Training data of 2-dimensional (2D) images were generated from the T1W images using the following procedures: the original T1W image was reoriented perpendicular to the anterior–posterior commissure line and the transaxial section at the basal ganglia level was located manually at the center of the image domain. Thereafter, the image was rotated in the right / left direction with angles from −10 to +10°, up / right direction with angles from −10 to +10° and cephalad / caudal direction with angles from −5 to +5° with step size of 1°. In the rotated image, 5 transaxial sections were sampled as 2D images at distances of −4, −2, 0, 2, 4 mm from the central transaxial section. Furthermore, the 2D images were flipped in the right/left direction. The total number of 2D images was 2 328 480 (48 510 images / original T1W image).
Generation of Artificial Brain Images Using Deep Convolutional GAN
We applied deep convolutional GAN (DCGAN) to generate artificial brain images on the basis of a previously described procedure with minor modifications (7). The DCGAN generator consisted of a fully connected layer projecting input of a 100-dimensional uniform distribution to the following layers of 4 fractionally strided convolutions with filter sizes of 256, 128, 64, and 32 and a kernel size of 5 × 5. The rectified linear unit activation functions were used except for the output layer, which used the tanh function. Batch normalization was performed in each layer. As discriminators, leaky rectified linear unit functions were used in all layers with a slope of leak of 0.2. Batch normalization was performed except for the output layer. Training and testing were implemented using NVIDIA (R) CUDA 9.0, Google (R) TensorFlow 1.7.0, and Python 3.6, and performed with 35 epochs on a personal computer with CPU, Intel (R) Core i7-2600 CPU 3.4 GHz; memory, 8 Gbytes; GPU, NVIDIA (R) GeForce GTX 1080; OS, CentOS 7.4.
Human Visual Assessment
The likelihood of images having been created by the DCGAN was evaluated visually by 5 radiologists: 2 neuroradiologists (NRs, board-certified NRs with >15 years of experience in reading MR scans) and 3 non-NRs (NNRs, including 2 body radiologists (<10 years of experience) and 1 resident (<4 years of experience in reading MR scans). A visual evaluation of the images (single 2D axial T1W images) was performed independently, and the readers were blinded to the diagnosis and any further clinical information (other than knowing that the patients had undergone imaging because of a history of cerebrovascular accident and that healthy volunteers had been included). Before the investigation, the readers gained familiarity with the software tool by reading a training set of 10 cases. In total, 50 real and created images were randomly selected from the data set. The number of created images in the provided data set randomly varied from 40% to 60%, and thus, the readers did not necessarily read the exact same MR images. To evaluate the entire data set, 1 session per reader was performed and the authors had 10 seconds to judge each image. The conditions did not change for any of the readers throughout the assessment. After the images had been displayed, binary reporting (real vs. created image) was performed visually by both interpreters.
Statistical Analysis
Most of the observations described in this feasibility study are of a descriptive nature. Quantitative values were expressed as percentages, as appropriate. The true-positive (TP), false-negative (FN), false-positive (FP), and true-negative (TN) rates were recorded, and the accuracy was computed as follows: (TP + TN) / (TP + FN + FP + TN).
Results
None of the investigated images was selected by an interpreter as unknown. Of the created images, the NRs rated 45% and 71% as real magnetic resonance imaging (MRI) images (NNRs, 24%, 40%, and 44%). In contradistinction, 44% and 70% of the real images were rated as generated images by the NRs (NNRs, 10%, 17%, and 27%). Figure 1B displays selected artificial and acquired brain MR images, which were included in the evaluations by both interpreters. This figure is provided as an interactive quiz, that is, figure panels identifying artificial MR images are given in the figure legend, which enables the reader to rate the images based on her or his own impression before viewing the answer key.
The accuracy for all observers was as follows: NR1, 0.55; NR2, 0.30; NNR1, 0.83; NNR2, 0.72, and NNR3, 0.64. Table 1 gives an overview about all obtained results (including TP, FN, FP, and TN).
Table 1.
Overview of Obtained Results from All 5 Readers (2 NRs and 3 NNRs)
| Reader | TP | FN | FP | TN | Accuracy |
|---|---|---|---|---|---|
| NR1 | 56 | 44 | 45 | 55 | 0.55 |
| NR2 | 30 | 70 | 71 | 29 | 0.30 |
| NNR1 | 90 | 10 | 24 | 76 | 0.83 |
| NNR2 | 83 | 17 | 40 | 60 | 0.72 |
| NNR3 | 73 | 27 | 44 | 56 | 0.64 |
True-positive (TP), false-negative (FN), false-positive (FP), and true-negative (TN) rates for each reader are displayed.
Discussion
In the present feasibility study, we generated artificial brain MR images using a neural network–based algorithm (DCGAN) and further validated the created images in a visual assessment by readers with different levels of experience.
As technological advances or novel concepts are introduced in the field of radiology (10–12), reader studies (eg, interobserver agreement studies or comparison with human visual assessment as a gold standard) are indispensable before testing a potential clinical benefit in a real-world scenario (13, 14). Hence, if DCGAN for the improvement of machine learning should be applied more broadly (eg, to assist the radiologist in diagnosis by identifying target lesions) (9), quality control studies to validate the integrity of DCGAN-derived images should be performed. Of note, after creating brain MR images using DCGAN, the herein presented results show that the created artificial images may even convince an experienced NR that he or she is looking at real brain MR images. As shown in the created images, the GAN technique reproduced characteristic features of brain MR images, such as distribution of gray and white matter regions and the configuration of lateral ventricles, which appeared to mimic real human anatomy (Figure 1B (b)). In addition, high-intensity regions around the basal ganglia such as hemorrhage or edema were delineated in the training images including patients status post cerebrovascular accidents (Figure 1B (f)). Although the structure of the neural networks used in the present study was based on a previously reported technique with minor modifications for the matrix size of the output images (7), NRs rated 45% and 71% of the created images as real. Some of the created images were more obviously artificial owing to reasons that included, but were not limited to, distortion (Figure 1B (d)), inappropriate small spaces of the ventricles (Figure 1B (c)), or blurring of boundaries between gray and white matter regions (Figure 1B (h)). Nonetheless, as human assessment served as gold standard in the present study, the GAN technique may be applicable to generate artificial MR images to imitate real brain images. As machine-based learning is seen as a “data-hungry” technology, novel advances are needed to provide large data sets that would otherwise be difficult to obtain through traditional collation in a database (15). In this regard, the present quality control study shows that DCGAN can help meet the need to generate large data sets of MR images in a high-quality manner, such that even experienced NRs may be misled. Interestingly, the NRs did not perform better than the NNRs (range of accuracy, NRs, 0.30–0.55 vs. NNRs, 0.64–0.83). This may be caused by the realness of the created images, by the small sample size, or the low number of included readers. Thus, future studies addressing this issue, preferably by indicating reasons for the radiologist's decisions, are definitely warranted.
Nonetheless, GAN-derived brain MR images can be made readily applicable to be implemented to augment existing data, which in turn may improve machine learning algorithms. Current medical applications include simple differentiation of benign vs. malignant lesions (9), but DCGAN could also potentially be applied to more complex evaluations of multiple target lesions. In recent years, several reporting and data systems (RADS) have been described for imaging of the breast (BI-RADS), thyroid (TI-RADS), prostate (PI-RADS, PSMA-RADS), lung (LUNG-RADS), neuroendocrine tumors (SSTR-RADS), or liver (LI-RADS) (10, 11, 16–21). Those in-depth RADS-based evaluations can communicate important information to treating clinicians, provide a framework for understanding indeterminate findings, and allow for standardized data to be collected from large clinical trials. However, many types of lesions that might be classifiable by such a system are rare and would not be encountered to the extent that an effective machine learning algorithm could be trained from real images (10–12, 14, 21), indicating the need for effective data augmentation to automate RADS reporting. Other applications include augmentation of brain MRI for extremely rare pediatric genetic disorders, for example, the Hutchinson–Gilford progeria syndrome (prevalence, 1 in 18 million) or the Aicardi syndrome (1 in 557,000), which may provide both radiologists and machine learning algorithms with brain MR images for training purposes (22–25).
This feasibility study has several limitations: future studies generating 3-dimensional MR brain images, which include a larger data set with more interpreting radiologists with different levels of experience, are needed. Additionally, obvious limitations in the created images (eg, distortion) have to be addressed. Moreover, the feasibility of this technique should be tested in different imaging modalities, such as PET, or even hybrid techniques such as PET/MRI. In addition, the reader's confidence, preferably on a 5-point Likert scale, and reasons for false negatives and false positives should be assessed in future studies (26). Moreover, the number of created images in the provided data set randomly varied from 40% to 60%, and thus, the readers may have read cases with slightly different levels of difficulty. Future studies may also always provide the same data set for every reader.
In the present quality control study, DCGAN-created brain MR images were able to convince NRs that they were viewing true images instead of artificial brain MR images. Thus, DCGAN may be nearing readiness to be implemented for synthetic data augmentation for data-hungry technologies, such as supervised machine learning, which, in turn, can pave the way to incorporate AI in even highly complex medical imaging cases.
Acknowledgments
Equal Contribution: “K.K. and R.A.W” contributed equally to this work.
This work was supported by the Competence Network of Heart Failure funded by the Integrated Research and Treatment Center (IFB) of the Federal Ministry of Education and Research (BMBF), and German Research Council (DFG grant HI 1789/3-3). This project has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 701983.
Conflict of Interest: The authors have no conflict of interest to declare.
Disclosure: Nothing to disclose.
Footnotes
- GAN
- Generative adversarial networks
- DCGAN
- deep convolutional GAN
- MR
- magnetic resonance
- NRs
- neuroradiologists
- NNRs
- non-neuroradiologists
- PET
- positron emission tomography
- T1W
- T1-weighted
- 2D
- 2-dimensional
- MRI
- magnetic resonance imaging
References
- 1. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 2. Greenspan H, van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35:1153–1159. [Google Scholar]
- 3. Ullrich NJ, Gordon LB. Hutchinson-Gilford progeria syndrome. Handb Clin Neurol. 2015;132:249–264. [DOI] [PubMed] [Google Scholar]
- 4. Perez L, Wang J. The effectiveness of data augmentation in image classification using deep learning. ArXiv e-prints [Internet]. 2017. December 01, 2017. Available from: https://ui.adsabs.harvard.edu/#abs/2017arXiv171204621P.
- 5. Simard PY, Steinkraus D, Platt JC, editors. Best Practices for Convolutional Neural Networks Applied to Visual Document Analysis. ICDAR; 2003. [Google Scholar]
- 6. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. Adv Neural Inf Process Syst. 2014. [Google Scholar]
- 7. Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:151106434 2015. [Google Scholar]
- 8. Costa P, Galdran A, Meyer MI, Niemeijer M, Abramoff M, Mendonca AM, Campilho A. End-to-end adversarial retinal image synthesis. IEEE Trans Med Imaging. 2018;37:781–791. [DOI] [PubMed] [Google Scholar]
- 9. Ben-Cohen A, Klang E, Raskin SP, Amitai MM, Greenspan H. Virtual PET images from CT data using deep convolutional networks: initial results. In: Tsaftaris S, Gooya A, Frangi A, Prince J, eds. Simulation and Synthesis in Medical Imaging. New York: Springer, Cham; 2017. pp. 49–57. [Google Scholar]
- 10. Rowe SP, Pienta KJ, Pomper MG, Gorin MA. Proposal for a structured reporting system for prostate-specific membrane antigen-targeted PET imaging: PSMA-RADS Version 1.0. J Nucl Med. 2018;59:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Werner RA, Solnes LB, Javadi MS, Weich A, Gorin MA, Pienta KJ, Higuchi T, Buck AK, Pomper MG, Rowe S, Lapa C. SSTR-RADS Version 1.0 as a reporting system for SSTR PET imaging and selection of potential PRRT candidates: a proposed standardization framework. J Nucl Med. 2018;59:1085–1091. [DOI] [PubMed] [Google Scholar]
- 12. Rowe SP, Pienta KJ, Pomper MG, Gorin MA. PSMA-RADS Version 1.0: a step towards standardizing the interpretation and reporting of PSMA-targeted PET imaging studies. Eur Urol. 2018;73:485–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gennaro G. The “perfect” reader study. Eur J Radiol. 2018;103:139–146. [DOI] [PubMed] [Google Scholar]
- 14. Shenderov E, Gorin MA, Kim S, Johnson PT, Allaf ME, Partin AW, Pomper MG, Antonarakis ES, Pienta KJ, Rowe SP. Diagnosing small bowel carcinoid tumor in a patient with oligometastatic prostate cancer imaged with PSMA-Targeted [(18)F]DCFPyL PET/CT: value of the PSMA-RADS-3D designation. Urol Case Rep. 2018;17:22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. [DOI] [PubMed] [Google Scholar]
- 16. Orel SG, Kay N, Reynolds C, Sullivan DC. BI-RADS categorization as a predictor of malignancy. Radiology. 1999;211:845–850. [DOI] [PubMed] [Google Scholar]
- 17. Tessler FN, Middleton WD, Grant EG. Thyroid imaging reporting and data system (TI-RADS): A user's guide. Radiology. 2018;287:29–36. [DOI] [PubMed] [Google Scholar]
- 18. Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany CM, Thoeny HC, Verma S. PI-RADS prostate imaging – reporting and data system: 2015, Version 2. Eur Urol. 2016;69:16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKee BJ, Regis SM, McKee AB, Flacke S, Wald C. Performance of ACR lung-RADS in a clinical CT lung screening program. J Am Coll Radiol. 2016;13(2 Suppl):R25–R29. [DOI] [PubMed] [Google Scholar]
- 20. Purysko AS, Remer EM, Coppa CP, Leao Filho HM, Thupili CR, Veniero JC. LI-RADS: a case-based review of the new categorization of liver findings in patients with end-stage liver disease. Radiographics. 2012;32:1977–1995. [DOI] [PubMed] [Google Scholar]
- 21. Werner RA, Bundschuh RA, Bundschuh L, Javadi MS, Higuchi T, Weich A, Sheikhbahaei S, Pienta KJ, Buck AK, Pomper MG, Gorin MA, Lapa C, Rowe SP. Molecular imaging reporting and data systems (MI-RADS): a generalizable framework for targeted radiotracers with theranostic implications. Ann Nucl Med. 2018;32:512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ullrich NJ, Kieran MW, Miller DT, Gordon LB, Cho YJ, Silvera VM, Giobbie-Hurder A, Neuberg D, Kleinman ME. Neurologic features of Hutchinson-Gilford progeria syndrome after lonafarnib treatment. Neurology. 2013;81:427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ullrich NJ, Gordon LB. Hutchinson–Gilford progeria syndrome. In: Islam MP and Roach ES, eds. Handbook of Clinical Neurology, Neurocutaneous Syndromes, 3rd Series Amsterdam, Netherlands; 2015;132: pp. 249–264. [DOI] [PubMed] [Google Scholar]
- 24. Hopkins B, Sutton VR, Lewis RA, Van den Veyver I, Clark G. Neuroimaging aspects of Aicardi syndrome. Am J Med Genet A. 2008;146A:2871–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kroner BL, Preiss LR, Ardini MA, Gaillard WD. New incidence, prevalence, and survival of Aicardi syndrome from 408 cases. J Child Neurol. 2008;23:531–535. [DOI] [PubMed] [Google Scholar]
- 26. Keeble C, Baxter PD, Gislason-Lee AJ, Treadgold LA, Davies AG. Methods for the analysis of ordinal response data in medical image quality assessment. Br J Radiol. 2016;89:20160094. [DOI] [PMC free article] [PubMed] [Google Scholar]