Abstract
Background: Although methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene promoter predicts response to temozolomide in patients with glioblastoma, no consensus exists as to which assay is best for its detection. Materials and Methods: Methylation of MGMT promoter was examined by methylation-specific polymerase chain reaction (MSP), quantitative real-time MSP, methylation-sensitive high-resolution melting analysis, and two commercial pyrosequencing (PSQ) kits. Survival was compared among 48 patients with glioblastoma according to assay results. Results: Only PSQ and MSP significantly separated patients who benefited from temozolomide, with PSQ being the superior method. For PSQ analysis, the cut-off value that best correlated with prognostic outcome was 7% methylation of MGMT. Median survival in patients with MGMT promoter methylation above this cut-off value was 7.8 months longer compared to those with less than 7% methylation. Two-year overall survival for the two groups was 42% and 7.4%, respectively. Conclusion: PSQ is the method of choice for MGMT promoter methylation analysis in routine clinical practice.
Keywords: MGMT gene promoter, methylation, pyrosequencing, methylation-specific PCR, glioblastoma, overall survival
Glioblastoma multiforme (GBM) is one of the most common and malignant tumors of the central nervous system. Median overall survival (OS) for fit patients with GBM receiving standard treatment ranges between 12 and 18 months, with a 2-year survival rate of 27% (1,2). Standard management of patients with GBM involves tumor-resective surgery and radiotherapy with concomitant temozolomide, followed by six cycles of adjuvant temozolomide (2,3). Alkylating agents, such as temozolomide, exert their cytotoxic effect through alkylation of the O6 position of guanine (4). Such alkyl adducts are rapidly removed by the DNA-repair enzyme O6-methylguanine-DNA methyltransferase (MGMT), thereby counteracting the cytotoxic effect of temozolomide (5). The expression level of the MGMT enzyme, is therefore, inversely related to the outcome of patients with GBMs treated with temozolomide. Expression of the MGMT protein is highly regulated by hypermethylation of the MGMT promoter, whereby hypermethylation provokes transcriptional silencing (6). Thus, MGMT promoter methylation has become a clinically relevant prognostic and predictive marker in patients with GBM treated with alkylating agents (1,7).
Although different diagnostic methods for determining the extent of MGMT promoter methylation are available, all assays measure the methylation status of cytosines only in a fraction of the 5’→cytosine→phosphate→guanine→3’ (CpG) dinucleotides located in the MGMT promoter. The MGMT promoter contains 98 CpG sites. Methylation of two CpG regions, CpGs 25-50 [differentially methylated region 1 (DMR1)] and CpGs 73-90 (DMR2), has been demonstrated to correlate with transcriptional silencing (8,9). Although analysis of MGMT promoter methylation status is now standard practice in newly-diagnosed GBM, there is no consensus as to which molecular test is the optimal one.
The most popular techniques used to assess MGMT promoter methylation are based on methylation-specific polymerase chain reaction (MSP) and pyrosequencing (PSQ). MSP is a qualitative technique using methylation-specific primers to amplify separately fully methylated and unmethylated sequences of the MGMT promoter (10). GBM with detectable amplification of a methylated sequence is scored as methylation-positive. Variants of the MSP techniques are methylation-sensitive high-resolution melting analysis (MS-HRM) and quantitative MSP (qMSP). MS-HRM is a semi-quantitative PCR technique which, based on comparison of melting profiles of PCR products from tumor with melting profiles of a standard, measures the percentage of methylated amplicons (11). The percentage of methylated amplicons detected in an unmethylated control sample is defined as the cut-off value in order to separate unmethylated GBMs from methylated GBMs. qMSP is a quantitative assay that determines the number of copies of a methylated MGMT promoter element. The number of copies of methylated MGMT promoter is thereafter normalized to the number of copies of a housekeeping gene to give the relative level of methylated amplicons (12-15). qMSP only recognizes a fully methylated sequence. The relative level of methylated MGMT promoter detected in an unmethylated control sample is, therefore, used to define the cut-off value in order to separate unmethylated GBMs from methylated GBMs. Other MSP assays used are methylation-specific multiplex ligation-dependent probe amplification (MS-MPLA), where restriction of an unmethylated CpG in the recognition site results in amplification of only the methylated sequence (16), and MethyLight qMSP, a qMSP technique using a methylation-specific probe (17).
PSQ is a semi-quantitative technique which quantifies in real time each added nucleotide during sequencing to give the percentage of methylation at each CpG present in the sequence measured (18,19). Usually the average percentage of methylation for all CpGs measured in a PSQ assay is used to score samples as unmethylated or methylated. The optimal cut-off value for prognostication is the most critical issue in the use of PSQ. To date, reported cut-off values have ranged from 2.7-35% and the number of CpGs measured from four to 62 (20).
MGMT promoter methylation, regardless of whether measured by MSP, qMSP, PSQ, MS-HRM, MS-MLPA, or MethyLight qMSP, has been demonstrated to predict clinical outcome (1,13,21-33). PSQ has been claimed to be best technique to identify patients with GBM likely to benefit from therapy with temozolomide (21-24). For MS-HRM, MS-MLPA, and MethyLight qMSP, on the other hand, some studies showed a weak predictive value (21,24), whereas other reports concluded that these methods are equivalent to PSQ or MSP (or better than PSQ) when it comes to quantitation of methylation levels (24-26,31,32). In this report, we compared two commercial PSQ kits, MSP, MS-HRM and qMSP to determine which technique had the best predictive power for survival in patients with GBM.
Materials and Methods
Patients and samples. Tumor samples from 48 patients with primary GBM and four patients with meningioma who underwent surgery at the Department of Neurosurgery (Oslo University Hospital) between January 2005 and January 2009 were included in this study. The four meningioma samples served as negative controls for MGMT promoter methylation (28). All patients with GBM received radiochemotherapy with 25 of them also receiving 1-6 courses of adjuvant temozolomide. All GBM (N=48) and meningioma cases (N=4) used in this study were included in a previous publication comparing survival in methylated and unmethylated cases using one commercial PSQ kit (PyroMark Q96 CpG MGMT kit; cat. number 972032; Qiagen, Hilden, Germany) and qMSP (34). For each patient, the histological diagnosis was reviewed by an expert neuropathologist (34). Written, informed consent was obtained from living patients whereas permission to include deceased patients was obtained from the National Health Authorities. The study was approved by the Regional Ethics Committee (S-06046).
DNA isolation and bisulfite conversion. The protocols for DNA extraction and quality measurements were described previously (34). In that study, 58 GBM cases were included in the OS analysis (34). For 10 of the samples in our previous study, DNA or frozen tissue material was not available, leaving a total of 48 GBM cases to be analyzed in this study. Spare DNA was available from our previous study for 45 of the GBM cases and all four meningiomas, while for three of the GBM cases, new DNA had to be extracted from frozen samples. Extraction was performed using Maxwell® 16 Tissue DNA Purification Kit (Promega, Madison, WI, USA) followed by quality measurements as described elsewhere (34). Unmethylated cytosine residues were converted to uracil by bisulfite treatment of 500 ng DNA using EpiTect Bisulfite Kit (Qiagen) and QiaCube automated purification system (Qiagen) according to the manufacturer’s procedures. After conversion, the final concentration of DNA was 12.5 ng/μl.
Methylation-specific PCR. For MSP, melting curve analysis was used to detect PCR products in our samples (35). The forward and reverse primers targeting methylated and unmethylated exon 1 of the human MGMT gene are listed in Table I and correspond to those described by Esteller et al. (36). The PCR product amplified by the primers analyzed methylation of CpG numbers 76-80 and 84-87 (see Figure 1). The 20 μl reaction volume contained 1× Precision Melt Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 200 nM of each forward and reverse primer, and 25 ng of bisulfite-converted DNA. Three replicates of each sample were used to ensure statistical representativity. Real-time PCR followed by melting curve analysis was run on a CFX96 Touch™ Real-Time PCR Detection system (Bio-Rad Laboratories). The thermal cycling included an initial step at 95˚C for 2 min followed by 40 cycles of 50 s at 95˚C, 50 s at 59˚C, and 50 s at 72˚C. Following the last cycle, PCR products were incubated for 10 s at 95˚C before the melting curve was generated by heating from 65˚C to 95˚C in increments of 0.5˚C/5 s while continuously measuring the fluorescence. The melting curves were analyzed using Bio-Rad CFX Manager Software (Bio-Rad Laboratories). Melting peaks determined for methylated and unmethylated controls, respectively, were used to identify methylated and unmethylated PCR products in the samples (EpiTect PCR Control DNA Set, cat. number 59695; Qiagen). Samples having only methylated PCR products and samples having both methylated and unmethylated PCR products were both scored as methylation-positive.
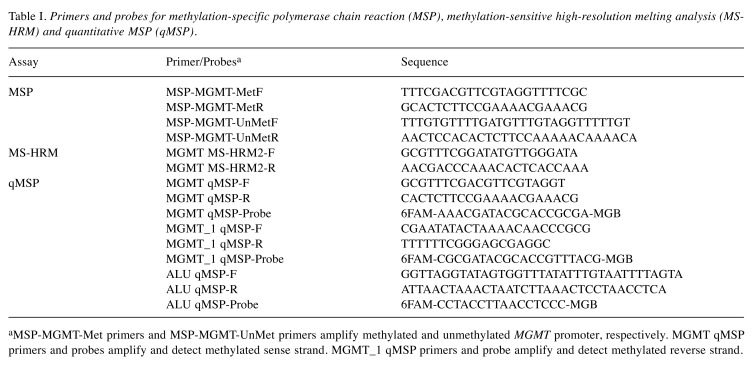
Table I. Primers and probes for methylation-specific polymerase chain reaction (MSP), methylation-sensitive high-resolution melting analysis (MSHRM) and quantitative MSP (qMSP).
aMSP-MGMT-Met primers and MSP-MGMT-UnMet primers amplify methylated and unmethylated MGMT promoter, respectively. MGMT qMSP primers and probes amplify and detect methylated sense strand. MGMT_1 qMSP primers and probe amplify and detect methylated reverse strand.
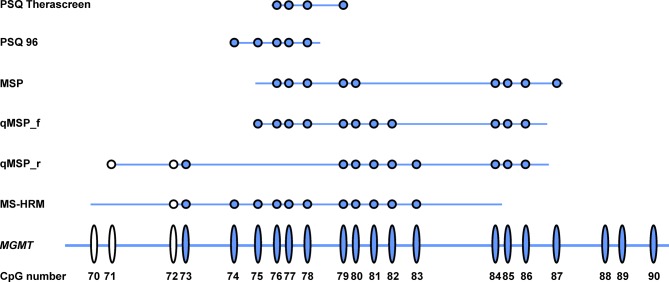
Figure 1. CpGs analyzed by the pyrosequencing PSQ Therascreen, PSQ 96, methylation-specific polymerase chain reaction (MSP), quantitative MSP (qMSP) and methylation-sensitive high-resolution melting analysis (MS-HRM). Each ellipse represents a CpG site in the O6-methylguanine-DNA methyltransferase (MGMT) promoter. Each dot represents the CpGs analyzed by the different assays. Dots and ellipses in blue are CpGs of the MGMT promoter’s DMR2 region. Only CpGs in the primer and probe regions were analyzed by the MSP and qMSP assays. Note that CpGs in the primer region of MS-HRM were not analyzed because the primers for MS-HRM amplify MGMT promoter both when methylated and unmethylated at the primer binding sites with the same efficiency. qMSP_f, qMSP of reading strand; qMSP_r, qMSP of reverse strand.
Methylation-sensitive high-resolution melting. The primers used for MS-HRM and targeting exon 1 of the human MGMT gene were the MGMT MS-HRM2 primer set described by Wojdacz and Dobrovic with some modification (see Table I) (11). The PCR amplified a 110-bp fragment which analysed 12 CpGs (CpG numbers 72-83, see Figure 1) (11). The position of the amplified fragment on the University of California, Santa Cruz Genome Browser, February 2009 (GRCh37/19) was: chr10:131,265,469-131,265,578 (see Figure 1) (11). Three replicates of each sample were used. The 20 μl reaction volume contained 1× Precision Melt Supermix (Bio-Rad Laboratories), 200 nM of each forward and reverse primer, and 20 ng of bisulfite-converted DNA. Real-time PCR followed by a melting curve step was run on a CFX96 Touch Real-Time PCR Detection system (Bio-Rad Laboratories). The thermal cycling included an initial step at 95˚C for 2 min, followed 50 cycles of 10 s at 95˚C and 30 s at 64˚C. The melting curve step was performed according to the company’s recommendation (Bio-Rad Laboratories): an initial step of 10 s at 95˚C followed by temperature ramping from 65˚C to 95˚C rising by 0.2˚C/10 s and fluorescence acquisition. The data files generated by the CFX96 system were imported using Precision Melt Analysis software (Bio-Rad Laboratories) and further analyzed.
In order to create standards for the assay, the sense strand of a 400 bp DNA fragment was used, which corresponds to the sequence chr10:131,265,281-131,265,680 (GRCh37/hg19 Assembly). The 400-bp sequence was analyzed with Methyl Primer Express Software v1.0 (Applied Biosystems, Thermo Fisher Scientific Inc., Waltham, MA, USA) and both methylated and unmethylated sequences were obtained. The respective methylated and unmethylated DNA fragments were commercially synthesized and cloned into two different PEX-A plasmids (Europhins, Brussels, Belgium). The two plasmids were mixed in order to obtain a range of methylated and unmethylated allele dilutions. Each of the experimental runs included methylated/unmethylated dilutions corresponding to 100%, 70%, 50%, 20%, 10%, 5%, 1%, 0.1%, and 0% methylated allele. For all standards, 6,000 copies of plasmid were used as template. The calculations were based on the assumption that 3.3 pg of human genomic DNA contains one copy of MGMT gene and that 20 ng of bisulfite-converted DNA was used per assay. Thus, 20 ng of human genomic DNA contained 6000 copies of MGM gene (http:// www6.appliedbiosystems.com/support/tutorials/pdf/quant_pcr.pdf).
Quantitative methylation-specific qPCR. Quantitation of MGMT promoter methylation assessed by qMSP is described in (34). The data produced by Håvik et al. (34) were re-analyzed for OS in this study for 48 of the patients with GBM and the four with meningiomas. A total of 15 CpG sites were covered by the qMSP assay (Figure 1). Eleven CpGs of the 12 CpG sites 75-86 were covered by qMSP for the sense strand (primers and probes named MGMT qMSP) and 11 CpGs of the 14 CpG sites 71-86 were covered by qMSP for the opposite strand (primers and probes named MGMT_1 qMSP). qMSP results of the four meningiomas were used to calculate the threshold value for scoring methylation-positive samples (34). Only samples where both qMSP assays scored above the threshold value were scored as methylation-positive (34).
Pyrosequencing. Two PSQ kits were used to analyze MGMT promoter methylation. The analysis using the PyroMark Q96 CpG MGMT kit (cat. number 972032; Qiagen) and the PyroMark MD system (Qiagen) was described elsewhere (34). For analysis of MGMT promoter methylation using PyroMark therascreen MGMT kit (cat. number 971061; Qiagen) and the PyroMark Q24 system (Qiagen), 50 ng of bisulfite-converted DNA was used. Bisulfite-converted DNA was amplified in a PCR reaction according to the manufacturer’s recommendation (PyroMark Therascreen kit; Qiagen) on a C-1000 Thermal cycler (Bio-Rad Laboratories). Subsequent sample preparation and PSQ were also performed according to the manufacturer’s procedure (PyroMark Therascreen kit; Qiagen).
The PyroMark Q96 CpG MGMT kit detected five CpG sites located in exon 1 of the human MGMT gene (CpG 74-78; for CpG numbering and localization, see Figure 1), whereas the PyroMark Therascreen MGMT kit detected four CpG sites in the same location (CpG 76-79) (Figure 1).
Statistical analysis. All GBM cases (N=48) were included in the OS analysis. However, for MS-HRM, the data were obtained for only 37 out of the 48 cases. Survival data were calculated from the date of surgery. Information about the patients’ date of surgery, death or last follow-up is described elsewhere (34).
Receiver operating characteristic (ROC) curve analysis was used to estimate the optimal cut-off value for the two PSQ assays, using the mean percentage MGMT methylation for the CpGs covered by the two assays. The area under the ROC curve (AUROC) was calculated after fitting ordinary logistic regressions with the dependent variable indicating if a patient lived at least 18 months after diagnosis or not. Methylation was included as an independent variable. Hazard ratios (HR) were estimated using Cox proportional hazards regression with a dummy variable for methylation level. The AUROC and HR calculations were performed using Stata v15.1 software (StataCorp LLC, College Station, TX, USA).
OS analysis was performed using the Kaplan–Meier procedure. The survival curves were compared using the log-rank test. For PSQ, methylation was dichotomized according to the ROC analysis described above. OS calculations were performed using IBM SPSS Statistics v23 software (IBM Corporation, Armonk, NY, USA) and GraphPad Prism v7 (GraphPad Software, Inc. La Jolla, CA, USA). p-Values of less than 0.05 were considered statistically significant.
Results
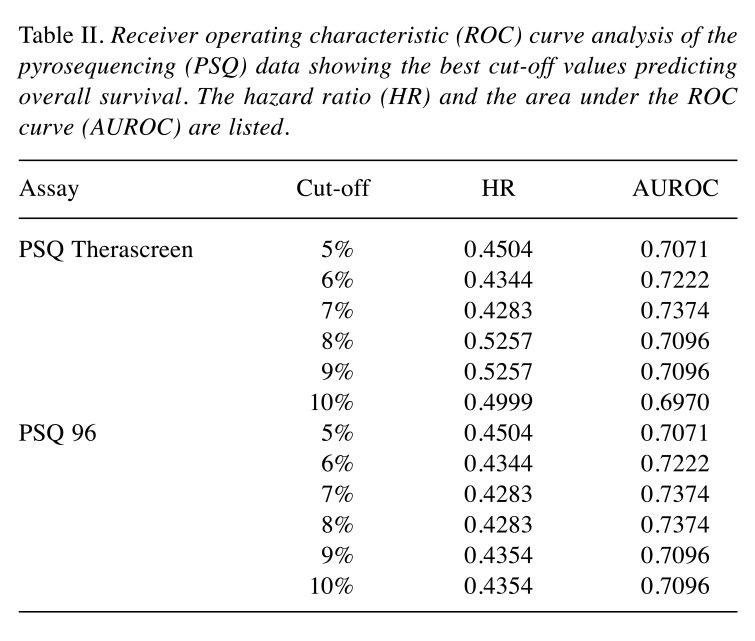
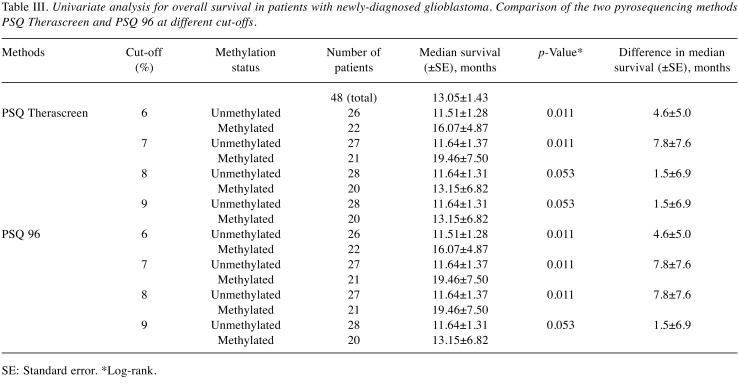
Determining the cut-off value for PSQ. In order to compare the prognostic ability of the different methods, the optimal cut-off value for PSQ needed to be identified. ROC curve analysis is the method of choice for predicting optimal cut-off values (37,38). The mean percentage methylation of the CpGs analyzed in the two PSQ assays were used in our ROC curve analysis, where methylation cut-off scores (1-15%) were plotted to identify the optimum cut-off value for the prediction of OS of 18 months or more after surgery. The AUROC results, including HR values, are listed in Table II. The highest values for AUROC were at a cut-off of 7% for PSQ Therascreen and 7 and 8% for PSQ 96. The AUROC values for both PSQ kits were 0.7374, with an HR of 0.4283 (confidence interval=0.2187-0.8390, p=0.013). The univariate analysis for OS in patients with newly-diagnosed GBM for the two PSQ kits, with cut-offs in the range from 6-9%, verified cut-offs at 7% for PSQ Therascreen and 7 and 8% for PSQ 96, as being optimal (Table III).
Table II. Receiver operating characteristic (ROC) curve analysis of the pyrosequencing (PSQ) data showing the best cut-off values predicting overall survival. The hazard ratio (HR) and the area under the ROC curve (AUROC) are listed.
Table III. Univariate analysis for overall survival in patients with newly-diagnosed glioblastoma. Comparison of the two pyrosequencing methods PSQ Therascreen and PSQ 96 at different cut-offs.
SE: Standard error. *Log-rank.
Comparison of PSQ, MSP, MS-HRM, and qMSP. For the two PSQ kits, MSP, MS-HRM and qMSP, univariate analyses were performed to measure OS. The variables were dichotomized as methylated and unmethylated according to their optimized cut-off values and OS was calculated.
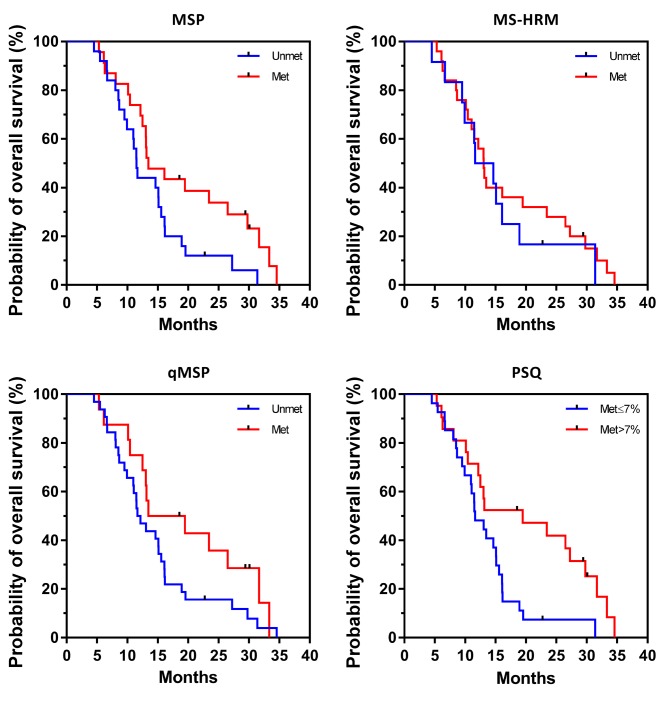
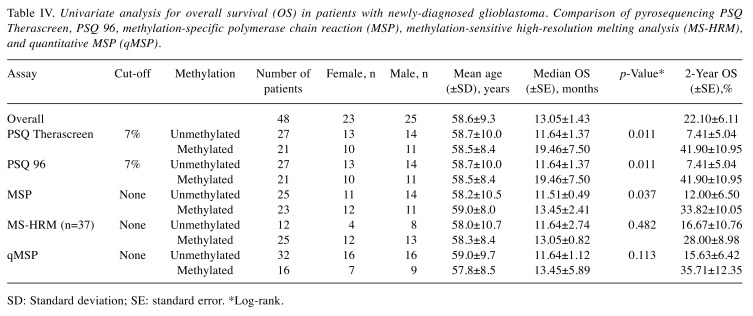
Unadjusted Kaplan–Meier plots of OS for the four methods are shown in Figure 2. Log-rank tests showed significant differences in OS between the groups with methylated and unmethylated MGMT promotor for the two PSQ kits (p=0.011) and MSP (p=0.037), but not for MS-HRM and qMSP (p=0.482 and p=0.113, respectively) (Table IV). Median OS using MSP was 11.5 months for the group with unmethylated MGMT and 13.5 months for the group with methylated MGMT, whereas 2-year OS was 12% and 33.8%, respectively. For PSQ, median OS was 11.6 months in the group with unmethylated MGMT and 19.5 months in the group with methylated MGMT, whereas 2-year OS was 7.4% and 41.90%, respectively.
Figure 2. Kaplan–Meier plots of overall survival according to O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status as analyzed by methylation-specific polymerase chain reaction (MSP), quantitative MSP (qMSP), methylation-sensitive high-resolution melting analysis (MS-HRM) and pyrosequencing (PSQ). For PSQ, the results of only PSQ Therascreen are shown. Met: Methylated; Unmet: unmethylated.
Table IV. Univariate analysis for overall survival (OS) in patients with newly-diagnosed glioblastoma. Comparison of pyrosequencing PSQ Therascreen, PSQ 96, methylation-specific polymerase chain reaction (MSP), methylation-sensitive high-resolution melting analysis (MS-HRM), and quantitative MSP (qMSP).
SD: Standard deviation; SE: standard error. *Log-rank.
Discussion
In this study, we identified PSQ as being the best technique for prognostication based on tumor MGMT promoter methylation analysis in patients with GBM. The difference in median OS between the groups with unmethylated and methylated MGMT was approximately 8 months (p=0.011) by PSQ, whereas the difference in median OS was only 2 months when MSP was used; this, however, was still significant (p=0.037). The two other techniques assessed in our study, MS-HRM and qMSP, did not reveal significant survival differences (p=0.482 and 0.113, respectively).
The different results produced by the four methods compared in this study might be explained by variances in the CpGs analyzed and differences in methodology. Of the 98 CpG sites in the MGMT promoter, methylation of two CpG regions, CpGs 25-50 (DMR1) and CpGs 73-90 (DMR2), seems to correlate with transcriptional silencing (8,9). DMR2 in particular plays a critical role in controlling the MGMT promoter (8). MSP, qMSP, and MS-HRM cover CpGs 76-80 and 84-87, CpGs 71-86 (except CpG 74), and CpGs 72-83, respectively. The two PSQ assays performed, PSQ 96 and PSQ Therascreen, analyze five and four CpGs, respectively, namely CpGs 74-78 and 76-79 in DMR2. Unlike MSP and qMSP, presumably important sites such as CpG 86 and 87 are not evaluated by the two PSQ kits (8). Despite this and the low number of CpGs analyzed as compared to the other three methods, PSQ is superior when it comes to prognostication based on separation of patients into groups with and without MGMT promotor methylation. A major disadvantage with using MSP and qMSP is that both these techniques only detect fully methylated MGMT promoter at the annealing sites of primers and probe (see Figure 1). This may lead to a problem in sensitivity; however, both these methods have in previous studies been found to be of prognostic value (1,21-30,39).
PSQ overcomes the problem with heterogenous methylation patterns by providing information on the extent of methylation at each individual CpG site with high sensitivity and specificity (19). The most critical issue in the use of PSQ is to identify a threshold value for dichotomizing the original continuous data, that is, to define the optimal prognostication cut-off value (9,34,40). The cut-off value can be set based on negative controls, arbitrarily, or as the median value of the whole patient group tested. Another and probably better method for defining an optimal threshold value from continuous test results is use of the ROC curve (37,41). We used an approach to measure AUROC by defining 18 months’ survival as a point for separating patients into long-term and short-term survivors. Based on this, ROC analysis identified 7% methylation for PSQ Therascreen and 7-8% methylation for PSQ 96 as optimal cut-off values. The AUROC value for both PSQ kits was 0.76, with an HR of 0.4283 (CI=0.2187-0.8390, p=0.013), indicating it to be a reliable assay and confirming the suitability of the cut-off value for discriminating methylated from unmethylated promoter. Unadjusted Kaplan–Meier plots for the two PSQ kits with cut-offs in the range from 6% to 9% verified optimal cut-offs at 7% and 7 and 8% for PSQ Therascreen and PSQ 96, respectively (Table III). These results are on a par with findings by Quillien et al. (24), Kim et al. (42), and Quillien et al. (43) who used ROC analysis to find optimal cut-off points for the PSQ 96 kit at 6%, 9%, and 8%, respectively. Linear regression showed good correlation between the two PSQ kits (R2=0.9226, N=48, data not shown), in agreement with the findings of Quillien et al. (33). Additionally, there was 100% correlation between cases defined as methylated or unmethylated when comparing the two PSQ kits, supporting the notion that these two PSQ assays are interchangeable (33).
MSP has been clinically validated for analysis of MGMT promoter methylation status (2,3) and is a purely qualitative assay that analyzes methylation of nine CpGs encompassing CpGs 76-80 and CpGs 84-87, covering a large part of DMR2. Primers are generated to either amplify unmethylated or fully methylated PCR products, that is, PCR products either unmethylated or fully methylated at CpG sites covered by the primers. As melting point analyses were used to detect methylated and unmethylated PCR products, the melting point of the PCR products might be affected by heterogenous methylation of the three CpGs in the sequence between the two primers. Therefore, heterogeneous methylation of CpG sites may lead to lower sensitivity and specificity, and failure of detection by MSP. In spite of these drawbacks, MSP was able to generate groups with methylated and unmethylated MGMT promotor with significantly different median OS (p=0.037), in accordance with several previous reports (1,21-24,26-29,39).
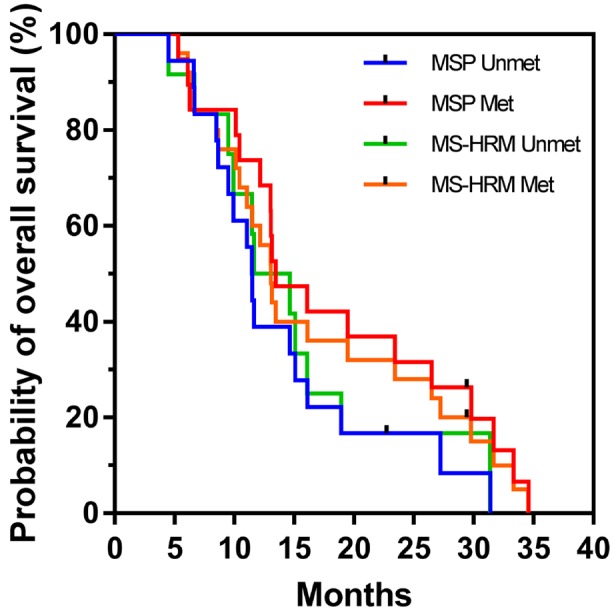
Our MS-HRM assay, which analyzed CpG site 72-83, was not useful for prognostication in this patient cohort. An explanation for this discrepancy compared with results in previous studies might be the low number of patients (N=37) included in our MS-HRM analysis. In support of this, when looking at only the 37 cases analyzed by MS-HRM, MSP did not give significant differences in OS for patients with methylated and unmethylated MGMT promoter (p=0.096, N=37) (Figure 3).
Figure 3. Kaplan–Meier plot of overall survival according to methylation, comparing methylation-specific polymerase chain reaction (MSP) and methylation-sensitive high-resolution melting analysis (MSHRM) when N=37. Log-rank p-values were 0.096 and 0.482 for MSP and MS-HRM, respectively.
The reason why the qMSP assay used here was not useful for prognostication might be its highly restrictive nature based on analysis of both the sense and reverse strand, as well as the variation in CpGs analyzed on the sense and reverse strand (see Figure 1). As can be seen in Table IV, only 16 patients out of 48 were scored as having MGMT promoter methylation by qMSP. When comparing MSP and qMSP, only 16 out of the 23 cases scored as methylated by MSP were scored as methylated by qMSP, indicating that our qMSP setup was too restrictive when compared to previous studies (22,30).
In our hands, only MSP and PSQ were able to significantly separate patients with GBM into two prognostic groups based on MGMT promoter methylation status. The superiority of PSQ for such analysis (21-24) makes it the method of choice for analysis of MGMT promoter methylation status in routine clinical practice.
Competing Interests
The Authors declare that they have no competing interests in regard to this study.
Acknowledgements
The Authors thank Dr. Annette Bentsen Håvik for providing us with the earlier published data for the PSQ 96 and qMSP analyses (34). This work was supported by grants from the Radiumhospitalets Legater and Larvik kreftforening.
References
- 1.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozo-lomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, for the European Organisation for Research Treatment of Cancer Brain Tumor and Radiotherapy Groups and the National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO, on behalf of the European Organisation for Treatment of Cancer Brain Tumour and Radiation Oncology Groups and the National Cancer Institute of Canada Clinical Trials Research Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-Year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Stevens MF, Bradshaw TD. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5:102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 6.Cabrini G, Fabbri E, Lo Nigro C, Dechecchi MC, Gambari R. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review) Int J Oncol. 2015;47:417–428. doi: 10.3892/ijo.2015.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, Hegi ME. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine. Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 8.Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121:651–661. doi: 10.1007/s00401-011-0803-5. [DOI] [PubMed] [Google Scholar]
- 9.Everhard S, Tost J, El Abdalaoui H, Criniere E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Thillet J. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): A new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. doi: 10.1093/nar/gkm013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, Moreau F, Hamou MF, Renard I, Delorenzi M, Flamion B, DiGuiseppi J, Bierau K, Hegi ME. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parrella P, la Torre A, Copetti M, Valori VM, Barbano R, Notarangelo A, Bisceglia M, Gallo AP, Balsamo T, Poeta ML, Carella M, Catapano D, Parisi S, Dallapiccola B, Maiello E, D'Angelo V, Fazio VM. High specificity of quantitative methylation-specific PCR analysis for MGMT promoter hypermethylation detection in gliomas. J Biomed Biotechnol. 2009;2009:531692. doi: 10.1155/2009/531692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ida CM, Butz ML, Jenkins RB, Sarkaria JN, Kitange GJ, Giannini C, Kipp BR. Real-time methylation-specific polymerase chain reaction for MGMT promoter methylation clinical testing in glioblastoma: an alternative detection method for a heterogeneous process. Am J Clin Pathol. 2017;148:296–307. doi: 10.1093/ajcp/aqx073. [DOI] [PubMed] [Google Scholar]
- 15.Maxwell JA, Johnson SP, Quinn JA, McLendon RE, Ali-Osman F, Friedman AH, Herndon JE 2nd, Bierau K, Bigley J, Bigner DD, Friedman HS. Quantitative analysis of O6-alkylguanine-DNA alkyltransferase in malignant glioma. Mol Cancer Ther. 2006;5:2531–2539. doi: 10.1158/1535-7163.MCT-06-0106. [DOI] [PubMed] [Google Scholar]
- 16.Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): Simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- 19.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 20.Bienkowski M, Berghoff AS, Marosi C, Wohrer A, Heinzl H, Hainfellner JA, Preusser M. Clinical Neuropathology practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clin Neuropathol. 2015;34:250–257. doi: 10.5414/NP300904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christians A, Hartmann C, Benner A, Meyer J, von Deimling A, Weller M, Wick W, Weiler M. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS One. 2012;7:e33449. doi: 10.1371/journal.pone.0033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu CY, Ho HL, Lin SC, Chen MH, Hsu SP, Yen YS, Guo WY, Ho DM. Comparative assessment of 4 methods to analyze MGMT status in a series of 121 glioblastoma patients. Appl Immunohistochem Mol Morphol. 2017;25:497–504. doi: 10.1097/PAI.0000000000000331. [DOI] [PubMed] [Google Scholar]
- 23.Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, Etcheverry A, Hamlat A, Loussouarn D, Campion L, Campone M, Vallette FM, Gratas-Rabbia-Re C. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 24.Quillien V, Lavenu A, Karayan-Tapon L, Carpentier C, Labussiere M, Lesimple T, Chinot O, Wager M, Honnorat J, Saikali S, Fina F, Sanson M, Figarella-Branger D. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118:4201–4211. doi: 10.1002/cncr.27392. [DOI] [PubMed] [Google Scholar]
- 25.Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin Epigenetics. 2016;8:49. doi: 10.1186/s13148-016-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metellus P, Coulibaly B, Nanni I, Fina F, Eudes N, Giorgi R, Barrie M, Chinot O, Fuentes S, Dufour H, Ouafik L, Figarella-Branger D. Prognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: a prospective patient cohort. Cancer. 2009;115:4783–4794. doi: 10.1002/cncr.24546. [DOI] [PubMed] [Google Scholar]
- 27.Reifenberger G, Hentschel B, Felsberg J, Schackert G, Simon M, Schnell O, Westphal M, Wick W, Pietsch T, Loeffler M, Weller M, German Glioma N. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131:1342–1350. doi: 10.1002/ijc.27385. [DOI] [PubMed] [Google Scholar]
- 28.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 29.Hsu CY, Ho HL, Lin SC, Chang-Chien YC, Chen MH, Hsu SP, Yen YS, Guo WY, Ho DM. Prognosis of glioblastoma with faint MGMT methylation-specific PCR product. J Neurooncol. 2015;122:179–188. doi: 10.1007/s11060-014-1701-1. [DOI] [PubMed] [Google Scholar]
- 30.Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12:116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Wu X, Yao Y, Zhou LF, Mao Y. Direct, real-time PCR (MethyLight) assay for methylation of O6-methylguanine-DNA methyltransferase promoter in glioma. Chin Med J. 2009;122:1342–1345. [PubMed] [Google Scholar]
- 32.Shah N, Lin B, Sibenaller Z, Ryken T, Lee H, Yoon JG, Rostad S, Foltz G. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6:e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quillien V, Lavenu A, Ducray F, Meyronet D, Chinot O, Fina F, Sanson M, Carpentier C, Karayan-Tapon L, Rivet P, Entz-Werle N, Legrain M, Zalcman EL, Levallet G, Escande F, Ramirez C, Chiforeanu D, Vauleon E, Figarella-Branger D. Clinical validation of the CE-IVD marked Therascreen MGMT kit in a cohort of glioblastoma patients. Cancer Biomark. 2017;20:435–441. doi: 10.3233/CBM-170191. [DOI] [PubMed] [Google Scholar]
- 34.Håvik AB, Brandal P, Honne H, Dahlback HS, Scheie D, Hektoen M, Meling TR, Helseth E, Heim S, Lothe RA, Lind GE. MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J Transl Med. 2012;10:36. doi: 10.1186/1479-5876-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorente A, Mueller W, Urdangarin E, Lazcoz P, von Deimling A, Castresana JS. Detection of methylation in promoter sequences by melting curve analysis-based semiquantitative real time PCR. BMC Cancer. 2008;8:61. doi: 10.1186/1471-2407-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 37.Greiner M, Pfeiffer D, Smith RD. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med. 2000;45:23–41. doi: 10.1016/s0167-5877(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 38.Yuan G, Niu L, Zhang Y, Wang X, Ma K, Yin H, Dai J, Zhou W, Pan Y. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J Neurooncol. 2017;133:193–201. doi: 10.1007/s11060-017-2433-9. [DOI] [PubMed] [Google Scholar]
- 39.Uno M, Oba-Shinjo SM, Camargo AA, Moura RP, Aguiar PH, Cabrera HN, Begnami M, Rosemberg S, Teixeira MJ, Marie SK. Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma. Clinics. 2011;66:1747–1755. doi: 10.1590/S1807-59322011001000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, Wong H, Liloglou T, Haylock B, Walker C. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DC, Kim KU, Kim YZ. Prognostic role of methylation status of the MGMT promoter determined quantitatively by pyrosequencing in glioblastoma patients. J Korean Neurosurg Soc. 2016;59:26–36. doi: 10.3340/jkns.2016.59.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quillien V, Lavenu A, Ducray F, Joly MO, Chinot O, Fina F, Sanson M, Carpentier C, Karayan-Tapon L, Rivet P, Entz-Werle N, Legrain M, Zalcman EL, Levallet G, Escande F, Ramirez C, Chiforeanu D, Vauleon E, Figarella-Branger D. Validation of the high-performance of pyrosequencing for clinical MGMT testing on a cohort of glioblastoma patients from a prospective dedicated multicentric trial. Oncotarget. 2016;7:61916–61929. doi: 10.18632/oncotarget.11322. [DOI] [PMC free article] [PubMed] [Google Scholar]