Abstract
Pectinase catalyzed the degradation of pectin substances and has been used in various biotechnological industries. In the current study, 23 bacterial strains were isolated from rotten vegetables, soil and air. The isolated bacterial strains were qualitatively screened for pectinase production on pectin agar medium and only three strains HR 4, HR 21 and HR 23 were observed to produce extracellular pectinase. These strains were further screened quantitatively for pectinase production through submerged fermentation technology in pectin containing fermentation medium. Strain HR 4 from rotten brinjal (Solanum melongena) was found to produce higher pectinase as compared to others. The maximum pectinase producing bacterial strain was identified as Bacillus licheniformis on the basis of morphological, physiological and biochemical characteristics. For further confirmation of identification, 16S rDNA sequence analysis was performed. The 16S rDNA sequences were aligned and the phylogenetic tree was constructed. The phylogenetic tree confirmed that the strain was belonging to B. licheniformis. The 16S rDNA sequences of this new strain were submitted to GenBank and designated as B. licheniformis KIBGE-IB21 with the GenBank accession number JQ 411812. The newly isolated pectinase producing B. licheniformis used apple pectin as carbon and yeast extract as nitrogen source for maximum pectinase production.
Keywords: Identification, 16S rDNA sequences, Bacillus licheniformis, Pectinase production
1. Introduction
Pectinase has different applications in industries such as clarification and filtration of fruits juices, winemaking process for increasing the quality of wine, coffee and tea fermentation, degumming of plant fibers, oil extraction, textile, papermaking, protoplasm isolation and purification of plant virus as well as pre-treatment of pectin containing wastewaters [1], [2]. Pectinase is one of the important factors in the plant pathological process, plant–microbe symbiosis and in the decomposition of plant matter decay. The microbial pectinase is playing an important role in nature by contributing to natural recycling of carbon in the environment. Different microorganisms are known to produce pectinase with different molecular mass and catalytic properties [3], [4], [5]. Aspergillus niger is mostly used for the industrial production of pectinase [6]. Pectinases from fungus sources are usually acidic in nature and only can work in acidic conditions. Production of alkaline pectinase remains under developed, as only few reports are available on the production of alkaline pectinase by bacterial strains [7], [8], [9]. Alkaline pectinase can be used for the treatment of pectinous substances containing wastewater from vegetables and food processing industries [10]. Bacillus is one of the large genera of bacterial strains. It is a rod shaped, endospore bearing bacteria and belong to the family Firmicutes. The genus Bacillus covered a great diversity of strains and some of them are strictly aerobic, while others are facultative anaerobic. The Bacillus especially Bacillus subtilis and Bacillus licheniformis are excellent candidates for large-scale production of commercially important enzymes [11]. B. licheniformis is a saprophytic bacterium found in nature and commonly found in soil and other natural environments. The B. licheniformis is capable of growing on a large diversity of nutrient sources because of synthesizing and secreting different hydrolytic enzymes and this quality makes the B. licheniformis an industrially important microorganism [12], [13]. In the current study, different bacterial strains were isolated from indigenous sources and screened for pectinase production. Finally, maximum pectinase producing bacterial strain was identified using conventional and molecular techniques.
2. Material and methods
2.1. Isolation of pure bacterial strains
Several bacterial strains were isolated from rotten vegetables and soil samples collected from vegetative fields of KIBGE, University of Karachi, Pakistan. Soil and rotten vegetable samples (1.0 g) were mixed in 100 ml normal saline, afterward serially diluted from 10−1 to 10−6 ratio with normal saline. 100 μl of each diluted sample was inoculated in nutrient agar medium and incubated at 37 °C for 24 h. The isolated colonies were selected to obtain pure bacterial cultures. Cultures were maintained in nutrient agar slants.
2.2. Qualitative screening of bacterial strains for pectinase production
Pure bacterial isolates were grown at 37 °C for 24 h onto the pectin agar medium described by Soares et al. [14] for the screening of pectinolytic activity. Medium was composed of citrus pectin (1.0%), ammonium sulfate (0.14%), di-potassium hydrogen phosphate (0.6%), potassium dihydrogen phosphate (0.20%), magnesium sulfate (0.01%) and agar-agar (2.0%) of pH 6.0. A clear zone around the colony was detected by overlying with potassium–iodide solution [15].
2.3. Quantitative screening of bacterial strains for pectinase production
The bacterial strains showing the maximum zone of hydrolysis on pectin agar medium were screened for pectinase production using submerged fermentation in the above mentioned medium without agar-agar. The pure cultures were inoculated in 10% inoculums at 37 °C for 24 h, then transferred into 90.0 ml fermentation medium, and incubated at 37 °C for 24 h. After incubation, biomass was separated by centrifugation at 10,000 rpm for 15 min. The supernatant was used to evaluate pectinase activity.
2.4. Enzyme assay
Pectinase activity was measured by the estimation of the amount of galacturonic acids through the DNS method [16] using 1.0% citrus pectin as a substrate and mono-d-galacturonic acid as a standard. One unit of pectinase was defined as the “amount of enzyme required to generate 1 μmole of galacturonic acid under standard assay conditions”.
2.5. Estimation of total protein
Total protein was estimated through Lowry’s method using BSA (Bovine serum albumin) as a standard [17].
2.6. Identification of bacterial strain
The identification of a selected bacterial strain was performed on the basis of morphological, biochemical and molecular characteristics.
2.6.1. Morphological characterization
Morphological characteristics such as colony morphology (color, shape, margin, elevation, and surface) and cell morphology (shape, gram reaction, and arrangement) of the selected bacterial strain were studies for identification [18].
2.6.2. Biochemical characterization
The bacterial strain was subjected to different biochemical tests including catalase test, methyl red (MR) and Voges Proskauer (VP) tests, gas production from glucose and mannitol, anaerobic growth, growth in 7.0% NaCl, SDA, starch hydrolysis, citrate utilization and nitrate reduction test [18].
2.6.3. Molecular characterization
16S rDNA sequence analysis was performed for molecular based identification of selected isolates. The genomic DNA was extracted using the method reported by Chen and Kuo with slight modifications [19]. The extraction of DNA was confirmed by running in 1.0% agarose electrophoresis gel containing ethidium bromide and visualized under UV light. PCR amplification and 16S rDNA sequence analysis were performed [20]. The 16S rDNA gene was PCR-amplified by using bacterial universal primers set 16SF: GAGTTTGATCCTGGCTCAG and 16SR: AGAAAGGAGGTATCCAG CC. The PCR reaction mixture was prepared in a final 50 μl of reaction volume consisting of 5.0 μl PCR reaction buffer, 1.0 μl of dNTPs mix, 1.0 μl of 10 pM primers, 2.5 μl genomic DNA template and 2.5 U of DNA polymerase. The amplification was carried out in an Applied Biosystem GenAmp PCR system 2700. The PCR amplified production was analyzed by 1.0% agarose gel electrophoresis and after visualization using UV light the PCR amplified product was purified using Wizard SV gel and PCR Clean-Up system (promega), according to manufacturer’s instructions. The sequence analysis of amplified DNA fragment was done by DNA sequencer ABI3130 Genetic Analyzer (Applied BioSystem). Phylogenetic and molecular evolutionary analyses were done using MEGA 4 software [21].
2.7. Effect of carbon and nitrogen sources on pectinase production
The effect of carbon and nitrogen sources on pectinase production was analyzed by incubating B. licheniformis in production media containing different carbon and nitrogen sources through a one variable at a time approach.
3. Results and discussion
Several bacterial strains were isolated from rotten vegetables, air and soil samples collected from different areas of Karachi, Pakistan using pure culture study (Table 1). Out of these strains, 1.0–3.0 were from cauliflower, 4.0 and 5.0 were from brinjal, 6.0 and 7.0 were from cabbage, 8.0–10 were from beetroot, 11–21 from soil, and 22 and 23 were isolated from air. All strains were maintained on nutrient agar slants.
Table 1.
Bacterial strains isolated from indigenous sources.
| S. No | Bacterial strains | Sources |
|---|---|---|
| 1 | HR 1 | Cauliflower (Brassica oleracea) |
| 2 | HR 2 | Cauliflower (Brassica oleracea) |
| 3 | HR 3 | Cauliflower (Brassica oleracea) |
| 4 | HR 4 | Brinjal (Solanum melongena) |
| 5 | HR 5 | Brinjal (Solanum melongena) |
| 6 | HR 6 | Cabbage (Brassica oleracea) |
| 7 | HR 7 | Cabbage (Brassica oleracea) |
| 8 | HR 8 | Beetroot (Beta vulgaris) |
| 9 | HR 9 | Beetroot (Beta vulgaris) |
| 10 | HR 10 | Beetroot (Beta vulgaris) |
| 11 | HR 11 | Soil |
| 12 | HR 12 | Soil |
| 13 | HR 13 | Soil |
| 14 | HR 14 | Soil |
| 15 | HR 15 | Soil |
| 16 | HR 16 | Soil |
| 17 | HR 17 | Soil |
| 18 | HR 18 | Soil |
| 19 | HR 19 | Soil |
| 20 | HR 20 | Soil |
| 21 | HR 21 | Soil |
| 22 | HR 22 | Air |
| 23 | HR 23 | Air |
After the pure culture study, the purified bacterial isolates were subjected to pectinolytic activity by growing on pectin agar medium at 37 °C for 24 h [14]. The pectinolytic activity was detected by visualizing a clear zone around the colony using potassium–iodide flooding method [15]. Among these isolates only three strains, HR 4, HR 21 and HR 23 showed pectinolytic activity on pectin agar medium (Fig. 1).
Figure 1.
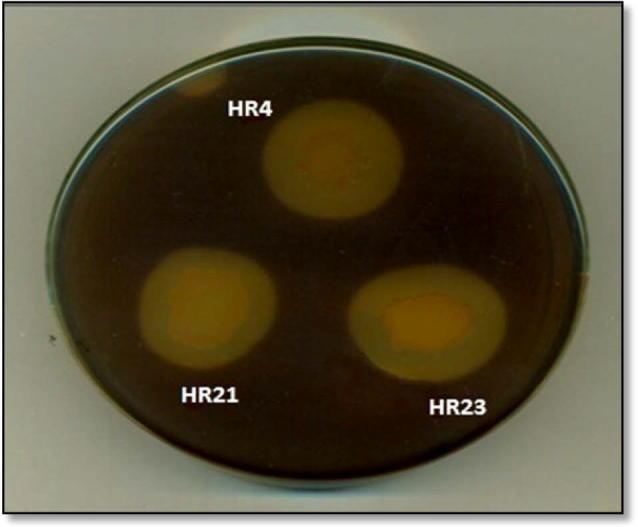
Qualitative screening of bacterial strains for pectinolytic activity on pectin agar medium.
The bacterial strains were further screened for pectinase production using batch fermentation by growing in a production medium containing 1.0% pectin as a sole source of carbon at 37 °C for 24 h. It was observed that the pectinase production varied from strain to strain, HR 4 produced 341 U mg−1, HR 21 produced 160 U mg−1 and HR 23 produced 140 U mg−1 pectinase isolated from rotten vegetable (Brinjal), soil and air, respectively (Table 2). The strain HR 4 which produced high pectinase compared to HR 21 and HR 23 was selected for further studies.
Table 2.
Production of pectinase from different bacterial strains.
| Bacterial strains | Pectinase activity (U mg−1) | Relative activity (%) |
|---|---|---|
| HR 4 | 341 | 100 |
| HR 21 | 160 | 46.92 |
| HR 23 | 140 | 41.05 |
The identification of the selected bacterial strain was done on the basis of morphological, physiological and biochemical characteristics as well as 16S rDNA sequence analysis. On the basis of morphology, physiology and biochemical tests, the selected strain was identified as B. licheniformis. It showed oxidase and catalase activities and was able to produce nitrogen gas from nitrate and hydrolyzed starch. It utilized various sugars including glucose, maltose and galactose to produce acid and was unable to produce acid from adonitol, sorbitol, mannose and sucrose. Its unutilized urea and citrate did not exhibit decarboxylase activity, as well as indole and H2S production. The details of biochemical characteristics of pectinase producing strain are given in Table 3.
Table 3.
Morphological, physiological and biochemical characteristics of maximum pectinase producing bacterial strain.
| Cellular characteristics | |
| Gram’ staining | Positive |
| Morphology | Rods with rounded ends |
| Motility | Motile |
| Spore | Central to para-central, ellipsoidal to cylindrical in shape |
| Size | 0.6–1.0 μm in length |
| Colonial morphology | |
| Nutrient agar | Finely wrinkled, dull, opaque, adherent colonies |
| Fermentation reaction | |
| Glucose | Acid |
| Maltose | Acid |
| Galactose | Acid |
| Adonitol | – |
| Mannose | – |
| Sucrose | – |
| Biochemical reaction | |
| Indole | Negative |
| Methyl red | Negative |
| Voges Proskauer | Positive |
| Citrate | Negative |
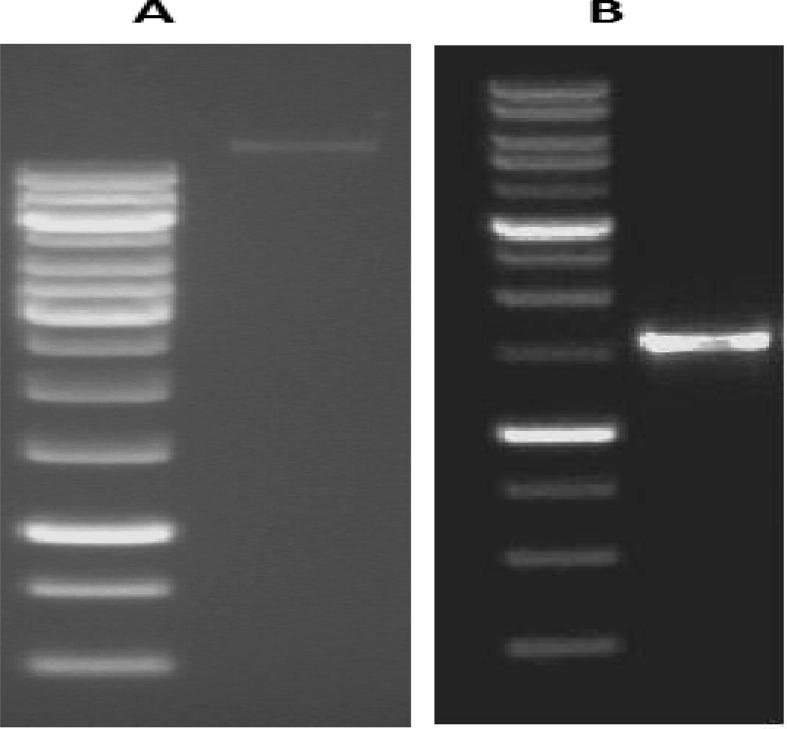
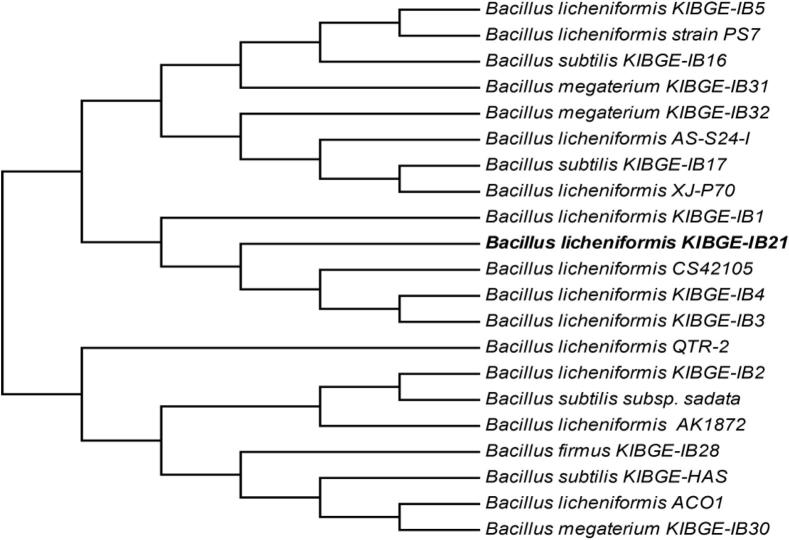
The genomic DNA of the microorganism was extracted and 16S rDNA was amplified by PCR using genomic DNA of the microorganism as template. The PCR product was examined by agarose gel electrophoresis (Fig. 2). The PCR product was purified and sequencing was done in triplicate in order to get the correct sequence. The nucleotide sequence of 16S rDNA (Fig. 3) was submitted in GenBank database under the accession number JQ 411812. The 16S rDNA sequence of the bacterial strain was aligned with all available 16S rDNA sequences in GenBank database and finally the phylogenetic tree was constructed (Fig. 4). The phylogenetic tree indicated that this bacterial isolate belonged to the genus Bacillus and the pattern of the tree determined that the strain is closely related to other B. licheniformis strains with 99% 16S rDNA similarity.
Figure 2.
Agarose gel electrophoresis. (A) DNA extracted from Bacillus licheniformis KIBGE-IB21. (B) PCR product of 16S rDNA of B. licheniformis KIBGE-IB21.
Figure 3.
Nucleotide sequence of 16S rDNA from Bacillus licheniformis KIBGE-IB21.
Figure 4.
Phylogentic tree showing the relation of Bacillus licheniformis KIBGE-IB21 with other species.
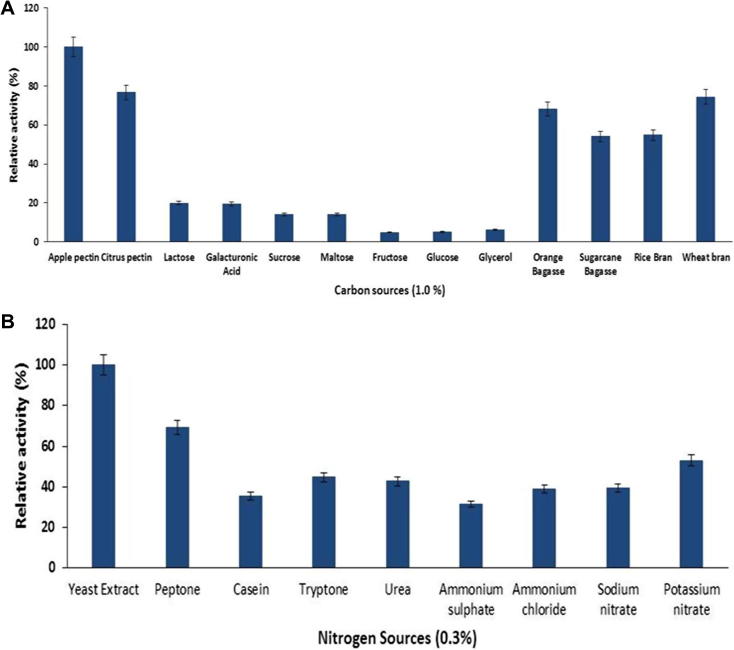
Carbon and nitrogen are the major elements required for structural and metabolic activities of living organisms. So the sources of these elements have a profound effect on the production of enzymes by microorganisms. The effect of carbon sources on the production of pectinase was determined by performing the fermentation in a production media containing different carbon sources. B. licheniformis produced higher concentrations of pectinase in the medium containing apple pectin as a sole source of carbon, followed by citrus pectin and wheat bran (Fig. 5A). The utilization of B. licheniformis KIBGE-IB21 at commercial level can reduce the cost of pectinase production by using wheat bran as a sole source of carbon. The production of pectinase was decreased in the media containing lactose, glucose, sucrose, maltose, and galacturonic acid which may be due to catabolic repression effect of these metabolites.
Figure 5.
Effect of carbon (A) and nitrogen (B) sources on pectinase production by Bacillus licheniformis. (mean ± S.E., n = 6).
The sources of nitrogen cause significant impact on microbial enzyme production. The influence of various nitrogen sources on pectinase production by B. licheniformis was determined by performing the fermentation in a production media with various organic and inorganic nitrogen sources. I observed that the B. licheniformis produced more pectinase in the medium with complex organic nitrogen sources like yeast extract and peptone as compared to medium with inorganic nitrogen sources (Fig. 5B). In terms of pectinase production yeast extract supported B. licheniformis to produce higher pectinase as compared to peptone. Yeast is a complex nutrient source with unknown composition containing essential amino acids, vitamins and other trace elements which may be responsible for enhancing pectinase production.
4. Conclusion
Several bacterial strains were isolated from indigenous sources and screened for pectinase production. Among these strains only three strains showed pectinolytic activity on pectin agar medium. These three (03) strains were further screened for pectinase production by using submerged fermentation technology and HR 4 was observed to produce higher pectinase. On the basis of physiological, morphological and biochemical characteristics the strain was identified as B. licheniformis. 16S rDNA sequence analysis was performed to finalize the identification at molecular level. The sequences of the current strain were found to be 99% similar to 16S rDNA sequences of B. licheniformis from various sources in GenBank database. B. licheniformis used apple pectin and wheat bran as sole carbon sources, and yeast extract as a nitrogen source to produce maximum pectinase. Further biochemical characterization of pectinase from B. licheniformis KIBGE-IB21 will be required to enhance its industrial feasibility.
Acknowledgment
All authors gratefully acknowledged the Karachi Institute of Biotechnology and Genetic Engineering (KIBGE) for providing financial support.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Hoondal G.S., Tiwari R.P., Tiwari R., Dahiya N., Beg Q.K. Appl. Microbiol. Biotechnol. 2002;59:409–418. doi: 10.1007/s00253-002-1061-1. [DOI] [PubMed] [Google Scholar]
- 2.Kashyap D.R., Vohra P.K., Chopra S., Tewari R. Bioresour. Technol. 2001;77:215–227. doi: 10.1016/s0960-8524(00)00118-8. [DOI] [PubMed] [Google Scholar]
- 3.Esquivel J.C.C., Voget C.E. J. Biotechnol. 2004;110:21–28. doi: 10.1016/j.jbiotec.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Kaur G., Kumar S., Satyanarayana T. Bioresour. Technol. 2004;94:239–243. doi: 10.1016/j.biortech.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Gummadi S.N., Panda T. Process Biochem. 2003;38:987–996. [Google Scholar]
- 6.Maldonado M.C., Cáceres S., Galli E., Navarro A.R. Folia Microbiologica (Praha) 2002;47:409–412. doi: 10.1007/BF02818699. [DOI] [PubMed] [Google Scholar]
- 7.Beg Q.K., Bhushan B., Kapoor M., Hoondal G.S. J. Ind. Microbiol. Biotechnol. 2000;24:396–402. [Google Scholar]
- 8.Hayashi K., Inoue Y., Shiga M., Sato S., Tokano R., Hirayae K., Hibi T., Hara S. Phytochemistry. 1997;45:1359–1363. doi: 10.1016/s0031-9422(97)00191-x. [DOI] [PubMed] [Google Scholar]
- 9.Cao J., Zheng L., Chen S. Enzyme Microb. Technol. 1992;14:1013–1016. [Google Scholar]
- 10.Tanabe H., Kobayashi Y., Akamatsu T. Agric. Biol. Chem. 1988;52:1855–1856. [Google Scholar]
- 11.Schallmey M., Singh A., Ward O.P. Can. J. Microbiol. 2004;50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- 12.Neves M.A.D., Kimura T., Shimizu N., Shiiba K. Braz. Arch. Biol. Technol. 2006;49:481–490. [Google Scholar]
- 13.Mojović L., Nikolić S., Rakin M., Vukasinović M. Fuel. 2006;85:1750–1755. [Google Scholar]
- 14.Soares M.M.C.N., Da Silva R., Carmona E.C., Gomes E. World J. Microbiol. Biotechnol. 2001;17:79–82. [Google Scholar]
- 15.Fernandes-Salomao T.M., Amorim A.C.R., Chaves-Alves V.M., Coelho J.L.C., Silva D.O., Araujo E.F. Rev. Microbiol. 1996;27:15–18. [Google Scholar]
- 16.Miller G.L. Anal. Chem. 1959;31:426–428. [Google Scholar]
- 17.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Holt J.G., Krieg N.R., Sneath P.H.A., Staley J.T., William S.T. nineth ed. Williams and Walkins; Baltimore: 1994. Bergey′s Manual of Determinative Bacteriology. pp. 787. [Google Scholar]
- 19.Chen W., Kuo T. Nucleic Acids Res. 1993;21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansari A., Aman A., Siddiqui N.N., Iqbal S., Qader S.A.U. Pak. J. Pharm. Sci. 2012;25:195–201. [PubMed] [Google Scholar]
- 21.Tamura K., Dudley J., Nei M., Kumar S. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]