Abstract
The study confirms the role of the two Arabidopsis hemoglobin genes (Glb1 and Glb2) during somatic embryogenesis and proposes the involvement of ethylene in the regulation of embryo development. Suppression of both Glb1 and Glb2 results in accumulation of nitric oxide (NO) and a different embryogenic response. Compared to WT tissue, down-regulation of Glb1 (Glb1 RNAi line) compromises the embryogenic process, while repression of Glb2 (Glb2−/− line) increases the number of embryos. These differences were ascribed to the differential accumulation of NO in the two lines, as Glb1 is a more effective NO scavenger compared to Glb2. A high elevation of NO level [achieved pharmacologically using the NO donor sodium nitroprusside (SNP), or genetically using the Glb1 suppressing line], activated the two ethylene biosynthetic genes 1-aminocyclopropane-1-carboxylate synthase (ACC synthase) and 1-aminocyclopropane-1-carboxylate oxidase (ACC oxidase). Ethylene accumulation repressed embryogenesis, as shown by the decreased embryo number observed in tissue treated with the ethylene releasing agent Ethephon (ETH), as well as by the increased embryo production obtained with the two ethylene insensitive mutant lines (ein2-1 and ein3-1). A repression in ethylene level increased the expression of many auxin biosynthetic genes and favored the accumulation of the auxin indole-acetic acid (IAA) at the sites of the explants where embryogenic tissue will form. Collectively these data reveal that high levels of NO, generated by the Glb1 suppressing line, but not by the Glb2 suppressing line, might increase the level of ethylene, which represses the production of auxin. Auxin is the inductive signal required for the formation of the embryogenic tissue.
Keywords: Auxin, Ethylene, Hemoglobin, Somatic embryogenesis
1. Introduction
Plant hemoglobins (Hb) were discovered in the early 20th century [1], and are mainly involved in oxygen transport and nitric oxide (NO) scavenging. Hemoglobins can be classified into 3 main classes; class 1 includes non-symbiotic Hbs, class 2 symbiotic Hbs, and class 3 truncated Hbs. In Arabidopsis, two Hb genes have been characterized: Glb1 (class 1) and Glb2 (class 2) which are encoded by single genes [12], [51], [54]. Classes 1 and 2 are similar in structure to animal myoglobins and human globins [51], while class 3 globins are closer to truncated globins from prokaryotes [54]. Class 1 Hbs have high affinity for oxygen comparing to class 2 Hb, while class 3 Hbs have the weakest oxygen binding ability [12].
Hemoglobins are expressed in many organisms including bacteria, fungi and plants [26], [53], where they participate in many tasks, such as oxygen transport and NO scavenging [12], [15], [20]. Also, there are several recent studies showing that modulation of class 1 Hb levels may affect development and morphogenetic processes in plants [15], [14]. Plant Hbs are involved in dormancy breakage by modulating NO and ethylene that control abscisic acid (ABA) metabolism and signaling pathways [2]. Recently, it was reported that Hbs play also an effective role in somatic embryogenesis through auxin modulation [10].
Somatic embryogenesis is a process where somatic embryos, similar in morphology and structure to seed embryos, are produced by somatic cells in culture [60], [22], [5]. Somatic embryogenesis was first described almost 50 years ago by Steward et al. [48] who were able to produce viable embryos from isolated carrot cells. This system was recognized as a model to study the regulatory mechanisms underlying early events in plant embryogenesis [60], [5].
Auxin biosynthesis and distribution are critical for plant embryogenesis [7], [49], [3]. Quadruple mutations of YUCs, key enzymes in auxin biosynthesis, impair distribution of auxin resulting in severe developmental defects such as the absence of hypocotyls or root meristem. These studies indicate that depletion in auxin synthesis and/or transport compromises embryogenesis [7]. During the induction phase of Arabidopsis somatic embryogenesis, auxin polar transport, mediated by PIN1, is essential for the establishment of auxin gradients and the formation of somatic embryos [3].
Evidence indicates that non-symbiotic Hbs influence and modify the auxin signaling and subsequently somatic embryogenesis by modulating the endogenous NO levels [10]. Nitric oxide is tightly linked to many hormones such as auxin and ethylene [27], [59], [32], [34]. The two Arabidopsis Hbs: Glb1 and Glb2 have been shown to scavenge NO. Compared to Glb2, Glb1 is a more efficient NO scavenger, as shown by the high levels of NO accumulating in Glb1-suppressing tissue [18].
Ethylene regulates a number of developmental and physiological processes such as seed germination, root hair development, root nodulation, abscission, flower senescence and fruit ripening [31], [58], [57], [36]. Production of this plant growth regulator is tightly regulated by internal signals during development and in response to environmental stimuli triggered by biotic and abiotic stresses. Ethylene synthesis is also induced during the excision of plant explants utilized for the induction of somatic embryogenesis [23], [6], [28]. Hence, the role of this hormone has been examined in various culture systems. The regulation of ethylene biosynthesis and ethylene signaling influences the efficiency of de novo organogenesis. Ethylene was in fact shown to modulate the formation and development of somatic embryos [3], [35], [37]. The inhibition of ethylene production by AgNO3 or CoCl2 induces somatic embryogenesis in coffee [25] and Arabidopsis [3]. In line with these observations, Chen and Chang [7] observed that low levels of 1-aminoacyl cyclopropane-1-carboxylic acid (ACC), the precursor of ethylene, compromise the formation of embryos in Oncidium leaf cultures, whereas high levels elevated the frequency of somatic embryos.
Auxin and ethylene interaction is at the center of many developmental processes [11], [33], [29], [40], [46], [47], [13]. Exogenous auxin induced ethylene biosynthesis in root tips by activating several genes including those encoding 1-aminocyclopropane-1-carboxylate synthase (ACC synthase) [52]. Ethylene inhibited root growth by activating WEAK ETHYLENE INSENSITIVE 2, 7, I2 and I7 [47], genes encoding subunits of a rate-limiting enzyme in the biosynthesis of tryptophan, the precursor of auxin [42]. Genetic studies in Arabidopsis roots demonstrated that ethylene positively regulates auxin biosynthesis in the apex, and enhances the auxin response in the elongation zone resulting in changes in cell expansion [47], [44], [50].
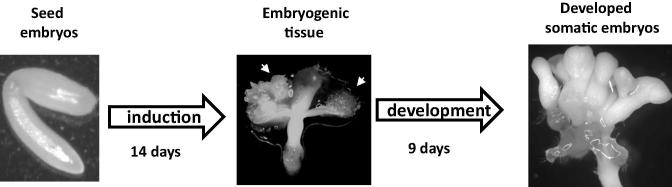
The present study tries to define the relationship between Glbs, NO and ethylene in the regulation of somatic embryogenesis by using the Arabidopsis system. Arabidopsis somatic embryos are generated by a two-step process: an induction step where embryogenic tissue forms from the zygotic embryos used as explants, and a developmental phase required for the development and growth of the embryos in a medium devoid of growth regulators (Fig. 1) [10], [4]. Our results confirm that suppression of the Arabidopsis class 1 Hb (Glb1) and class 2 Hb (Glb2) has different effects on the formation of somatic embryos, and places ethylene as a possible downstream component of the Glb-regulation of embryogenesis.
Figure 1.
The Arabidopsis somatic embryogenic system. Seed embryos are dissected and placed on an auxin-containing induction medium for 14 days. During this time the embryogenic tissue (arrows) forms from the cotyledons of the explants. Transfer of the tissue on an auxin-free development medium for 9 days results in the formation of fully mature somatic embryos.
2. Materials and methods
2.1. Plant materials
Two Arabidopsis (Columbia) lines suppressing Glb1 or Glb2 were donated by Dr. Hebelstrup [16]. They included the Glb1 RNAi line where Glb1 (class 1 Hb) was partially suppressed by RNAi mediated mechanisms, and the Glb2−/− knock out line where Glb2 (class 2 HB) was completely repressed. The pASA1::GUS reporter line (CS16701) and the ein2-1 and ein3-1 mutant lines were obtained from the Arabidopsis Biological Resource Center (ABRC). The following lines were received as gifts: the pYUC4::GUS line [8], the pPDF1.2::GUS line [24] and the pEBS::GUS [30].
2.2. Growth conditions and induction of somatic embryogenesis
Arabidopsis seeds were sterilized in a solution containing 70% ethanol and 0.5% Triton X-100 for 5 min, followed by a wash in 90% ethanol for 10 min. The sterilized seeds were germinated on half-strength MS medium [39] in a tissue culture cabinet (20–22 °C, 16 h light/8 h dark photoperiod). Following the procedure described by Bassuner et al. [4], developing siliques were harvested from the plants and the zygotic embryos were excised and used as explants for somatic embryogenesis. The explants were initially placed on the 2,4-dichlorophenoxyacetic acid (2,4-D) containing induction medium for 14 days and then transferred onto a development medium devoid of growth regulators. Fully developed embryos were counted 9 days after transfer onto the development medium (Fig. 1).
2.3. Chemical treatments
Modulations of NO levels were performed using the NO donor sodium nitroprusside (SNP) or the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide) (cPTIO), at the concentrations reported by Elhiti et al. [10]. The ethylene donor Ethephon (ETH) and the ethylene biosynthetic blocker O-(carboxymethyl)hydroxylamine hemihydrochloride (AOA) were also applied at the concentrations indicated in the text. Both compounds were dissolved in water. All treatments were performed during the 14 days on induction medium.
2.4. Total RNA isolation and quantitative real-time PCR analysis
TRIzol reagent (Invitrogen) was used to extract the total RNA from tissue harvested 7 days on induction medium. The extracted RNA was first treated with DNase I RNase-free (Promega) and these used for cDNA synthesis using the cDNA Reverse Transcription Kit (Applied Biosystems).
Quantitative real-time PCR was performed as described by Elhiti et al. [9]. All primers are listed in Table. 1. The relative expressions of the target genes were calculated with the 2−ΔΔCt method using UBQ10 (AT4G05320) as a reference [19].
Table 1.
Primers used for the quantitative (q)RT-PCR results.
| Gene | Sequence | Acc. No. | Name |
|---|---|---|---|
| UBQ10-F | AACTTTGGTGGTTTGTGTTTTGG | AT4G05320 | UBIQUITIN 10 |
| UBQ10-R | TCGACTTGTCATTAGAAAGAAAGAGATAA | AT4G05320 | UBIQUITIN 10 |
| ACCOX-F | CAAACTCTCTCGGTACACAATGA | AT2G19590 | ACC OXIDASE |
| ACCOX-R | GGATGAATGCGAGGCCAATA | AT2G19590 | ACC OXIDASE |
| ACCSYN-F | GCGCTTTGGCGAGTTATTATC | AT3G61510 | ACC SYNTHASE |
| ACCSYN-R | GGAGTGTGTCTTCGTCCATATT | AT3G61510 | ACC SYNTHASE |
| ERF1-F | CCGCTCCGTGAAGTTAGATAAT | AT3G23240 | ETHYLENE RESPONSE FACTOR 1 |
| ERF1-R | TCTTTCACCAAGTCCCACTATTT | AT3G23240 | ETHYLENE RESPONSE FACTOR 1 |
| ERF10-F | CGAGTTTGTCCTGACCAGTTT | AT1G03800 | ETHYLENE RESPONSE FACTOR 10 |
| ERF10-R | GGTTCCATTCGCAGCTTACA | AT1G03800 | ETHYLENE RESPONSE FACTOR 10 |
| ASA1-F | ACAAGGATGCTAACAAACGGCGTG | AT1G19920 | ATP SULFURYLASE ARABIDOPSIS 1 |
| ASA1-R | TCTGGCACTCACAGTGTTCGTCTT | AT1G19920 | ATP SULFURYLASE ARABIDOPSIS 1 |
| Yuc4-F | CTAACGGATGGAAAGGAGAGAAG | AT4G32540 | YUCCA 4 |
| Yuc4-R | GCGATCTTAACGGCGTCATA | AT4G32540 | YUCCA 4 |
| AMI1-F | ATCTCGTCGGTGAAGCCAGAGTTT | AT1G08980 | AMIDASE 1 |
| AMI1-R | CCGAGCAAAGTTGAAAGAGCCGTT | AT1G08980 | AMIDASE 1 |
| IGPS-F | TCTTGGAGGAGATCACATGG | AT2G04400 | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE |
| IGPS-R | GGAGGAGCATCCTCTACAGC | AT2G04400 | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE |
| PAI3-F | ACACAACACCTTTCAAACCCGTGG | AT1G29410 | PHOSPHORIBOSYLANTHRANILATE ISOMERASE 3 |
| PAI3-R | CAAAGCACTGCACTGAGCCATGAT | AT1G29410 | PHOSPHORIBOSYLANTHRANILATE ISOMERASE 3 |
| CYP79B2-F | ATGCTCGCGAGACTTCTTCAAGGT | AT4G39950 | CYTOCHROME P450, FAMILY 79, SUBFAMILY B, POLYPEPTIDE 2 |
| CYP79B2-R | AGATGCTCCGGCAATCTAAGGTCA | AT4G39950 | CYTOCHROME P450, FAMILY 79, SUBFAMILY B, POLYPEPTIDE 2 |
2.5. β-GUS assays
GUS staining was carried out exactly as described by Sieburth and Meyerowitz [45]. A minimum of 20 samples were used per treatment. A dissecting microscope equipped with a Leica DC500 digital camera was used for capturing the images.
2.6. IAA immunolocalization
IAA localization was performed exactly as described by Elhiti et al. [10]. Material at day 7 on induction medium was first pre-fixed in freshly prepared 4% aqueous 1-ethyl-3-(3-dimethyl-aminopropyl)-carbodiimide hydrochloride at 4 °C for 2 h, and then post-fixed in FAA (10% formalin, 5% acetic acid, and 50% ethanol) overnight at 4 °C. The fixed tissue was dehydrated in ethanol series, embedded in paraplast, sectioned (10 μM), and deparaffinized in xylene. The sections were incubated in blocking solution [1× phosphate-buffered saline (PBS) solution pH 7, 0.1% Tween 20, 1.5% glycine, and 5% bovine serum albumin (BSA)] at room temperature for 1 h. 150 μl of monoclonal primary IAA-antibodies (1 mg/ml, Sigma) diluted 1:200 in 10 mM PBS containing 0.8% BSA were applied to the sections and incubated in a high humidity chamber for 4 h at room temperature. The slides were washed first in 10 mM PBS containing 0.88 g/L NaCl, 0.1% Tween 20, and 0.8% BSA for 5 min, and then in 10 mM PBS with 0.8% BSA for 5 min in order to remove any excess of Tween 20. The slides were incubated in 200 μl secondary antibodies [anti-mouse IgG alkaline phosphatase conjugate (1 mg/ml), Promega, USA] overnight in a high humidity chamber, washed 2 times in 1× PBS containing 0.88 g/L NaCl, 0.1% Tween 20, and 0.8% BSA for 10 min, and then incubated in water for 15 min to remove the excess of secondary antibodies. Samples were stained using 250 μl Western blue (Promega) for 40 min.
2.7. Statistical analysis
Tukey’s post hoc test for multiple variances was used to compare differences among samples. All experiments were carried out using three biological replicates.
3. Results
3.1. Embryo number is differentially affected by manipulations in NO and ethylene levels
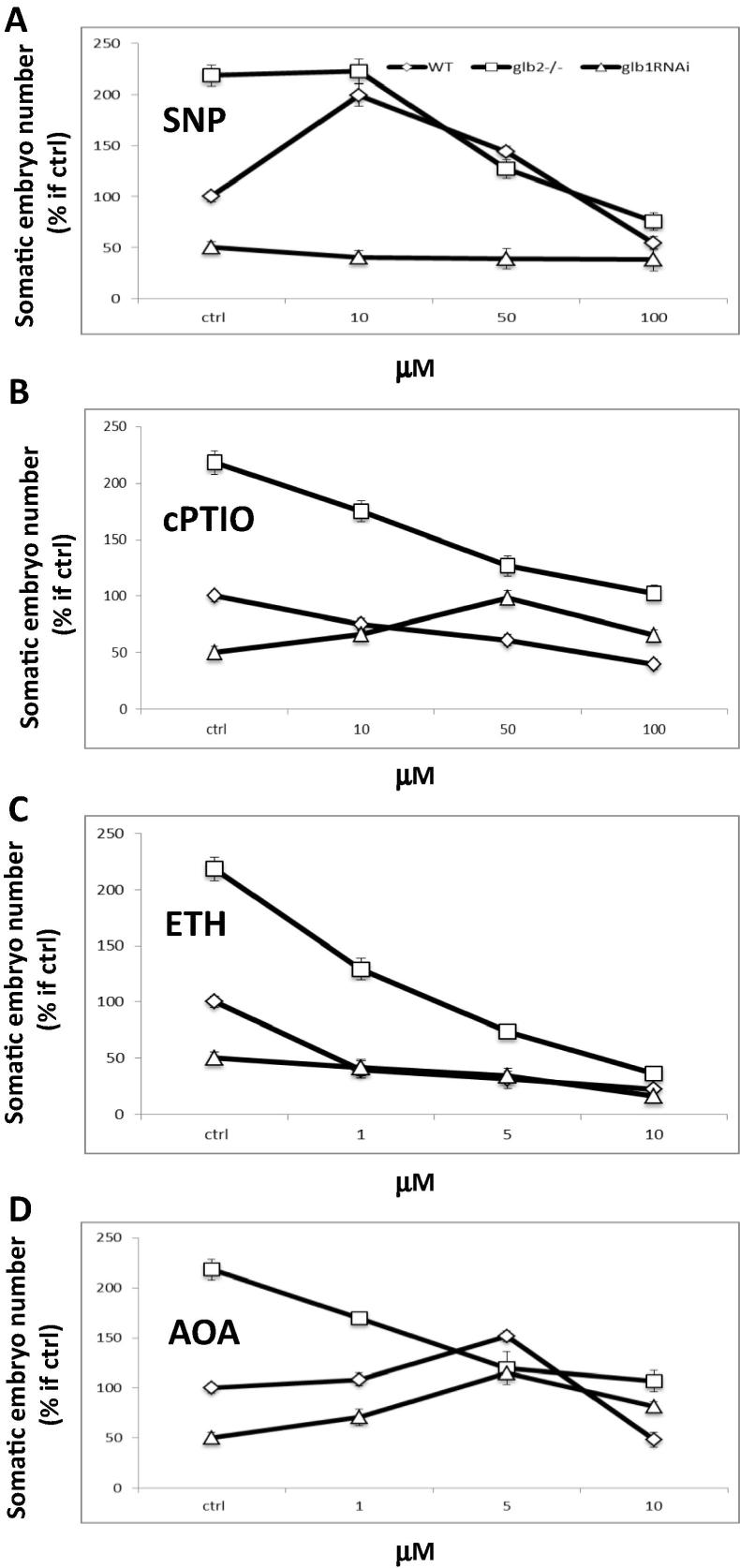
Arabidopsis somatic embryogenesis is a two-step process including an induction phase where 2,4-D is used to stimulate the formation of the embryogenic tissue (arrows in Fig. 1) from the cotyledons of the explant, and a development phase required for the growth of the embryos in an environment devoid of growth regulators. The number of WT embryos harvested after 9 days on development medium was increased by applications of the NO donor SNP at a concentration of 10 μM. Higher concentrations of the donor repressed embryogenesis in both the WT and the Glb2−/− mutant line. Inclusions of SNP had no effects on the embryogenic output of the Glb1 RNAi line which was consistently lower than that of the other lines (Fig. 2A).
Figure 2.
Number of somatic embryos [expressed as a percentage of WT control (ctrl)] collected from the WT, Glb1 RNAi and Glb2−/− mutant lines treated with increasing levels of NO donor sodium nitroprusside (SNP) (A), the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) (B), the ethylene releasing agent Ethephon (ETH) (C), and the ethylene biosynthetic inhibitor O-(carboxymethyl)hydroxylamine hemihydrochloride) (AOA) (D). Compounds were added in the induction medium. Values are means ± SE of at least three biological replicates.
Scavenging of NO by (2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide) (cPTIO) reduced the embryo number in both WT and Glb2−/− knock out line and this effect was concentration dependent. The embryo number in the Glb1 RNAi line increased following applications of cPTIO reaching a maximum value at a concentration of 50 μM, before declining (Fig. 2B).
Inclusions of the ethylene donor Ethephon (ETH) repressed embryogenesis in the WT and Glb2−/− mutant line, and this repression increased with elevated levels of the compound. Ethephon had no significant effects on the number of embryos produced by the Glb1 RNAi line (Fig. 2C).
The number of embryos in the Glb2−/− mutant line declined significantly following applications of the ethylene biosynthetic inhibitor O-(carboxymethyl)hydroxylamine hemihydrochloride (AOA). This was in contrast to the WT and Glb1 RNAi lines, which showed an increase in embryogenesis up to a concentration of 5 μM (AOA). Higher levels of the inhibitor decreased the embryo number in these two cell lines (Fig. 2D).
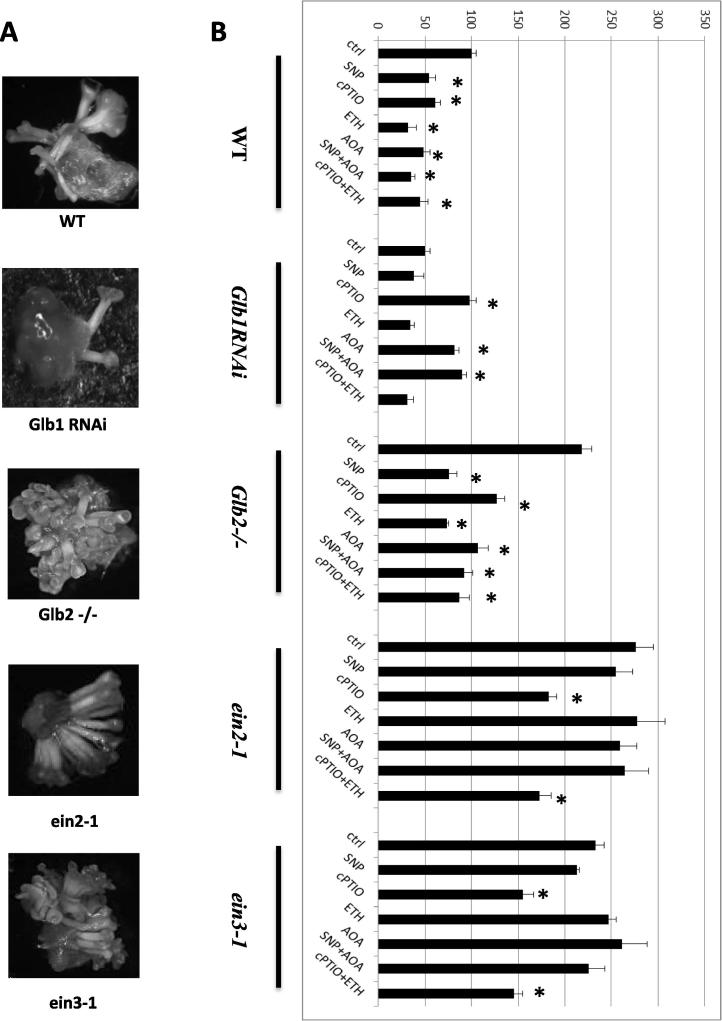
To further confirm the involvement of ethylene in somatic embryogenesis two ethylene insensitive lines: ethylene-insensitive 2 (ein2-1) and 3 (ein3-1) were used. Compared to WT, embryo number was significantly increased in these lines, and an increase in NO levels by SNP had no significant effect (Fig. 3).
Figure 3.
(A) Micrographs showing embryo production after 9 days on development medium in the WT, Glb1 RNAi, Glb2−/− mutant, and the two ethylene insensitive mutant lines ein2, ein3. (B) Number of embryos expressed as a percentage of WT control (ctrl) in the different lines subjected to different treatments in the induction medium: SNP (100 μM), ETH (10 μM), cPTIO (50 μM), or AOA (10 μM). Values are means ± SE of at least three biological replicates. ∗ indicates statistically significant differences (P < 0.005) from the WT (ctrl) value.
3.2. Nitric oxide (NO) alters the expression of ethylene biosynthetic genes
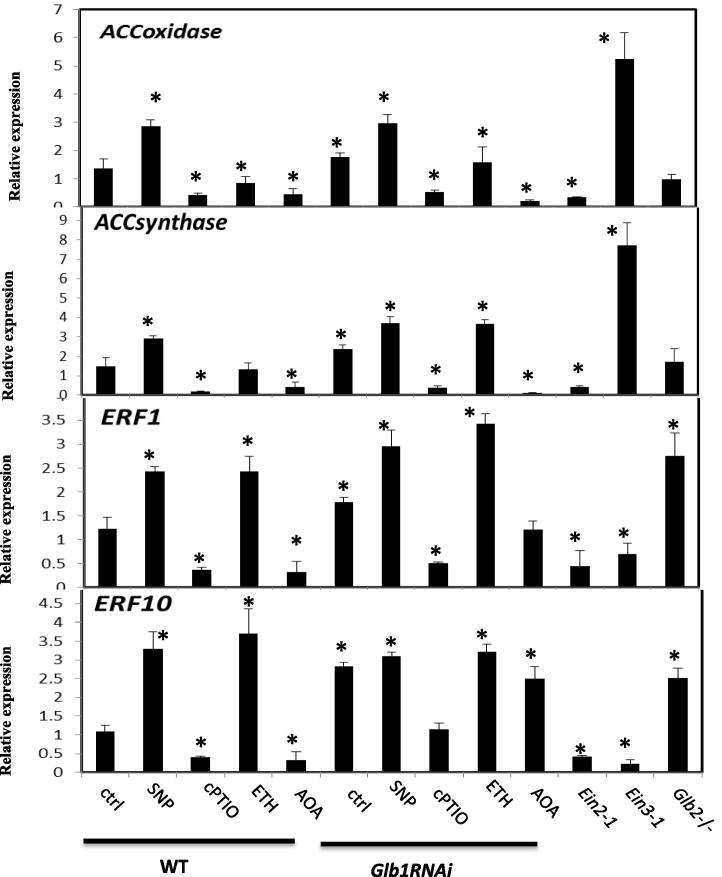
Ethylene synthesis was analyzed as a transcriptional level by measuring the expression of the two ethylene biosynthetic genes: 1-amino-cyclopropane-1-carboxylate synthase (ACC synthase) which synthesizes the ethylene precursor 1-aminoacyl cyclopropane-1-carboxylic acid (ACC) from S-adenosyl-l-methionine, and 1-amino-cyclopropane-1-carboxylate oxidase (ACC oxidase) which oxidizes ACC to form ethylene. At day 7 on the induction medium, the expression of ACC oxidase and ACO synthase was increased in NO enriched environments, i.e. (SNP-treated WT line and Glb1 RNAi line) (Fig. 4). Consistent with these results, a reduction in NO level by cPTIO in both WT and Glb1 RNAi line reduced the expression of both genes (Fig. 4). The changes in expression levels of the ethylene responsive factor 1 (ERF1) and 10 (ERF10) also suggest that NO and ethylene affect the overall mechanisms of ethylene response (Fig. 4).
Figure 4.
Expression level of the two ethylene biosynthetic aminocyclopropane-1-carboxylic acid oxidase (ACC oxidase) and aminocyclopropane-1-carboxylic acid synthase (ACC synthase), and the ethylene responsive factor 1 (ERF1), 10 (ERF10). Treatments were conducted as described in Fig. 3. Values are means ± SE of at least three biological replicates. ∗ indicates statistically significant differences (P < 0.005) from the WT (ctrl) value set at 1.
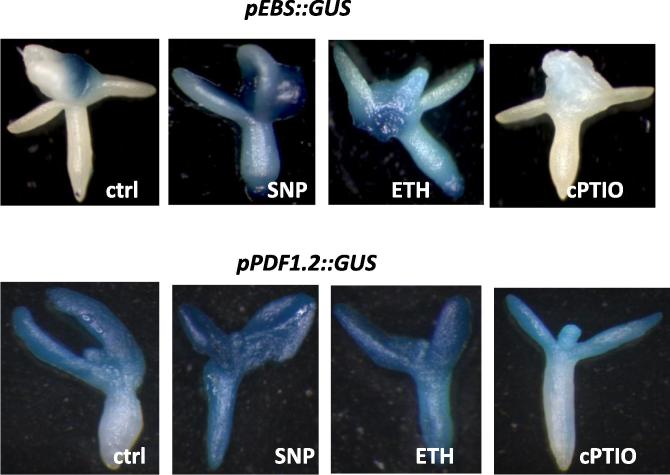
The effect of NO on ethylene level was further confirmed using two reporter lines (pEBS::GUS) (an ethylene reporter construct in which the GUS reporter gene is driven by a synthetic EIN3-responsive promoter) and (pPDF1.2::GUS) (in which the GUS reporter gene is driven by ethylene responsive promoter PDF1.2) (Fig. 5). In both lines the GUS signal increased with high levels of NO (by SNP) and ethylene (by ETH), while it decreased following depletion of NO (by cPTIO).
Figure 5.
GUS localization in the pEBS::GUS and pPDF1.2::GUS reported lines. Embryos were cultured for 7 days on induction medium before being harvested and stained for GUS. Concentrations of the treatments were identical to those reported in Fig. 3.
3.3. Ethylene response affects the expression of IAA biosynthesis genes
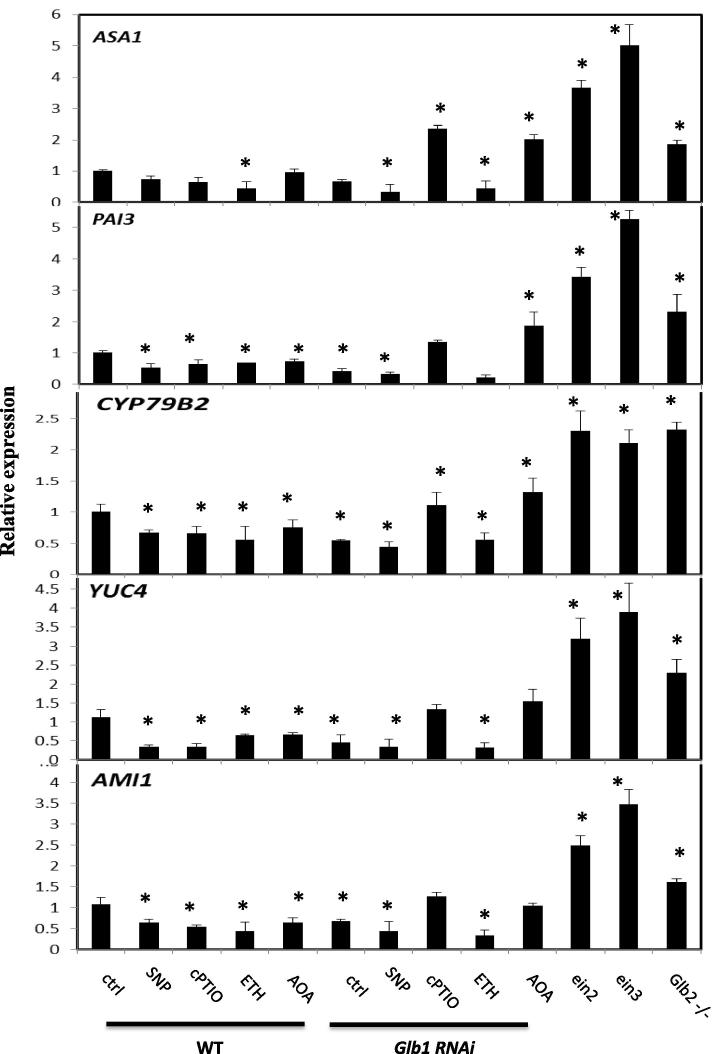
Auxin is the main inductive signal promoting the re-differentiation of the somatic cells and formation of embryogenic tissues [43]. Previous studies showed that suppression of Glb2 increased IAA production at the site of embryo formation [10]. To further expand whether IAA biosynthesis was regulated through an ethylene response, the expression of several key genes involved in IAA biosynthesis was measured at day 7 on induction medium. The expression of anthranilate synthase α–subunit (ASA1), anthranilate isomerase (PAI3), cytochrome P450 CYP79B2 (CYP79B2), YUCCA4 (YUC4), and amidase1 (AMI1) increased in the ethylene mutants ein2 and 3 (Fig. 6). Their expression was generally lower in the Glb1 RNAi line compared to the WT and the Glb2−/− mutant lines. Furthermore, in the Glb1 RNAi line treatments depleting NO (by cPTIO) and ethylene (by AOA) increased the expression level of all the genes analyzed.
Figure 6.
Expression level by quantitative (q) RT-PCR of the auxin biosynthetic genes anthranilate synthase α-subunit (ASA1), anthranilate isomerase (PAI3), amidase1 (AMI1), cytochrome P450 CYP79B2 (CYP79B2) and YUCCA4 (YUC4). Values are means ± SE of at least three biological replicates and normalized to WT (control, ctrl) set at 1. ∗ indicates statistically significant differences (P < 0.005) from the WT (control, ctrl) value.
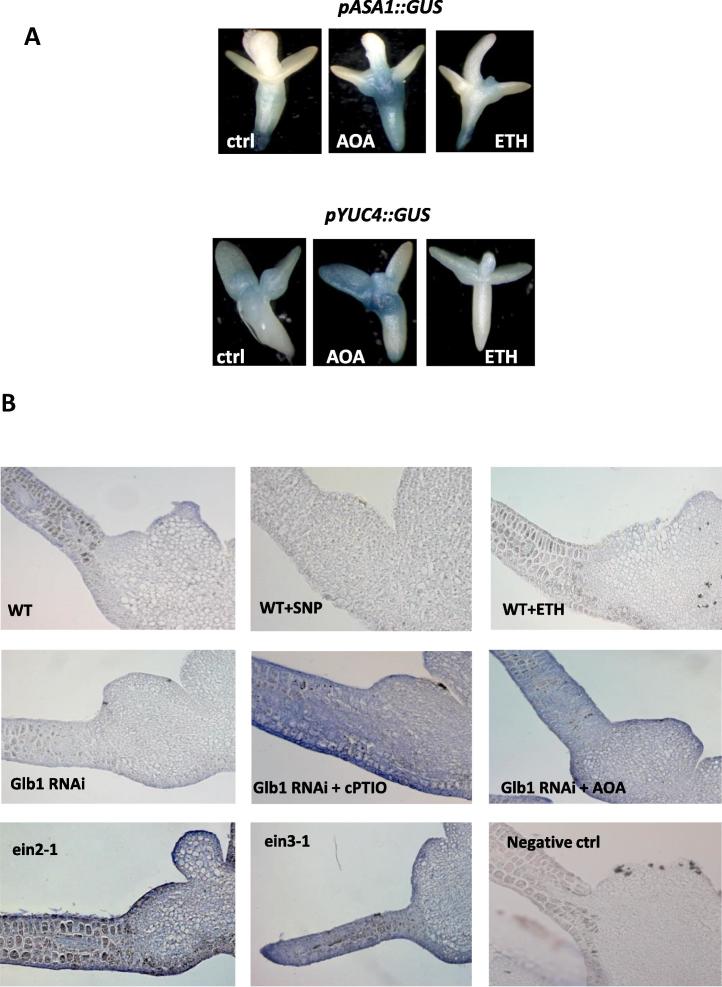
These expression patterns were confirmed using 2 different promoter lines (pASA1::GUS) and (pYUC4::GUS). In both lines GUS signal was increased when ethylene level was depleted with AOA, and decreased when ethylene level was augmented with ETH (Fig. 7A).
Figure 7.
(A) GUS localization in the pASA1::GUS and pYUC4::GUS reported lines. Embryos were cultured for 7 days on induction medium before being harvested and stained for GUS. Concentrations of the treatments were identical to those reported in Fig. 3. (B) Immunolocalization of IAA along the cotyledons of the zygotic explants. Tissue was collected at 7 days on induction medium. Concentrations of the treatments were identical to those reported in Fig. 3.
Immunolocalization of the auxin IAA in the embryogenic tissue arising from the cotyledons of the zygotic explants was also performed (Fig. 7B). Wild type (WT) tissue treated with SNP or ETH, as well as Glb1 RNAi tissue had low IAA signals. A strong IAA signal was detected in tissues with depleted NO level (Glb1 RNAi tissue treated with cPTIO) or ethylene (Glb1 RNAi tissue treated with AOA), and in the ethylene mutants (ein2-1 and ein3-1).
4. Discussion
Nitric oxide (NO) plays a vital role in different developmental processes ranging from leaf expansion, root growth and senescence [55], [56]. Besides being involved in wounding and pathogen interactions [38], NO has proven to contribute to ethylene biosynthesis [17]. However, some studies found that NO represses ethylene biosynthesis [41]. The level of cellular NO is mediated by several factors, including the presence of Hbs, which are effective regulators of NO. As reviewed by Hill [18], both the Arabidopsis Glb1 and Glb2 are able to reduce NO levels, although Glb1 is a stronger scavenger due to its higher affinity to NO. Therefore, tissue suppressing Glb1 accumulates more NO than tissue suppressing Glb2 [18]. Small differences in NO levels might have divergent effects on embryogenesis, with high levels decreasing embryo production and intermediate levels improving the process.
The relationship between NO and ethylene during somatic embryogenesis has not been studied before. Here we show that excessive NO levels, produced pharmacologically using SNP or genetically using the Glb1 RNAi line, reduce somatic embryo number, possibly by elevating the level of ethylene, and this response might be integrated in the Hb-regulation of embryogenesis.
Effects of exogenous application of ethylene during somatic embryogenesis are contradictory. While ethylene improves production of embryogenic callus in spinach [21], it represses the embryogenic competence in Arabidopsis [3]. Applications of the ethylene blocker (O-(carboxymethyl) hydroxylamine hemihydrochloride) (AOA) enhance the embryo production in the Glb1 down regulating line (Glb1 RNAi line) (Fig. 3) characterized by the highest NO level [18], but not in the Glb2−/− mutant line which has intermediate NO levels between the WT and the Glb1 RNAi values. These effects were independent from increasing the levels of NO by SNP, thus suggesting that NO is upstream of the ethylene response. An experimental increase of ethylene by ETH repressed embryogenesis in both the Glb1 down regulating line and the Glb2−/− mutant line (Fig. 3). Compared to WT tissue, embryogenesis was significantly increased in lines insensitive to ethylene: ein2-1 and ein3-1. Embryo production in these lines remained higher than WT even following manipulation in NO (by SNP and cPTIO) (Fig. 3).
In a high NO environment, such is the case of SNP-treated tissue or tissue suppressing Glb1, the expression level of the two ethylene biosynthetic genes ACC oxidase and ACC synthase is up-regulated (Fig. 4). Furthermore, this up-regulation can be reversed if NO level is experimentally reduced by cPTIO. This trend was observed in both WT and GLB1 line. The expression of both genes was reduced in the ein2-1 line, but increased in the ein3-1 line. No significant differences in the expression levels of both genes were measured in the Glb2−/− mutant line (Fig. 4), which accumulated less NO compared to the Glb1 RNAi line. Generally similar expression profile patterns were also observed for the ethylene responsive factor 1 (ERF1) and 10 (ERF10), thus suggesting that NO and ethylene affect the overall mechanisms of ethylene response (Fig. 4).
Compared to WT, the expression of these two genes is not altered in the Glb2−/− mutant line, thus suggesting that both ACC oxidase and ACC synthase are only responsive to high levels of NO. Activation of the two biosynthetic genes most likely results in increased ethylene synthesis, which in Arabidopsis compromises somatic embryogenesis. This is demonstrated both pharmacologically, by the fact that application of the ethylene releasing agent ETH reduces the number of embryos in the WT line (Fig. 3), and genetically, using the two ethylene insensitive mutants ein2-1 and ein3-1. Production of somatic embryos in these two mutants is highly favored (Fig. 3). Furthermore, ethylene appears to be downstream of NO as an increase in NO production by SNP in the ein2-1 and ein3-1 has no effects on the embryogenic output of the lines. This notion is also supported by the observation that the two ethylene responsive promoters pEBS and pDF1.2 are activated in a high NO environment (SNP) and repressed in a NO depleted environment (cPTIO) (Fig. 5).
The elevated levels of ethylene triggered by excessive NO might be linked to the repression of auxin, the inductive signal triggering the de-differentiation of the cotyledon cells and the production of embryogenic tissues [43]. The expression of all IAA biosynthetic genes investigated: ASA1, PAI3, AMI1, CYP79B2, and YUC4 was suppressed when ethylene level was increased by ETH but increased in the ethylene insensitive mutants ein2-1 and ein3-1 (Fig. 6). The effect of ethylene was particularly apparent on ASA1 and YUC4, the expression of which was induced when ethylene biosynthesis was blocked by AOA (Fig. 7A). Immunolocalization studies of IAA further confirm that IAA localizes within the cotyledons of the zygotic explants producing the embryogenesis tissue, and that this localization is affected by levels of NO and ethylene consistent with a model in which NO and ethylene regulate IAA accumulation through ethylene (Fig. 7B).
Collectively, these data suggest that ethylene is integrated in the Glb1 and NO regulation of in vitro embryogenesis. In the proposed model (Fig. 8), the excessive level of NO generated in the Glb1 down regulating line (or by pharmacological treatments) induces the expression level of the ethylene biosynthetic genes ACC synthase and ACC oxidase, and possibly increases the endogenous ethylene content. Accumulation of ethylene down regulates several auxin biosynthetic genes leading to the depletion of endogenous auxin, the inductive signal for the formation of the embryogenic tissues. Depletion of IAA reduces somatic embryo production in the line suppressing Glb1. These events compromising the embryogenic output, do not occur in the Glb2−/− mutant line as Glb2 is not an effective NO scavenger as Glb1. As a result, the level of NO accumulated as a result of Glb2 suppression is lower and possibly not sufficient to activate ethylene biosynthesis (Fig. 8).
Figure 8.
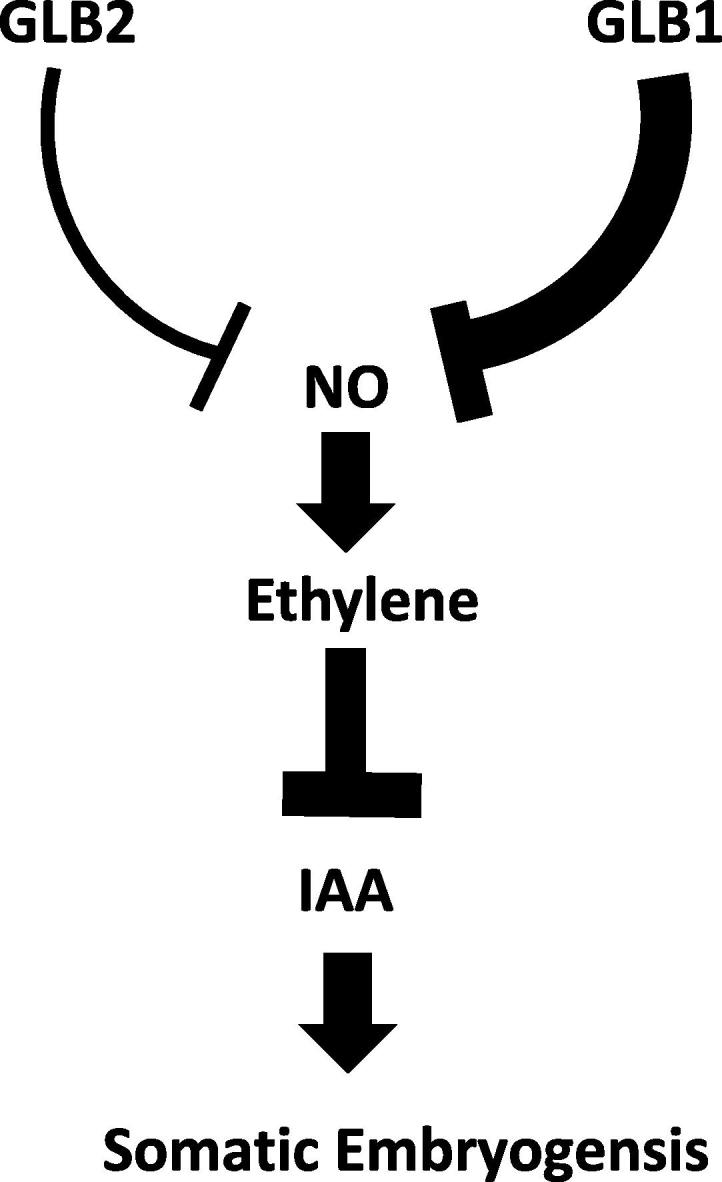
Proposed model regulating Arabidopsis somatic embryogenesis. Glb1 is a more effective NO scavenger of Glb2, therefore, their suppression would result in a differential accumulation of NO. The Glb1 RNAi line accumulates more NO than the Glb2−/− line. Accumulation of NO activates the transcription of the two ethylene biosynthetic genes (ACC synthase and ACC oxidase) and would possibly increase ethylene level. Ethylene transcriptionally down-regulates many IAA biosynthetic genes and reduces the IAA signal on the zygotic explants. Since IAA is the inductive signal for somatic embryogenesis, a depletion in IAA reduces the ability of the Glb1 RNAi line to produce embryos.
Future studies will be undertaken to investigate the precise mode of action of ethylene response in the modulation of auxin biosynthesis, and possibly in the identification of transcription factors directly influencing the expression of genes encoding auxin biosynthetic enzymes.
Acknowledgments
This work was supported by NSERC Discovery Grants to CS and by a Fellowship to MM granted by the Egypt’s Ministry of Higher Education and Research. The authors thank Doug Durnin for technical support and editing of the manuscript. The following people are also acknowledged for providing seed material. Dr. Kim Hebelstrup, University of Aarhus, Denmark (Glb2−/− line; Glb1 RNAi), Prof. Eva Sundberg, Department of Plant Biology and Forest Genetics, Swedish University of Agriculture Science (pYUC4::GUS), Prof. Corné M.J. Pieterse, Department of Plant Microbe Interactions, Utrecht University (pPDF1.2::GUS), Prof. Jose M. Alonsoof, Department of Plant and Microbial Biology, North Carolina State University (pEBS::GUS), Prof. Brad Mark Binder, Biochemistry and Cellular and Molecular Biology Department, University of Tennessee, Knoxville, USA (ein2-1; ein3-1), the Arabidopsis Biological Resource Center (ABRC) (pASA1::GUS).
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Appleby C.A. Sci. Prog. 1992;76:365–398. [Google Scholar]
- 2.Arc E., Galland M., Godin B., Cueff G., Rajjou L. Front. Plant Sci. 2013;4:346. doi: 10.3389/fpls.2013.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai B., Su Y.H., Yuan J., Zhan X.S. Mol. Plant. 2012;6(4):1247–1260. doi: 10.1093/mp/sss154. [DOI] [PubMed] [Google Scholar]
- 4.Bassuner B.M., Lam R., Lukowitz W., Yeung E.C. Plant Cell Rep. 2007;26:1–11. doi: 10.1007/s00299-006-0207-5. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum K.D., Alvarado A.S. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleecker B., Kende H. Annu. Rev. Cell Dev. Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Chen J.T., Chang W.C. Biol. Plant. 2003;46:455–458. [Google Scholar]
- 8.Eklund D.M., Ståldal V., Valsecchi I., Cierlik I., Eriksson C., Hiratsu K. Plant Cell. 2010;22:349–363. doi: 10.1105/tpc.108.064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhiti M., Tahir M., Gulden R.H., Khamiss K., Stasolla C. J. Exp. Bot. 2010;61:4069–4085. doi: 10.1093/jxb/erq222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elhiti M., Hebelstrup K.H., Wang A., Li C., Cui Y., Hill R.D., Stasolla C. Plant J. 2013;74:946–958. doi: 10.1111/tpj.12181. [DOI] [PubMed] [Google Scholar]
- 11.Gazzarrini S., McCourt P. Ann. Bot. 2003;91:605–612. doi: 10.1093/aob/mcg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta K.J., Hebelstrup K.H., Mur L., Igamberdiev A.U. FEBS Lett. 2011;585:3843–3849. doi: 10.1016/j.febslet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 13.He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebelstrup K.H., Jensen E.O. Planta. 2008;227:917–927. doi: 10.1007/s00425-007-0667-z. [DOI] [PubMed] [Google Scholar]
- 15.Hebelstrup K.H., Hunt P., Dennis E., Jensen S.B., Jensen E.O. Physiol. Plant. 2006;127:157–166. [Google Scholar]
- 16.Hebelstrup K.H., Ostergaard-Ensen E., Hill R.D. Methods Enzymol. 2008;437:595–604. doi: 10.1016/S0076-6879(07)37030-4. [DOI] [PubMed] [Google Scholar]
- 17.Hebelstrup K.H., Van Zanten M., Mandon J., Voesenek L.A., Harren F.J., Cristescu S.M., Moller I.M., Mur L.A. J. Exp. Bot. 2012;63:5581–5591. doi: 10.1093/jxb/ers210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill R.D. AoB Plants. 2012;10(1093):1–13. doi: 10.1093/aobpla/pls004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong M., Bahn C., Lyu A., Jung S., Ahn H. Plant Cell Physiol. 2010;51:1606–1694. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]
- 20.Igamberdiev A.U., Stoimenova M., Serege’lyes C., Hill R.D. Planta. 2006;223:1041–1046. doi: 10.1007/s00425-005-0145-4. [DOI] [PubMed] [Google Scholar]
- 21.Ishizaki T., Komai F., Masuda K., Megumi C. J. Am. Soc. Hort. Sci. 2000;125:21–24. [Google Scholar]
- 22.Jiménez V.M., Bangerth F. Physiol. Plant. 2001;111:389–395. doi: 10.1034/j.1399-3054.2001.1110317.x. [DOI] [PubMed] [Google Scholar]
- 23.Johnson R., Ecker R. Annu. Rev. Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- 24.Koornneef A., Leon-Reyes A., Ritsema T., Verhage A., Den Otter F.C., Van Loon L.C., Pieterse C.M.J. Plant Physiol. 2008;147:1358–1368. doi: 10.1104/pp.108.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar V., Ramakrishna A., Ravishankar G.A. In Vitro Cell. Dev. Biol. Plant. 2007;43:602–607. [Google Scholar]
- 26.Kundu S., Premer S.A., Hoy J.A., Trent J.T., Hargrove M.S. Biophys. J. 2003;84:3931–3940. doi: 10.1016/S0006-3495(03)75121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamattina L., Garcia-Mata C., Graziano M., Pagnussat G. Annu. Rev. Plant Biol. 2003;54:109–136. doi: 10.1146/annurev.arplant.54.031902.134752. [DOI] [PubMed] [Google Scholar]
- 28.Li H., Guo H. J. Plant Growth. 2007;26:106–117. [Google Scholar]
- 29.Li H., Johnson P., Stepanova A., Alonso M., Ecker R. Dev. Cell. 2004;7:193–204. doi: 10.1016/j.devcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Li G., Li B., Dong G., Feng X., Kronzucker H., Shi W. J. Exp. Bot. 2013;10(1093):1–13. doi: 10.1093/jxb/ert019. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman N. Annu. Rev. Plant Physiol. 1979;30:533–591. [Google Scholar]
- 32.Liu Y., Yu X., Cui Y., Sun H., Sun N., Tang C. Cell Res. 2007;17:638–649. doi: 10.1038/cr.2007.34. [DOI] [PubMed] [Google Scholar]
- 33.Lorenzo O., Piqueras R., Sánchez-Serrano J., Solano R. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manjunath L., Ramadug C., Majil M., Williams S., Irey M., Lee F. Plant Dis. 2010;94(6):781. doi: 10.1094/PDIS-94-6-0781A. [DOI] [PubMed] [Google Scholar]
- 35.Mantiri F.R., Kurdyukov S., Lohar D.P., Sharopova N., Saeed N.A., Wang X.D., Vandenbosch K.A., Rose R.J. Plant Physiol. 2008;146:1622–1636. doi: 10.1104/pp.107.110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattoo K., Suttle C. CRC Press; Boca Raton, Florida: 1991. The Plant Hormone Ethylene. (337 p. 0-8493-4566-9) [Google Scholar]
- 37.Mauri P.V., Manzanera J.A. Acta Physiol. Plant. 2011;33:717–723. [Google Scholar]
- 38.Mur L., Prats E., Pierre S., Hall M., Hebelstrup K. Front. Plant Sci. 2013;4:1–7. doi: 10.3389/fpls.2013.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murashige T., Skoog F. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 40.Nemhauser L., Mockle C., Chory J. PLoS Biol. 2004;2:1460–1471. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parani M., Rudrabhatla S., Myers R., Weirich H., Smith B., Leaman D.W. Plant Biotechnol. J. 2004;2:359–366. doi: 10.1111/j.1467-7652.2004.00085.x. [DOI] [PubMed] [Google Scholar]
- 42.Radwanski R., Last L. Plant Cell. 1995;7:921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raghavan V. Am. J. Bot. 2004;91:1743–1756. doi: 10.3732/ajb.91.11.1743. [DOI] [PubMed] [Google Scholar]
- 44.Růžička K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. Plant Cell. 2007;19:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sieburth L.E., Meyerowitz E.M. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stepanova N., Hoyt M., Hamilton A., Alonso M. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepanova N., Robertson-Hoyt J., Yun J., Benavente M., Xie Y., Doležal K., Schlereth A., Jürgens G., Alonso M. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 48.Steward F.C., Marion O.M., Mears K. Am. J. Bot. 1958;45:705–708. [Google Scholar]
- 49.Su H., Zhao Y., Liu B., Zhang L., O’Neill D., Zhang S. Plant J. 2009;59:448–460. doi: 10.1111/j.1365-313X.2009.03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swarup R., Perry P., Hagenbeek D., Straeten D., Beemster S., Sandberg G., Bhalerao R., Ljung K., Bennett J. Plant Cell. 2007;19:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trevaskis B., Watts R.A., Andersson C., Llewellyn D., Hargrove M.S., Olson J.S., Dennis E.S., Peacock W.J. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12230–12234. doi: 10.1073/pnas.94.22.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuchisaka A., Theologis A. Plant Physiol. 2004;136:2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinogradov S.N., Hoogewijs D., Bailly X., Arredondo-Peter R., Gough J., Dewilde S., Moens L., Vanfleteren J.R. BMC Evol. Biol. 2006;6:31. doi: 10.1186/1471-2148-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watts R.A., Hunt P.W., Hvitved A.N., Hargrove M.S., Peacock W.J., Dennis E.S. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10119–10124. doi: 10.1073/pnas.191349198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wendehenne D., Pugin A., Klessig F., Durner J. Trends Plant Sci. 2001;6:177–183. doi: 10.1016/s1360-1385(01)01893-3. [DOI] [PubMed] [Google Scholar]
- 56.Yadav S., David A., Baluska F., Bhatla S.C. Plant Signal Behav. 2013;8(3):e231961–e231968. doi: 10.4161/psb.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang F. HortScience. 1985;20(1):41–45. [Google Scholar]
- 58.Yang F., Hoffman E. Annu. Rev. Plant Physiol. 1984;35:155–189. [Google Scholar]
- 59.Zhu C., Gan L., Shen Z., Xia K. J. Exp. Bot. 2006;57:1299–1308. doi: 10.1093/jxb/erj103. [DOI] [PubMed] [Google Scholar]
- 60.Zimmerman J.L. Plant Cell. 1993;5(10):1411–1423. doi: 10.1105/tpc.5.10.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]