Abstract
Genetic variability and identification of some molecular markers were studied in twenty promising lines of wheat using agronomic traits, ISSR (inter simple sequences repeats) and RAPD (random amplified polymorphic DNA) markers. Significant variation was evidenced in all agronomic traits. The lines proved to be superior to the check cultivar Sahel1 in yield and its component traits. Lines L2, L7 and L8 were the best in most yield component traits in both seasons. Moreover, Lines L2, L4, L5, L7 and L8 showed drought tolerance by which they displayed high performance in agronomic traits as well as a low drought susceptibility index. The percentage of polymorphism was 39.3% and 53.2% for ISSRs and RAPDs, respectively. UBC-881 belonged to penta-nucleotide repeat sequences (GGGTG) that produced the highest level of polymorphism, while UBC-846 belonged to di-nucleotide repeat sequences (CA) that produced the lowest level of polymorphism. Genetic similarities among wheat lines based on ISSR and RAPD markers ranged from 0.81 to 1.00 and from 0.86 to 0.98, respectively. There was a low average of PIC (polymorphism information content) values which were 0.10 (ISSR) and 0.15 (RAPD). The RAPD technique exhibited a higher marker index (MI = 0.69) compared to ISSR (MI = 0.43). There was insignificant correlation between ISSR and RAPD data (0.168, p > 0.05). There were two markers (UBC-881450bp and OPF-10540bp), on each of which two traits regressed significantly. The associated markers each explained a maximum regression of 18.92–34.95% of the total available variation for individual associated traits.
Keywords: Drought, Genetic diversity, Single marker analysis, Triticum aestivum L, Yield traits
1. Introduction
Bread wheat (Triticum aestivum L.) is considered to be one of the most important cereal crops in the world as well as in Egypt. Increasing wheat production is an important national goal in order to decrease the great gap between production and human consumption especially under the yearly increase in the population with a greater rate than production. Expansion of the wheat area in reclaimed lands faces the problem of drought stress conditions because of the dominance of sandy soil and limited moisture from rainfall. Therefore, great efforts have been directed by plant breeder toward developing suitable wheat genotypes which have high production and tolerance for drought conditions.
Drought tolerance is defined as the ability of a plant to live, grow and reproduce satisfactorily with limited water supply or under periodic conditions of water deficit [59]. Crop plants should not only have the ability to survive under drought but also the ability to produce a harvestable yield. Research into the molecular aspects of drought tolerance has tended to focus on plant survival at the expense of yield. However, severe water deficits are rare in viable agriculture, and asking how crops respond to or survive extreme drought is unlikely to have much practical impact [40]. Drought tolerance is a quantitative trait, with complex phenotype and genetic control [32]. Understanding the genetic basis of drought tolerance in crop plants is a prerequisite for developing superior genotypes through conventional breeding. Given the complexity of the genetic control of drought tolerance (multigenic, low-heritability, and high G × E interactions), marker assisted selection (MAS) has not contributed significantly to cultivar improvement for dry environments and breeding has relied on direct phenotypic selection.
Over the years, the methods for detecting and assessing genetic diversity have extended from analysis of discrete morphological to biochemical and molecular traits. Several PCR-based markers were developed and applied to assess the genetic variation among populations and genetic resources. These marker systems are different in technical principle, type of inheritance, reproducibility, amount of polymorphism and in their costs [42]. Molecular markers provide a direct measure and go beyond indirect diversity measures based on agronomic traits or geographic origin. Applying molecular markers and recognition of polymorphic nucleotide sequences dispersed throughout the genome have provided new possibility for evaluating genetic diversity and determining of inter and intra-species genetic relationships [22], [25]. The right choice of a technique, proper sampling procedure and judicious interpretation, these laboratory methods can provide reliable and accurate results for variety identification and genetic purity testing in a considerably short period of time [51]. Efforts have been made to characterize wheat genotypes with molecular markers [23]. Among the available methods, ISSR and RAPD markers have been widely used for diversity studies, DNA fingerprinting, map construction and linkage analysis among wheat genotypes. Marker association studies have been conducted, which not only allow mapping of genes/QTLs (QTLs, for quantitative trait loci) with higher levels of confidence, but also allow detection of genes/QTLs, which would otherwise escape detection in linkage-based studies [9], [38]. Moreover, molecular markers are widely used to detect the location of drought-induced genes. Because molecular markers are not subject to environmental influence they are considered superior to morphological markers [31]. Different molecular markers are currently available for genome mapping and tagging of different traits which is useful for marker assisted breeding technique in wheat in stress conditions [5]. The present study aimed to: (1) determine the performance and behavior of 20 bread wheat lines for yield traits under normal and drought stress conditions, (2) evaluate the usefulness of molecular markers viz. ISSRs and RAPDs, in assessing and analyzing the nature and the extent of genetic diversity among tested inbred lines and (3) identify molecular markers strictly associated with important agronomic traits.
2. Materials and methods
2.1. Plant materials
Seeds of twenty Egyptian lines of bread wheat (T. aestivum L.) in the F10 generation were selected and developed from some crosses (Table 1), at experimental Farm of the Faculty of Agriculture, Sohag University, Egypt. In addition Sahel1 was used as a check cultivar.
Table 1.
Brief description of the origin of lines (L1–L20) of bread wheat used in phenotypic and molecular marker analyses.
| Lines | Codes | Parents | Lines | Codes | Parents |
|---|---|---|---|---|---|
| L1 | F5-sp69 | (Sakha-69 × C.B-6) | L11 | F5-sp4 | (Gemaza-1 × C.B-15) |
| L2 | F5-Dr56 | (Sakha-8 × C.B-8) | L12 | DI-H7 | (Sakha-206 × Gemaza-3) |
| L3 | F5-Dr12 | (Gemaza-3 × Sakha-69) | L13 | DI-H25 | (Gemaza-3 × Sakha-8) |
| L4 | DII-H12 | (Giz-163 × Sedes-8) | L14 | DII-H8 | (Giza-163 × Giza-167) |
| L5 | DI-H19 | (Giz-164 × Sakha-8) | L15 | DI-H17 | (Giza-155 × C.B-C6) |
| L6 | F5-Dr33 | (Gemaza-3 × C.B-8) | L16 | DI-H13 | (Gemaza-3 × Sedes-1) |
| L7 | DI-H14 | (Giza-155 × Giza-164) | L17 | DI-H19 | (Sakha-8 × Giza-164) |
| L8 | DII-H20 | (Giz-163 × Sedes-6) | L18 | DII-H6 | (Sakha-610 × C.B139) |
| L9 | DI-H28 | (C.B-261 × Gemaza-3) | L19 | DI-H22 | (Gemaza-3 × Giza-164) |
| L10 | DII-H28 | (Sedes-8 × Giza-167) | L20 | F5-Dr73 | (C.B-8 × Sakha-69) |
2.2. Phenotypic evaluation
Split plot design with four replicates was used in this study. The main plot was occupied with the irrigation treatments. Genotypes were allocated in the subplots. Each plot consisted of 15 rows (20 cm spacing) of 3.5 meter length, i.e., 10.5 m2 (1/400 feddan) and then converted to hectare. The first experiment was grown under supplemental water applied regularly as recommended (Normal “N”) and the second did not receive any irrigation after the heading stage (drought stress “D”). Planting date was 18th and 20th November in 2011/2012 and 2012/2013 seasons, respectively. The experimental field soil was sandy-clay in texture. Normal agronomic practices of growing wheat were carried out until harvest.
2.3. Data recorded
Plant height (cm), number of spikes/m2, spike length (cm) and 1000-kernel weight (gm) were measured. Grain and biological yields (ton/hectares) were determined for the plot area and then converted to yield per hectare. Thus, harvest index (grain yield/total biological yield) was determined.
2.4. Statistical analysis
The combined analysis of two season data was conducted according to Snedecor and Cochran [52]. Mean values of the recorded data were compared by using the least significant differences (L.S.D.0.05) according to Wynne et al. [60]. Drought susceptibility Index (DSI): was calculated according to the method of Fischer and Maurer [19]. Stress tolerance index (STI), for grain yield was computed as a formula used by Farshadfar et al. [18].
2.5. DNA extraction, ISSR and RAPD assays
Lines seeds were planted into pots containing peat moss and placed under greenhouse conditions at 22 °C (Laboratory of plant reproduction and development (RDP), ENS of Lyon, France). Total Genomic DNA was extracted from young leaf pieces (approximately 1 cm2) using the BioSprint 96 Workstation (RDP, ENS of Lyon, France) and the DNA Plant Kit (Qiagen), according to the instructions of the supplier. PCR assay was performed in a 20 μl volume containing 2 μl of 10 × PCR blue buffer, 0.4 μl of dNTPs (PRomyga, Madison, USA) 2 μl of primer, 0.2 μl of GoTaq DNA polymerase Core System mixture (Promega), 13.4 μl of sterile ultrapure deionized water and 2 μl of 100 ng DNA template. A negative-DNA control was obtained by adding 1μ of sterile ultrapure deionized water. The Thermal Cycler was programed by: 1 cycle (an initial denaturing step) of 5 min at 95 °C, 40 cycles of 30 s at 95 °C (denaturation step), 30 s at 33–55 °C (annealing step, optimized for each primer), 1 min 30 s at 72 °C (elongation step) and 5 min at 72 °C (final extension), then kept at 20 °C. PCR products were visualized by conventional agarose gel electrophoresis [49]. For ISSR markers, 26 primers were tested as single primers for the amplification of genomic DNA. Of these, 16 primers produced polymorphic band patterns. Twenty single primers of RAPD markers were screened across the 20 wheat lines, and twelve of them were polymorphic.
2.6. Data analysis
DNA banding patterns generated by ISSR and RAPD were analyzed by computer program Gene Profiler (version 4.03). The presence (1) or absence (0) of each band was recorded for each line for all tested primers. To measure the informativeness of the ISSR and RAPD markers in differentiating among 20 wheat lines, polymorphism information content (PIC) was calculated according to the formula proposed by Ghislain [21], as PIC = 1 − [(p)2 + (q)2], where p is the frequency of the allele band present and q is frequency of the allele band absent across wheat lines. The marker index (MI) was also calculated for each ISSR and RAPD primer as MI = PIC × ηβ, where PIC is the mean PIC value, η the number of bands, and β is the proportion of polymorphic bands, based on the method of Powell et al. [43]. Analysis of variance (ANOVA) was conducted using the 1–0 data. The association analysis was conducted using simple linear regression. For this, data on individual phenotypic trait were regressed on whole 1–0 binary marker data for each individual marker using Excel program. The coefficient of determination (R)2 was calculated as R2 = 1 − (SSE/SST), where SSE is the sum of squares of error and SST is the total sum of squares. Genetic similarity estimates for ISSR and RAPD markers were determined using Nei and Li’s method [39]. Dendrograms were generated with the unweighted pair group method with arithmetic mean (UPGMA) using the computational package MVSP version 3.1. A cophenetic matrix was derived from each matrix to test goodness of fit of the clusters by comparing the matrices using the Mantel test [30]. Finally, the correlation between ISSR and RAPD distances was calculated using NTSYS-pc version 2.2 [45].
3. Results and discussion
3.1. Phenotypic evaluation
3.1.1. Analysis of variance
The combined analysis of variance for tested traits (Table 2) revealed highly significant differences affected by years, water stress treatments and genotypes except harvest index. These results showed that the wheat inbred lines responded differently when they were grown under water stress conditions.
Table 2.
Mean squares of the combined analysis of variance for all studied traits over two years.
| S.O.V | D.F | Mean squares |
||||||
|---|---|---|---|---|---|---|---|---|
| Plant height | No. of spikes/m2 | 1000-kernel weight | Spike length | Biological yield | Grain yield | Harvest index | ||
| Year (Y) | 1 | 293.19⁎⁎ | 48807.20⁎⁎ | 373.96⁎⁎ | 9.91⁎⁎ | 82.24⁎⁎ | 7.08⁎⁎ | 14.18 |
| Drought (D) | 1 | 10487.06⁎⁎ | 351655.20⁎⁎ | 8346.37⁎⁎ | 38.85⁎⁎ | 3547.28⁎⁎ | 373.35⁎⁎ | 0.35 |
| Y × D | 1 | 3.81 | 2497.61⁎⁎ | 11.75⁎⁎ | 0.01 | 2.25 | 0.82 | 17.47 |
| Error a | 12 | 37.57 | 1111.90 | 3.31 | 0.07 | 13.41 | 2.26 | 11.59 |
| Genotype (G) | 19 | 501.07⁎⁎ | 26766.17⁎⁎ | 69.45⁎⁎ | 5.73⁎⁎ | 40.11⁎⁎ | 7.47⁎⁎ | 251.70⁎⁎ |
| Y × G | 19 | 0.75 | 39.15 | 1.14 | 0.03 | 0.90 | 0.01 | 12.60 |
| D × G | 19 | 80.12⁎⁎ | 766.88⁎⁎ | 19.75⁎⁎ | 0.14⁎⁎ | 9.31⁎⁎ | 1.87⁎⁎ | 68.68⁎⁎ |
| Y × D × G | 19 | 0.77 | 25.98 | 0.90 | 0.03 | 0.24 | 0.01 | 1.92 |
| Error b | 228 | 11.53 | 249.88 | 1.99 | 0.06 | 1.15 | 0.18 | 8.05 |
Significant at 5% and 1% levels of probability, respectively.
3.1.2. Effect of wheat genotypes
Table 2 showed the differences between the tested wheat lines in yield and its components. It was appeared that some introduced lines were superior to the check cultivar (Sahel1) in yield and its component traits. Within the studied lines, L2, L7 and L8 surpassed all other lines and Sahel1 in most yield component traits in both seasons. It could be concluded that wheat yield and its components were adjusted a lot due to the wheat gene effect since lines L2, L7 and L8 were more effective in predicting more yield and its components. These results were in agreement with [15], [62], [35], they found there are significant differences among wheat cultivars in yield and its components.
3.1.3. Effect of drought stress conditions
The response in terms of wheat yield and its components by exposing plants to skipping irrigation at the heading stage is presented in Table 3. Normal irrigation for wheat plants significantly results in higher yield and its components as compared to drought stress. Preventing irrigation at the heading stage induced a decrease in the wheat yield and its components. Preventing irrigation at the heading stage may cause a great decrease in some important processes affecting yield production such as cell division and enlargement mainly that of spikes, flowering, fertilization, kernel formation and food translocation to the formed kernels. Similar results were obtained by Emel [17], Reiad et al. [44], Amin [3], Mohamed and Said [33], Anwar et al. [4], they concluded that drought stress conditions decreased wheat yield significantly and most of its components.
Table 3.
The range and mean values for all studied traits under well-watered and drought stress conditions over two years.
| Traits | Well-watered |
Drought stress |
||
|---|---|---|---|---|
| Range | Means ± S.E | Range | Means ± S.E | |
| Plant height (cm) | 91.20–112.49 | 102.94 ± 1.57 | 76.76–103.34 | 91.49 ± 1.67 |
| No. of spikes/m2 | 232.88–390.13 | 317.17 ± 10.75 | 185.13–301.63 | 250.87 ± 8.45 |
| 1000-kernel weight (gm) | 34.04–42.36 | 38.88 ± 0.59 | 24.16–31.93 | 28.67 ± 0.75 |
| Spike length (cm) | 10.85–12.90 | 11.87 ± 0.14 | 10.25–12.34 | 11.17 ± 0.12 |
| Biological yield (ton/hec.) | 9.971–18.051 | 14.998 ± 0.991 | 6.525–10.045 | 8.339 ± 0.321 |
| Grain yield (ton/hec.) | 3.050–6.151 | 4.776 ± 0.589 | 1.980–4.071 | 2.694 ± 0.111 |
| Harvest index (%) | 24.64–39.40 | 32.51 ± 0.99 | 22.42–43.70 | 32.44 ± 0.77 |
3.1.4. Effect of the interaction between genotype x drought
The different studied lines exposed to drought effect, showed depression in the mentioned studied characters (Table 3). Line L7 showed superiority mainly in tested characters by 97.53 cm, 295.88, 11.30 cm, 31.51 gm, 3.13 ton/hec., 10.05 ton/hec. and 31.27% for plant height, number of spikes/m2, spike length, 1000-kernel weight, grain yield, biological yield and harvest index respectively, as compared with check cultivar under drought stress (Table 4). These results indicated that L7 is considered as the most promising line under drought stress conditions in yield and its components. Thus the variation in genotypes over the environments could provide scope for breeding drought tolerant cultivars [10], [50].
Table 4.
Mean performance of all traits recorded for 20 bread wheat lines under normal (N) and drought stress (D), drought susceptibility index (DSI) and stress drought index (STI) over two years.
| Lines | Plant height |
No. of spikes/m2 |
Spike length (cm) |
1000-kernel weight |
Grain yield (ton/hec.) |
DSI | STI | Biological yield (ton/hec.) |
Harvest index% |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | D | N | D | N | D | N | D | N | D | N | D | N | D | |||
| L1 | 102.49 | 92.69 | 324.75 | 264.63 | 11.71 | 11.21 | 38.23 | 28.68 | 5.02 | 2.78 | 1.03 | 55.44 | 14.31 | 7.91 | 35.09 | 39.26 |
| L2 | 100.15 | 89.60 | 367.75 | 301.63 | 11.33 | 10.65 | 39.89 | 31.65 | 6.15 | 4.07 | 0.78 | 66.17 | 15.95 | 9.45 | 39.40 | 43.70 |
| L3 | 103.96 | 96.14 | 335.50 | 273.88 | 11.20 | 10.69 | 39.87 | 30.90 | 5.80 | 3.10 | 1.07 | 53.54 | 16.19 | 9.45 | 36.34 | 36.49 |
| L4 | 98.75 | 89.14 | 337.00 | 266.75 | 12.04 | 11.51 | 36.66 | 28.31 | 4.46 | 3.150 | 0.68 | 70.55 | 11.94 | 8.38 | 37.68 | 37.58 |
| L5 | 106.78 | 97.44 | 298.88 | 233.63 | 12.73 | 12.06 | 37.13 | 28.61 | 5.20 | 3.43 | 0.78 | 66.00 | 16.85 | 9.52 | 30.83 | 36.06 |
| L6 | 111.53 | 99.04 | 378.88 | 288.75 | 12.40 | 11.71 | 39.70 | 27.78 | 4.63 | 2.46 | 1.08 | 53.03 | 15.30 | 8.48 | 30.31 | 32.04 |
| L7 | 104.19 | 97.53 | 390.13 | 295.88 | 12.18 | 11.30 | 40.45 | 31.51 | 5.32 | 3.130 | 0.95 | 58.81 | 17.99 | 10.05 | 38.30 | 31.27 |
| L8 | 112.49 | 103.34 | 324.13 | 266.50 | 12.75 | 12.16 | 36.36 | 28.76 | 5.01 | 3.21 | 0.83 | 63.95 | 15.15 | 9.29 | 32.25 | 34.53 |
| L9 | 110.18 | 94.64 | 251.13 | 207.00 | 11.99 | 10.88 | 36.20 | 26.43 | 3.74 | 2.24 | 0.92 | 59.92 | 15.14 | 7.22 | 24.64 | 31.63 |
| L10 | 96.43 | 88.19 | 356.50 | 287.63 | 11.63 | 10.83 | 38.16 | 30.14 | 4.56 | 2.83 | 0.87 | 62.19 | 14.49 | 8.86 | 31.03 | 31.98 |
| L11 | 103.54 | 92.64 | 324.88 | 252.13 | 11.50 | 11.04 | 40.65 | 31.93 | 4.89 | 2.70 | 1.03 | 55.22 | 17.07 | 8.69 | 28.89 | 31.12 |
| L12 | 111.00 | 98.19 | 239.75 | 187.50 | 12.63 | 11.95 | 40.61 | 28.54 | 4.25 | 1.98 | 1.23 | 46.55 | 14.36 | 6.53 | 29.15 | 30.37 |
| L13 | 97.99 | 89.69 | 349.88 | 274.50 | 12.15 | 11.21 | 41.13 | 30.76 | 5.79 | 2.549 | 1.29 | 43.98 | 17.23 | 8.51 | 33.58 | 29.99 |
| L14 | 109.28 | 81.84 | 232.88 | 185.13 | 11.68 | 10.59 | 42.36 | 25.06 | 3.05 | 1.93 | 0.85 | 63.31 | 9.97 | 6.60 | 30.68 | 29.37 |
| L15 | 101.40 | 91.01 | 359.75 | 293.75 | 12.90 | 12.34 | 41.53 | 29.81 | 5.81 | 2.39 | 1.36 | 41.14 | 18.05 | 9.92 | 32.13 | 24.17 |
| L16 | 91.20 | 76.76 | 281.88 | 223.63 | 11.90 | 11.33 | 36.23 | 26.31 | 4.44 | 2.03 | 1.25 | 45.69 | 17.25 | 9.05 | 25.87 | 22.42 |
| L17 | 100.08 | 91.11 | 285.63 | 228.50 | 11.43 | 10.68 | 36.64 | 24.34 | 4.16 | 2.24 | 1.06 | 53.92 | 13.90 | 6.83 | 30.30 | 32.85 |
| L18 | 101.99 | 86.74 | 282.00 | 189.88 | 11.18 | 10.44 | 34.04 | 24.16 | 3.71 | 2.09 | 1.01 | 56.28 | 11.94 | 7.24 | 31.04 | 30.19 |
| L19 | 96.38 | 85.96 | 305.00 | 233.63 | 10.85 | 10.25 | 40.45 | 28.68 | 4.66 | 2.15 | 1.24 | 46.25 | 12.30 | 6.70 | 39.33 | 32.36 |
| L20 | 98.96 | 88.08 | 317.13 | 262.50 | 11.25 | 10.64 | 41.40 | 31.04 | 4.85 | 2.56 | 1.09 | 52.70 | 14.60 | 8.16 | 33.24 | 31.40 |
| Mean | 102.94 | 91.49 | 317.17 | 250.87 | 11.87 | 11.17 | 38.88 | 28.67 | 4.86 | 2.65 | 14.99 | 8.34 | 32.51 | 32.44 | ||
| Check | 104.28 | 94.65 | 334.65 | 274.38 | 11.64 | 11.31 | 39.50 | 29.88 | 4.78 | 3.12 | 15.23 | 8.16 | 29.66 | 30.69 | ||
| LSD0.05 | 2.37 | 11.01 | 0.18 | 0.98 | 0.29 | 1.98 | 0.75 | |||||||||
3.1.5. Drought susceptibility index (DSI) and stress tolerance index (STI)
DSI is derived from the yield difference between stress and non stress environments [6]. Moreover, many authors used the DSI [19], [18], [29] to characterize the yield stability between two environments. Results of DSI and STI (Table 3), indicated that lines L2, L4, L5, L7 and L8 which had a relatively high grain yield and low drought susceptibility index under drought stress, can be used in breeding programs to produce varieties or hybrids having high grain yield ability and high tolerance to drought stress.
3.2. Diversity analysis
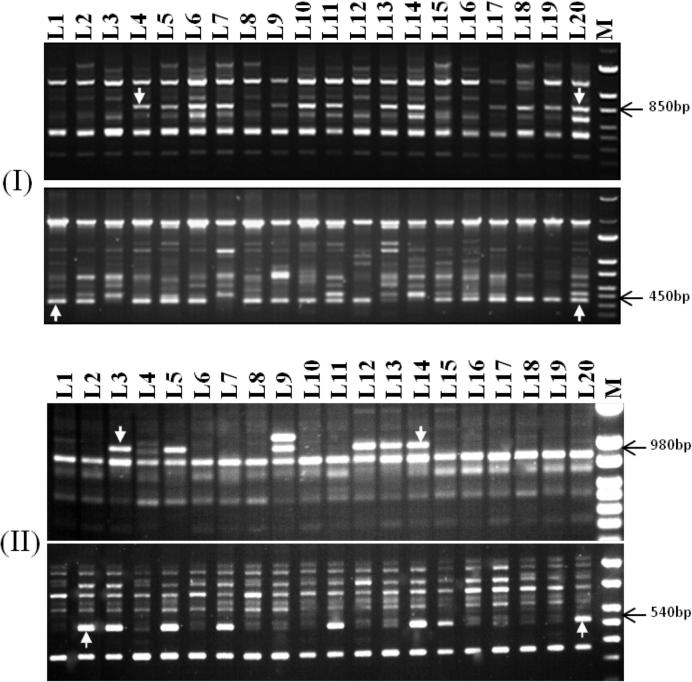
The sixteen ISSR primer sets utilized to analyze wheat lines generated 4–12 bands with approximately an average of 7 bands per primer. A total of 117 bands were scored, of which 46 (39.3%) were polymorphic and 71 (60.7%) were monomorphic (Table 5). The highest percentage of polymorphism (%P) was 75.0%, generated by UBC-881 (Fig. 1-I), while the lowest was 11.1%, generated by UBC-846. The band size ranged from 440 to 2000 bp to that generated by UBC-811 and UBC-833, respectively. The study result of %P was in agreement with that (47.3%) obtained by El Siddig et al. [12]. In this regard, Emel [17] found a higher %P than that herein reported (76.07%). Also, Carvalho et al. [8] documented a very high %P (98.5%) using 18 ISSR primers in 99 wheat accessions. Contrary, Tok et al. [58] showed, the highest percent of polymorphic loci was very low (17.59%) among wheat genotypes. Compared to other molecular markers used for genetic diversity in wheat, our %P was about similar to those of SSR (50.3%, [31] TRAP (40%, [2], RAPD (52.6%, [60], AFLP (50.2%, [11], and SRAP (54.81%, [18].
Table 5.
Primers used in ISSR analysis, their sequences, AB, PB,%P, PIC, MI and FS for wheat lines (L1–L20).
| Primer | Sequence | AB | PB | %P | PIC | MI | FS (bp) |
|
|---|---|---|---|---|---|---|---|---|
| Larger | Smaller | |||||||
| UBC-808 | (AG)8C | 6 | 2 | 33.3 | 0.06 | 0.12 | 940 | 560 |
| UBC-811 | (GA)8AC | 8 | 5 | 62.5 | 0.29 | 1.45 | 1200 | 440 |
| UBC-812 | (GA)2GG(AG)4AA | 5 | 1 | 20.0 | 0.04 | 0.04 | 1000 | 775 |
| UBC-815 | (TC)8A | 5 | 1 | 20.0 | 0.04 | 0.04 | 920 | 450 |
| UBC 818 | G(GGGGT)3 | 7 | 3 | 42.9 | 0.10 | 0.30 | 1635 | 620 |
| UBC-819 | (GT)8A | 5 | 1 | 20.0 | 0.04 | 0.04 | 1200 | 725 |
| UBC-826 | (AC)8C | 6 | 2 | 33.3 | 0.06 | 0.12 | 1370 | 535 |
| UBC-833 | (GA)8TT | 11 | 5 | 45.5 | 0.10 | 0.50 | 2000 | 635 |
| UBC-834 | (AG)8YT | 7 | 2 | 28.6 | 0.13 | 0.26 | 1850 | 880 |
| UBC-840 | (CT)8TT | 12 | 7 | 58.3 | 0.23 | 1.61 | 1950 | 725 |
| UBC-846 | (CA)8RT | 9 | 1 | 11.1 | 0.04 | 0.04 | 1000 | 530 |
| UBC-849 | (GT)8YA | 8 | 2 | 25.0 | 0.06 | 0.12 | 1025 | 590 |
| UBC-852 | (GATA)2(GACA)2 | 5 | 1 | 20.0 | 0.04 | 0.04 | 1345 | 420 |
| UBC-876 | (GATA)2(GACA)2 | 7 | 3 | 42.9 | 0.10 | 0.30 | 1840 | 770 |
| UBC-880 | (TC)8AA | 4 | 1 | 25.0 | 0.12 | 0.12 | 1250 | 550 |
| UBC-881 | (GGGTG)3 | 12 | 9 | 75.0 | 0.20 | 1.80 | 1485 | 535 |
| Total | 117 | 46 | ||||||
| Means | 7.3 | 2.9 | 0.10 | 0.43 | ||||
AB, amplified bands; PB, polymorphic bands;%P, percent of polymorphism; PIC, polymorphism information content; MI, marker index; FS, fragment size and bp, base pair.
Figure 1.
(I), ISSR fingerprinting with primers UBC-811, B: UBC-849 and C: UBC-881 and (II), RAPD fingerprinting with primers OPAM-01 and B: OPF-10 of tested wheat lines (L1–L20), the arrows indicate the different molecular markers.
ISSR primers used in this study were composed of di-, tetra- and penta-nucleotide repeat sequences. These varied primers were found to show different levels of polymorphism. The UBC-881 belonged to penta-nucleotide repeat sequences (GGGTG) that produced the highest level of polymorphism, while the UBC-846 belonged to di-nucleotide repeat sequences (CA) that produced the lowest level of polymorphism (Table 5). Likely, [54], [53], they proposed that the polymorphism rates were higher when the motifs comprise three to five nucleotides of microsatellite primers in wheat. In agreement with results of Najaphy et al. [37], the results showed, UBC-852 that belonged to tetra-nucleotide repeat sequences (GATA) (GACA) produced a low level of polymorphism (20%).
The polymorphism information content (PIC) index has been used extensively in many genetic diversity studies [56], [57]. Moreover, the PIC value of markers indicated the usefulness of DNA markers for gene mapping, molecular breeding and germplasm evaluation [41]. In this study, PIC values for the 16 ISSR primers varied from 0.04 to 0.23 with an average of 0.10 (Table 5). In this regard, Saleh [48] obtained similar results to that ranging from 0.05 to 0.27 with an average of 0.195 using the AFLP assay. Also, Muthusamy et al. [36] documented an average of 0.20 per ISSR primer among rice beans. The moderate values of PIC for the ISSR primers could be attributed to the diverse nature of wheat accessions and/or highly informative ISSR markers [37]. In this study, MI values ranged from 0.04 to 1.80 for the primers (UBC-812, UBC-815, UBC-819, UBC-846 and UBC-852), and UBC-881, respectively. Our MI values were smaller than those, from 0.41 to 3.36, reported by Najaphy et al. [37].
Twenty RAPD primers were used for initial screening with 20 representative wheat lines, only 12 primers amplified polymorphic patterns. Amplification products yielded a total of 95 scorable bands, between them 50 (53.2%) were polymorphic with an average of 4.2 per primer (Table 3 and Fig. 1-II). The highest number of bands (15) was obtained with primer OPF-4, while the lowest number (5) was obtained with primers OPG-05 and OPA-15 (Table 6). In accordance with [14], [13], they reported a %P of 52.6%, while, the %P of the present study (53.2%) was higher than (18.8%), obtained by Saleh [48]. The %P obtained in the present study based on the RAPD technique varied from those obtained in previous studies on different plant species, like barley (77.06%, [24]; rice bean (70.30%, [8], Vigna species (48%, [1] and maize (76.14%, [27].
Table 6.
Primers used in RAPD analysis, their sequences, AB, PB,%P, PIC, MI and FS for wheat lines (L1-L20).
| Primer | Sequences | AB | PB | %P | PIC | MI | FS (bp) |
|
|---|---|---|---|---|---|---|---|---|
| Larger | Smaller | |||||||
| OPF-4 | GAATGCGGAG | 15 | 11 | 73.3 | 0.21 | 2.31 | 2100 | 500 |
| OPF-10 | GGGCCACTCA | 9 | 2 | 22.2 | 0.10 | 0.20 | 1150 | 360 |
| OPA-17 | GACCGCTTGT | 8 | 4 | 50.0 | 0.16 | 0.64 | 2000 | 375 |
| OPG-05 | CTGACGTCAC | 5 | 4 | 80.0 | 0.14 | 0.56 | 1700 | 400 |
| OPAM-01 | TCACGTACGG | 7 | 3 | 42.8 | 0.13 | 0.39 | 2000 | 490 |
| OPP-05 | CCCCGGTAAC | 8 | 3 | 37.5 | 0.10 | 0.30 | 1900 | 635 |
| OPA-04 | AATCGGGCTG | 6 | 3 | 50.0 | 0.14 | 0.42 | 1350 | 440 |
| OPA-06 | GGTCCCTGAC | 8 | 6 | 75.0 | 0.18 | 1.08 | 1650 | 540 |
| OPA-09 | GGGTAACGCC | 6 | 2 | 33.3 | 0.14 | 0.28 | 1975 | 375 |
| OPA-12 | TCGGCGATAG | 10 | 5 | 50.0 | 0.16 | 0.80 | 2050 | 380 |
| OPA-14 | TCTGTGCTGG | 8 | 6 | 75.0 | 0.21 | 1.26 | 1890 | 410 |
| OPA-15 | TTCCGAACCC | 5 | 1 | 20.0 | 0.10 | 0.10 | 1100 | 300 |
| Total | 95 | 50 | ||||||
| Means | 7.9 | 4.2 | 0.15 | 0.69 | ||||
AB, amplified bands; PB, polymorphic bands;%P, percent of polymorphism; PIC, polymorphism information content; MI, marker index; FS, fragment size and bp, base pair.
In this investigation, the PIC values for the 12 RAPD primers ranged from 0.10 (OPF-10, OPP-05 and OPA-15) to 0.21 (OPF-4 and OPA-14) with an average of 0.15 (Table 6). The average of PIC was smaller than 0.34 which was obtained by Khavarinejad and Karimov [28] for wheat spring genotypes. MI values for the RAPD technique were between 0.10 (OPA-15) and 2.31 (OPF-4).
3.3. Single marker analysis
The present study involved a set of 20 bread wheat lines, which constitute important and diverse inbred lines of Egyptian bread wheat, exhibiting moderate to high genetic variability for phenotypic traits analyzed during this work. Ninety-six polymorphic molecular markers (ISSR = 46; RAPD = 50) were screened using single marker analysis method across the means of 7 phenotypic traits (Table 3), and 4 of these markers were significantly associated with 5 phenotypic traits. Results showed that there were two markers (UBC-881450bp and OPF-10540bp), on each of which two traits regressed significantly, and individual traits (grain yield) which regressed significantly on more than one marker. In analysis, the associated markers each explained a maximum regression from 18.92% to 34.95% of the total available variation for individual associated traits (Table 7). Roy et al. [47] showed, the associated markers each explained a maximum of 8.12% and 29.38% for tiller numbers and florets per spike traits analyzing a total of 99 and 133 polymorphic SSR and AFLP bands, respectively. Recently, Khaled and Hamam [20] obtained a maximum regression of 10.74 (days to heading) to 11.60% (spike length) of the total available variation for individual associated traits. In this investigation, the ISSR markers UBC-811850bp and UBC-881450bp (Table 7 and Fig. 1, I) were regarded probably as candidate markers which were linked to spike length and yield traits, respectively. Our findings were supported by the findings of Nei and Li [39] who demonstrated that five ISSRs were regarded as candidate markers, linked to spike length per plant trait in wheat genotypes.
Table 7.
Detailed analyses of variances (ANOVA) involving simple linear regression (R2) for traits using 46 ISSR and 50 RAPD polymorphic bands.
| Marker/type | Trait | Condition | SV | df | SS | MS | R2 | p-Value |
|---|---|---|---|---|---|---|---|---|
| UBC-811850bp (ISSRs) | Spike length | Normal | Genotypes | 1 | 0.92 | 0.92⁎ | 24.09 | 0.028 |
| Error | 18 | 2.93 | 0.16 | |||||
| Total | 19 | 3.86 | ||||||
| UBC-881450bp (ISSRs) | Grain yield | Normal | Genotypes | 1 | 5.54 | 5.54⁎⁎ | 34.95 | 0.006 |
| Error | 18 | 10.31 | 0.57 | |||||
| Total | 19 | 15.85 | ||||||
| Biological yield | Normal | Genotypes | 1 | 24.02 | 24.02⁎ | 26.02 | 0.022 | |
| Error | 18 | 68.27 | 3.79 | |||||
| Total | 19 | 92.29 | ||||||
| Drought | Genotypes | 1 | 1434.61 | 1434.61⁎ | 19.39 | 0.051 | ||
| Error | 18 | 5960.72 | 331.15 | |||||
| Total | 19 | 7395.33 | ||||||
| OPAM-01980bp (RAPDs) | Number of spikes/m2 | Normal | Genotypes | 1 | 9054.73 | 9054.73⁎ | 23.55 | 0.030 |
| Error | 18 | 29379.50 | 1632.19 | |||||
| Total | 19 | 38434.23 | ||||||
| OPF-10540bp (RAPDs) | 1000-kernel weight | Normal | Genotypes | 1 | 31.01 | 31.01⁎⁎ | 30.33 | 0.011 |
| Error | 18 | 71.22 | 3.96 | |||||
| Total | 19 | 102.23 | ||||||
| Drought | Genotypes | 1 | 25.94 | 25.94⁎ | 23.66 | 0.029 | ||
| Error | 18 | 83.69 | 4.65 | |||||
| Total | 19 | 109.63 | ||||||
| Grain yield | Normal | Genotypes | 1 | 3.03 | 3.03⁎ | 18.92 | 0.053 | |
| Error | 18 | 12.82 | 0.71 | |||||
| Total | 19 | 15.85 |
Significant at 0.05 and 0.01 probability levels, respectively.
RAPD markers OPAM-01980bp and OPF-10540bp (Fig. 1, II) were regarded may be as candidate markers which were linked to the number of spikes per plant and 1000-kernel weight and grain yield, respectively. A significant negative correlation was observed between the number of spikes/m2 trait and the presence of the OPAM-01980bp marker. In other words, by increasing the number of spikes/m2, the probability of observing the OPAM-01980bp marker decreases (Fig. 1,II). Similar results were obtained by Mohammadi et al. [34].
Under normal conditions, results in Table 7 showed significant and highly significant regressions (0.92∗, p = 0.028), (5.54∗∗ p = 0.006) and (24.02∗, p = 0.022) on spike length, grain and biological yield traits, respectively. Moreover, under drought conditions, results showed significant regressions (1434.61∗, p = 0.051) and (25.94∗, p = 0.029) on biological yield and 1000-kernel weight traits, respectively. Recently, El-Rawy and Youssef [16] used SRAP markers to evaluate bread wheat genotypes under drought stress, they reported that SRAP molecular markers were able to generate some unique and specific bands for certain genotypes related to drought tolerance. Determining the genetic basis of tolerance involves correlating the incidence of molecular markers with phenotypic scores to predict DNA genomic regions that harbor a factor influencing the plant’s response [46]. Our results demonstrated the importance of understanding which ISSR and RAPD markers function in the environment in which the markers may be deployed. Molecular markers that respond most consistently and to the greatest extent in the target environment are the prime candidates for marker-assisted selection. Therefore, ISSR and RAPD markers identified during the present study need to be subjected to validation and/or functional analysis of respective traits.
3.4. Similarity coefficient analysis
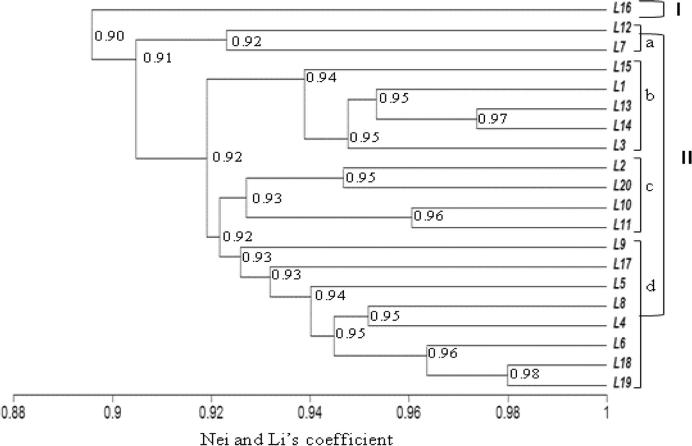
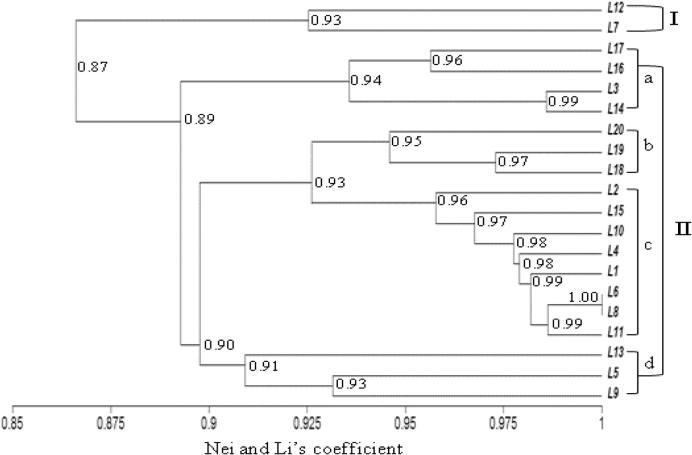
The similarity coefficient for ISSR markers ranged from 0.86 to 0.98 (Table 8, below diagonal). The UPGMA cluster analysis based on the ISSR marker separated the tested lines into two different clusters based on a similarity coefficient of 0.90 (Fig. 2). The line “L16” was placed alone in the first cluster. The second main cluster was separated into four sub-branches: I-a with lines “L12 and L7”, I-b with lines “L15, L1, L13, L14 and L3”, I-c which contained lines “L2, L20, L10 and L11” and I-d with other studied lines (Fig. 2). In this main cluster, lines “L19 and L8” gathered at a high similarity coefficient of 0.98%. Based on RAPD data, the similarity coefficient ranged from 0.81 to 1.00 (Table 8, above diagonal). On the Dendrogram, the cluster analysis separated the tested lines into two main clusters (Fig. 3). The first cluster includes lines L12, and L7. The second cluster was sub-divided into four sub-clusters, whereas the first (I-a) had lines L17, L16, L3, and L14 but the second (I-b) included L20, L19 and L18. The third sub-cluster (I-c) contained 11 lines (Fig. 3) which gathered at 0.90 similarity coefficient with the fourth sub-cluster (I-d: L13, L5 and L9). In accordance with Zar and Ahmadi [61] the dendrogram constructed by the combined RAPD and ISSR data gave a relatively different clustering pattern.
Table 8.
Similarity percent of ISSR (below diagonal) and RAPD (above diagonal) markers among wheat lines (L1–L20).
| Lines | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 | L10 | L11 | L12 | L13 | L14 | L15 | L16 | L17 | L18 | L19 | L20 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | 0.94 | 0.89 | 0.97 | 0.91 | 0.99 | 0.90 | 0.99 | 0.89 | 0.97 | 0.97 | 0.89 | 0.89 | 0.88 | 0.96 | 0.90 | 0.92 | 0.95 | 0.95 | 0.92 | |
| L2 | 0.93 | 0.94 | 0.94 | 0.90 | 0.96 | 0.90 | 0.96 | 0.91 | 0.97 | 0.97 | 0.85 | 0.89 | 0.93 | 0.96 | 0.93 | 0.94 | 0.92 | 0.89 | 0.92 | |
| L3 | 0.95 | 0.92 | 0.89 | 0.90 | 0.90 | 0.84 | 0.90 | 0.91 | 0.91 | 0.92 | 0.85 | 0.89 | 0.99 | 0.90 | 0.93 | 0.94 | 0.86 | 0.83 | 0.86 | |
| L4 | 0.91 | 0.91 | 0.90 | 0.93 | 0.99 | 0.90 | 0.99 | 0.91 | 0.97 | 0.97 | 0.88 | 0.91 | 0.87 | 0.96 | 0.90 | 0.91 | 0.94 | 0.92 | 0.92 | |
| L5 | 0.92 | 0.90 | 0.91 | 0.95 | 0.92 | 0.86 | 0.92 | 0.93 | 0.9 | 0.93 | 0.87 | 0.90 | 0.92 | 0.92 | 0.83 | 0.88 | 0.88 | 0.85 | 0.88 | |
| L6 | 0.94 | 0.93 | 0.93 | 0.95 | 0.92 | 0.91 | 1.00 | 0.90 | 0.99 | 0.99 | 0.90 | 0.90 | 0.89 | 0.97 | 0.91 | 0.93 | 0.96 | 0.93 | 0.90 | |
| L7 | 0.91 | 0.92 | 0.90 | 0.92 | 0.89 | 0.91 | 0.91 | 0.81 | 0.90 | 0.93 | 0.93 | 0.81 | 0.83 | 0.91 | 0.82 | 0.84 | 0.87 | 0.85 | 0.85 | |
| L8 | 0.93 | 0.93 | 0.91 | 0.95 | 0.94 | 0.94 | 0.92 | 0.90 | 0.99 | 0.99 | 0.90 | 0.90 | 0.89 | 0.97 | 0.91 | 0.93 | 0.96 | 0.93 | 0.90 | |
| L9 | 0.90 | 0.90 | 0.90 | 0.92 | 0.91 | 0.93 | 0.92 | 0.93 | 0.91 | 0.89 | 0.85 | 0.91 | 0.93 | 0.87 | 0.87 | 0.91 | 0.86 | 0.83 | 0.86 | |
| L10 | 0.93 | 0.91 | 0.89 | 0.95 | 0.91 | 0.95 | 0.90 | 0.94 | 0.92 | 0.97 | 0.88 | 0.89 | 0.90 | 0.96 | 0.93 | 0.94 | 0.94 | 0.92 | 0.92 | |
| L11 | 0.93 | 0.93 | 0.92 | 0.92 | 0.91 | 0.95 | 0.89 | 0.91 | 0.92 | 0.96 | 0.89 | 0.89 | 0.90 | 0.99 | 0.90 | 0.92 | 0.95 | 0.92 | 0.92 | |
| L12 | 0.90 | 0.90 | 0.90 | 0.90 | 0.86 | 0.90 | 0.92 | 0.92 | 0.91 | 0.94 | 0.91 | 0.85 | 0.84 | 0.90 | 0.84 | 0.82 | 0.86 | 0.86 | 0.8 | |
| L13 | 0.96 | 0.93 | 0.94 | 0.94 | 0.93 | 0.94 | 0.91 | 0.96 | 0.90 | 0.93 | 0.93 | 0.91 | 0.87 | 0.87 | 0.84 | 0.86 | 0.94 | 0.92 | 0.92 | |
| L14 | 0.95 | 0.92 | 0.95 | 0.93 | 0.93 | 0.94 | 0.90 | 0.93 | 0.90 | 0.91 | 0.92 | 0.89 | 0.97 | 0.89 | 0.91 | 0.96 | 0.85 | 0.82 | 0.85 | |
| L15 | 0.93 | 0.94 | 0.95 | 0.91 | 0.94 | 0.92 | 0.92 | 0.94 | 0.92 | 0.90 | 0.91 | 0.91 | 0.94 | 0.94 | 0.91 | 0.90 | 0.93 | 0.93 | 0.9 | |
| L16 | 0.90 | 0.90 | 0.91 | 0.88 | 0.90 | 0.90 | 0.85 | 0.90 | 0.87 | 0.90 | 0.91 | 0.87 | 0.93 | 0.92 | 0.91 | 0.96 | 0.87 | 0.87 | 0.85 | |
| L17 | 0.90 | 0.91 | 0.91 | 0.92 | 0.94 | 0.94 | 0.89 | 0.91 | 0.92 | 0.92 | 0.94 | 0.89 | 0.92 | 0.93 | 0.91 | 0.92 | 0.89 | 0.86 | 0.86 | |
| L18 | 0.92 | 0.92 | 0.90 | 0.94 | 0.94 | 0.97 | 0.93 | 0.95 | 0.94 | 0.93 | 0.93 | 0.88 | 0.92 | 0.92 | 0.92 | 0.88 | 0.95 | 0.97 | 0.95 | |
| L19 | 0.91 | 0.93 | 0.90 | 0.96 | 0.95 | 0.96 | 0.92 | 0.94 | 0.93 | 0.92 | 0.91 | 0.87 | 0.91 | 0.91 | 0.92 | 0.87 | 0.93 | 0.98 | 0.95 | |
| L20 | 0.92 | 0.95 | 0.91 | 0.90 | 0.92 | 0.92 | 0.93 | 0.92 | 0.91 | 0.93 | 0.94 | 0.91 | 0.92 | 0.90 | 0.93 | 0.90 | 0.93 | 0.93 | 0.93 |
Figure 2.
UPGMA-Dendrogram of genetic similarities using ISSR data based on Nei and Li’s coefficient among tested wheat lines (L1–L20).
Figure 3.
UPGMA-Dendrogram of genetic similarities using RAPD data based on Nei and Li’s coefficient among tested wheat lines (L1–L20).
3.5. Association of ISSR and RAPD data
The correlation (r) and the Mantel test statistic (Z) were calculated to measure the degree of relationship between the similarity matrices obtained with ISSR and RAPD data. Results showed that this correlation was insignificant (0.168, p > 0.05). The main reason for the difference between RAPD and ISSR results is that the two marker techniques targeted different parts of the genome [55]. But, Kaul et al. [26] reported a high correlation (r = 0.86) between RAPD and ISSR markers in Jatropha curcas plants. Finally, the optimal strategies of the breeding system require extensive knowledge of the breeding materials employed. Results presented here will be useful to understand the current status of genetic diversity between Egyptian bread wheat lines. Because all PCR-based markers applied in the present study were randomly selected from the entire wheat genome, exploring genetic variation for specific traits could not be expected [58] with advances in mapping of quantitative trait loci (QTL) for many agronomic important traits in wheat [7], it becomes possible to detect the allelic variation at these loci among accessions by using marker haplotypes.
4. Conclusion
This study gave some important clues that each genetic trait under study responded differently to drought stress. Genotypes also showed a wide variation in their response to drought stress tolerance. Lines L2, L4, L5, L7 and L8 showed drought tolerance by showing a high performance in agronomic traits as well as a low drought susceptibility index, indicating that due to their drought tolerance they can be employed in breeding programs in stress environments. RAPD and ISSR markers could be efficient for the determination of the genetic diversity of bread wheat lines. These molecular markers can be used for the selection of diverted wheat lines which would increase the efficiency and precision of breeding programs. The results of single marker analysis showed that 4 out of 96 molecular markers were significantly associated with 5 phenotypic traits. We believe that at least one of the markers identified during the present study would be validated and used for marker-assisted selection programs involving Egyptian bread wheat lines.
Acknowledgments
The authors thank Prof. Dr. Galal A.R. El-sherbeny, Head of the Genetic Department, Faculty of Agriculture, Sohag University, Egypt for providing seeds. I am thankful to the Laboratory of Plant Reproduction and Development (RDP) in Lyon, France for experimental chemicals and equipment. Finally I would like thank the Egyptian Government for providing the short scientific mission.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Abd El-Hady E.A.A., Haiba A.A.A., Abd El-Hamid N.R., Al-Ansary A.M.F., Mohamed A.Y. N. Y. Sci. J. 2010;3:120–128. [Google Scholar]
- 2.Al-Doss A.A., Saleh M., Moustafa K.A., Elshafei A.A., Barakat M.N. Afr. J. Agric. Res. 2011;5:3065–3074. [Google Scholar]
- 3.Amin E.H.M., Ibrahim A.A., Saleh M.E., Ali A.G.A. Zagazig J. Agric. Res. 2010;37:803–828. [Google Scholar]
- 4.Anwar J., Ahmad A., Khaliq T., Mubeen M., Sultana S.R. Crop Environ. 2011;2:24–27. [Google Scholar]
- 5.Ashraf M. Biotechnol. Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Blum A., Shipler L., Golan G., Mayer J. Field Crops Res. 1989;22:289–296. [Google Scholar]
- 7.Börner A., Schumann E., Furste A., Coster H., Leithold B., Roder S., Weber E. Theor. Appl. Genet. 2002;105:921–936. doi: 10.1007/s00122-002-0994-1. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho A., Lima-Brito J., Maçãs B., Guedes-Pinto H. Biochem. Genet. 2009;47:276–294. doi: 10.1007/s10528-009-9227-5. [DOI] [PubMed] [Google Scholar]
- 9.Darvasi A., Weintreb A., Minke V., Weller S., Soller M. Genetics. 1993;134:943–951. doi: 10.1093/genetics/134.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhanda S.S., Sethi G.S., Behl R.K. J. Agron. Crop Sci. 2004;190:6–12. [Google Scholar]
- 11.Eivazi A.R., Naghavi M.R., Hajeidari M., Pirseyedi S.M., Ghaffari M.R., Mohammadi S.A., Majidi I., Salekedeh G.H., Mardi M. Ann. Appl. Biol. 2007;152:81–91. [Google Scholar]
- 12.El Siddig M.A., Dweikat I., Baenziger S., El Hussein A.A., Elbasyoni I. Middle East J. Sci. Res. 2013;14:1135–1142. [Google Scholar]
- 13.El-Assal S.E.D., Gaber A. Am. J. Appl. Sci. 2012;9:724–735. [Google Scholar]
- 14.El-Mouhammady A.A., Rady M.R., El-Seidy E.H. World Appl. Sci. J. 2014;29:506–516. [Google Scholar]
- 15.El-Murshedy W.A. J. Agric. Res. Kafer El sheikh Univ. 2008;34:25–42. [Google Scholar]
- 16.El-Rawy M.A., Youssef M. J. Crop Sci. Biotechnol. 2014;17:183–189. [Google Scholar]
- 17.Emel S. Pak. J. Bot. 2010;42:2755–2763. [Google Scholar]
- 18.Farshadfar E., Ghandha M., Zahravi M., Sutka J. Acta Agron. Hung. 2001;49:59–66. [Google Scholar]
- 19.Fischer R.A., Maurer R. Aust. J. Agric. Res. 1978;29:897–912. [Google Scholar]
- 20.Khaled A.G.A., Hamam K.A. Egypt. J. Genet. Cytol. 2015;44(1) In press. [Google Scholar]
- 21.Ghislain M., Zhang D., Fajardo D., Hanuman Z., Hijmans R. Genet. Resour. Crop Eval. 1999;46:547–555. [Google Scholar]
- 22.Gostimsky S.A., Kokaeva Z.G., Konovalov F.A. Russ. J. Genet. 2005;41:480–492. doi: 10.1007/s11177-005-0101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta P.K., Roy J.K. Plant Cell Tissue Organs Cult. 2002;70:229–234. [Google Scholar]
- 24.Hou Y., Yan Z., Wei Y., Zheng Y. Barley Genet. Newsl. 2005;35:9–22. [Google Scholar]
- 25.Karakas O., Gurel F., Uncuoglu A.A. Genet. Mol. Biol. 2010;33:719–730. doi: 10.1590/S1415-47572010005000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaul V.K., Kachhwaha S., Kothari S.L. Indian J. Biotechnol. 2012;11:54–61. [Google Scholar]
- 27.Khaled A.G.A., El-sherbeny G.A.R., Elsayed H.M.A. Egypt. J. Genet. Cytol. 2013;42:73–88. [Google Scholar]
- 28.Khavarinejad M., Karimov M. Afr. J. Biotechnol. 2012;11:14724–14731. [Google Scholar]
- 29.Kilic H., Yagbasanlar T. Not. Bot. Horti. Agrobo. 2010;38:164–170. [Google Scholar]
- 30.Mantel N. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 31.Maric S., Boleric S., Martinicic J., Petic I., Kozumplic V. Plant Breeding. 2004;123:366–369. [Google Scholar]
- 32.McWilliam J. In: Baker F., editor. CAB International; Wallingford, UK: 1989. pp. 1–11. (Drought Resistance in Cereals). [Google Scholar]
- 33.Mohamed N.E.M., Said A.A. Egypt J. Agron. 2014;36:123–146. [Google Scholar]
- 34.Mohammadi A., Majidi-Heravan A.E., Bihamta M., Heidari-Sharifabad H., Ahmadi H. Iran. J. Genet. Plant Breeding. 2010;1:59–64. [Google Scholar]
- 35.Morsy A.M., Abd El-Hameed I.M. Egypt J. Agron. 2012;34:227–247. [Google Scholar]
- 36.Muthusamy S., Kanagarajan S., Ponnusamy S. Electron. J. Biotechnol. 2008;11:1–10. [Google Scholar]
- 37.Najaphy A., Parchina R.A., Farshadfara E. Biotechnol. Biotechnol. Equip. 2011;25:2634–2638. [Google Scholar]
- 38.Neale D.B., Savolainen O. Trends Plant Sci. 2004;9:325–330. doi: 10.1016/j.tplants.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Nei M., Li W.H. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Passioura J.B. Funct. Plant Biol. 2002;29:537–546. doi: 10.1071/FP02020. [DOI] [PubMed] [Google Scholar]
- 41.Peng J.H., Lapitan N.L.V. Funct. Integr. Genomic. 2005;5:80–96. doi: 10.1007/s10142-004-0128-8. [DOI] [PubMed] [Google Scholar]
- 42.Powell W., Orozco-Castillo C., Chalmers K.J., Provan J., Waugh R. Electrophoresis. 1995;16:1726–1730. doi: 10.1002/elps.11501601285. [DOI] [PubMed] [Google Scholar]
- 43.Powell W., Morgante M., Andre C., Hanafey M., Vogel J., Tingey S., Rafalsky A. Mol. Breeding. 1996;2:225–238. [Google Scholar]
- 44.Reiad M.Sh., Yasein M., Tolba A.M., Abd-El-Samie F.S. Egypt J. Agron. 2007;29:69–83. [Google Scholar]
- 45.Rohlf F.J. Exeter Software; Setauket, USA: 2000. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System. Version 2.1. [Google Scholar]
- 46.Roy S.J., Tucker E.J., Tester M. Plant Biol. 2011;14:232–239. doi: 10.1016/j.pbi.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Roy J.K., Bandopadhyay R., Rustgi S., Balyan H.S., Gupta P.K. Curr. Sci. 2006;90:683–689. [Google Scholar]
- 48.Saleh B. J. Plant Biol. Res. 2012;1:1–11. [Google Scholar]
- 49.Sambrook J., Fritsch E.F., Maniatis T. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. A Laboratory Manual. [Google Scholar]
- 50.Shafeeq S., Rahman M., Zafar Y. Pak. J. Bot. 2006;38:1671–1678. [Google Scholar]
- 51.Silvanacrieste S.M., Vallis M.A., Jose F.M., Gimense A., Lopes C.R. Genet. Resour. Crop Eval. 2005;52:1079–1086. [Google Scholar]
- 52.G.W. Snedecor, W.G. Cochran, 7th ed., Lowa State Univ. Press., Ames., Lowa, U.S.A. (1980).
- 53.Sofalian O., Chaparzadeh N., Javanmard A., Hejazi M.S. Int. J. Agric. Biol. 2008;10:466–468. [Google Scholar]
- 54.Song Q.J., Fichus E.W., Cregan P.B. Theor. Appl. Genet. 2002;104:286–293. doi: 10.1007/s001220100698. [DOI] [PubMed] [Google Scholar]
- 55.Souframanien J., Gopalakrishna T. Theor. Appl. Genet. 2004;109:1687–1693. doi: 10.1007/s00122-004-1797-3. [DOI] [PubMed] [Google Scholar]
- 56.Tatikonda L., Wani S.P., Kannan S., Beerelli N., Sreedevi T.K., Hoisington D.A., Devi P., Varshney R.A. Plant Sci. 2009;176:505–513. doi: 10.1016/j.plantsci.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Thudi M., Manthena R., Wani S.P., Tatikonda L., Hoisington D.A., Varshney R.A. J. Plant Biochem. Biotechnol. 2010;19:209–216. [Google Scholar]
- 58.Tok D., Senturk-Akfirat F., Sevinc D., Aydin Y., Altinkut-Uncuoglua A. Turk. J. Field Crops. 2011;16:157–165. [Google Scholar]
- 59.Turner N.C. In: Mussell H., Staples C.R., editors. John Wiley & Sons; New York: 1979. pp. 343–372. (Stress Physiology in Crop Plants). [Google Scholar]
- 60.Wynne J.C., Emery D.A., Rice P.W. Crop Sci. 1970;10:713–715. [Google Scholar]
- 61.Zar M., Ahmadi J. Genet. Plant Physiol. 2011;1:45–55. [Google Scholar]
- 62.Zeidan E.M., Abbd El-Hameed I.M., Bassiouny A.H., Waley A.A. Faculty of Agriculture Cairo, University; Egypt: 2009. 4th Conf. Technologies in Agriculture. [Google Scholar]