Abstract
The aim of the present study was to assess the association of single nucleotide polymorphisms (SNPs) of Calpain (CAPN) gene with birth weight (BW), final weight (FW) and average daily gain (ADG) in three Egyptian sheep breeds: Barki, Rahmani and Ossimi. Blood samples were collected from 108 animals representing the three breeds. DNA was isolated using salting out procedure and then the quality and quantity of DNA extracted were measured. A 190 bp of CAPN was amplified by PCR using specific primers. The allele and genotype frequencies for all the identified SNPs were calculated. The PCR products corresponding to each genotype were sequenced to identify SNPs associated with the traits in question. Two SNPs (C→T) were detected in the nucleotides 44 and 154. For each SNP, the two mentioned alleles were named C and T, respectively. The sequenced CAPN segments were subjected to nucleotide blast at NCBI, which revealed 99% identity with that reported for sheep in Genbank. The TT was the least common genotype, whereas frequencies of CT and CC genotypes were fluctuated in the three sheep breeds under study. Animal carrier TT genotype had higher BW, FW and ADG than those with CT genotype, while the lowest values were associated with CC genotype. For the three traits under study, Rahmani had the highest estimates followed by Ossimi and Barki. Males exhibited heavier BW and FW as well as higher ADG compared with females. The results generated provide preliminary indication of the functional diversity present in Barki, Rahmani and Ossimi sheep and the possibility of using this polymorphism in Egyptian sheep genetic improvement.
Keywords: Egyptian sheep, Polymorphism, Calpain, PCR-RFLP, SNP
1. Introduction
Recent developments in molecular biology and statistics have provided the possibility of using genomic variation to accelerate the rate of genetic improvement of livestock. Application of marker-assisted selection (MAS) can be more effective in traits that are expressed late in the life of the animal or controlled by a few genes [1].
Calpains (CAPN) encode intracellular calcium-activated cysteine proteases that have been involved in several physiological and pathological processes [2]. Calpains are regulated by a variety of factors, including a 30-kDa small subunit [3], calcium and phospholipids [4], and calpastatin, a widely distributed calpain-specific inhibitor [5] and [6]. In living muscle, calpains are responsible for remodeling proteins that maintain the structure of skeletal muscle (myofibrillar linkage proteins, MLP) such as titin, nebulin and desmin [7]. The calpain activity is basically required for myoblast fusion as well as cellular proliferation and growth [8]. Polymorphism in CAPN gene has been identified in different breeds of sheep [9], [10], [11]. Page et al. [12] proposed CAPN as a potential candidate gene for meat tenderness in cattle, causing degradation in myofibrillar proteins postmortem. Calpain substrates include a variety of enzymes such as cytoskeletal proteins [13], kinases and phosphatases [14], and epidermal growth factor receptors [15]. The extent of physiological cleavage of these and other proteolytic proteins depends mainly on the presence and the activity of specific cell inhibitors.
The most common sheep breeds in Egypt are Barki, Ossimi and Rahmani. Barki is originated from the Libyan province Barka, and is spread along the coastal Mediterranean zone form west of Alexandria to the eastern provinces in Libya. Ossimi name is derived from a village near to Cairo, called Ossim, Giza governorate. It is the major breed in the Nile Valley and Delta zones, where the breed is generally more productive in middle Egypt rather than in Upper Egypt. Rahmani breed originates mainly from Northern Syria and Southern Turkey. The breed is most popular in the North and Middle of the Nile Delta. All breeds are classified as fat tailed animals producing coarse wool, well adapted to harsh environmental conditions and are raised basically for lamb production, followed by wool and milk. Generally animals belonging to the three sheep breeds are usually kept by small holders where herd size is of 1–3 heads, fed on low quality diets, bred through natural service, herd books and breed registration are generally not available [16].
The objective of the present study is to determine the polymorphism of calpain gene in three Egyptian sheep breeds and to evaluate the association of these SNPs with birth and final weights as well as average daily gain.
2. Material and methods
2.1. Animals
The present study was performed on 108 animals (males and females), representing the three Egyptian sheep breeds (Barki, Rahmani and Ossimi), animals were reared in the Agricultural Experiment Station, belonging to Faculty of Agriculture, Cairo University. Animals were classified into heavy and light final body weight (56 animals per breed per phenotype). A 10-ml blood sample was collected through vein puncture from each animal in a tube containing ethylenediaminetetraacetic acid (EDTA) as anticoagulant, and sent to the laboratory under cooled conditions. Traits measured were birth weight (BW), final weight at slaughter (FW) and average daily gain (ADG).
2.2. DNA extraction
Genomic DNA was extracted from the whole blood using salting out procedure described by Miller et al. [17]. Ultraviolet Spectrophotometer and 0.8 per cent agarose gel electrophoresis were used to check the quantity and quality of DNA.
2.3. Polymerase chain reaction (PCR)
The DNA fragment of the studied gene was amplified through PCR technique. This amplified fragment covered a part of exon 2, intron 2, exon 3 and a part of intron 3. The primer sequence of CAPN was the previously designed by Nassiry et al. [18].
A 20 μl PCR cocktail consisted of 20 pmol forward (AACATTCTCAACAAAGTGGTG) and reverse primers (ACATCCATACAGCCACCAT), 0.2 mM dNTPs and 1.25U of Taq DNA polymerase. The cocktail was aliquoted into PCR tubes with 100 ng of ovine DNA. The reaction was cycled with the following conditions; initial denaturation for 5 min at 94 °C followed by 35 cycles of denaturation at 94 °C, annealing at 60 °C and extension at 72 °C, each step for 1 min and the final extension for 5 min at 72 °C. The amplification was verified by electrophoresis on 2% agarose gel (w/v) in 1X TBE buffer. The size of PCR product was measured using Gene Ruler of 100-bp ladder as a molecular weight marker. The gel was stained with ethidium bromide and visualized under UV transilluminator.
2.4. Sequence analysis
The PCR products corresponding to distinct patterns were purified and got sequenced by Macrogen Incorporation (Seoul, South Korea). Sequence analyses and alignment to reveal nucleotide substitutions were carried out using nucleotide blast at NCBI (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi) for sequence homology and comparison searches in public databases [19].
2.5. Statistical analyses
Animals were assumed to be unrelated. Deviations from the Hardy–Weinberg equilibrium were tested by the χ2-test. The genotypic and allelic frequencies were calculated using XLSTAT [20]. The program was used to test the significance of the fixed effects on the studied traits (BW, FW and ADG). Marker genotypes were considered to be independent in all analyses. Association analysis of the single genotypes at different loci in CAPN gene was performed individually for the three studied traits in Barki, Rahmani and Ossimi breeds.
Statistical analyses were performed for BW, FW and ADG. The model used to analyze the data was as follows:
where,
Yijklm = the trait measured on each of the ijklmth animal,
μ = the overall population mean,
Gi = the fixed effect associated with ith genotype,
Bj = the fixed effect of jth breed (j = 1, 2, 3), where 1 = Barki, 2 = Ossimi and 3 = Rahmani,
Sk = the fixed effect of kth sex (k = 1, 2), where 1 = female and 2 = male,
Ilm = the effect of interaction between any two factors,
eijklm = random residual error.
Effects associated with year of birth and season of birth are not included in the linear model, as the preliminary statistical analyses revealed that these effects did not affect significantly the traits studied. The effects of farm, feeding regime and age of animals were not built into the linear model, because all animals were raised on the same farm.
3. Results
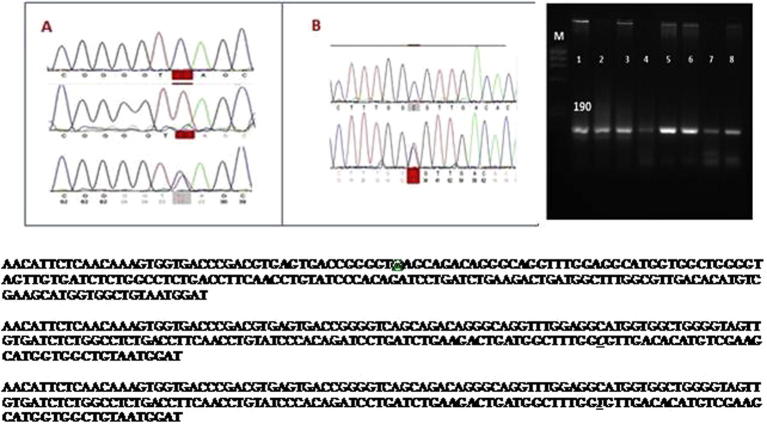
PCR amplification of the CAPN gene produced a 190 bp fragment in the samples genotyped of Barki, Rahmani and Ossimi breeds (Fig. 1).
Figure 1.
(A) PCR products of CAPN4 gene after running on 2% agarose gel electrophoresis. Fragment size for the CAPN4 was about 190 bp. Nucleotide sequence of 190 bp CAPN4 amplified fragment in high and low performance animals of Barki, Rahmani and Ossimi sheep. Intron 5 is in bold. “@”at nt-44 is nucleotide variation (C/T) is in italic and underlined. (c) (A) Demonstrations of sequenced results; the red color indicates the mutation sites. (B) Demonstrations of sequenced results; the red color indicates the mutation sites. (c). Nucleotide sequence of 190 bp CAPN4 amplified fragment in high and low performance animals of Barki, Rahmani and Ossimi sheep. Exon 6 is in bold. “C” is detected at nt-154 is in italic and underlined. (c). Nucleotide sequence of 190 bp CAPN4 amplified fragment in high and low performance animals of Barki, Rahmani and Ossimi sheep. Exon 6 is in bold. “T” is detected at nt-154 is in italic and underlined.
The PCR amplicons were purified and sequenced for SNP identification. Sequences obtained were aligned with the Genbank data to detect differences in nucleotide sequences. Variation in nucleotide sequences between the investigated samples and those of sheep published in Genbank were investigated using NCBI/Blast sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Moreover the obtained sequences were submitted and accepted at the international gene bank and got accession numbers: KT377442.1; KT377441.1; KT377440.1; KT377439.1; KT377438.1; KT377437.1; KT377436.1. The sequence analysis revealed two nucleotide substitutions at nt 44 in a part of intron 5 (C→T) and nt 154 in the part of exon 6 (C→T) (SNP position), this mutation is considered from the transition type. Accordingly, two alleles were detected (C and T) and subsequently three genotypes are expected (CC, CT and TT).
The obtained DNA sequence of the genotypes CC, CT and TT at nt 44 are shown in Fig. 1.
Results of the genotype and allele frequencies for SNP at the nt 44 are presented in Table 1. The C allele was the most frequent allele and subsequently the CC genotype was the most frequent genotype. Regarding the results of the genotype and allele frequencies for SNP at the nt 154 are presented in Table 2. It is noticed also that the C allele was more frequent than allele T, and subsequently the CC genotype was the most frequent followed by the CT then the TT genotypes.
Table 1.
Genotypic and allelic frequencies of Calpain4 gene for the SNPs at the position of nt 44.
| Breed | Genotype frequency |
Allele Frequency |
|||
|---|---|---|---|---|---|
| CC | CT | TT | C | T | |
| Barki | 0.50 | 0.50 | 0.00 | 0.75 | 0.25 |
| Rahmani | 0.37 | 0.44 | 0.19 | 0.59 | 0.41 |
| Osseimi | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
Table 2.
Genotypic and allelic frequencies of Calpain4 gene for the SNPs at the position of nt 154.
| Breed | Genotype frequency |
Allele Frequency |
|||
|---|---|---|---|---|---|
| CC | CT | TT | C | T | |
| Barki | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| Rahmani | 0.81 | 0.19 | 0.00 | 0.91 | 0.9 |
| Osseimi | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
In respect to the SNP at nt 154, two alleles were found (C and T), where C allele was highly dominant in Barki, Rahmani and Ossimi breeds. The TT genotype was absent in the three breeds, as shown in Fig. 1. Therefore, the SNPs and genotypes at nt 154 were not included in the association analysis.
Among the two SNPs examined in the CAPN gene, position 154 was monomorphic (CC genotype) in all the animals included in the present study, except three belonging to Rahmani breed that showed CC and CT genotypes. So, the result focused the final analysis only on the polymorphic and significant regions after the primary analysis. Samples selected to be sequenced were solely corresponding to position 44 of CAPN gene and included three genotypes (CC, CT and TT) for Rahmani and Ossimi; and only two genotypes (CC and CT) for Barki.
Least squares means and standard deviations as well as the minimum and maximum values for BW, FW and ADG are illustrated in Table 3. Moreover, least squares means of these traits for the three sheep breeds are presented in Table 4.
Table 3.
Summary statistics for phenotypic data.
| Trait | Minimum | Maximum | Mean | Std. deviation |
|---|---|---|---|---|
| Birth weight (kg) | 2.40 | 4.100 | 3.211 | 0.352 |
| Final weight (kg) | 34 | 63 | 47.026 | 6.281 |
| Average daily gain (g/day) | 68 | 142 | 93.450 | 15.980 |
Table 4.
Least squares means of birth weight, final weight and average daily gain (ADG) for Rahmani, Ossimi and Barki sheep breeds.
| Breeds | Birth weight (kg) | Final weight (kg) | ADG (g/day) | Breeds |
|---|---|---|---|---|
| Rahmani | 3.513 | 53.792 | 104.875 | Rahmani |
| Ossimi | 3.389 | 48.952 | 104.862 | Ossimi |
| Barki | 3.288 | 48.189 | 100.386 | Barki |
Table 4 shows the standardized regression coefficients of the studied variables including nucleotides in CAPNgene (nt 44-CC, nt 44-CT and nt 44-TT), breed of animal (Barki, Ossimi and Rahmani),sex of animal (females and males)and nucleotide by sex interaction on the three growth performance traits involved in the current study. Both CC and CT genotypes had negative effects on BW, FW and ADG. On the other hand, no assocaition (relationship) between TT genotype and the three traits studied has been established probably due to the limited number of samples and low frequency of T allele.
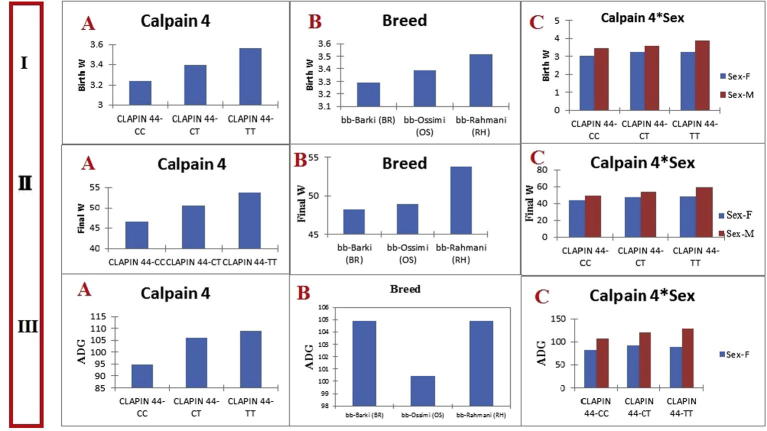
The TT genotype was associated with higher BW compared to CT and CC genotypes (Fig. 2IIA). The three sheep breeds exhibited variation in BW, where Rahmani had heavier BW than Ossimi and Barki breeds (Fig. 2IIB). Birth weight was significantly (P < 0.05) affected by sex of animal. Males with genotypes TT, CT and CC were heavier at birth than females with the corresponding genotypes (Fig. 2IIC).
Figure 2.
(I) A: The relation between the genotypes CC, CT and TT and birth weight for the CAPN4 gene. B: The relation between the sheep breeds (Barki, Ossimi and Rahmani) and birth weight for the CAPN4 gene. C: The relation between the genotypes (CC, CT and TT) by sex (male and female) interaction and birth weight for the CAPN4 gene. (II) A: The relation between the genotypes CC, CT and TT and final weight. B: The relation between the sheep breeds (Barki, Ossimi and Rahmani) and final weight for the CAPN4 gene. C: The relation between the genotypes (CC, CT and TT) by sex (male and female) interaction and final weight for the CAPN4 gene. (III) A: The relation between the genotypes CC, CT and TT and ADG for the CAPN4 gene. B: The relation between the sheep breeds (Barki, Ossimi and Rahmani) and ADG for the CAPN4 gene. C: The relation between the genotypes (CC, CT and TT) by sex (male and female) interaction and ADG for the CAPN4 gene.
Regarding the final weight, the standardized regression coefficients of genotype, breed, sex of animal and interaction of genotype with sex of animal on the three traits studied are given in Table 5. As shown in Fig. 3, animals with TT genotype had heavier FW, followed by those with CT and CC genotypes. Breed of animal affected significantly (P < 0.05) FW. Rahmani were significantly heavier at FW than Ossimi and Barki. Also, sex of animal had a significant (P < 0.05) effect on FW; where male carrier TT, CT and CC genotypes showed heavier FW compared to females with the same genotypes.
Table 5.
Standardized regression coefficients on birth weight, final weight and average daily gain (ADG).
| Source | Birth weight | Final weight | ADG |
|---|---|---|---|
| nt 44-CC | −0.575 | −10.395 | −0.649 |
| nt 44-CT | −0.429 | −6.000 | −0.284 |
| nt 44-TT | 0.000 | 0.000 | 0.000 |
| Barki | −0.323 | −4.937 | 0.061 |
| Ossimi | 0.072 | −2.679 | 0.000 |
| Rahmani | 0.000 | 0.000 | −0.897 |
| Sex-F | −0.789 | 14.012 | 0.000 |
| Sex-M | 0.000 | 0.000 | 0.411 |
| nt 44-CC*Sex-F | 0.233 | 6.325 | 0.000 |
| nt 44-CC*Sex-M | 0.000 | 0.000 | 0.360 |
| nt 44-CT*Sex-F | 0.358 | 5.623 | 0.000 |
| nt 44-CT*Sex-M | 0.000 | 0.000 | 0.000 |
| nt 44-TT*Sex-F | 0.000 | 0.000 | 0.000 |
| nt 44-TT*Sex-M | 0.000 | 0.000 | −0.649 |
Figure 3.
(I) Breed differences in birth weight, final weight and ADG for CAPN4 gene. (II) The effect of the genotypes (CC, CT and TT) by sex (male and female) interaction on birth weight, final weight and ADG for the CAPN4 gene.
Considering ADG, the standardized regression coefficients of independent factors and their pair-wise interactions on ADG are illustrated in Table 4. Fig. 2IIIA presents the effects of different subclasses of genotype, sex of animal and genotype by sex interaction on ADG. Genotype had a significant effect on ADG; where animals with TT genotype had higher ADG compared to those with CT and CC genotypes. The sheep breeds varied in their ADG; both Rahmani and Barki breeds exhibited significantly (P < 0.05) higher ADG than Ossimi breed. In addition, ADG was significantly (P < 0.05) affected by sex, where males with genotypes TT, CT and CC had higher ADG than females with the corresponding genotypes.
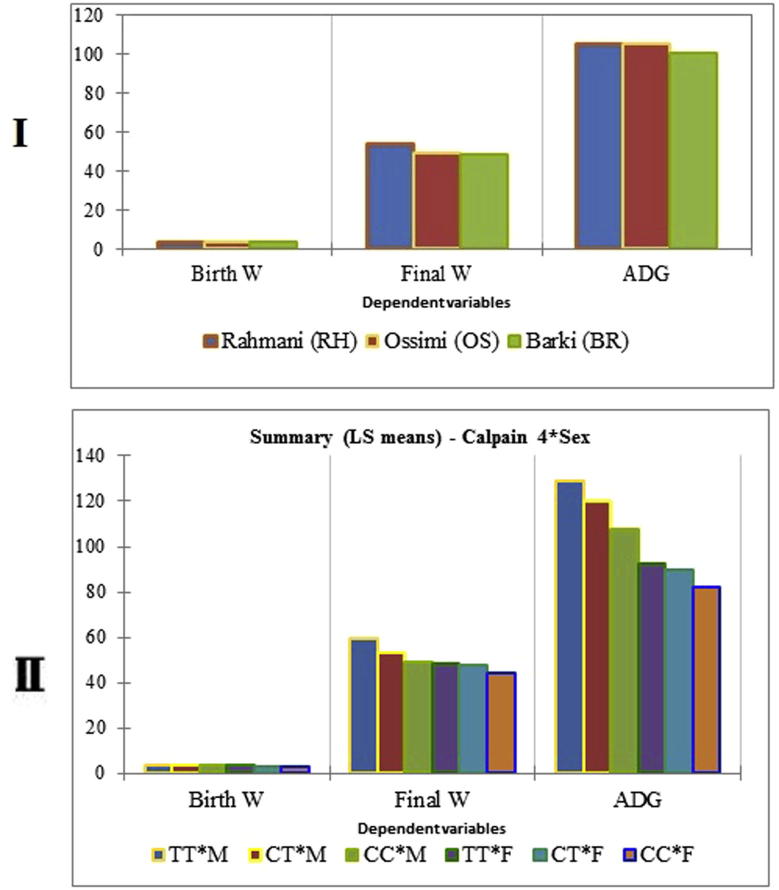
Summary of the different estimates of BW, FW and ADG for each breed are shown in Table 5 and Fig. 3I. In all traits studied, Rahmani breed had the best performance followed by Ossimi and Barki, respectively.
The cumulative effects of different genotypes of CAPN gene and sex of animal on BW, FW and ADG are shown in Table 7 and Fig. 3II. In general, males with any given genotype had heavier weight and higher ADG than their counterpart females.
Table 7.
Least squares means of genotypes (CC, CT and TT) by sex (male and female) interaction on birth weight, final weight and ADG for CAPN4 gene.
| Source | Birth weight | Final weight | ADG |
|---|---|---|---|
| TT*M | 3.872 | 59.443 | 128.673 |
| CT*M | 3.572 | 53.443 | 119.673 |
| CC*M | 3.461 | 49.048 | 107.644 |
| TT*F | 3.248 | 48.126 | 92.713 |
| CT*F | 3.219 | 47.748 | 89.360 |
| CC*F | 3.010 | 44.055 | 82.183 |
4. Discussion
The CAPNgene was investigated as a potential candidate gene for growth and meat tenderness as reported in earlier studies of Koohmaraie [21], Chung et al. [9], Arora et al. [10] and Naveen et al. [11]. The exon 5 and 6 as well as intron 5 in ovine CAPN gene were amplified and produced a 190 bp fragment. Part of this fragment from nt-1 to nt-28 corresponds to exon 5, from nt-29 to nt-127 corresponds to intron 5 and from nt-128 to nt-190 is part of exon 6 [9]. Our results of sequence analysis revealed two nucleotide substitutions at nt 44 in a part of intron 5 and nt 154 in a part of exon 6 (SNP position) and non-synonymous substitution was observed at the two position C→T (transition). The substitutions at exon 6 nt154 C→T, this SNP is a silent mutation {GGC (Glycine) → GGT (Glycine)}, which creates no substitution effect for amino acid sequence of Calpain protein. Arora et al. [10] identified nine novel SNPs in four candidate genes, including CAPN4, for mutton quality traits across 11 Indian sheep breeds. Of these SNPs, six transitions and three transversions were observed. The primers for CAPN4 were in exons 4 and 5 and also in intron 4. In contrast to those results obtained in the present study, Naveen et al. [11] amplified by PCR-SSCP 192 bp of exons 5 and 6 of ovine CAPN gene and found genotype frequency of 0.672 and 0.295 for AA and AB, and allele frequency of 0.820 and 0.180 for A and B, respectively. Analysis of sequences revealed addition/insertion of one nucleotide ‘A’ in B allele.
Three different patterns (CC, CT and TT) were found at the SNP position nt 44, while at nt 154 there were two patterns (CC and CT) detected in the three sheep breeds studied. With regard to allelic frequencies, the C allele was the predominant one in all breeds. The highest frequency for T allele was found in Rahmani, while the lowest one was observed in Barki. In all the three breeds studied the frequncy of CC and CT genotypes were comparable, while the frequncy of TT genotype was always the lowest. The identification of two genotypes in CAPN gene has been demonstrated by Nassiry et al. [18] and Naveen et al. [11] in Iranian Kurdi sheep and in Indian Bandur sheep.
Contradicting the current study, Dehnavi et al. [22] using single strand conformational polymorphism (SSCP) found two alleles (A and B) which produced only two genotypes (AA and AB), with frequencies of 0.69and 0.31, respectively. Similar results were previously reported by Tahmoorespour et al. [23] who found A and B alleles with frequencies of 0.56 and 0.44 for CAPN in Baluchi sheep. In Karakul sheep, Chung et al. [9] found allelic frequencies of 0.69 for A and 0.31 for B, while the genotypic frequencies of AA, AB and BB were 0.70, 0.30 and 0.00,respectively.
Females carry CT genotype for CAPN gene was heavier at birth than those with CC genotype. On the other hand, females with CC genotype had heavier FW compared to female carrier CT genotype. The results obtained suggest that specific nucleotides can be used as genetic markers in early selection process for heavier birth or final weights.
Males with CT genotype had significantly heavier BW, FW and ADG compared to females with the same genotype (Table 6). Similarly, males showed higher estimates than females for the three traits in the three breeds investigated. Sex of animal influenced significantly birth weight. Superiority in growth performance of males compared to females in various sheep breeds was reported by De Zegher et al. [24], Loos et al. [25] and Cruickshank et al. [26].
Table 6.
Summary of least squares means of Rahmani, Ossimi and Barki breeds on birth weight, final weight and ADG for CAPN4 gene.
| Breed | Birth weight | Final weight | ADG |
|---|---|---|---|
| Rahmani | 3.513 | 53.792 | 104.875 |
| Ossimi | 3.389 | 48.952 | 104.862 |
| Barki | 3.288 | 48.189 | 100.386 |
Esenbuga and Dayıoğlu [27] reported that sex had highly significant effect on BW of Awassi and Red-Karman lambs. Babar et al. [28] indicated that BW in Lohi sheep was significantly influenced by sex of the lamb. In contrast, Sahani et al. [29] and Guevara et al. [30] reported that sex of animal had no significant effect on BW of Marwari and Pelibuey X Wiltshire Horn lambs. Differences in sexual chromosomes, probably in the position of genes related to growth, physiological characteristics and difference in endocrinal system (type and measure of hormone secretion, especially sexual hormones) lead to difference in animal growth. In relation to endocrinal system, estrogen hormone has a limited effect on the growth of long bones in females. This could be one of the reasons for which females have smaller body size and lighter body weight in comparison to males [31], [32], [33]. Compbella et al. [34] reported significant effects of the breed on live weight gains of sheep from birth to weaning. Also, Taiwo et al. [35] reported significant differences between sheep breeds in post-weaning gain.
Breed of animal affected significantly traits studied. Rahmani breed had the heaviest birth and final weightsfollowed by Ossimi and Barki, respectively. Considering ADG, Rahmani and Barki had higher ADG than Ossimi breed. The results obtained in this study demonstrated the importance of breed, sex and genotypes as sources of variation in BW, FW and ADG in Egyptian sheep breeds.
The existence of major genes associated with growth performance can accelerate the rate of genetic improvement in such quantitative traits through the integration of information provided by genes/markers into traditional selection schemes in so called marker-assisted selection (MAS). Subsequently, this would provide excellent opportunities for improving meat quality and quantity.
One of the aims of the present study is to determine the polymorphism of calpain gene in Egyptian sheep breeds. Another aim was to associate the detected polymorphism in CAPN gene with growth performance. The preliminary findings indicate that the genotypes detected in CAPN gene can be used as a candidate gene marker for body weight and average daily gain. In order to substantiate these observations, future studies will be needed to ascertain and establish the effects of reported SNPs in the Egyptian sheep breeds.
5. Conclusion
The present study was conducted to determine the polymorphism of calpain gene in Egyptian sheep. Of the two alleles detected in the gene, C allele was always predominant over T allele in Barki, Rahmani and Ossimi breeds. The obtained results indicated that the TT was the least common genotype in all breeds, while frequencies of CT and CC genotypes were fluctuated between nucleotide positions (44 vs. 154) in Rahmani breed. Male animal carrier of TT genotype had significantly heavier birth and final weights as well as higher average daily gain than those with CT genotype, while the lowest values were associated with CC genotype. Also, TT genotype in females was associated with the best performance in the three traits under study, followed by CT and CC genotypes. Rahmani breed had heavier birth and final weights than Barki, which was preceeded by Ossimi. Both Rahmani and Ossimi had on average higher ADG than Barki breed. Because the CAPN gene is considered as candidate gene for growth and meat quality traits, additional studies may be conducted to ascertain the association of those reported polymorphisms with meat production traits in the Egyptian sheep breeds.
Acknowledgements
This work was funded by National Research Center, Dokki, Giza, Egypt. The authors are grateful to the staff of Agricultural Experiment Station, Faculty of Agriculture, Cairo University for the help provided during sample collection.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Montaldo H.H., Meza-Herrera C.A. Use of molecular markers and major genes in the genetic improvement of livestock. Electron. J. Biotechnol. 1998;1(2):83–89. [Google Scholar]
- 2.Goll D.E., Thompson V.F., Taylor R.G., Zalewska T. Is Calpain activity regulated by membranes and autolysis or by calcium and Calpastatin. BioEssays. 1992;14:549–556. doi: 10.1002/bies.950140810. [DOI] [PubMed] [Google Scholar]
- 3.Sorimachi H., Shoichi I., Suzuki K. Structure and physiological function of calpin. Biochem. J. 1997;328:721–732. doi: 10.1042/bj3280721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saido T.C., Nago S., Shiramine M., Tsukaguchi M., Yoshizawa T., Sorimachi H., Ito H., Tsuchiya T., Kawashima S., Suzuki K. Distinct kinetics of subunit autolysis in mammalian m-calpain activation. FEBS Lett. 1994;346:263–267. doi: 10.1016/0014-5793(94)00487-0. [DOI] [PubMed] [Google Scholar]
- 5.Kawasaki H., Emori Y., Suzuki K. Calpastatin has two distinct sites for interaction with calpain effect of calpastatin fragments on the binding of calpain to membranes. Arch. Biochem. Biophys. 1993;305(2):467–472. doi: 10.1006/abbi.1993.1448. [DOI] [PubMed] [Google Scholar]
- 6.Croall D.E., McGrody K.S. Domain structure of calpain: mapping the binding site for Calpastatin. Biochemistry. 1994;33:13223–13230. doi: 10.1021/bi00249a008. [DOI] [PubMed] [Google Scholar]
- 7.Huff-Lonergan E., Mitsuhashi T., Beekman D.D., Parrish F.C., Olsonand D.G., Robson R.M. Proteolysis of specific muscle structural proteins by m calpain at low pH and temperature is similar to degradation in postmortem bovine muscle. J. Anim. Sci. 1996;74:993–1008. doi: 10.2527/1996.745993x. [DOI] [PubMed] [Google Scholar]
- 8.Kuryl J., Kapelanski W., Pierzchala M., Grajewska S., Bocian M. Preliminary observation on the effect of calpastation gene (CAST) polymorphism on carcass traits in pigs. J. Anim. Sci. 2003;21:87–95. [Google Scholar]
- 9.H.Y. Chung, M.S. Davis, H.C. Hines. The Ohio State University. Midwestern Section. Am. Soc. Anim. Sci., Supplement 2.79, 32–110, (2001) 37.
- 10.Arora R., Yadav H.S., Yadav D.K. Identification of novel single nucleotide polymorphisms in candidate genes for mutton quality in Indian sheep. Anim. Mol. Breed. 2014;4(1):1–5. [Google Scholar]
- 11.Naveen K.S., Jayashankar M.R., Nagaraja R., Nagaraja C.S., Nadeem F., Satyanarayana K. Genetic polymorphism of ovine Calpine gene in Bander sheep. Int. J Sci. Environ. Technol. 2015;4(3):804–812. [Google Scholar]
- 12.Page B.T., Casas E., Heaton M.P., Cullen N.G., Hyndman D.L., Morris C.A., Crawford A.M., Wheeler T.L., Koohmaraie M., Keele J.W., Smith T.P.L. Evaluation of single-nucleotide polymorphisms in CAPN1 for association with meat tenderness in cattle. J. Anim. Sci. 2002;80:3077–3085. doi: 10.2527/2002.80123077x. [DOI] [PubMed] [Google Scholar]
- 13.Schoenwaelder S.M., Yuan Y., Cooray P., Salem H.H., Jackson S.P. Calpain of focal adhesion proteins regulates the cytoskeletal attachment of integrin aIIbß3 (Platelet Glycoprotein IIb/IIIa) and the cellular retraction of fibrin clots. J. Biol. Chem. 1997;272:1694–1702. doi: 10.1074/jbc.272.3.1694. [DOI] [PubMed] [Google Scholar]
- 14.McGinnis M.K., Whitton M.M., Gnegy M.E., Wang K.W. Calcium/calmodul independent protein kinase IV is cleaved by caspase-3 and calpain in SH-SY5Y human neuroblastoma cells undergoing apoptosis. J. Biol. Chem. 1998;273:19993–20000. doi: 10.1074/jbc.273.32.19993. [DOI] [PubMed] [Google Scholar]
- 15.Glading A., Chang P., Lauffenburger D.A., Wells A. Epidermal growth factor receptor activation of calpain is required for fibroblast motility and occurs via an ERK/MAP kinase signaling pathway. J. Biol. Chem. 2000;275(4):2390–2398. doi: 10.1074/jbc.275.4.2390. [DOI] [PubMed] [Google Scholar]
- 16.Galal S. Farm animal genetic resources in Egypt: factsheet. Egypt. J. Anim. Prod. 2007;44:1–23. [Google Scholar]
- 17.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl. Acids Res. 1988;16:12–15. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nassiry M.R., Shahroudi F.E., Tahmoorespur M., Javadmanesh A. Genetic variability and population structure in beta-lactoglobulin, calpastatin and calpain loci in Iranian Kurdi sheep. Pak. J. Biol. Sci. 2007;10(7):1062–1067. doi: 10.3923/pjbs.2007.1062.1067. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.XLSTAT, Statistical and data analysis software packages for Microsoft Excel. 224 Centre Street, 3rd Floor, New York, NY 10013, USA, 2014.
- 21.Koohmaraie M. Ovine skeletal muscle multicatalytic proteinase complex (proteasome): purification, characterization, and comparison of its effects on myofibrils with mu-calpains. J. Anim. Sci. 1992;70:3697–3708. doi: 10.2527/1992.70123697x. [DOI] [PubMed] [Google Scholar]
- 22.Dehnavi E., AhaniAzari M., Hasani S., Nassiry M.R., Mohajer M., Ahmadi A.K., Shahmohamadi L., Yousefi S. Polymorphism of Myostatin Gene in Intron 1 and 2 and Exon 3, and Their Associations with YearlingWeight, Using PCR-RFLP and PCR-SSCP Techniques in Zel Sheep. Biotechnol. Res. Int. 2012;12:133–138. doi: 10.1155/2012/472307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. Tahmoorespour, M.R. Nassiry, A. Javadmanesh, in: Proceedings of the 1st Agricultural Biotechnology Conference of Iran. Iran, 51, 2005.
- 24.De Zegher F., Devlieger H., Eeckels R. Fetal growth: boys before girls. Horm. Res. 1999;51:258–259. doi: 10.1159/000023382. [DOI] [PubMed] [Google Scholar]
- 25.Loos R.J., Derom C., Eeckels R., Derom R., Vlietinck R. Length of gestation and birthweight in dizygotic twins. Lancet. 2001;358(9281):560–561. doi: 10.1016/S0140-6736(01)05716-6. [DOI] [PubMed] [Google Scholar]
- 26.Cruickshank J.K., Mzayek F., Liu L., Kieltyka L., Sherwin R., Webber L.S., Srinavasan S.R., Berenson G.S. Origins of the ‘black/white’ difference in blood pressure: roles of birth weight, postnatal growth, early blood pressure, and adolescent body size: the Bogalusa heart study. Circulation. 2005;111:1932–1937. doi: 10.1161/01.CIR.0000161960.78745.33. [DOI] [PubMed] [Google Scholar]
- 27.Esenbuga N., Dayıoğlu H. Effect of some environmental factors on grown traits of Awassi and Red Karaman Lambs. Turk. J. Vet. Anim. Sci. 2002;26:145–150. [Google Scholar]
- 28.Babar M.E., Ahmad Z., Nadeem A., Yaqoob M. Environmental factors affecting birth weight in Lohi sheep. Pak. Vet. J. 2004;24(1):5–8. [Google Scholar]
- 29.Sahani M.S., Bapra D.L., Singh M. Influence of various non-genetic factors on pre- and post-weaning body weight of Marwari lambs under hot arid region. Livestock Adviser. 1989;14(11):41–45. [Google Scholar]
- 30.Guevara V.G., Cero R.A., Patao G.Y., Isaela A.T., Quiroz V.O., Llano P.N. Preweaning growth in 3/4 Pelibuey-114 Wiltshire Horn lambs. Revista de Production Animal, Cuba. 1993;7(3):173–175. [Google Scholar]
- 31.Vaez-torshizi V.R., Jomeh E.N., Nik-Khah A., Hejazi M. A study of pre-weaning traits in a Baluchi Sheep flock. Iran. J. Agric. Sci. 1992;23:33–42. [Google Scholar]
- 32.Shahroudi E.F., Shiri A., Twakolyan J., DaneshMesgaran M. Estimation of maternal effects on growth traits of Kurdish lam in north of khorasan. Pajouhesh Sazandegi. 2003;50:62–66. [Google Scholar]
- 33.Rashidi A., Mokhtari M.S., Jahanshahi A.S., Abadi M.R.M. Genetic parameter estimates of pre-weaning growth traits in Kermani sheep. Small Ruminant Res. 2008;74(1):165–171. [Google Scholar]
- 34.Cambella J., Martinaz N., Gonalez J. Study of some factors affecting birth and weaning weights of lambs. Anim. Breed. Abstr. 1990;48:853. [Google Scholar]
- 35.Taiwo B.B.A., Ngere L.O., Adebayo I.O.A. Comparative growth performance of Nigerian dwarf sheep and its crosses with Permer, Uda and Yankassa. World Rev. Animal Prod. 1992;18:57–63. [Google Scholar]