Abstract
Because of its high case fatality rate, listeriosis locates among the most frequent causes of death due to food-borne illness. In this study, a total of 150 processed meat samples were collected from Giza Governorate, Egypt. Phenotypic and genotypic identification of Listeria monocytogenes was performed using PCR incorporating listeriolysin O virulence gene hlyA followed by DNA sequence analysis. L. monocytogenes was confirmed in 4% of each of beef burger, minced meat, and luncheon samples. Phylogenetic analysis showed that all the six Egyptian isolates have high homology with Colombian isolate (EF030606), except one Egyptian isolate which showed high homology with Indian isolate (EU840690). The public health significance of these pathogens as well as recommended sanitary measures were discussed.
Keywords: L. monocytogenes, Processed meat, Egypt, Phylogenetic analysis
1. Introduction
The genus Listeria includes only two pathogenic strains, Listeria monocytogenes is a well-known cause of abortion, encephalitis and septicemia in man and animals. Listeria ivanovii is of major veterinary importance as a cause of abortions, still births and neonatal septicemias in sheep and cattle, and a rare human infection [24].
L. monocytogenes has become remarkably important as a food-borne pathogen. The ability to persist in diverse conditions such as low temperature and pH, high salt concentrations and multiplication under refrigeration temperatures makes it a serious threat to public health [7], [12]. Because of its high case fatality rate, listeriosis ranks among the most frequent causes of death due to food-borne illness with the highest hospitalization rates (91%) and mortality rate up to 30% [13].
The detection of this pathogen in food by routine culture methods is difficult by the sporadic or low levels of contamination, by the presence of a high level of background microflora [17]. Moreover, these methods are laborious and time consuming [1], while immediate action should be taken in case of contamination especially in the case of foods having short shelf-lives, such as meat or dairy products. In general, DNA-based typing approaches are recognized as simple and cost-effective methods that have better discriminatory power than phenotypic approaches and being better for investigating L. monocytogenes outbreaks [5]. The classification based on the nucleic acid sequence has become more common. The gene hly encoding listeriolysin O is one such target gene for specific detection of L. monocytogenes [21].
Molecular fingerprinting is by far considered the most precise method for studying the epidemiology of foodborne diseases and allows prediction of the relationships between Listeria isolates, regardless of the origin or the geographical location [9].
Usually, L. monocytogenes is susceptible to a wide range of antibiotics, but resistance to multiple antibiotics was recorded [18]. The presence of multiple key virulence factors (Virulence markers) such as listeriolysin O (LLO encoded by hlyA) significantly regulates the virulence and pathogenicity of L. monocytogenes [21]. Moreover, food and host environments may present variable expression of virulence genes, resulting in varied infectivity [4]. Therefore, it is essential to study the epidemiological significance and distribution of virulence determinants within these isolates [23]. Thus the aim of this study was to determine the prevalence and phylogenetic characterization of L monocytogenes isolated from processed meat collected from markets in Egypt.
2. Materials and methods
2.1. Samples
A total of 150 meat samples were collected from Giza Governorate over the period of October 2013 to September 2014. These samples included 25 minced meats, 25 luncheons, 50 sausages, 50 beef burger.
2.2. Isolation and identification of L. monocytogenes
L. monocytogenes was isolated from the examined food samples according to the International Organization for Standardization procedure [11]. Suspected colonies were transferred to tryptic soya agar plates with 0.6% yeast extract (TSA-YE) for further biochemical identification using Listeria Microbact 12L (Oxoid, UK). For DNA extraction, colonies were suspended in 500 μl of PBS, pH 7.2, washed 3 times in PBS. The cell suspension was centrifuged for 10 min at 800g, then the supernatant was discarded carefully and the pellet was dried and stored at −20 °C till use.
2.3. Polymerase chain reaction (PCR) analysis and DNA sequencing
2.3.1. DNA extraction
For extraction of DNA, bacterial pellets were re-suspended with 200 μl PBS. DNA was extracted from L. monocytogenes isolates using the DNA extraction Kits (GF-1, Vivantis Co., Malaysia) according to manufacturer’s instructions.
2.3.2. PCR
This PCR amplifies a 234 bp region of the hlyA gene (ᾳ-Hemolysin, listeriolysin O) encoding listeriolysin O. PCR was performed according to Furrer et al. [8]. 50 μl volume containing 2 μl of each primer (10 μM), 25 μl of 2X Taq Master Mix (Cat. No. PLMM01,Vivantis Co., Malaysia), Primers, LMA: CGGAGG TTCC GCAAAAGATG and LMB: CCTCCAGAGTGATCGATGTT. Polymerase chain reaction (PCR) amplification conditions were: 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 45 s at 55 °C, 45 s at 72 °C and a final extension of five min at 72 °C. The PCR products were analyzed using 1% agarose gel electrophoresis.
The positive PCR products were then sequenced in MACROGEN Company (Korea) on 3730 × L sequencers (Applied Biosystem, USA). The accuracy of data was confirmed by two-directional sequencing with the forward and reverse primers used in PCR.
The nucleotide sequences obtained in this study were analyzed using the BioEdit 7.0.4.1 and ClustalW2 (http://www.clustal.org/) programs. The resulting sequences were aligned with hlyA gene of reference sequences of Liateria spp. using a neighbor-joining analysis of the aligned sequences implemented in the program CLC Sequence Viewer 6.
3. Results and discussion
Contamination of the meat with L. monocytogenes generally occur after the slaughter and may come from the skin of the animals, the hands of the workers, the equipment and the tools used [16]. It is also important to comment that the presence of any Listeria spp. may be indicative of poor hygiene and cross-contamination scenarios which cold favor the persistence of L. monocytogenes [2].
In the present study, PCR results showed that L. monocytogenes were confirmed in 8 out of 25 of beef burgers examined (4%), one out of 25 of minced meat (4%), and one out of 25 of Luncheon (4%) from the total 100 examined food samples. The sausage samples were totally negative.
This suggests the presence of a significant public health hazard linked to the consumption of this meat sold in Giza Governorate contaminated with L. monocytogenes.
3.1. Nucleotide sequence accession numbers
Six sequences PCR samples (Giza 1–6) used in this study have been deposited in the Gene Bank database under accession no: KR812472, KR812473, KR81274, KR534875, KR812475 and KR812476 respectively. Phylogenetic analysis showed that all the six Egyptian isolates have high homology with Colombian isolate (EF030606), except one Egyptian isolate which showed high homology with Indian isolate (EU840690). This may be due to the importation of animals and raw meat from the Latin America and India. This suggests the presence of a significant public health hazard linked to the consumption of this meat sold in Egypt contaminated with L. monocytogenes.
From Fig. 1, the differences in the sequences of the six Egyptian isolates (Giza 1–6) which we examined in comparison with the other Gene Bank isolates from different isolation sources depending on the analysis of the hlyA gene may reflect sequencing artifacts or may be true differences. Sallen et al. [20] reported the common presence of sequence polymorphism between different isolates of the same Listeria species.
Figure 1.
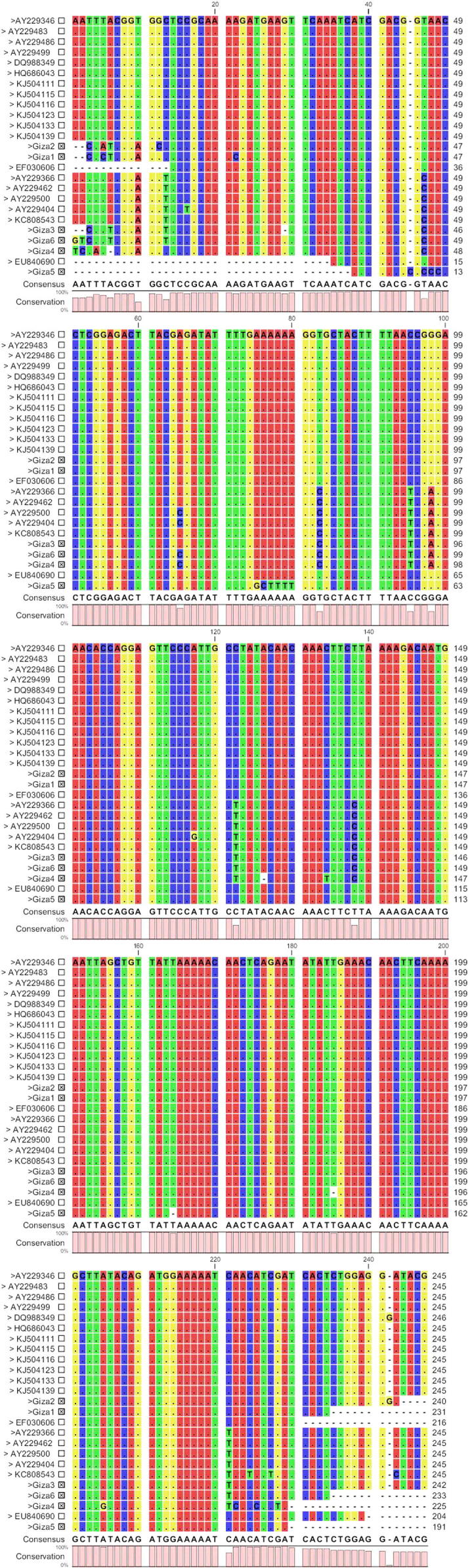
Sequence alignment of Listeria monocytogenes (present study) isolated from processed meat (Giza 1–6) with the closely related isolates from different sources. Only variable sites are shown with different color. Dashes in the middle indicate gaps.
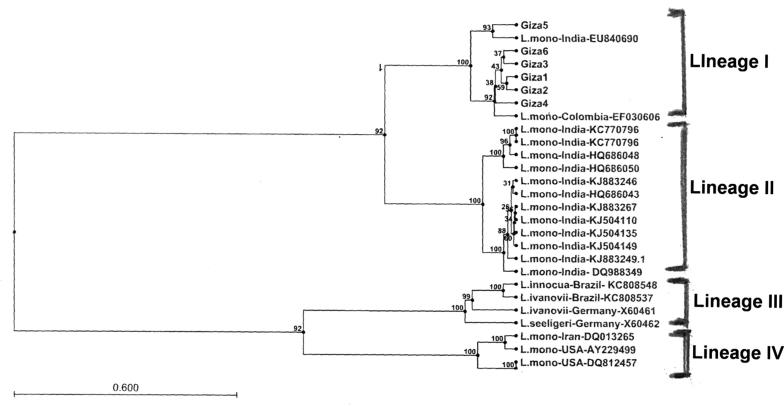
DNA sequencing of representative samples confirmed the identification of the Listeria species. Blasting the obtained sequences with those in database and the deduced phylogenetic analysis (Figure 2, Figure 3) were induced. Clear sequences of hlyA gene were obtained from the 6 isolates. Homology results (98–99%) proved that the six isolates were L. monocytogenes. The neighbor-Joining (NJ) phylogenetic analysis based on the hlyA gene (Fig. 2) showed that all the six Egyptian isolates were clustered with relevant sequences of L. monocytogenes EU840690 and EF030606 isolates in the same group (Lineage 1) as shown in Fig. 2.
Figure 2.
Phylogenetic relationship of selected strains of Listeria spp. from different geographical areas, representing the four distinct lineages, based on the listeriolysin gene. The Gene Bank accession numbers of the isolates used are given.
Figure 3.
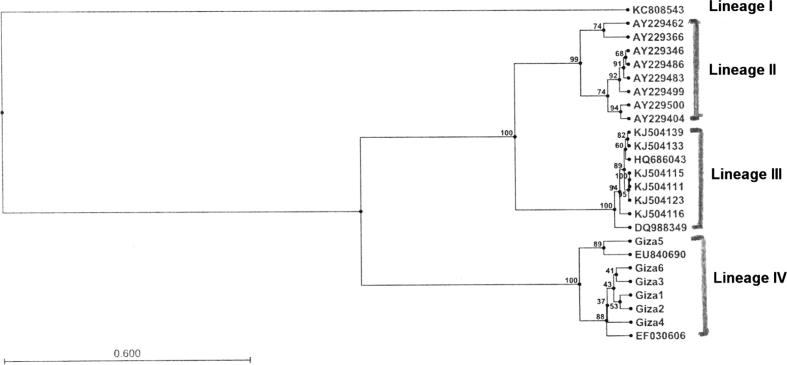
Phylogenetic relationship of selected strains of Listeria spp. from different sources, representing the four distinct lineages, based on the listeriolysin gene. The Gene Bank accession numbers of the isolates used are given.
Concerning the geographical distribution (Fig. 2), hlyA gene analysis of our isolates in comparison with other L. monocytogenes isolates from India, Colombia, Brazil and USA showed that all the Egyptian isolates have high homology with Colombian isolate (EF030606), Except Egypt 5 isolate which showed high homology with Indian isolate (EU840690). This may be due to the importation of animals and raw meat from the Latin America and India.
This was confirmed by Phylogenetic analysis based on the hly gene of L. monocytogenes applied by Headley et al. [10] which revealed the identification of L. monocytogenes isolates from cattle and small ruminants in Brazil with encephalitic listeriosis. This finding is of significant importance to understand the epidemiology of L. monocytogenes in Egypt.
The phylogenetic tree based on hlyA gene sequence clearly differentiates between the L. monocytogenes, Listeria innocua, L. ivanovii and Listeria seeligeri. It was noticed from Fig. 2 that our isolates were in a different cluster (lineage I) from that of the other Listeria spp. as shown in Lineage III.
This was different when using the 16S rRNA gene for phylogenetic analysis as reported by Soni and Dubey [21] and Soni et al. [22], that noticed that due to the close relationship (>99%) between the members of Listeria species, other Listeria sp. (L. innocua, L. ivanovii, L. seeligeri and Listeria welshimeri) are also clustered in same L. monocytogenes group because of the highly conserved nature of the 16S rRNA gene among its species.
In order to ascertain the presence of pathogens in their respective environment, the detection of one of the major virulence factors is a better option. Among the various virulence factors, LLO (a 58 kDa hemolysin protein encoded by hly gene) is the main virulence factor and pathogenic marker for the detection of Listeria sp. [3].
In the present study, we identified 6 isolates of L. monocytogenes originating from processed meat based on hlyA gene sequence. On the basis of sequence similarity, all the Egyptian Isolates (except Egypt 5) were found to be closely related to L. monocytogenes isolate EF030606, isolated from human clinical samples and from food processing plants and food (Hams and chicken) in Colombia while Egyptian isolate5 was closely related to isolate EU840690 separated from raw and packaged meat and meat products in India as shown in the lineage (4) of phylogenetic tree (Fig. 3). The details of isolate, strain isolation source, country and accession numbers are shown in Table 1.
Table 1.
Details of L. monocytogenes isolates used in the present study with the available ones in Gene Bank from different habitats and country.
| Ser. no | Isolate | Strain | Serotype | Source of Isolation | Country | Access number |
|---|---|---|---|---|---|---|
| 1 | Giza 1 | L. monocytogenes | – | processed meat | Egypt | KR812472 |
| 2 | Giza 2 | L. monocytogenes | – | processed meat | Egypt | KR812473 |
| 3 | Giza 3 | L. monocytogenes | – | processed meat | Egypt | KR812474 |
| 4 | Giza 4 | L. monocytogenes | – | processed meat | Egypt | KR534875 |
| 5 | Giza 5 | L. monocytogenes | – | processed meat | Egypt | KR812475 |
| 6 | Giza 6 | L. monocytogenes | – | processed meat | Egypt | KR812476 |
| 7 | 72 | L. monocytogenes | 4d | Clinical and environmental (poultry meat) | USA | AY229346 |
| 8 | 57 | L. monocytogenes | 3b | Clinical and environmental (poultry meat) | USA | AY229366 |
| 9 | 193 | L. monocytogenes | 3a | Clinical and environmental (poultry meat) | USA | AY229404 |
| 10 | 126 | L. monocytogenes | 1/2b | Clinical and environmental (poultry meat) | USA | AY229462 |
| 11 | 101 | L. monocytogenes | 1/2c | Clinical and environmental (poultry meat) | USA | AY229483 |
| 12 | 88 | L. monocytogenes | 4a | Clinical and environmental (poultry meat) | USA | AY229486 |
| 13 | 168 | L. monocytogenes | 4b | Clinical and environmental (poultry meat) | USA | AY229499 |
| 14 | 172 | L. monocytogenes | 1/2a | Clinical and environmental (poultry meat) | USA | AY229500 |
| 15 | – | L. monocytogenes | – | Poultry meat | India | DQ988349 |
| 16 | LMO6 | L. monocytogenes | – | Human clinical samples and from food processing plants and food (Hams and chicken) | Colombia | EF030606 |
| 17 | 62TP_BRSL3 | L. monocytogenes | – | Raw and packaged meat and meat products | India | EU840690 |
| 18 | W1 | L. monocytogenes | – | River gangs water | India | HQ686043 |
| 19 | Lm4 | L. monocytogenes | – | Swine slaughter house & meat market | Brazil | KC808543 |
| 20 | Pb1 | L. monocytogenes | – | Human placental bit | India | KJ504111 |
| 21 | VS1 | L. monocytogenes | – | Human vaginal swab | India | KJ504115 |
| 22 | M2 | L. monocytogenes | – | Cow milk | India | KJ504116 |
| 23 | VB1 | L. monocytogenes | – | Vegetables | India | KJ504123 |
| 24 | VT1 | L. monocytogenes | – | Vegetables | India | KJ504133 |
| 25 | VC1 | L. monocytogenes | – | Vegetables | India | KJ504139 |
The existence of the isolate KC808543 in separate group (cluster1) may be due to that it was isolated from swine not bovine slaughter house, while cluster 2 was isolated from human clinical samples and poultry meat. Finally, the cluster 3 was isolated from different sources (clinical samples, river water, poultry meat, cow’s milk and vegetables). This explains why our isolates were grouped in the cluster 4 which were isolated from raw, packaged meat and food processing plants.
However, we have found that strains from lineage 4 may be more likely to survive thermal inactivation than strains from the other three lineages of L. monocytogenes, indicating that lineages 4 may be better adapted to the food-processing environment than the other three lineage isolates, this was agreed by De Jesus and Whiting [6], which explains that with the development of unique ecological adaptations for these isolates such as enhanced psychrotolerance and growth at refrigeration temperatures.
The close relation between our isolates and Colombian isolate (EF030606) that was isolated from some human clinical samples demonstrate the close relationship between the disease in human and animals. Finally this proof that our Egyptian isolates are more frequently isolated in food-borne epidemics of listeriosis and in sporadic cases of human and animal listeriosis. This was agreed by Liu et al. [14] and Roberts et al. [19].
Finally, in order to minimize human listeriosis, foods should be cooked to an internal temperature of 70 °C for more than 20 min to ensure distraction of L. monocytogenes. Reheat cooked food thoroughly (70 °C) immediate aseptic packaging of the finished product to avoid post processing environmental contamination. Proper cold storage of meat and meat products (freezing −18 °C) and proper personal hygiene of food handlers is advisable as reported by Mahmoud et al. [15].
In conclusion, the hlyA gene sequence information could be the reliable option to indicate the presence of L. monocytogenes, tracking the source of infection and identifying the geographical distribution and divergence. More work, however, is required on L. monocytogenes in order to ascertain its presence in clinical and environmental samples in Egypt as there is limited information available on such aspects for tropical countries including Egypt. The molecular technologies such as the next generation sequencing are likely to be helpful in rapid acquisition of sequence data to facilitate detection and characterization of other pathogenic strains of L. monocytogenes.
Acknowledgements
The authors wish to express their gratitude to the National Research Centre, Cairo, for providing and financial support of this study.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Anon . 1996. Rapporti ISTISAN ISSN; pp. 1123–3117. 96/35. [Google Scholar]
- 2.Azevedo I., Regalo M., Mena C., Almeida G., Luisa Carneiro L., Teixeira P., Hogg T., Gibbs P. Food Control. 2005;16(2):121–124. [Google Scholar]
- 3.Barbuddhe S.B., Chaudhari S.P., Malik S.V.S. J. Vet. Med. B. 2002;49:181–184. doi: 10.1046/j.1439-0450.2002.00527.x. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee S.S., Hossain H., Otten S., Kuenne C., Kuchmina K., Machata S., Domann E., Chakraborty T., Hain T. Infect. Immun. 2006;74(2):1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B., Pyla R., Kim T., Silva J.L., Jung Y. Food Microbiol. 2010;27:645–652. doi: 10.1016/j.fm.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 6.De Jesus A.J., Whiting R.C. J. Food Prot. 2003;66:1611–1617. doi: 10.4315/0362-028x-66.9.1611. [DOI] [PubMed] [Google Scholar]
- 7.Fayol L., Beizig S., Monnier Le A., Lacroze V., Simeoni U. Arch. Pediatr. 2009;164:353–356. doi: 10.1016/j.arcped.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Furrer B., Candrian U., Hoefelein C., Luethy J. J. Appl. Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 9.Gendel S.M., Ulaszek J. J. Food Prot. 2000;63:179–185. doi: 10.4315/0362-028x-63.2.179. [DOI] [PubMed] [Google Scholar]
- 10.Headley S.A., Lívia Bodnar, Juliana T.T., Fritzen D.E., Bronkhorst, Alice F., Alfieri W., Okano A.A., Alfieri Braz. J. Microbiol. 2013;44(3):889–896. doi: 10.1590/s1517-83822013000300036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ISO . International Organization for Standardization; Geneva, Switzerland: 2004. ISO 11290-1: 1996/Amd 1. [Google Scholar]
- 12.Jemmi T., Stephan R. Rev. Sci. Tech. Off. Int. Epiz. 2006;25(2):571–580. [PubMed] [Google Scholar]
- 13.Kyoui D., Takahashi H., Miya S., Kuda T., Kimura B. BMC Microbiol. 2014:14–15. doi: 10.1186/1471-2180-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D., Lawrence M.L., Austin F.W., Ainsworth A.J. J. Microbiol. Methods. 2007;71:133–140. doi: 10.1016/j.mimet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Mahmoud M.S., Ahmed A.N., Hussain I. Pak. J. Nutrition. 2003;2(6):346–349. [Google Scholar]
- 16.Marinsek J., Grebenc S. Slovenian. Slovenian Vet. Res. 2002;39(2):131–136. [Google Scholar]
- 17.Norton D.M., McCamey M.A., Gall K.L., Scarlett J.M., Boor K.J., Wiedmann M. Appl. Environ. Microbiol. 2001;67(1):198–205. doi: 10.1128/AEM.67.1.198-205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nwachukwu N.C., Orji F.A., Iheukwumere I., Ekeleme U.G. Aust. J. Basic Appl. Sci. 2010;4:1571–1576. [Google Scholar]
- 19.Roberts A., Nightingale K., Jeffers G., Fortes E., Kongo J.M., Weidman M. Microbiology. 2006;152:685–693. doi: 10.1099/mic.0.28503-0. [DOI] [PubMed] [Google Scholar]
- 20.Sallen B., Rajoharison A., Desvarenne S., Quinn F., Mabilat C. Intern. J. Syst. Bacteriol. 1996;7:669–674. doi: 10.1099/00207713-46-3-669. [DOI] [PubMed] [Google Scholar]
- 21.Soni D.K., Dubey S.K. Mol. Biol. Rep. 2014 doi: 10.1007/s11033-014-3724-2. [DOI] [PubMed] [Google Scholar]
- 22.Soni D.K., Singh M., Singh D.V., Dubey S.K. BMC Microbiol. 2014;14:241–251. doi: 10.1186/s12866-014-0241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedmann M., Nightingale K.K. Food Technol. 2009;63(4):44–49. [Google Scholar]
- 24.Yadav M.M., Roy A. Zoonoses Public Health. 2009;56:515–524. doi: 10.1111/j.1863-2378.2008.01201.x. [DOI] [PubMed] [Google Scholar]