Abstract
The objective of this research was to find the basic data on genetic diversity of mtDNA D-Loop in Aceh cattle and its association with Bhutanese, Chinese, and Indian cattle. There were sixty samples of DNA which had been sequenced; i.e. Banda Aceh (11), Saree (20), and Indrapuri (29). To the best of our knowledge this is the first published data on the complete mitochondrial D-Loop sequence of Aceh cattle. Results show that Aceh cattle have the closest relationship to Bos indicus and have been influenced by Bos taurus. The closest genetic ranges among Aceh cattle, Bhutanese, Chinese, Indian and Zebu were Aceh–Zebu (0.0138), Aceh–Bhutanese (0.0156), Aceh–Chinese (0.0190) and Aceh–Indian (0.0193). D-Loop mtDNA analyses showed that there were 27 haplotypes in which twenty-one samples spread in haplotype 1, two samples were in haplotype 2, and the other four haplotypes had various samples in the range of three to seventeen samples. One sample of Aceh cattle from Saree has a closest maternal genetic with B. taurus. One of the four mutations among the star-shaped clusters on median joining network was a new specific haploid-group in Aceh cattle. From this finding it could be assumed that Aceh cattle form a specific haplotype and it can be conclude that Aceh cattle are animal genetic resources from Aceh in Sumatera Island that have to be preserved.
Keywords: Aceh cattle, Mitochondrial D-Loop, Polymorphism, Haplotype
1. Introduction
Cattle are one of the importance domestic animals in Indonesia. They are also regarded as excellent meat producers. To increase the demand for their products, attention has been focused on the genetic improvement of these species. The mitochondrial genome (mtDNA) of vertebrates has become a common tool for resolving phylogenetic relationships at different evolutionary depths due to its peculiar properties [5] that include predominantly maternal mode of inheritance [6]. The mtD-loop sequences have provided significant insight into the domestication and past migration history of cattle [10], [9]. The information of mtDNA D-Loop varieties in Aceh cattle and other domestic cattle in Indonesia had been studied previously [1], but the information about Aceh and Zebu cattle from other countries (Bhutan, China, and India) had never been reported. Therefore, the objective of this research was to find the basic data of the mtDNA D-Loop genetic varieties of Aceh cattle and its association with the Bhutanese, Chinese, and Indian cattle to complete the information about Aceh cattle genetic traits which had been provided before. It is hoped that this research is able to give thorough information which is beneficial for the progress and development policies in improving the genetic quality of Aceh cattle.
2. Material and methods
2.1. Sample collection
Sixty fresh blood samples were collected from different locations in Aceh. The DNA samples used for sequencing were taken from Banda Aceh (11), Saree (20), and Indrapuri (29). The process of isolation, extraction, and purification of DNA were carried out in Genetic and Animal Breeding Laboratory, Faculty of Animal Science Bogor Agricultural Institute. The research of mtDNA D-Loop was conducted in Molecular and Genetic Laboratory CAAS (Chinese Academy of Agricultural Sciences) Beijing-China. The sequencing of DNA was conducted in Laboratory Sunbio Beijing (Primer F: 5′-ATGCCGCGTGAAACCAGCAA-3′ and R: 5′CCTGAAGAAAGAACCAGATGC-3′ [3]. The comparison sequences of cattle used were taken from Gen Bank. The blood sample of Aceh cattle was taken using venoject (EDTA) 5 ml on vena jugularis.
2.2. Extraction of DNA genome
The extraction of DNA genome was conducted using the method of Sambrook et al. [15] which had been modified using buffer lyses cell (400 μl 1× STE, and 40 μl 10% SDS and 10 μl proteinase-K. DNA was purified using phenol–chloroform that was added with 40 μl 5 M NaCl and 400 μl phenol and chloroform iso amyl alcohol (CIAA). The DNA was precipitated using 40 μl 5 M NaCl and 800 μl ethanol absolute. Precipitation was then washed by adding 800 μl 70% ethanol, centrifuged at 12,000 rpm speed for 5 min, the ethanol was discarded and evaporated. Then, the DNA precipitation was dissolved in 10 μl 80% TE (Elution buffer).
2.3. Amplification of mtDNA D-Loop
Each PCR reaction was made with the volume of 50 μl with the composition of 5 μl 1× buffer PCR; 4 μl dNTP; 1 μl Taq DNA Polymerase; 1 μl Primer Forward and 1 μl Primer Reverse; 4 μl DNA; and 34 μl dH2O. PCR engine used was GeneAmp PCR System 9700 Applied Biosystem. The initial denaturation at 95 °C for 5 min was done once and then repeated 31 times, each with a denaturation step at 94 °C for 30 s, 30 s annealing at the temperature of 60 °C, extension at 72 °C in 30 s, and continued with extension at 72 °C for 5 min. PCR product was stored at the temperature of 4 °C for 25 min. The primer used is shown in Table 1.
Table 1.
Primer bases D-Loop.
| Primer Ref. | Temperature annealing | PCR product | Primer sequence |
|---|---|---|---|
| Loftus et al. [10] | 60 °C | 980 bp | F:5′-CTGCAGTCTCACCATCAACC-3′ |
| R:5′-GCTGGGACCAAACCTATGTC-3′ |
2.4. Electrophoreses of PCR product
The PCR product was electrophoresed by a machine of Applied Bio system 3130x Genetic Analyzer and analyzed by Gene Mapper version 4.0 software.
2.5. Analysis of data
The mtDNA D-Loop data analysis was done by parallelizing nucleotide D-Loop using software MEGA program version 4.1 [16], by comparing the sequence of Gen-Bank. DnaSP program was used to find NJ tress and identification of haplotype, then using software NETWORK 4.510.
3. Results and discussion
3.1. Phylogenetic analysis of mtDNA D-Loop sequences
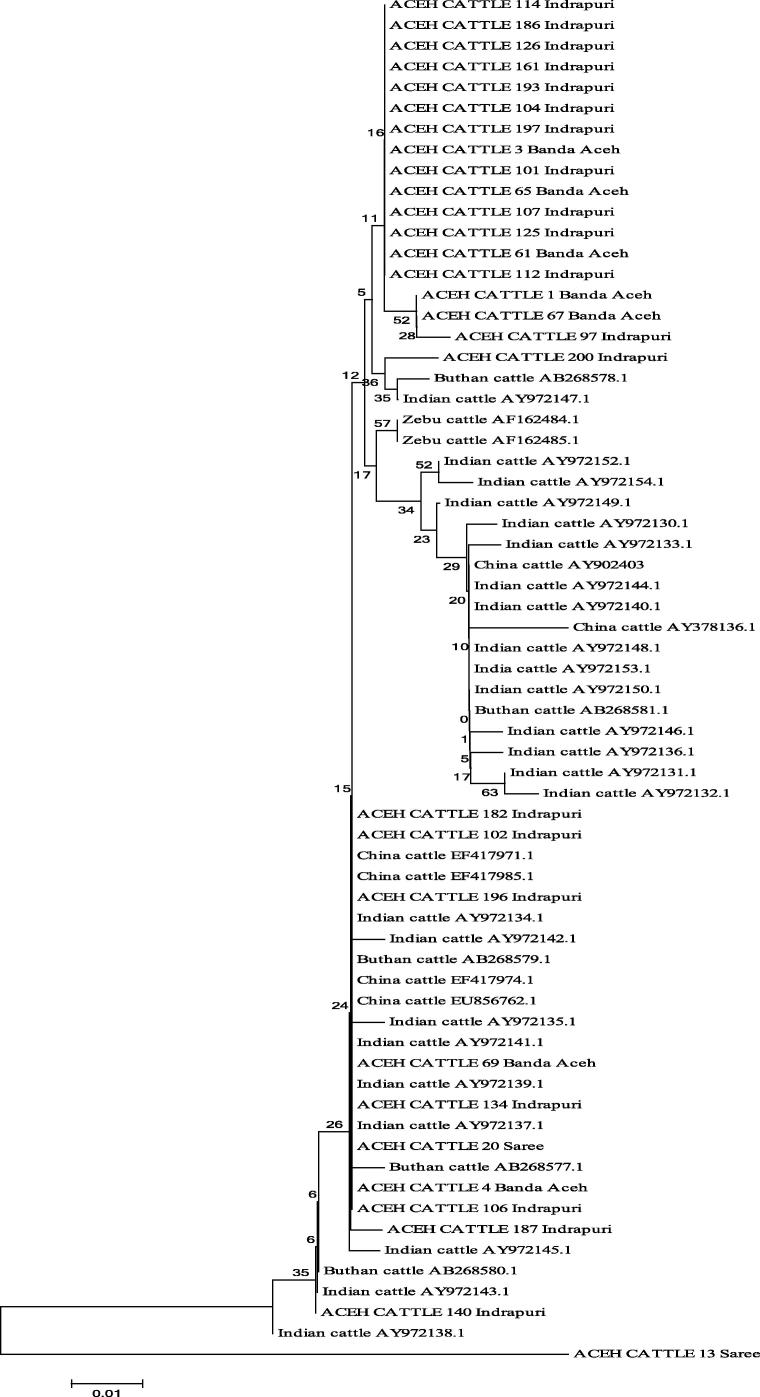
The analysis of nucleotide sequence varieties was made after the sequence of Aceh cattle was parallelized with the standard sequence from Gene Bank (V00645) by Anderson et al. [2]. The sixty samples represented 27 haplotypes of the mtDNA D-Loop complete sequence in Aceh cattle, Bhutanese, Chinese, Indian and Zebu cattle. The Neighbor-Joining (NJ) phylogenetic tree of haplotypes in Aceh cattle, Bhutanese, Chinese and Zebu are shown in Fig. 1.
Figure 1.
Dendogram Neighbor-Joining based on the 2 parameter-Kimura method (230 bp) of Aceh, Bhutanese, Chinese, Indian and Zebu cattle by applying bootstrap 1000 times.
Based on the analysis using Neighbor Joining (NJ) tree, it was shown that all samples were divided into two clusters; Bos taurus and Bos indicus. On NJ tree, individuals having the same maternal origin would form a cluster together. The NJ tree indicated that Aceh cattle population (Banda Aceh, Indrapuri, and Saree) widely spread among Bhutanese, Chinese, Indian, and Zebu cattle (Fig. 1). Meanwhile, one sample of Aceh cattle from Saree formed a separate cluster which had the maternal origin of B. taurus. The result showed that most Aceh cattle had maternal origin of B. indicus. This result supported previous researches on Aceh cattle [11], [1]). The NJ tree also indicated that Aceh cattle from Indrapuri form a specific haplotype which indicates that Aceh cattle from Indrapuri are animal genetic resources that have to be preserved.
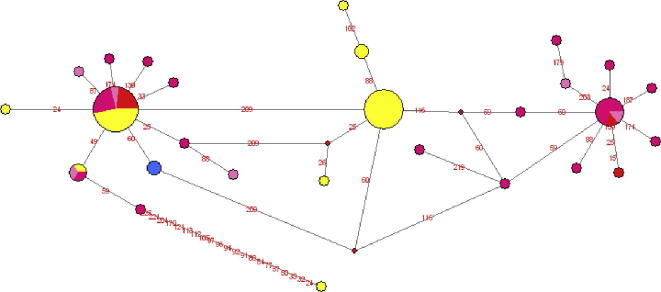
The analysis of D-Loop sequence showed that there were 27 haplotypes in which twenty-one samples spread in haplotype 1, two samples were in haplotype 2, and the other four haplotypes had various samples in the range of three to seventeen samples. An observation toward the correlation of phylogenetic on NJ tree could be formed and built using Median Joining Network (Fig. 2) from 230 bp mtDNA D-Loop of Aceh, Bhutanese, Chinese, Indian, Zebu cattle and referent sample from Gen Bank.
Figure 2.
Construction of median joining network from 230 bp D-Loop sequence; Aceh cattle (Yellow), Indonesian Zebu (blue), Chinese Zebu (red), Indian (pink) and Bhutanese (light pink).
Sequence of 230 bp was a very variable fragment of mtDNA of control region that has been applied in other researches [12], [7], [4], [10], [11]. The result of mtDNA D-Loop sequence formed two star-shaped clusters in which haplotype predominant was in the middle of the two star-shaped clusters (Fig. 2). Four mutations and one unique haplotype of sixty samples of Aceh cattle separated the two star-shaped clusters. Zebu cattle from some countries were in both star-shaped clusters and there was no significant difference on the maternal origin of Aceh cattle. The other research showed that 230 bp from mtDNA control region of B. indicus was divided in two star-shaped clusters which were separated by four mutation points [4]. In this research, Zebu sequence was divided into two clusters and in the middle of the two clusters there were haplotypes separated by four mutations and it was the special haplotype of Aceh cattle. Lai et al. [8] found five mutation points between two clusters, it showed that the number of mutation in two clusters had different value. Based on the previous researches, it is shown that one of the mutations was the new haplotypes.
3.2. Composition of mtDNA D-Loop nucleotide
The number of nucleotide of all samples had no similar value. It is caused by some nucleotide deletion and insertion on Aceh, Bhutanese, Chinese, Indian, and Zebu cattle (Table 2). This research is also supported by the results of Abdullah et al. [1]. Based on the range of the analyzed 230 bp nucleotide, the result of nucleotide parallelization of Aceh, Bhutanese, Chinese, Indian and Zebu cattle is shown on the Table 3.
Table 2.
Average composition of D-Loop nucleotide of Aceh, Bhutanese, Chinese, Indian, and Zebu cattle.
| Breed | Number of sample | T | C | A | G | A + T | G + C |
|---|---|---|---|---|---|---|---|
| Aceh | 29 | 27.1 | 26.0 | 32.3 | 14.6 | 59.4 | 40.6 |
| Bhutanese | 5 | 26.8 | 26.3 | 32.5 | 14.5 | 59.3 | 40.8 |
| Chinese | 6 | 26.5 | 26.6 | 32.4 | 14.6 | 58.9 | 41.2 |
| Indian | 24 | 26.3 | 26.8 | 32.5 | 14.5 | 58.8 | 41.3 |
| Zebu | 2 | 26.3 | 26.8 | 32.5 | 14.5 | 58.8 | 41.3 |
| B. taurus | 1 | 29.0 | 24.2 | 33.3 | 13.4 | 62.3 | 37.6 |
Table 3.
The genetic distance of Aceh, Bhutanese, Chinese, Indian, and Zebu based on 2 parameter Kimura methods.
| Aceh | Bhutanese | Chinese | Indian | Zebu | |
|---|---|---|---|---|---|
| Aceh | 0.0000 | ||||
| Bhutanese | 0.0156 | 0.0000 | |||
| Chinese | 0.0190 | 0.0184 | 0.0000 | ||
| Indian | 0.0193 | 0.0179 | 0.0171 | 0.0000 | |
| Zebu | 0.0138 | 0.0113 | 0.0103 | 0.0134 | 0.0000 |
The difference in the nucleotide between Aceh cattle and B. taurus of Gene Bank (access code V 00645) was T = 1.9%; C = 1.8; A = 1%; and G = 1.2%. The average rate of nucleotide composition A + T and G + C between Aceh cattle and B. taurus was 2.9% and 3%. Aceh had the highest frequency of Nucleotide T (0.271) among the Bhutanese, Chinese, Indian, and Zebu cattle. Followed by Bhutanese (0.268), Chinese (0.265), Indian and Zebu (0.263). Nucleotide C had the lowest frequency in Aceh cattle (0.260), followed by Bhutanese (0.263) and Chinese (0.266). The same frequency was found in Indian and Zebu cattle (0.268).
Based on Table 3 it can be concluded that the composition of nucleotide ATGC in Indian and Zebu cattle had the same value. It shows that Indian and Zebu cattle had the same maternal origin. This result is supported by Nozawa [13] which stated that the central of Zebu cattle (B. indicus) gene was found in Indian. Patricia et al. [14] stated that genealogy of mtDNA mammal had high substitution rank. Aceh cattle had a various range of bases. It showed that the area of D-Loop mtDNA of Aceh cattle had mutated.
3.3. Genetic distance of Aceh, Bhutanese, Chinese, Indian and Zebu cattle
The analysis of Pairwise Distance Calculation with the model of parameter Kimura was applied to analyze the genetic distance or the closeness of genetic association of Aceh, Bhutanese, Chinese, Indian and Zebu cattle which were from the gene bank. The genetic distance between Aceh cattle and the standard cattle from the gene bank was 0.0000–0.0165. The genetic distance of Aceh cattle was 0.0040–0.1500. This result had a higher value than that of Abdullah et al. [1] which found the genetic distance in Aceh cattle to be about 0.0025–0.0075. The genetic distance of Bhutanese cattle was about 0.0005–0.028, Chinese cattle was 0.014–0.033, Indian was 0.005–0.029, and Zebu was 0.00.
The lowest genetic distance belonged to Aceh cattle with the standard cattle (Table 3), was about 0.0138, and was between Aceh and Zebu cattle. Meanwhile, the highest genetic distance was between Aceh cattle and Indian cattle, about 0.0193. The sequence of genetic closeness of all samples from the closest to the furthest were Chinese–Zebu (0.0103); Bhutanese–Zebu (0.0113); Indian–Zebu (0.0134); Aceh–Zebu (0.0138); Aceh–Bhutanese (0.0156); Chinese–Indian (0.0171); Bhutanese–Indian (0.0179); Bhutanese–Chinese (0.0184); Aceh–Chinese (0.0190) and Aceh–Indian (0.0193).
The value of genetic distance of some samples of cattle applied was less than 0.5 which means that generally, the cattle were close genetically. Nijman et al. [12] stated that based on the control of mitochondrial DNA of Madura and Bali-Malaysia cattle, allele from Zebu cattle (B. indicus) was found.
4. Conclusion
Based on the research of mtDNA D-Loop region, it is shown that Aceh cattle have a closest relationship to B. indicus and have been influenced by B. taurus. One of the four mutations among the star-shaped clusters on median joining network was a new specific haploid-group in Aceh cattle, and Aceh cattle from Indrapuri are animal genetic resources that have to be preserved.
Acknowledgement
This research was undertaken with the research funding from DIKTI Sandwich Program 2009/2010.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Abdullah Man, Noor R.R., Handiwirawan E. J. Indonesian Trop. Anim. Agric. 2008:1–10. [Google Scholar]
- 2.Anderson S., de Brujin M.H., Coulson A.R., Eperon I.C., Sanger F., Young I.G. J. Mol. Biol. 1982;154(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 3.Bailey J.F., Richards M.B., Macaulay V.A., Colsob I.B., James I.T., Bradley D.G., Hedges R.E.M., Sykes B.C. Proc. R. Soc. London, Ser. B. 1996;263:1463–1473. doi: 10.1098/rspb.1996.0214. [DOI] [PubMed] [Google Scholar]
- 4.Baig M., Pereira A.B., Mohammad R., Kulkarni K., Farah S., Luikari G. Curr. Sci. 2005;89(1):38–40. [Google Scholar]
- 5.Carmela G., Reyes A., Pesole G., Saccone C. Mol. Biol. Evol. 2000;17:1022–1031. doi: 10.1093/oxfordjournals.molbev.a026383. [DOI] [PubMed] [Google Scholar]
- 6.Gyllensten U., Wharton D., Wilson S.C. Nature. 1985;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 7.Kim K.I., Heon Lee J., Soo Lee S., Hoon Yan Y. Biochem. Genet. 2003;41:91–98. doi: 10.1023/a:1022021900205. [DOI] [PubMed] [Google Scholar]
- 8.Lai S.J., Liu Y.P., Liu Y.X., Li X.W., Yao Y.G. Mol. Phylogenet. Evol. 2006;38:146–154. doi: 10.1016/j.ympev.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Lenstra J.A., Bradley D.G. In: The Genetics of Cattle. Fries R., Ruvinsky A., editors. CAB Int.; Wallingford: 1999. pp. 1–14. [Google Scholar]
- 10.Loftus R.T., MacHugh D.E., Bradley D.G., Sharp P.M., Cunningham P. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2757–2761. doi: 10.1073/pnas.91.7.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uggla C.M. Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences; Box 7080, 75007 Uppsala, Sweden: 2008. Investigating genetic variability within specific indigenous Indonesian cattle breeds. (Dissertation) [Google Scholar]
- 12.Nijman I.J., Ruiter C., Hanekamp E., Verkaar E.L.C., Ochieng J., Shamshad S.B.M., Rege J.E.O., Hanotte O., Barwegen M., Susilawati T., Lenstra J.A. Heredity. 2003;90:10–16. doi: 10.1038/sj.hdy.6800174. [DOI] [PubMed] [Google Scholar]
- 13.K. Nozawa, Phylogenetics studies on the native domestic animals in East and Southeast Asia, in: Proc. Workshop Animals Genetic Resources in Asia and Oceania, Tsakuba, 3–7 September 1979, Society for the Advancement of Breeding Researches in Asia and Oceania (SABRAO), Tsukuba, 1979, pp. 23–43.
- 14.Patricia M.M., Garcia P.P., Dulout F. Genet. Mol. Biol. 2002;25(1):1–7. [Google Scholar]
- 15.Sambrook J., Fritsch E.F., Maniatis T. second ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Molecular Cloning: A laboratory Manual. [Google Scholar]
- 16.Tamura K., Dudley J., Nei M., Kumar S. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]