ABSTRACT
Maintaining hepatic functional characteristics in-vitro is considered one of the main challenges in engineering liver tissue. As hepatocytes cultured ex-vivo are deprived of their native extracellular matrix (ECM) milieu, developing scaffolds that mimic the biomechanical and physicochemical properties of the native ECM is thought to be a promising approach for successful tissue engineering and regenerative medicine applications. On the basis that the decellularized liver matrix represents the ideal design template for engineering bioinspired hepatic scaffolds, to derive quantitative descriptors of liver ECM architecture, we characterised decellularised liver matrices in terms of their biochemical, viscoelastic and structural features along with porosity, permeability and wettability. Together, these data provide a unique set of quantitative design criteria which can be used to generate guidelines for fabricating biomaterial scaffolds for liver tissue engineering. As proof-of-concept, we investigated hepatic cell response to substrate viscoelasticity. On collagen hydrogels mimicking decellularised liver mechanics, cells showed superior morphology, higher viability and albumin secretion than on stiffer and less viscous substrates. Although scaffold properties are generally inspired by those of native tissues, our results indicate significant differences between the mechano-structural characteristics of untreated and decellularised hepatic tissue. Therefore, we suggest that design rules - such as mechanical properties and swelling behaviour - for engineering biomimetic scaffolds be re-examined through further studies on substrates matching the features of decellularized liver matrices.
KEYWORDS: decellularisation, design criteria, ECM-mimicking scaffold, hepatic cells, liver, tissue engineering
INTRODUCTION
The liver, by way of its central role in both endogenous and exogenous metabolism, is one of the most widely studied organs in the human body. Hepatic tissue and its derivatives are used in many applications ranging from in-vitro liver models for investigating hepatic drug metabolism and diseases, to tissue regeneration and incorporation into bio-artificial liver devices.1–3 One of the main challenges in engineering liver tissue is to maintain the native hepatic functional characteristics in-vitro. Indeed, conventional monolayer hepatocyte cultures often fail to remain functional, and this is thought to be due to the absence of the native three-dimensional (3D) multi-parametric, multi-stimuli in-vivo milieu, that is not recapitulated in-vitro.4–6 Reproducing the 3D features of native liver is crucial since the in-vivo environment affects cell behaviour and function through a variety of soluble and insoluble signalling factors, including physical, chemical and mechanical cues (e.g. oxygen concentration, substrate stiffness).7,8 These environmental signals are transmitted within the cell, inducing a cascade of interactive events at various levels (i.e. molecular, subcellular and cellular levels).9,10 Beyond cell-cell communications, cell-ECM (extra-cellular matrix) interactions are of primary importance since they play a fundamental role in hepatocyte growth,11 cell organisation,12 liver organ development,13 tissue regeneration,14 wound healing14 and liver diseases.15 The ECM is in a state of dynamic reciprocity with these cells in response to changes in the microenvironment and has been shown to provide cues that affect cell migration, proliferation and differentiation.16,17 In addition to physicochemical properties, mechanical cues are also known to play a key role in liver pathophysiology. Several hepatic pathologies, such as fibrosis and non-alcoholic fatty liver disease, are indeed associated with a significant degree of ECM remodelling, which involves dynamic changes in matrix stiffness, flexibility and density related to the dysregulation of predominant collagen and elastic fibres as well as minor components with both structural and signalling properties.18 In fact, both parenchymal and non-parenchymal cells exhibit mechano-sensitive behaviour, suggesting that the mechanics of liver ECM facilitates normal regeneration or paves the way for liver disease.19–27 As the 3D ultrastructure, surface topology, mechanical properties and composition of the native ECM all contribute to hepatic homeostasis, the current consensus is that an optimal scaffold for liver tissue engineering should mimic these architectural and physicochemical features to provide cells with an appropriate supportive framework and micro-environment.28–30
This study is therefore aimed at deriving a unique set of quantitative descriptors of decellularised liver which can be used to define an ideal hepatic tissue scaffold in terms of interfacial and mechanical properties, viscoelasticity, 3D microarchitecture and adhesion ligands.31 Since human primary hepatocytes exhibit good metabolic competency when cultured on porcine liver-derived ECM scaffolds, either in the form of 3D porous matrices.32 or hydrogels,33 decellularised porcine liver matrices (dECMs) may represent an attractive candidate for deriving design criteria for engineering hepatic scaffolds.34
Unlike healthy human livers which are not only difficult to obtain but also highly variable,35 decellularized porcine liver has proved to be a reproducible material with unique mechano-structural and physicochemical features.36–38 In a previous study focused on the identification of a standardised matrix conserving method for eliminating cells from hepatic tissue, we investigated several immersion and agitation protocols to decellularise porcine liver samples.38 We demonstrated that a 3 days long immersion and agitation procedure based on non-ionic detergents (i.e. Triton X-100) ensured complete cell removal while preserving the native ECM features. Increasing treatment duration (up to 5 days) or using more aggressive detergents (i.e. sodium dodecyl sulphate, SDS) resulted in matrix degradation, while complete cell removal was not achieved in the absence of detergents.
In this paper, we extend and complete the systematic characterisation of porcine liver dECMs described in Mattei et al.,38 by quantifying biochemical and viscoelastic properties along with porosity, permeability, wettability and other parameters relevant to water-scaffold interaction. This unique dataset can be used to generate guidelines for fabricating bioinspired tissue engineered livers. As a proof concept, given that tissue mechanics are known to play a critical role in liver pathophysiology, a brief study on hepatic cell response to substrate viscoelasticity was conducted on collagen hydrogels with mechanical properties either matching those of liver dECMs or not. Cell viability, metabolism and morphology were evaluated up to 7 days of culture.
RESULTS
Immunohistochemistry
The immuno-histochemical analysis showed the presence of the three proteins investigated both in the fresh and in the decellularised samples (Figure 1).
Figure 1.
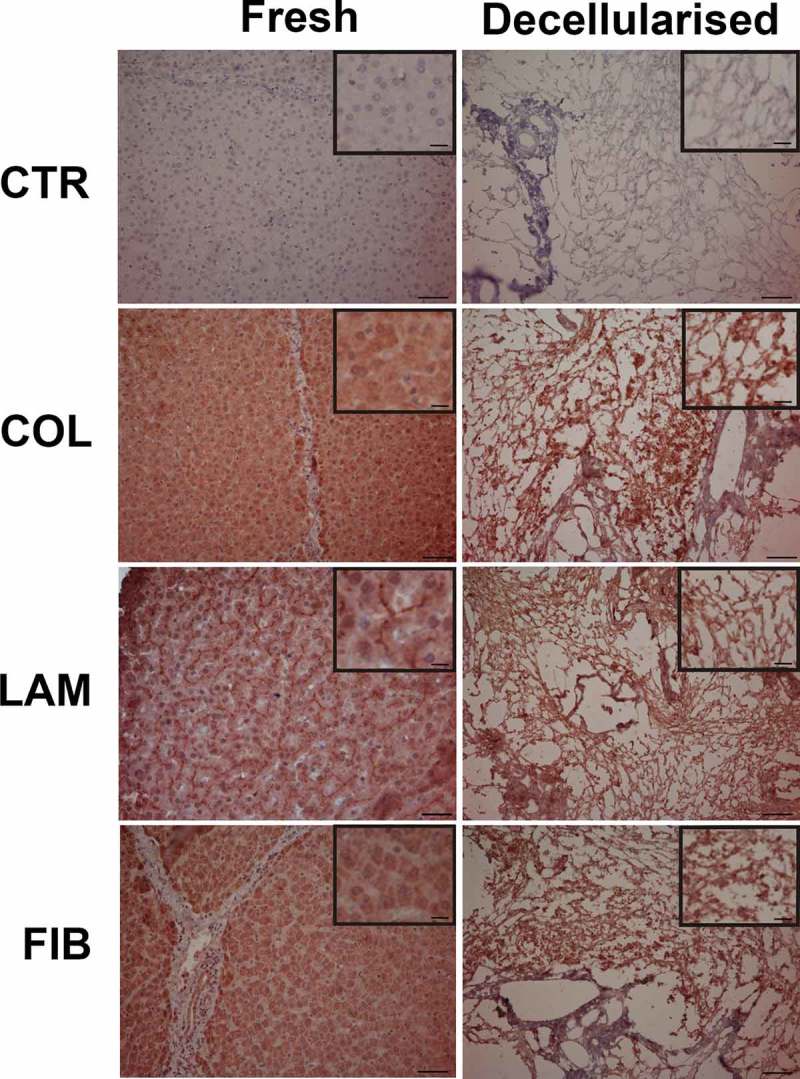
Immuno-histochemical staining of the fresh (left column) and decellularized (right column) liver. Insets within each micrograph show a magnification to better display collagen, laminin and fibronectin localization. In both fresh and decellularised negative controls (CTR) where the primary antibodies were omitted, immunostaining was not detected. Collagen (COL) immunoreactivity was evenly distributed in the fresh liver parenchyma, laminin (LAM) was mainly localised in the extracellular spaces, while fibronectin (FIB) was found in the hepatocyte cytoplasm, membranes and extracellular spaces. The decellularized samples also showed positivity to collagen, laminin and fibronectin. Scale bars = 50 µm in the main micrographs and 10 µm in the insets.
In fresh liver, collagen and fibronectin were mainly distributed in the cells and in the extracellular spaces. In particular, collagen positivity was evenly localised in the liver parenchyma while fibronectin immunolabeling was found in the hepatocyte cytoplasm, membranes and extracellular spaces. On the other hand, laminin displayed a marked extracellular localization. Moreover, fibronectin, collagen and laminin were maintained in the decellularised samples when compared to that observed in the fresh liver.
The average dimension of the dECM fibres (likely consisting of aggregates of multiple protein fibrils) was estimated from control histological images of decellularised liver samples and found to be 1.09 ± 0.32 μm.
3D pore size
Confocal imaging of NI3 samples in the equilibrium swollen state showed a rich intra-lobular network with pores of 22.02 ± 0.65 μm diameter, estimated with a seeded region growing algorithm (Figure 2).
Figure 2.
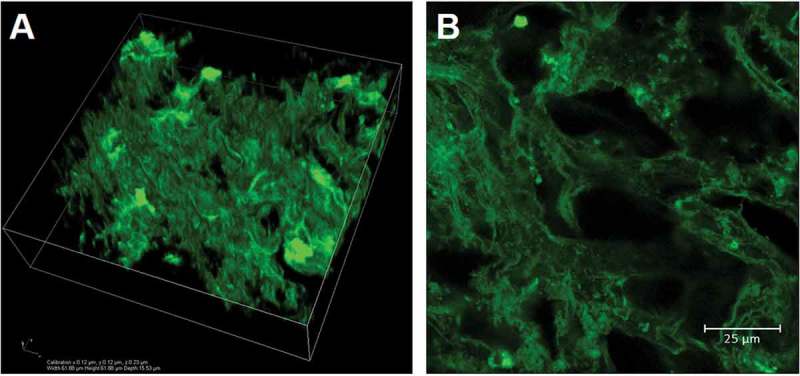
Confocal acquisitions of an eosin-stained NI3 liver decellularised matrix. A) 3D rendering of a 62 x 62 x 16 μm volume and B) detail of a central slice of the confocal scan showing a rich intra-lobular network with an average (equilibrium swollen) pore size of about 22 μm.
It is worth noting that, unlike the dry liver dECM pore size previously obtained via micro-CT imaging,38 the wet pore size obtained from confocal imaging of equilibrium swollen samples represents that of cell culture conditions. Other methods such as mercury intrusion porosimetry as well as micro-, meso- and macro-pore analysis by gas adsorption also require dry samples, and again do not generally allow for wet pore size characterization. Moreover, confocal imaging is able to resolve the fine intra-lobular reticular network observed with histological analyses, overcoming the resolution limitations encountered using micro-CT scans, which returned an average pore size of about 500 μm.
Swelling behaviour
Swelling ratio values () over time obtained for NI3 decellularised liver matrices are shown in Figure 3. Although one-way ANOVA analysis showed that only at 12 h is significantly lower than values obtained from 24 to 48 h (which are not statistically different from each other), it is more advisable to consider the swelling plateau reached after 36 h, taking the equilibrium mass swelling ratio equal to the average between values at 36 and 48 h (i.e. = 17.82 ± 0.98).
Figure 3.
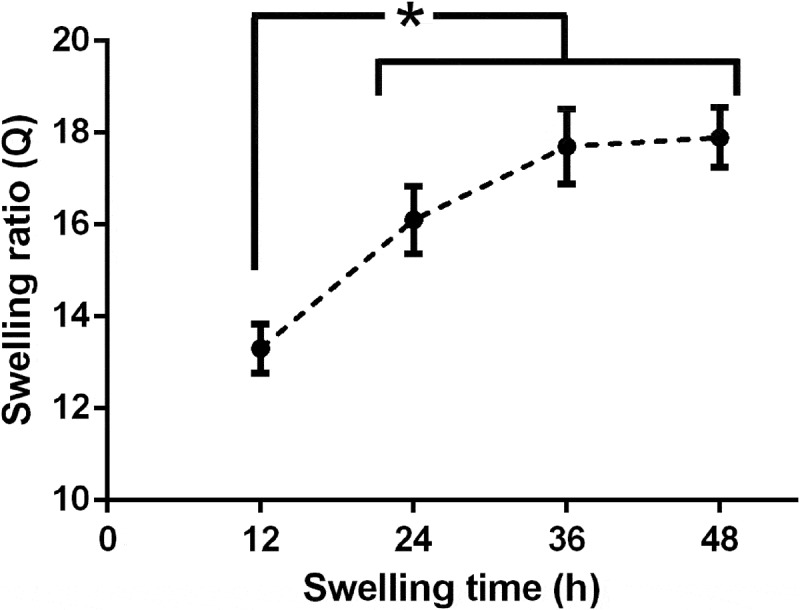
Mass swelling behaviour of NI 3 decellularised liver matrices. Black circles represent mean swelling ratio values (Q) at different time points, while error bars denote respective standard deviation. * = significant differences between samples (one-way ANOVA, p < 0.05).
Sorptivity
For both decellularised and untreated liver samples, sorptivity values were derived considering measurements in the first 100 s of acquisition, where was found to be linear with . In particular, the sorptivity coefficient for decellularised samples, = (1.25 ± 0.26)∙10–1 cm/s1/2 was found to be significantly higher than that obtained for untreated liver, = (2.52 ± 0.30)∙10–3 cm/s1/2 as expected since higher sample pore size and porosity are known to result in higher sorptivity coefficients.39
Wet porosity and permeability estimation
Residual mass swelling ratio of NI3 liver dECMs after pore-filling water expulsion was equal to = 3.59 ± 0.44. This result was used along with = 17.82 ± 0.98 to calculate the wet porosity according to Equation 2, obtaining = 0.81 ± 0.08, which is in agreement with cells occupying about 80% of liver volume.40
Equilibrium and residual mass swelling ratios for untreated liver were found to be equal to = 3.55 ± 0.26 and = 2.21 ± 0.14, respectively. These results yield = 0.41 ± 0.09, which is significantly lower than that obtained for decellularised liver, as expected due to the denser and more compact structure of untreated liver, owing to the presence of cells. Notably, the wet porosity of the native hepatic tissue largely consists of the liver blood volume fraction.41,42
The Darcy’s permeability of NI3 liver dECMs, estimated from wet porosity and pore size data, was found to be equal to = (1.53 ± 0.28) ∙ 10 ∙ 10–10 m2.
Viscoelastic characterisation
Since soft biological tissues are typically regarded as viscoelastic materials,43 characterising elastic properties (i.e. stiffness, elastic modulus) only is generally over-reductive to describe their mechanical behaviour. In agreement with experimental results previously obtained for untreated liver samples,37 the Linear Viscoelastic Region (LVR) of liver dECMs extended up to 0.03 compressive strain range, independent of the applied strain rate (), with an increase in the apparent compressive modulus with (Figure 4).
Figure 4.
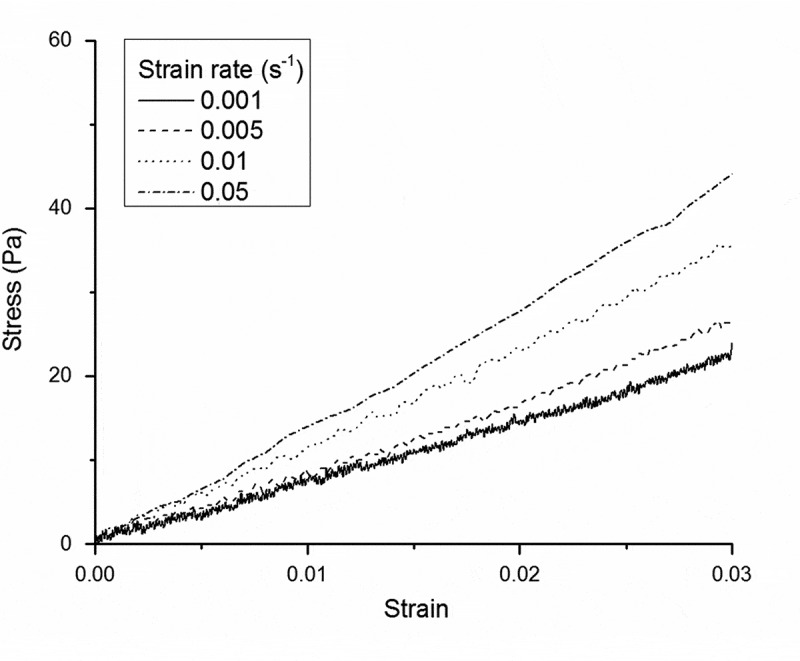
Examples of experimental LVR stress-strain data collected at various strain rates for NI3 liver dECMs. Decellularised liver LVR extended up to 3% strain and the apparent compressive modulus markedly increases with applied strain rate, as expected for viscoelastic materials.
Viscoelastic parameters obtained for liver dECMs are summarised in Table 1, where and represent the instantaneous and equilibrium elastic moduli, respectively, while is the characteristic relaxation time, defined as the Maxwell arm dashpot to spring constant ratio. For the sake of comparison, results are reported along with those previously obtained for untreated liver samples.37
Table 1.
Viscoelastic parameters of decellularised and untreated porcine liver estimated using the with both SLS and GM2 lumped models. Results are expressed as estimated parameter value ± standard error of estimation. The abbreviation n.s. stands for non-significant result
| Decellularised liver |
Untreated liver |
|||
|---|---|---|---|---|
| SLS | GM2 | SLS | GM2 | |
| 1.75 ± 0.01 | 1.75 ± (5.26 ∙ 104) n.s. | 2.04 ± 0.01 | 2.04 ± (3.21 ∙ 102) n.s. | |
| 0.76 ± 0.01 | 0.76 ± 0.01 | 0.91 ± 0.01 | 0.91 ± 0.01 | |
| 0.77 ± 0.01 | 0.77 ± (2.05 ∙ 105) n.s. | 1.10 ± 0.02 | 1.10 ± (3.05 ∙ 103) n.s. | |
| - | 0.77 ± (5.11 ∙ 104) n.s. | - | 1.10 ± (3.43 ∙ 102) n.s. | |
| 0.97 | 0.97 | 0.97 | 0.97 | |
As in the case of untreated liver samples, although a good fitting convergence was obtained for both SLS and GM2 models, the latter yielded non-significant results with very large standard errors, indicating model over-parameterisation. Therefore, the SLS model adequately represents the viscoelastic behaviour of both untreated and decellularised liver matrices within the investigated range of physiologically relevant strain rates. Decellularised matrices were found to be less rigid than untreated liver samples, in terms of both instantaneous and equilibrium moduli (p < 0.0001) and characterised by a shorter relaxation time (p < 0.0001). These differences in viscoelastic mechanical behaviour are likely linked to structural differences occurring upon cell removal, which results in a decrease of bulk sample stiffness and in a reduction of barriers to fluid flow. As a consequence, fluid is able to redistribute faster within the porous network of decellularised matrices, explaining the smaller characteristic relaxation time with respect to that of untreated tissue, in agreement with results reported by Evans et al.44
Maxwell SLS viscoelastic parameters of GTA-crosslinked collagen gels are reported in Table 2 as a function of GTA concentration. Notably, collagen samples not only get stiffer with increasing GTA concentration (as demonstrated by increased and values), but there is also a concomitant change in their viscoelastic behaviour from a more viscous towards a more elastic one (as indicated by the longer ), in agreement with data previously reported for gelatin samples.45 Collagen samples crosslinked with 1 mM GTA concentration closely mimic decellularised liver viscoelastic properties (Table 1), while those at higher GTA concentrations correlate well with liver fibrotic stiffening.46
Table 2.
Viscoelastic parameters of GTA-crosslinked collagen hydrogels estimated using the with a SLS lumped model. Results are reported as a function of GTA concentration and expressed as estimated parameter value ± standard error of estimation. GM2 fitting resulted in model over-parameterisation, as in the case of liver (data not shown)
| GTA [mM] | 1 | 2 | 20 | 100 | 200 |
|---|---|---|---|---|---|
| 1.88 ± 0.01 | 2.98 ± 0.02 | 4.72 ± 0.04 | 6.45 ± 0.03 | 7.95 ± 0.04 | |
| 0.81 ± 0.01 | 1.47 ± 0.01 | 2.63 ± 0.02 | 3.92 ± 0.02 | 5.12 ± 0.02 | |
| 0.70 ± 0.02 | 0.77 ± 0.02 | 0.85 ± 0.03 | 0.93 ± 0.02 | 0.99 ± 0.03 | |
| 0.98 | 0.98 | 0.97 | 0.97 | 0.98 |
Hepg2 response to collagen substrate viscoelasticity
Cell viability results at different time points are shown in Figure 5a, as percentages of the respective values obtained on the softest 1 mM GTA-crosslinked collagen gels (mimicking decellularised liver viscoelastic properties), here taken as 100% viability controls. A significant decrease in cell viability was observed on stiffer and less viscous substrates (crosslinked with 20, 100 and 200 mM GTA), independent of the culture time point.
Figure 5.
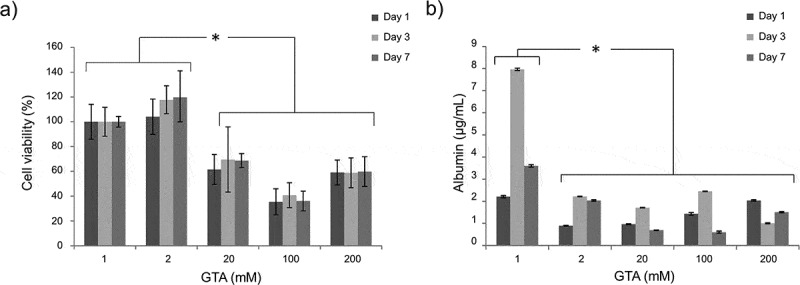
HepG2 viability (a) and albumin secretion (b) measured at different time points (day 1, 3, 7) on collagen scaffolds with different viscoelastic properties (here coded according to their GTA crosslinker concentration, Table 2). Statistical differences are denoted with an asterisk (p < 0.05).
Hepatic metabolic activity was evaluated at the same time points by measuring HepG2 albumin secretion in the cell culture medium. At each time point, albumin secretion on 1 mM GTA-crosslinked hydrogels was significantly higher than those measured on the other substrates (Figure 5b). Taken together, these results demonstrate that mimicking hepatic viscoelasticity is not only critical for maintaining cell viability, but also for promoting cell metabolic activities.
Cell morphology at day 3 and 7 was evaluated via immunofluorescence. Hepatocytes form a compact monolayer on substrates more closely mimicking hepatic viscoelasticity (i.e. 1 and 2 mM GTA). Conversely, as the substrate viscoelastic properties are changed towards more stiff and less viscous environments, the cells started forming clusters (Figure 6). This behaviour is in agreement with that reported in,47 where hepatocytes seeded on increasingly fibrotic-like substrates were less spread and suffered a decrease in albumin expression .
Figure 6.
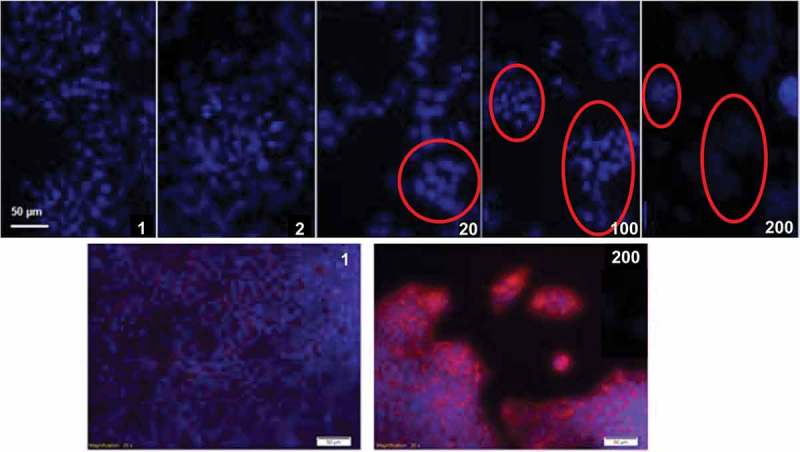
Immunofluorescent analyses of HepG2 cultured on collagen scaffolds with different viscoelastic properties (here coded according to their GTA crosslinker concentration, Table 2). Top row images show cell nuclei stained in blue (DAPI) at day 3. Cell clusters are highlighted with red circles. Bottom row images show HepG2 nuclei stained in blue (DAPI) and F-actin fibres stained in red (phalloidin) at day 7.
DISCUSSION
Tissue ECM is known to provide chemical, physical and biological cues affecting cell function and behaviour as a result of inside-out signalling.48 Therefore, developing scaffolds that mimic the mechanical and physicochemical properties of the native ECM is thought to be a promising approach for tissue engineering and regenerative medicine applications.49
Porcine liver dECMs may represent a suitable template for deriving hepatic scaffold design criteria, since they provide good support to primary hepatocytes.32,33 In light of the above considerations and building upon our previous studies on porcine liver decellularisation and characterisation,37,38 we performed an in-depth analysis of porcine liver decellularised matrices in terms of their biochemical, mechanical and structural features aimed at deriving a unique set of quantitative descriptors which can be used to fabricate ideal bioinspired hepatic scaffolds for liver tissue engineering. Beside in-vitro applications (such as pathophysiological organ models to study cell function and behaviour or to test new drugs and treatments), obtaining functional liver constructs could also help cope with the chronic shortage of donor organs.35,50
In order to investigate the presence and arrangement of hepatic ECM proteins, the histological analyses in38 were complemented with immunohistochemistry, while the 3D architecture of liver dECM was analysed using confocal imaging. Immunohistochemical staining for extracellular matrix moieties (laminin, collagen and fibronectin) in the decellularised samples demonstrated that liver architecture and biochemical features were largely conserved, In particular, the immunolabeled fibres observed in the histological images may represent the delicate network of connective tissue elements that supports the plates of hepatocytes within the lobules in native liver.34
The swelling behaviour was extensively characterised, while sorptivity index (i.e. the capacity of absorbing or desorbing liquid by capillarity), wet porosity and matrix permeability were quantified to provide robust design criteria for fabricating biomaterial scaffolds for liver. Finally, given the importance of matrix mechanics in the regulation of hepatic cell function and differentiation, we also tested untreated and decellularised liver samples at small physiologically relevant strains.38 Characterising decellularised liver tissue allows for the isolation of the extracellular matrix contribution to the overall mechanical behaviour of the liver and it is highly relevant for appropriate scaffold design given the known mechano-sensitivity of hepatic cells. Therefore, a complete viscoelastic characterisation of liver dECMs was performed using the , comparing results with those previously obtained for untreated hepatic tissue.37.
Table 3 summarises the quantitative parameters obtained in this study for both decellularised and untreated liver samples. It provides a quantitative dataset which can be used for defining the ideal design criteria and generating guidelines for the fabrication of hepatic ECM-mimicking scaffolds for application to liver tissue engineering.
Table 3.
Quantitative parameters obtained for decellularised and untreated liver samples. Results are expressed as mean ± standard deviation, with the only exception of those referred to viscoelastic properties that are reported as estimated parameter value ± standard error of estimation. N/A = not available. For each parameter investigated, results obtained for decellularised liver are significantly different than those of untreated liver (p < 0.05)
| Analysis | Parameter (units) | Decellularised liver | Untreated liver |
|---|---|---|---|
| Confocal imaging | Pore diameter (μm) | 22.02 ± 0.65 | N/A |
| Fibre diameter (μm) | 1.09 ± 0.32 | N/A | |
| Swelling behaviour | Equilibrium mass swelling ratio (w/w) | 17.82 ± 0.98 | 3.55 ± 0.26 |
| Residual mass swelling ratio (w/w) | 3.59 ± 0.44 | 2.21 ± 0.14 | |
| Wet porosity (v/v) | 0.81 ± 0.08 | 0.41 ± 0.09 | |
| Confocal imaging + swelling | Darcy’s permeability (m2) | (1.53 ± 0.28) ∙ 10–10 | N/A |
| Viscoelastic characterisation (Maxwell SLS) | Instantaneous elastic modulus (kPa) | 1.75 ± 0.01 | 2.04 ± 0.01 |
| Equilibrium elastic modulus (kPa) | 0.76 ± 0.01 | 0.91 ± 0.01 | |
| Characteristic relaxation time (s) | 0.77 ± 0.01 | 1.10 ± 0.02 |
Using one of the design parameters to illustrate an application of the data in Table 3, we generated a series of collagen hydrogels with different viscoelastic properties and examined the vitality, albumin production and morphology of hepatocytes seeded on the gels. The results, reported in Figures 5 and 6, show that hepatocyte vitality and metabolic function is higher on substrates with viscoelastic parameters which match those of liver dECM.
CONCLUSIONS
Most of the studies published to date seek to mimic untreated tissue properties to design optimal scaffolds for various cell applications, ranging from tissue engineering to in-vitro models. However, since it is widely accepted that an ideal scaffold should replicate native ECM features, the exemplar should be a (reproducible) decellularised matrix with highly preserved structure and constituents rather than the native untreated tissue. In this work, we extend and complete the systematic characterisation of porcine liver dECMs described in Mattei et al.38 Moreover, given that tissue mechanics are known to play a critical role in liver pathophysiology, a brief study on hepatic cell response to substrate viscoelasticity was conducted as a proof concept by seeding HepG2 cells on collagen hydrogels with mechanical properties either matching those of liver dECMs or not. Cell behaviour was evaluated up to 7 days of culture, showing better morphology, higher viability and albumin secretion on substrates mimicking dECMs mechanics.
In conclusion, we show that there are significant differences between the mechano-structural and swelling properties of untreated and decellularised hepatic tissue (summarised in Table 3). Moreover, cell behaviour on substrates mimicking liver dECM viscoelasticity is improved with respect to stiffer and less viscous substrates. As biomimetic hepatic scaffold design is generally inspired by the properties of native (untreated) tissue, we suggest that design rules for engineering liver scaffolds be re-examined through further studies on substrates matching the features of decellularized liver matrices.
MATERIALS AND METHODS
Liver decellularisation
Pig livers were collected from one year old healthy pigs as a slaughter by-product and decellularised using a 3-day long immersion and agitation procedure based on non-ionic detergents (NI3) described in Mattei et al.38 This method guarantees a good preservation of the architecture, structure and composition of the native hepatic ECM. Briefly, fresh pig livers were dissected into individual lobes and stored at −20 °C until use. Frozen livers were thawed at 4°C overnight and cut into 3 mm thick - 14 mm diameter discs, avoiding both the Glisson’s connective capsule and macroscopic vasculature. Liver discs were frozen at −20°C until use, then thawed and placed in 500 mL plastic bottles.
The decellularisation solution was added in a 20:1 v/w ratio with respect to the weight of liver disc samples, i.e. 200 mL solution per 10 g of liver samples (~ 20 discs). The bottles were placed on an orbital shaker (SO3, Stuart Scientific, Stone, UK) at 200 rpm in a cold room at 4°C, changing the decellularisation solution twice a day.
Immunohistochemistry
Liver samples were fixed by immersion in buffered formalin and processed for paraffin embedding. Immunohistochemistry was performed on 5 µm thick sections using the following primary antibodies: a goat polyclonal anti- Laminin β-1 (1:100, S. Cruz Biotech., Inc., sc-6018), a goat polyclonal anti- Fibronectin (1:100, S. Cruz Biotech., Inc., sc-6952), and a goat polyclonal anti- COL4A1/3: (1:100, S. Cruz Biotech., Inc., sc-9301). Epitope retrieval was carried out at 120°C in a pressure cooker for 5 min with a Tris/EDTA buffer, pH 9.0. Sections were pre-treated with 1% H2O2 in 0.1 M phosphate buffered saline (PBS 1x, pH 7.4) for 10 min to quench endogenous peroxidase activity, then rinsed with PBS 1x and blocked for 1 h with 5% normal horse serum (s-2000, Vector Labs, Burlingame, CA), 0.05% Triton X-100 in PBS. Sections were incubated overnight at 4°C with the primary antibodies.
Sections were then rinsed thrice in PBS 1x (10 min each), incubated with a biotinylated anti-goat IgG (BA-9500, Vector Labs) diluted 1:300 in PBS and then with the ABC system (PK-7200, Vector Labs). Immunostaining was visualized by incubating the sections in diaminobenzidine (sk-4105, Vector Labs) solution. Sections were counterstained with haematoxylin. The specificity of immune-histochemical staining was tested by substituting either the primary antibody, anti-goat IgG, or the ABC complex with PBS or non-immune serum. Under these conditions, staining was abolished.
Tissue sections were obtained from n = 3 independent liver samples. Each immunostaining was performed on n = 3 randomly selected sections, hence a total of 9 (immune-stained) + 3 (controls) = 12 sections per liver sample were analysed. Images of control and immuno-stained samples were acquired using a Leitz Diaplan light microscope (Leitz, Wetzlar, Germany) equipped with a 25x objective and connected to a digital camera (Nikon Digital Sight DS-U1, Nikon Instruments, Florence, Italy) interfaced with the NIS-Elements BR-4.13.00 software (Nikon Instruments). Control images of decellularised samples were used to characterize ECM fibre diameter. Briefly, images were firstly segmented using hisTOOLogy, an open-source software based on k-means clustering for quantitative analyses of digital colour images.51 Then, fiber diameters were obtained performing direct measurements on the segmented images using the “straight line” tool available in ImageJ software (NIH).
3D pore size characterization
The architectural analysis of decellularised liver matrices is fundamental to characterise their pore size, porosity and permeability. Confocal imaging was performed on eosin-stained equilibrium swollen matrices using a Nikon A1 confocal microscope (Nikon, Tokyo, Japan) equipped with a 10x objective. The scanning parameters were adjusted to obtain a pixel size less than 0.2 μm. In this study, n = 3 independent decellularised liver samples were imaged, acquiring n = 5 randomly selected volumes (62x62x16 μm) per sample investigated. Pores dimensions were derived using a seeded region growing algorithm,52 which was implemented as an ImageJ plug-in. Briefly, a seed was inscribed into each pore of the 3D structure acquired, then a sphere was grown from each seed until contacting the pixels representing the ECM network, thus returning the pore diameter.
Swelling behaviour
Decellularised liver samples were lyophilised for 24 h at −55°C/45 mTorr using a VirTis BenchTop Pro freeze-dryer (SP Scientific, PA, USA) to determine their dry weight (). Then, they were swollen in PBS 1x at room temperature and weighed every 12 h collecting values until a stable equilibrium swollen weight () was reached. For each time interval investigated, the mass swelling ratio was calculated as , until reaching an equilibrium value ().
Swelling experiments were performed testing n = 12 samples, which were weighed using an electronic balance (AS 220/C/2, Radwag, Poland) with an accuracy of 0.1 mg.
Sorptivity
Water absorption measurements were carried out to determine the sorptivity coefficient of both decellularised hepatic matrices and untreated liver specimens, used as control. Both samples were freeze-dried for 24 h at −55°C/45 mTorr, then pre-conditioned in the oven at 37°C until reaching a constant mass (i.e. a constant moisture level) and cut into 10 × 5 × 2 mm parallelepiped specimens to test. The lateral surface of each specimen was sealed with paraffin to avoid evaporation and maintain a uniaxial water flow during the test, while the upper and lower 5 × 2 mm opposite surfaces were left unsealed. Measurements were performed in an under-hook weighing configuration using a Radwag AS 220/C/2 electronic balance (Radwag, Poland) with an accuracy of 0.1 mg, mounted on a custom rack as shown in Figure 7. Weight data over time were recorded connecting the balance to a PC via the RS232 serial port.
Figure 7.
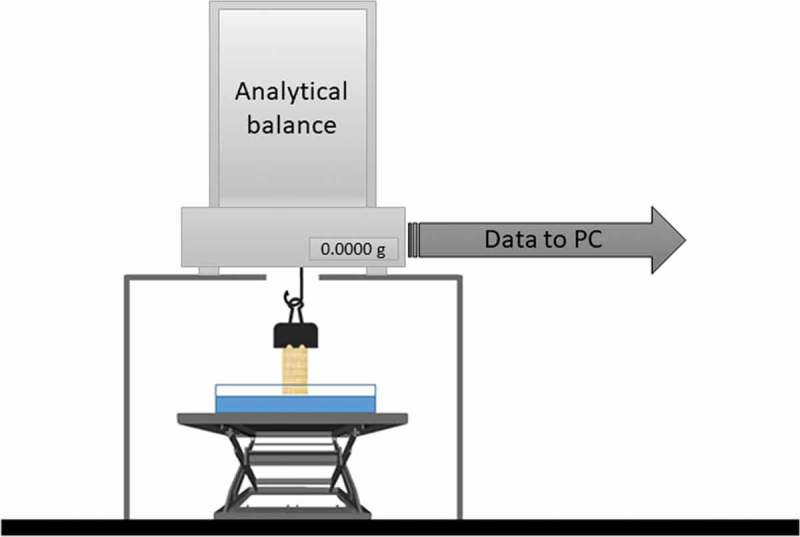
Experimental setup used for sorptivity measurements.
Dry samples were gently grasped with a paper clip and connected to the balance hook. A Petri dish filled with PBS 1x at room temperature was placed on a laboratory scissor jack positioned under the sample. Then data acquisition was started and the PBS-containing Petri dish slowly raised towards the bottom of the sample until an increase in the balance reading was observed. This increase is due to surface tension forces and was used as a reference point to identify sample contact with the PBS surface53: weight and time vectors were re-zeroed in correspondence to this point for sorptivity evaluation. The test surface was visually examined to ensure that no air was trapped under the specimen. If this occurred, or very occasionally, if the free water surface was significantly above the bottom surface of the specimen (indicated by a decrease in the balance reading due to buoyancy effects), the test was abandoned and the specimen reconditioned for a repeat test.
In the present study, weight measurements were recorded at 7 samples per second over a total test time of approximately 30 minutes. The sorptivity coefficient, (), is defined according to the following expression (Equation 1)39,54:
where represents the amount of PBS adsorbed (), denotes the cross-section area of the specimen that was in contact with PBS () and () is the time. To obtain the sorptivity coefficient, was plotted against the square root of time (), then was derived as the slope of the first linear portion of the curve.39 The density of PBS 1x was considered equal to that of water (i.e. 1 g/cm3) in the calculations.
Wet porosity and permeability estimation
Wet porosity for liver decellularised matrices was estimated adapting the method described by Weadock and colleagues.55 Briefly, liver dECMs were lyophilised as for swelling tests and their dry weight measured (). Then, they were swollen until equilibrium in PBS 1x at room temperature to determine the equilibrium swollen weight (). Finally, liver dECMs were sandwiched between two disks of filter paper (Whatman No. 3, United Kingdom) and placed under a 1 kg mass for 20 seconds to eliminate the pore-filling water, leaving only the portion which hydrates the dECM protein fibres, this giving the wet matrix weight (). The wet porosity (), defined as the volume of pore-filling water over the total liver dECM volume in the equilibrium swollen state, can be derived as follows (Equation 2).
where and are the density of PBS 1x (assumed as that of water, 1 g/cm3) and dry liver ECM (assumed as that of collagen, 1.34 g/cm3 56), respectively, while and represent the equilibrium mass swelling ratio (defined as ) and the residual mass swelling ratio (defined as ), respectively. Untreated liver samples were also tested and the results compared to those obtained for liver dECMs.
The wet porosity of liver dECMs was used along with the average pore diameter retrieved from 3D architectural analysis in the equilibrium swollen state to estimate the Darcy’s permeability () of decellularised liver matrices through the following relation proposed by Chor et al.57 (Equation 3).
Viscoelastic characterisation
Liver decellularised matrices were characterised using the epsilon dot method () to derive lumped viscoelastic parameters avoiding sample preload and deterioration during experiments.37,58 To ensure a repeatable sample testing state, essential to obtain meaningfully comparable data,27 samples were equilibrium swollen in PBS 1x at 4°C, then brought to room temperature and carefully measured using a calliper prior to testing.37,38,46,59 Hepatic tissue was considered as mechanically isotropic.36–38,60 Briefly, force and displacement time-series were recorded compressing samples at different strain rates (i.e. = 0.001, 0.005, 0.01 and 0.05 s−1) using a twin column Zwick/Roell ProLine Z005 uniaxial testing device (Zwick/Roell, Ulm, Germany) equipped with a 10 N load cell. The beginning of the sample compressive phase was identified as the instant at which the measured force crosses the abscissa towards monotonically increasing values with increasing displacement.37,45,58,61 Starting from this point, engineering stress () and strain () were calculated normalising measured force and displacement to sample’s cross-sectional area () and initial length (), respectively. The linear viscoelastic region (LVR) was identified as the region in which the stress varies linearly with the applied strain giving an of at least 0.995. Stress-time data within sample’s LVR (termed LVR stress-time data) obtained from measurements at different strain rates were globally fitted to derive viscoelastic constants for lumped parameter models as described in Tirella et al.58 A standard linear solid (SLS) and a 2-arm generalised Maxwell (GM2) model were used to fit experimental data.37 Six liver samples were tested at each strain rate, using a new sample for each trial to prevent permanent alterations due to repeated testing cycles (total number of specimens = 24).
Collagen hydrogels
Collagen hydrogels were obtained from type I collagen extracted from rat tails under sterile conditions following a standard protocol.62 After extraction, the collagen was freeze-dried and dissolved in 0.02 N acetic acid to obtain a 5.56 mg/mL solution. Then latter was then neutralised on ice by adding 10x concentrated M199 medium (Sigma-Aldrich) in a 9:1 collagen solution:M199 volume ratio, obtaining a 5 mg/mL neutralised collagen solution. Physically crosslinked hydrogels were obtained by pipetting 100 µL of the latter solution into the well of a 48-multiwell plate (Corning, Milan, IT), followed by 1 hour incubation at 37°C/5% CO2.63,64 After physical gelation, 100 µL of glutaraldehyde (GTA, Sigma-Aldrich) solution at different concentrations (i.e. 1, 2, 20, 100, 200 mM) were to each sample to modulate the substrate mechanical properties while keeping the other material parameters constant: this enables selective investigation of the role of substrate mechanics on cell response.59 Samples were incubated 1 hour at toom temperature (RT) to allow for GTA amino-crosslinking and then washed thrice with PBS 1x, soaked in a 0.1 M glycine solution for 2 h at room temperature to quench any residual aldehyde moiety and thus eliminate their potential toxicity.65 Finally, samples were rinsed twice with PBS and sterilised with 70% v/v ethanol (EtOH) aqueous solution for 2h at RT and placed under UV light for 30 minutes.59 Collagen samples were then incubated overnight with culture medium. The medium was then discarded and samples ready for either mechanical testing or cell seeding.
Hepg2 cell culture
The hepatocarcinoma cell line (HepG2) was purchased from ATCC (American Type Culture Collection ATCC, Manassas, VA). Cells were cultivated in Eagle’s Minimal Essential Medium (EMEM, 1 g/L glucose) (Sigma, Milan IT), supplemented with 10% inactivated fetal bovine serum (FBS) (Sigma, Milan IT), 1% non-essentials aminoacids (Lonza, Milan IT), 2 mM L-glutamine (Lonza, Milan IT), 100 U/mL penicillin and 100 μg/mL streptomycin (Lonza, Milan IT). All experiments were performed at passage 83, seeding cells at a density of 5 × 104 cells/well in a non-adhesive 48-well plate by carefully pipetting 500 μL of cell suspension on the samples. HepG2 cells were then cultured in a 37°C/5% CO2 humidified incubator up to 7 days, evaluating their viability and albumin secretion at different time points (i.e. day 1, 3 and 7) and assessing their morphology at day 7.
CellTiter-blue viability assay
CellTiter-Blue® Cell Viability Assay (Promega, Mannheim, Germany) was used according to the manufacturer’s instructions to assess cell viability at day 1, 3 and 7. Briefly, at each time point the medium in each well was removed (and collected for albumin secretion analyses) and replaced with 500 μL of fresh medium plus 50 μL of CellTiter-Blue® Reagent. Resofurin-mediated fluorescence was measured with a FLUOstar Omega microplate reader (BMG Labtech GmbH, Offenburg, Germany; excitation, 544 nm; detection, 590 nm) after 30 and 150 minutes of incubation. The slope of the fluorescence increase – representing the specific metabolic activity of cells – was calculated and expressed as percentage of that measured for cells cultured on softest collagen gel (i.e., the one crosslinked with 1 mM GTA) at the same time point.
Albumin secretion assay
Albumin secretion was quantified at day 1, 3 and 7 using an enzyme-linked immunosorbent assay (ELISA) kit (Bethyl Laboratories Inc., Montgomery, TX), according to manufacturer’s instructions. Absorbance readings at 450 nm were performed with a FLUOstar Omega microplate reader.
Cell morphology evaluation
HepG2 cells cultured were stained for F-actin and nuclei to assess their morphology after 3 and 7 days of culture on collagen hydrogels. For F-actin filaments staining, samples were incubated in the dark with rhodamine-conjugated phalloidin (Life-Technologies) diluted 1:400 in PBS containing 1% w/v BSA for 30 minutes at RT. After washing the samples three times with PBS, cell nuclei were stained incubating samples in the dark with 1 μg/mL DAPI (4ʹ,6-diamidino-2 phenylindole, Sigma-Aldrich) solution in PBS containing 1% w/v BSA for 30 minutes at RT. Then, samples were washed three times with PBS 1x and observed using a confocal microscope (Nikon A1, Nikon Co. Ltd., Tokyo, Japan).
Statistical analysis
All experiments were carried out at least in triplicate. Results are reported as mean ± standard deviation, unless stated otherwise. Comparisons between n groups of data (e.g. swelling ratio) referring to one factor only (e.g. time) were performed using one-way ANOVA followed by Tukey’s Multiple Comparison Test, while the Student’s t-test was used in case of two groups of data only (e.g. sorptivity coefficient of decellularised samples and untreated liver). Statistical significance was set at p < 0.05.
Acknowledgments
CM acknowledges funding from the Fondazione Umberto Veronesi under the Post-Doctoral Fellowship 2018. The Authors are grateful to the abattoir Desideri Luciano S.p.A. Via Abruzzi, 2 – Pontedera (Pisa), Italy, for kindly supplying fresh hepatic tissue.
References
- 1.Sbrana T, Ahluwalia A.. engineering quasi-vivo in vitro organ models. Adv Exp Med Biol. 2012. accessed 2012 December3;745:138–153. [Internet] http://www.ncbi.nlm.nih.gov/pubmed/22437817. [DOI] [PubMed] [Google Scholar]
- 2.Chang R, Emami K, Wu H, Sun W. Biofabrication of a three-dimensional liver micro-organ as an in vitro drug metabolism model. Biofabrication. 2010. accessed 2011 February15;2:045004 Internet. http://www.ncbi.nlm.nih.gov/pubmed/21079286. [DOI] [PubMed] [Google Scholar]
- 3.Naruse K, Tang W, Makuuch M. Artificial and bioartificial liver support: a review of perfusion treatment for hepatic failure patients. World J Gastroenterol. 2007;13:1516–1521. Internet. http://www.ncbi.nlm.nih.gov/pubmed/17461442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazzei D, Guzzardi MA, Giusti S, Ahluwalia A. A low shear stress modular bioreactor for connected cell culture under high flow rates. Biotechnol Bioeng. 2010. accessed 2012 November18;106:127–137. Internet. http://www.ncbi.nlm.nih.gov/pubmed/20091740. [DOI] [PubMed] [Google Scholar]
- 5.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger J, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87:1315–1530. Internet. http://link.springer.com/10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann A, Moll M, Gottwald E, Nies C, Zantl R, Wagner H, Burkhardt B, Sánchez J, Ladurner R, Thasler W, et al. 3D cultivation techniques for primary human hepatocytes. Microarrays. 2015;4:64–83. http://www.mdpi.com/2076-3905/4/1/64/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reilly GC, Engler AJ. Intrinsic extracellular matrix properties regulate stem cell differentiation. J Biomech. 2010. accessed 2012 November3;43:55–62. Internet. doi: 10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Nelson CM, Bissell MJ. Modeling dynamic reciprocity: engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin Cancer Biol. 2005. accessed 2012 March24;15:342–352. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2933210&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemir S, West JL. Synthetic materials in the study of cell response to substrate rigidity. Ann Biomed Eng. 2010. accessed 2010 July15;38:2–20. Internet. http://www.ncbi.nlm.nih.gov/pubmed/19816774. [DOI] [PubMed] [Google Scholar]
- 10.Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, Sc R, Fe W, Mp L. Elucidating the role of matrix stiffness in 3d cell migration and remodeling. Biophys J. 2011. accessed 2011 January20;100:284–293. Internet. http://www.ncbi.nlm.nih.gov/pubmed/21244824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond JS, Gilbert TW, Howard D, Zaitoun A, Michalopoulos G, Shakesheff KM, Beckingham IJ, Badylak SF. Scaffolds containing growth factors and extracellular matrix induce hepatocyte proliferation and cell migration in normal and regenerating rat liver. J Hepatol. 2011. accessed 2012 March1;54:279–287. Internet. http://www.ncbi.nlm.nih.gov/pubmed/21126791. [DOI] [PubMed] [Google Scholar]
- 12.Mattei G, Magliaro C, Giusti S, Ramachandran SD, Heinz S, Braspenning J, Ahluwalia A. On the adhesion-cohesion balance and oxygen consumption characteristics of liver organoids. PLoS One. 2017;12:e0173206 Internet. http://dx.plos.org/10.1371/journal.pone.0173206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semler EJ, Ranucci CS, Moghe PV. Tissue assembly guided via substrate biophysics: applications to hepatocellular engineering. Adv Biochem Eng Biotechnol. 2006. accessed 2012 October17;102:1–46. Internet. http://www.ncbi.nlm.nih.gov/pubmed/17089785. [DOI] [PubMed] [Google Scholar]
- 14.Jun J-I, Lau LF. Cellular senescence controls fibrosis in wound healing. Aging (Albany NY). 2010;2:627–631. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2984611&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanley KP, Oakley F, Sugden S, Wilson DI, Mann DA, Hanley NA. Ectopic SOX9 mediates extracellular matrix deposition characteristic of organ fibrosis. J Biol Chem. 2008. accessed 2012 April10;283:14063–14071. Internet. http://www.ncbi.nlm.nih.gov/pubmed/18296708. [DOI] [PubMed] [Google Scholar]
- 16.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006. accessed 2012 October22;22:287–309. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2933192&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002. accessed 2012 October22;14:608–616. Internet. http://www.ncbi.nlm.nih.gov/pubmed/12231357. [DOI] [PubMed] [Google Scholar]
- 18.Baiocchini A, Montaldo C, Conigliaro A, Grimaldi A, Correani V, Mura F, Ciccosanti F, Rotiroti N, Brenna A, Montalbano M, et al. Extracellular matrix molecular remodeling in human liver fibrosis evolution. PLoS One. 2016;11:e0151736 Internet. 10.1371/journal.pone.0151736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver W, Goldsmith EC. Regulation of tissue fibrosis by the biomechanical environment. Biomed Res Int. 2013. accessed 2013 October19;2013:1–10. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3679815&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells RG. The role of matrix stiffness in regulating cell behavior. Hepatology. 2008. accessed 2013 October19;47:1394–1400. Internet. http://www.ncbi.nlm.nih.gov/pubmed/18307210. [DOI] [PubMed] [Google Scholar]
- 21.Hansen LK, Wilhelm J, Fassett JT. Regulation of hepatocyte cell cycle progression and differentiation by type I collagen structure. Curr Top Dev Biol. 2006. accessed 2013 October20;72:205–236. Internet. http://www.ncbi.nlm.nih.gov/pubmed/16564336. [DOI] [PubMed] [Google Scholar]
- 22.Semler EJ, Ranucci CS, Moghe PV. Mechanochemical manipulation of hepatocyte aggregation can selectively induce or repress liver-specific function. Biotechnol Bioeng. 2000. accessed 2013 October20;69:359–369. Internet. http://www.ncbi.nlm.nih.gov/pubmed/10862674. [DOI] [PubMed] [Google Scholar]
- 23.Fassett J, Tobolt D, Hansen LK. Type I collagen structure regulates cell morphology and EGF signaling in primary rat hepatocytes through cAMP-dependent protein kinase A. Mol Biol Cell. 2006. accessed 2013 October20;17:345–356. [Internet] http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1345672&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Dranoff JA, Chan EP, Uemura M, Sévigny J, Wells RG. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007. accessed 2013 October20;46:1246–1256. [Internet] http://www.ncbi.nlm.nih.gov/pubmed/17625791. [DOI] [PubMed] [Google Scholar]
- 25.Olsen AL, Bloomer SA, Chan EP, Mda G, Georges PC, Sackey B, Uemura M, Janmey PA, Wells RG. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol. 2011. accessed 2013 November4;301:G110–8. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3129929&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozoya OA, Wauthier E, Turner RA, Barbier C, Prestwich GD, Guilak F, Superfine R, Lubkin SR, Reid LM. Regulation of hepatic stem/progenitor phenotype by microenvironment stiffness in hydrogel models of the human liver stem cell niche. Biomaterials. 2011. accessed 2013 November6;32:7389–7402. [Internet] http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3157321&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattei G, Ahluwalia A. Sample, testing and analysis variables affecting liver mechanical properties: A review. Acta Biomater. 2016;45:60–71. Internet. http://linkinghub.elsevier.com/retrieve/pii/S1742706116304603. [DOI] [PubMed] [Google Scholar]
- 28.Tirella A, La Marca M, Brace L-A, Mattei G, Aylott JW, Ahluwalia A. Nano-in-micro self-reporting hydrogel constructs. J Biomed Nanotechnol. 2015. accessed 2014 December16;11:1451–1460. Internet. http://openurl.ingenta.com/content/xref?genre=article&issn=1550-7033&volume=11&issue=8&spage=1451. [DOI] [PubMed] [Google Scholar]
- 29.Mattei G, Vozzi G. CFD modelling of a mixing chamber for the realisation of functionally graded scaffolds. Comput Chem Eng. 2016. accessed 2015 September2;84:43–48. Internet. http://www.sciencedirect.com/science/article/pii/S0098135415002823. [Google Scholar]
- 30.Ma PX. Biomimetic materials for tissue engineering. Adv Drug Deliv Rev. 2008. accessed 2013 November13;60:184–198. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2271038&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fathi I, Eltawila A. Whole-liver decellularization: advances and insights into current understanding [Internet]. In: Xenotransplantation - New Insights. InTech; 2017. http://www.intechopen.com/books/xenotransplantation-new-insights/whole-liver-decellularization-advances-and-insights-into-current-understanding
- 32.Lang R, Stern MM, Lang R, Smith L, Liu Y, Bharadwaj S, Liu G, Pm Baptista, Cr Bergman, Soker S, Jj Yoo, et al. Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials. 2011;1–11. accessed 2011 July14 Internet. http://www.ncbi.nlm.nih.gov/pubmed/21723601. [DOI] [PubMed] [Google Scholar]
- 33.Sellaro TL, Ranade A, Faulk DM, McCabe GP, Dorko K, Badylak SF, Strom SC. Maintenance of human hepatocyte function in vitro by liver-derived extracellular matrix gels. Tissue Eng Part A. 2010;16:1075–1082. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2863084&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barakat O, Abbasi S, Rodriguez G, Rios J, Wood RP, Ozaki C, Holley LS, Gauthier PK. Use of decellularized porcine liver for engineering humanized liver organ. J Surg Res. 2012. accessed 2012 November4;173:e11–25. Internet. http://linkinghub.elsevier.com/retrieve/pii/S0022480411007591. [DOI] [PubMed] [Google Scholar]
- 35.Mattei G, Magliaro C, Pirone A, Ahluwalia A. Decellularized human liver is too heterogeneous for designing a generic extracellular matrix mimic hepatic scaffold. Artif Organs. 2017;41:E347–55. Internet. doi: 10.1111/aor.12925. [DOI] [PubMed] [Google Scholar]
- 36.Marchesseau S, Heimann T, Chatelin S, Willinger R, Delingette H. Fast porous visco-hyperelastic soft tissue model for surgery simulation: application to liver surgery. Prog Biophys Mol Biol. 2010. accessed 2012 November2;103:185–196. Internet. http://www.ncbi.nlm.nih.gov/pubmed/20869382. [DOI] [PubMed] [Google Scholar]
- 37.Mattei G, Tirella A, Gallone G, Ahluwalia A. Viscoelastic characterisation of pig liver in unconfined compression. J Biomech. 2014. accessed 2014 October21;47:2641–2646. Internet. http://www.sciencedirect.com/science/article/pii/S0021929014003285. [DOI] [PubMed] [Google Scholar]
- 38.Mattei G, Di Patria V, Tirella A, Alaimo A, Elia G, Corti A, Paolicchi A, Ahluwalia A. Mechanostructure and composition of highly reproducible decellularized liver matrices. Acta Biomater. 2014. accessed 2014 January18;10:875–882. Internet. http://www.sciencedirect.com/science/article/pii/S1742706113005400. [DOI] [PubMed] [Google Scholar]
- 39.Tasdemir C. 2003. Combined effects of mineral admixtures and curing conditions on the sorptivity coefficient of concrete. Cem Concr Res. 33:1637–1642. doi: 10.1016/S0008-8846(03)00112-1. [DOI] [Google Scholar]
- 40.Nahmias Y, Berthiaume F, Yarmush ML. Integration of technologies for hepatic tissue engineering. Adv Biochem Engin/Biotechnol. 103;2006:309–329. [DOI] [PubMed] [Google Scholar]
- 41.Munk OL, Bass L, Roelsgaard K, Bender D, Hansen SB, Keiding S. Liver kinetics of glucose analogs measured in pigs by PET: importance of dual-input blood sampling. J Nucl Med. 2001;42:795–801. Internet. http://www.ncbi.nlm.nih.gov/pubmed/11337579 [PubMed] [Google Scholar]
- 42.Taniguchi H, Masuyama M, Koyama H, Oguro A, Takahashi T. Quantitative measurement of human tissue hepatic blood volume by C15O inhalation with positron-emission tomography. Liver. 2008;16:258–262. Internet. doi: 10.1111/j.1600-0676.1996.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 43.Fung YC. Biomechanics: mechanical properties of living tissues. 2nd New York: Springer; 1993. [Google Scholar]
- 44.Dw E, Ec M, Pm B, Soker S, Jl S. Scale-dependent mechanical properties of native and decellularized liver tissue. Biomech Model Mechanobiol. 2013. accessed 2013 September6;12:569–580. Internet. http://www.ncbi.nlm.nih.gov/pubmed/22890366. [DOI] [PubMed] [Google Scholar]
- 45.Mattei G, Cacopardo L, Ahluwalia A. Micro-mechanical viscoelastic properties of crosslinked hydrogels using the nano-epsilon dot method. Materials (Basel). 2017;10:889 Internet. http://www.mdpi.com/1996-1944/10/8/889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh W-C-C, Li P-C-C, Jeng Y-M-M, Hsu H-C-C, Kuo P-L-L, Li M-L-L, Yang P-M-M, Lee PH, Po HL. Elastic modulus measurements of human liver and correlation with pathology. Ultrasound Med Biol. 2002. accessed 2012 December8;28:467–474. Internet. http://linkinghub.elsevier.com/retrieve/pii/S0301562902004891. [DOI] [PubMed] [Google Scholar]
- 47.You J, Park S-A, Shin D-S, Patel D, Raghunathan VK, Kim M, Murphy CJ, Tae G, Revzin A. Characterizing the effects of heparin gel stiffness on function of primary hepatocytes. Tissue Eng Part A. 2013;19:2655–2663. Internet. http://online.liebertpub.com/doi/abs/10.1089/ten.tea.2012.0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010. accessed 2013 January24;123:4195–4200. Internet. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2995612&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim TG, Shin H, Lim DW. Biomimetic Scaffolds for Tissue Engineering. Adv Funct Mater.2012. accessed 2016 July27;22:2446–2468. Internet. Available from. doi: 10.1002/adfm.201103083. [DOI] [Google Scholar]
- 50.Adam R, Karam V, Delvart V, O’Grady J, Mirza D, Klempnauer J, Castaing D, Neuhaus P, Jamieson N, Salizzoni M, et al. Evolution of indications and results of liver transplantation in Europe. A report from the European Liver Transplant Registry (ELTR). J Hepatol. 2012;57:675–688. accessed 2014 July11 Internet. http://linkinghub.elsevier.com/retrieve/pii/S0168827812003364. [DOI] [PubMed] [Google Scholar]
- 51.Magliaro C, Tirella A, Mattei G, Pirone A, Ahluwalia A. HisTOOLogy: an open-source tool for quantitative analysis of histological sections. J Microsc. 2015. accessed 2015 November27;260:260–267. Internet. doi: 10.1111/jmi.12292. [DOI] [PubMed] [Google Scholar]
- 52.Adams R, Bischof L. Seeded region growing. IEEE Trans Pattern Anal Mach Intell. 1994;16:641–647. Internet. http://ieeexplore.ieee.org/document/295913/. [Google Scholar]
- 53.Sabir BB, Wild S, O’Farrell M. A water sorptivity test for martar and concrete. Mater Struct. 1998;31:568–574. Internet. http://www.springerlink.com/index/10.1007/BF02481540 [Google Scholar]
- 54.Bozkurt N, Yazicioglu S. Strength and capillary water absorption of lightweight concrete under different curing conditions. Indian J Eng Mater Sci. 17;2010:145–151. [Google Scholar]
- 55.Weadock K, Olson RM, Silver FH. Evaluation of collagen crosslinking techniques. Biomater Med Devices Artif Organs. accessed 2016 May25;11:293–318. [Internet] http://www.ncbi.nlm.nih.gov/pubmed/6091801. [DOI] [PubMed] [Google Scholar]
- 56.Chapman EW, Rodriguez F. Acrylic resin reinforcement of reconstituted collagen films. Polym Eng Sci. 1977. accessed 2014 January17;17:282–286. Internet. doi: 10.1002/pen.760170503. [DOI] [Google Scholar]
- 57.Chor MV, Li W. A permeability measurement system for tissue engineering scaffolds. Meas Sci Technol. 2007. accessed 2011 May18;18:208–216. Internet. http://stacks.iop.org/0957-0233/18/i=1/a=026?key=crossref.7d61d3e6027559b16d7b7a2fd1a34e04. [Google Scholar]
- 58.Tirella A, Mattei G, Ahluwalia A. Strain rate viscoelastic analysis of soft and highly hydrated biomaterials. J Biomed Mater Res Part A. 2014. accessed 2014 November28;102:3352–3360. Internet. http://www.ncbi.nlm.nih.gov/pubmed/23946054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mattei G, Ferretti C, Tirella A, Ahluwalia A, Mattioli-Belmonte M. Decoupling the role of stiffness from other hydroxyapatite signalling cues in periosteal derived stem cell differentiation. Sci Rep. 2015;5:10778 Internet. http://www.nature.com/doifinder/10.1038/srep10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pervin F, Chen WW, Weerasooriya T. Dynamic compressive response of bovine liver tissues. J Mech Behav Biomed Mater. 2011. accessed 2011 September23;4:76–84. Internet. http://www.ncbi.nlm.nih.gov/pubmed/21094481. [DOI] [PubMed] [Google Scholar]
- 61.Mattei G, Gruca G, Rijnveld N, Ahluwalia A. The nano-epsilon dot method for strain rate viscoelastic characterisation of soft biomaterials by spherical nano-indentation. J Mech Behav Biomed Mater. 2015;50:150–159. Internet. http://linkinghub.elsevier.com/retrieve/pii/S1751616115002088. [DOI] [PubMed] [Google Scholar]
- 62.Beken S, Vanhaecke T, De Smet K, Pauwels M, Vercruysse A, Rogiers V. Collagen-gel cultures of rat hepatocytes: collagen-gel sandwich and immobilization cultures. Methods Mol Biol. 1998. accessed 2014 January21;107:303–309. Internet. http://www.ncbi.nlm.nih.gov/pubmed/14577239. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi Y, Ishigaki T, Sano K, Miyamoto K, Nomura S, Horiuchi T. Three-dimensional invasion of epithelial-mesenchymal transition-positive human peritoneal mesothelial cells into collagen gel is promoted by the concentration gradient of fibronectin. Perit Dial Int. accessed 2014 January21;31:477–485. [Internet] http://www.ncbi.nlm.nih.gov/pubmed/21719684. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci U S A. 2012;109:9342–9347. accessed 2014 January21 [Internet] http://www.pnas.org/content/109/24/9342.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jayakrishnan A, Jameela SR. Glutaraldehyde as a fixative in bioprostheses and drug delivery matrices. Biomaterials. 1996. accessed 2014 March1;17:471–484. [Internet] http://www.sciencedirect.com/science/article/pii/0142961296827219. [DOI] [PubMed] [Google Scholar]


