Summary
Caspase-11, a cytosolic endotoxin (lipopolysaccharide: LPS) receptor, mediates pyroptosis, a lytic form of cell death. Caspase-11-dependent pyroptosis mediates lethality in endotoxemia, but it is unclear how LPS is delivered into the cytosol for the activation of caspase-11. Here we discovered that hepatocyte-released high mobility group box 1 (HMGB1) was required for caspase-11-dependent pyroptosis and lethality in endotoxemia and bacterial sepsis. Mechanistically, hepatocyte-released HMGB1 bound LPS and targeted its internalization into the lysosomes of macrophages and endothelial cells via the receptor for advanced glycation end-products (RAGE). Subsequently, HMGB1 permeabilized the phospholipid bilayer in the acidic environment of lysosomes. This resulted in LPS leakage into the cytosol and caspase-11 activation. Depletion of hepatocyte HMGB1, inhibition of hepatocyte HMGB1 release, neutralizing extracellular HMGB1, or RAGE deficiency prevented caspase-11-dependent pyroptosis and death in endotoxemia and bacterial sepsis. These findings indicate that HMGB1 interacts with LPS to mediate caspase-11-dependent pyroptosis in lethal sepsis.
Keywords: caspase-11, inflammasome, sepsis, endotoxemia, pyroptosis, HMGB1
Introduction
Elevated concentrations of circulating bacterial endotoxin(lipopolysaccharide [LPS]) are encountered in sepsis and removal of LPS is beneficial to patients with sepsis (Angus and van der Poll, 2013, Ronco et al., 2010). Endotoxemia-induced lethality depends largely on the activation of caspase-11, which is encoded by Casp11 (Hagar et al., 2013, Kayagaki et al., 2011, Kayagaki et al., 2013, Kayagaki et al., 2015, Wang et al., 1998). Activated caspase-11 cleaves gasdermin D (GSDMD) into pore-forming peptides that subsequently cause pyroptosis, a lytic form of cell death that releases eicosanoids, such as leukotriene B4 (LTB4), through cyclooxygenase (COX)-1 as well as alarmins, including interleukin(IL)-1α (Hagar et al., 2013). Deletion of Gasdmd or inhibition of COX-1 improves survival in endotoxemia (Hagar et al., 2013, Kayagaki et al., 2015). Caspase-11-mediated pyroptosis destroys the intracellular niche and activates local immune defenses by releasing chemoattractants and promoting vascular permeability (Kayagaki et al., 2011, Kayagaki et al., 2013, Shi et al., 2014, Hagar et al., 2013, Aachoui et al., 2013). Extracellular LPS triggers pyroptosis of immune cells and endothelial cellsonly after LPS has been delivered to caspase-11 inside the cell (Hagar et al., 2013, Kayagaki et al., 2013, Aachoui et al., 2013, Cheng et al., 2017). Uptake of intact gram-negative bacteria into macrophages with subsequent lysis of the phagolysosome can lead to activation of cytosoliccaspase-11 as does the uptake of LPS-containing outer membrane vesicles (OMVs) released by live extracellular bacteria (Meunier et al., 2014, Vanaja et al., 2016). It has been previously unknown, however, how extracellular or circulating LPS is delivered into the cytosol of cells for the activation of caspase-11.
High mobility group box-1 (HMGB1), a ubiquitous nuclear and cytosolic protein, is released into the circulation and mediates lethality during endotoxemia and sepsis (Wang et al., 1999, Wang et al., 2004, Lamkanfi et al., 2010, Lu et al., 2012). Genetic deletion of HMGB1 or neutralizing circulating HMGB1 confers protection in lethal endotoxemia and bacterial sepsis (Wang et al., 1999, Wang et al., 2004, Andersson and Tracey, 2011, Lamkanfi et al., 2010, Qin et al., 2006, Rittirsch et al., 2008). The biological activity of HMGB1 is dependent upon the redox status of its three cysteine residues (Lu et al., 2012, Kazama et al., 2008). Whereas the disulfide isoform activates toll-like receptor 4 (TLR4) and cytokineproduction, the fully reduced isoform does not (Lu et al., 2012). HMGB1 binds LPS (Youn et al., 2008), leading us to hypothesize that HMGB1 might deliver LPS into the cytosol of macrophages and endothelial cells to trigger caspase-11-dependent pyroptosis and lethality.
Here we show that HMGB1 enables extracellular LPS to activate cytosolic caspase-11 in macrophages and endothelial cells via receptor for advanced glycation end-products (RAGE)-dependent internalization of HMGB1-LPS complexes into lysosomes, where HMGB1 permeabilizes the phospholipid bilayer under acidic conditions. The destabilization of lysosomes and the leakage of LPS into the cytosol culminates in the activation of caspase-11. Hepatocytes are the major source of circulating HMGB1 during endotoxemia and bacterial sepsis, and deletion of Hmgb1in hepatocytes, prevention of hepatocyte HMGB1 release, administration of HMGB1 neutralizing antibody, or RAGE deficiency prevented caspase-11-dependent pyroptosis and death in endotoxemia and bacterial sepsis.
Results
HMGB1 Enables Extracellular LPS to Activate Caspase-11
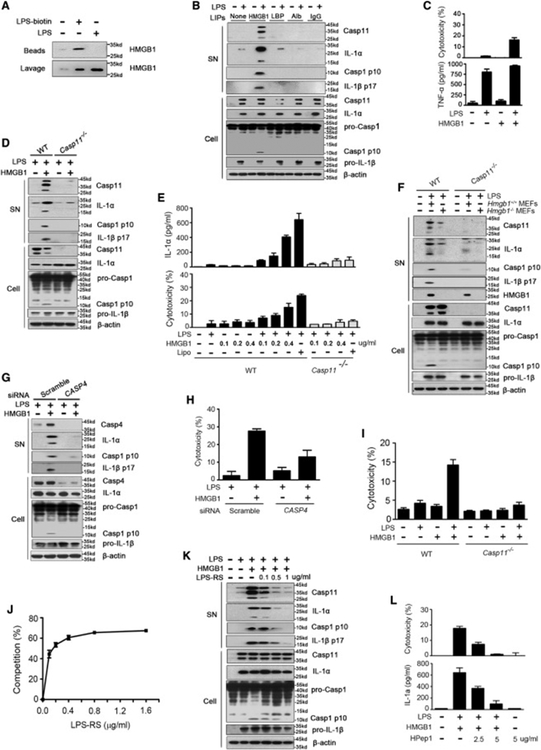
We first determined whether HMGB1 could physically interact with LPS. As shown by surface plasmon resonance, the reduced form of HMGB1 bound LPS in vitro (Figure S1A). To test whether HMGB1 binds LPS during endotoxemia, biotin-labeled LPS was administered into the peritoneal cavity of mice and soluble LPS binding molecules were isolated using streptoavidin-coated beads. The physical interaction between biotin-labeled LPS and HMGB1 during endotoxemia was demonstrated by both immunoblot and mass spectrometry (Figures 1A and S1B). In addition to HMGB1, we found that LPS-binding protein (LBP), albumin, and immunoglobin also interacted with LPS in vivo (Figure S1B). We next determined whether these LPS-interacting proteins enabled extracellular LPS to activate caspase-11. LPS failed to induce pyroptosis or the associated release of IL-1α in cultured macrophages in the presence of albumin, immunoglobin, or LBP (Figures 1B and S1C). In contrast, highly purified recombinant HMGB1 protein (reduced form) enabled LPS to induce macrophage pyroptosis, which largely depended on caspase-11 (Figures 1B–1E). HMGB1 alone failed to induce pyroptosis or tumor necrosis factor (TNF) production (Figure 1C). Importantly, endogenous HMGB1 released from necrotic Hmgb1+/+ mouse embryonic fibroblasts(Scaffidi et al., 2002) enabled LPS to induce the release and cleavage of IL-1α from wild-type but not caspase-11-deficient macrophages without enhancing TNF production (Figures 1F and S1D). Silencing of CASP4, one of the human homologs of Casp11 (Shi et al., 2014), blocked HMGB1-induced IL-1α and LDH release from human monocytic THP-1cells in the presence of LPS (Figures 1G and1H). Furthermore, HMGB1 enabled extracellular LPS to trigger pyroptosis in WT but not Casp11−/−mouse lung endothelial cells (Figure 1I).
Figure 1. HMGB1 Enables Extracellular LPS to Activate Caspase-11 through Physical Binding to LPS.
(A) Mice received i.p. biotin-labeled LPS (5 mg/kg) or normal saline (NS). Two hours later, peritoneal lavage fluid was incubated with streptavidin-coated beads. Pull down of HMGB1 was revealed by immunoblots.
(B) Immunoblots for caspase-11, IL-1α, caspase-1, IL-1β, and β-actin in the supernatants (SN) or cell lysates (cell) of mouse peritoneal macrophagesstimulated with the indicated LPS-interacting proteins (LIPs, 1 μg/mL) plus LPS (1 μg/mL) for 16 hr.
(C) LDH assay and ELISA for TNF in the supernatants of mouse peritoneal macrophages stimulated with LPS alone (1 μg/mL) or LPS (1 μg/mL)+HMGB1 (400 ng/mL) for 16 hr.
(D) Immunoblots for caspase-11, IL-1α, caspase-1, IL-1β, and β-actin in the supernatants (SN) or cell lysates (Cell) of WT or Casp11−/− peritoneal macrophages stimulated with LPS alone (1 μg/mL) or LPS (1 μg/mL)+HMGB1 (400 ng/mL) for 16 hr.
(E) ELISA for total IL-1α and LDH assay in the supernatants of WT or Casp11−/−peritoneal macrophages stimulated with LPS alone (1 μg/mL) or LPS (1 μg/mL)+HMGB1 of indicated concentration for 16 hr.
(F) Immunoblot to detect caspase-11, IL-1α, caspase-1, IL-1β, HMGB1, and β-actin in supernatants (SN) or cell lysates (Cell) of mouse peritoneal macrophages of indicated genotypes upon exposure to the necrotic Hmgb1−/− or Hmgb1+/+ MEFs (106 cells/mL) in the presence or the absence of LPS (1 μg/mL) for 16 hr (the ratio of necrotic cells to macrophages is 1:1).
(G) Immunoblot to detect caspase-4 (Casp4), IL-1α, caspase-1, IL-1β, and β-actin in supernatants (SN) or cell lysates (Cell) of human monocytic THP-1 cells transfected with scrambled siRNA or CASP4-specific siRNA upon HMGB1 (400 ng/mL) and LPS (1 μg/mL) stimulation for 16 hr.
(H) LDH assay in the supernatants of human monocytic THP-1 cells transfected with scrambled siRNA or CASP4-specific siRNA upon HMGB1 (400 ng/mL) and LPS (1 μg/mL) stimulation for 16 hr.
(I) LDH assay for WT or Casp11−/− mouse lung endothelial cells stimulated with LPS (1 μg/mL) alone or HMGB1 (400 ng/mL) alone or LPS (1 μg/mL)+HMGB1 (400 ng/mL) for 16 hr.
(J) The LPS-binding capacity of HMGB1 incubated with different concentrations of LPS-RS. Plates coated with recombinant HMGB1 (16 μg/mL) were incubated with biotin-labeled LPS (0.2 μg/mL) with indicated concentration of LPS-RS. Binding between plate-coated HMGB1 and biotin-labeled LPS was measured by using streptavidin-HRP. The percentage of binding competition by LPS-RS was determined.
(K) Mouse peritoneal macrophages were stimulated with HMGB1 (400 ng/mL) and LPS (1 μg/mL) in the presence of indicated doses of LPS-RS for 16 hr. Caspase-11, IL-1α, caspase-1, IL-1β, and β-actin were detected by immunoblot.
(L) LDH assay for cytotoxicity and ELISA for IL-1α in the supernatants of WT mouse peritoneal macrophages stimulated with LPS (1 μg/mL)+HMGB1 (400 ng/mL) in the presence of different concentrations of HPep1 for 16 hr. Graphs show the mean ± SD of technical replicates and are representative of at least three independent experiments. See also Figures S1 and S2
Caspase-11 induces inflammasome-mediated caspase-1 activation with pro-IL-1β cleavage and IL-1β release (Kayagaki et al., 2011). Consistent with these previous observations, deletion of Casp11 reduced HMGB1+LPS-induced IL-1β release, pro-IL-1β cleavage, and caspase-1 activation to the same amounts observed in Nlrp3−/− or Asc−/−macrophages (Figures 1B, 1D S2A, and S2B). Pharmacological inhibition of NLRP3 by MCC950 (Coll et al., 2015) reduced IL-1β release, pro-IL-1β cleavage, and caspase-1 activation in a concentration-dependent manner (Figures S2C and S2D). Furthermore, HMGB1+LPS-induced pro-IL-1β cleavage and IL-1β release largely depended on caspase-11 and the NLRP3 inflammasome (Figures S2A and S2B). Taken together, these observations indicate that HMGB1 enables extracellular LPS to activate caspase-11 and downstream caspase-1.
HMGB1-LPS Binding Is Important for Caspase-11-Dependent Pyroptosis
HMGB1 has been shown previously to bind LPS (Figure S1A; Youn et al., 2008). To determine whether a direct interaction between HMGB1 and LPS was required for caspase-11 activation, penta-acylated LPS from the Rhodobacter sphaeroides (LPS-RS), a competitive LPS antagonist that does not activate caspase-11 (Shi et al., 2014), was added to the assays. LPS-RS dose dependently blocked HMGB1-LPS interaction (Figure 1J) and inhibited HMGB1+LPS-induced pyroptosis in mouse peritoneal macrophages (Figures 1K and S2E). HPep1, a peptide known to specifically block the HMGB1-LPS binding (Youn et al., 2011), also dose dependently inhibited HMGB1+LPS-induced pyroptosis in mouse macrophages (Figure 1L).
HMGB1 contains two LPS-binding domains, one each in the A- and B-box domains. These two LPS-binding domains bind to the polysaccharide and lipid A moieties of LPS, respectively (Youn et al., 2011). However, truncated HMGB1 mutants containing either A-box or B-box could not bind LPS, suggesting that efficient HMGB1-LPS binding requires both LPS-binding domains (Youn et al., 2008, Youn et al., 2011). To further prove that HMGB1-LPS binding is important for HMGB1+LPS to induce pyroptosis, we used recombinant truncated forms of HMGB1 that included only A-box or B-box. In contrast to full-length HMGB1, the truncated HMGB1 form containing only A-box or B-box were unable to bind LPS efficiently (Figure S2F) and failed to induce pyroptosis in the presence of LPS (Figures S2G and 2H). Together, these findings establish that HMGB1 enables extracellular LPS to activate caspase-11 through physical interaction with LPS.
HMGB1 Delivers LPS into the Cytosol through RAGE-Mediated Internalization
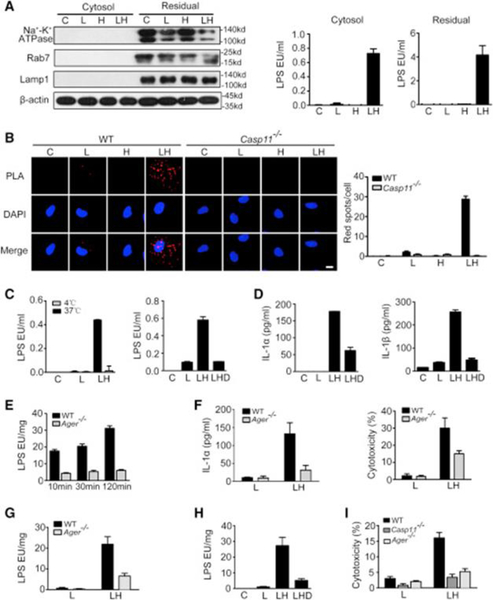
By using cholera toxin B or lipofectamine 3000, we found that HMGB1 is not required for the activation of caspase-11 by LPS delivered directly to the cytosol (Figures S2I–2L). To determine whether HMGB1 is essential for translocation of extracellular LPS to the cytosol, we isolated cytosol devoid of cytoplasmic membranes, endosomes, and lysosomes using low concentrations of digitonin (Vanaja et al., 2016) on macrophages treated with LPS with or without HMGB1 (Figure 2A). LPS was present in the cytosol of macrophages stimulated with HMGB1+LPS but not LPS alone. We next assessed the interaction between caspase-11 and LPS using a proximity ligation assay (PLA) and observed the interaction of LPS with caspase-11 in the cytosol of macrophages stimulated with HMGB1+LPS but not LPS alone (Figure 2B). Thus, HMGB1 mediates the translocation of LPS to the cytosol leading to LPS interaction with caspase-11.
Figure 2. HMGB1 Delivers Extracellular LPS into the Cytosol through RAGE-Mediated Internalization.
(A) Immunoblot for Na+-K+ ATPase, Rab7, Lamp1, and β-actin (left) and LPS activity assay (right) in the cytosolic and residual fraction (including cytoplasmicmembranes, endosomes, lysosomes, nuclei, etc.) from LPS (L, 1 μg/mL)- or LPS (1 μg/mL)+HMGB1 (400 ng/mL) (LH)-stimulated mouse peritoneal macrophages.
(B) The physical interaction between caspase-11 and LPS were visualized as the red spots by PLA in mouse peritoneal macrophages primed with 100 ng/mL LPS for 4 hr and then stimulated with LPS alone (L, 5 μg/mL), HMGB1 alone (10 μg/mL) (H), or LPS (5 μg/mL)+HMGB1 (10 μg/mL) (LH) for 2 hr. Scale bar: 10 μm.
(C and D) LPS activity assay in the cytosolic fraction (C) or ELISA for total IL-1αand IL-1β in cell culture medium (D) of LPS (L, 1 μg/mL) or LPS (1 μg/mL)+HMGB1 (400 ng/mL) (LH)-stimulated mouse peritoneal macrophages either placed at 4 or 37 degrees for 2 hr or pretreated with 20 μM dynasore (LHD) for 0.5 hr and washed away before LH treatment.
(E and F) LPS activity assay on the cytosolic fraction (E) or ELISA for IL-1α or LDH assay in cell culture medium (F) of LPS+HMGB1-stimulated peritoneal macrophages from WT or Ager−/− mice.
(G–I) LPS activity assay on the cytosolic fraction (G and H) or LDH assay in cell culture medium (I) of WT, Ager−/−, or Casp11−/− mouse lung endothelial cellspretreated with 20 μM dynasore for 0.5 hr stimulated with LPS or HMGB1 with or without pre-incubation of 20 μM dynasore for 0.5 hr.
Graphs show the mean ± SD of technical replicates and are representative of at least three independent experiments. See also Figure S3.
To assess how HMGB1 mediates LPS translocation to the cytosol, we used confocal microscopy to first localize LPS-bound HMGB1 in macrophages (Figure S3A) and found that HMGB1-LPS complexes co-localized with endosomal and lysosomal markers (Figure S3B). HMGB1 enhanced LPS internalization into endo-lysosomes (Figure 2A). We then tested whether internalization of HMGB1-LPS complexes into endo-lysosomes is a critical step for LPS translocation to the cytosol. Cooling cells to 4°C or addition of dynasore, both of which inhibit the active internalization processes (Thompson et al., 2012), prevented the cytosolic translocation of LPS and the release of IL-1α and IL-1β in mouse peritoneal macrophages stimulated with HMGB1+LPS (Figures 2C and 2D). HMGB1 internalization largely depends on RAGE, which is encoded by Ager (Liliensiek et al., 2004). Consequently, deletion of Ager or treatment with RAGE blocking peptide markedly reduced the number of HMGB1-LPS complexes in the cells (Figures S3C and S3D). Deletion of Ager blocked the cytosolic translocation of LPS, pyroptosis, and LPS-caspase-11 interaction by PLA in macrophages without altering the expression of caspase-11, pro-caspase-1, IL-1α, or pro-IL-1β (Figures 2E, 2F, S3E, and S3F). Like macrophages, endothelial cells express functional RAGE (Wautier et al., 1996, Hofmann et al., 1999, Liliensiek et al., 2004). Deletion of Ager impaired cytosolic delivery of LPS by HMGB1 in mouse lung endothelial cells (Figure 2G). Addition of dynasore also prevented the cytosolic translocation of LPS in endothelial cells (Figure 2H). Accordingly, RAGE deficiency prevented caspase-11-dependent pyroptosis in endothelial cells upon HMGB1+LPS stimulation (Figure 2I). Together, these results demonstrate that internalization of HMGB1-LPS complexes into endo-lysosomes through RAGE is a critical step for LPS translocation to the cytosol and caspase-11 activation.
HMGB1 Destabilizes Lysosomal Membranes Leading to LPS Release into the Cytosol for Caspase-11 Activation
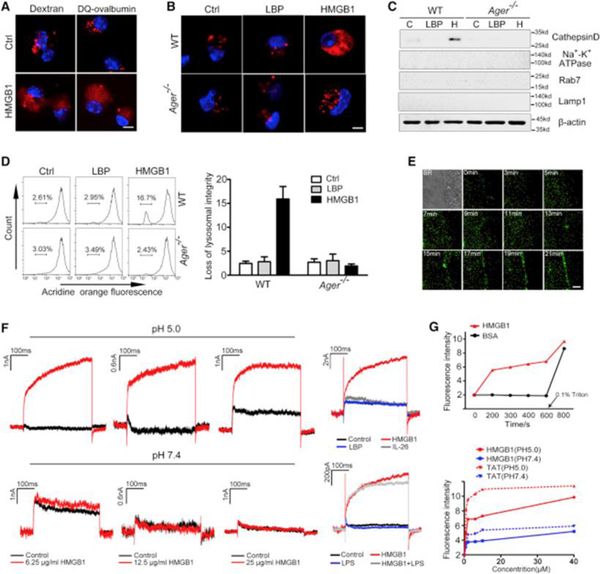
To assess the role of HMGB1 in the translocation of LPS from the endo-lysosomal compartment to the cytosol, mouse peritoneal macrophages were loaded with dextran, a fluorescent dye that accumulates in lysosomes and endosomes. Confocal microscopy revealed that exposure to HMGB1 but not LBP led to the transfer of dextran from the endosomes and lysosomes to the cytosol in WT but not Ager−/− macrophages (Figures 3A and 3B). Similar findings were obtained by using DQ-ovalbumin, another fluorescent dye that is normally quenched unless proteolytically processed into peptides in the lysosomes (Figure 3A). To confirm that HMGB1 induces lysosomal rupture with release of lysosomal contents into the cytosol, we isolated the cytosolic fraction of macrophages and measured the amounts of cathepsin D, a lysosomal protease. Notably, exposure to HMGB1 but not LBP resulted in the appearance of cathepsin D in the cytoplasmic compartment in a RAGE-dependent manner (Figure 3C). Next, we used acridine orange to quantitatively measure lysosomal integrity (Hornung et al., 2008). Exposure to HMGB1 but not LBP resulted in a disappearance of lysosomes in WT but not Ager−/−macrophages (Figure 3D). Lysosomal rupture triggers the activation of NLRP3 inflammasome in TLR agonist-primed macrophages independent of Caspase-11 (Hornung et al., 2008, Kayagaki et al., 2011). Accordingly, HMGB1 dose dependently induced IL-1β release from Casp11−/− but not Nlrp3−/− mouse macrophages primed with Pam3CSK4, a TLR2 agonist(Figure S3G). Taken together, these findings show that HMGB1 induces lysosomal rupture with release of lysosomal contents into the cytosol after RAGE-mediated internalization of HMGB1.
Figure 3. HMGB1 Destabilizes Lysosomal Membranes Leading to LPS Release into the Cytosol.
(A) Confocal microscopy of mouse peritoneal macrophages incubated with fluorescent dextran (red) or DQ ovalbumin (red) alone or together with HMGB1 (5 μg/mL) for 4 hr then fixed and stained with DAPI (blue). Scale bar: 10 μm.
(B) Confocal microscopy of WT or Ager−/− mouse peritoneal macrophages incubated with fluorescent dextran (red) alone or together with HMGB1 (5 μg/mL) or LBP (5 μg/mL) for 4 hr then fixed and stained with DAPI (blue). Scale bar: 10 μm.
(C) Immunoblot for Cathepsin D, Na+-K+ ATPase, Rab7, Lamp1, and β-actin in the cytosolic fraction from vehicle-treated (C) or HMGB1 (H, 5 μg/mL)- or LBP (5 μg/mL)-stimulated WT or Ager−/− mouse peritoneal macrophages.
(D) Flow cytometry of WT or Ager−/− mouse peritoneal macrophages stained with acridine orange and then treated with HMGB1 (5 μg/mL) or LBP (5 μg/mL) for 6 hr.
(E) Dynamic imaging of HMGB1 protein labeled with Alexa Fluor 488 (1 μg/mL) on living cell membranes. Scale bar: 2 μm.
(F) Whole-cell patch-clamp recording of HMGB1-induced inward current across the cytoplasmic membrane in proximity to the patch-clamp of HEK293 cells at neutral (pH = 7.4) or acidic (pH = 5.0) conditions.
(G) Fluorescent calcein dye was encapsulated into the liposomes, which were incubated with HMGB1, bovine serum albumin (BSA), or TAT at the indicated concentration for indicated time at neutral (pH = 7.4) or acidic pH (pH = 5.0). Liposome leakage was monitored by measuring calcein fluorescence intensity. Triton X-100 treatment was used to achieve 100% liposome leakage.
Graphs show the mean ± SD of technical replicates and are representative of three independent experiments. See also Figure S3.
Next, we investigated the mechanisms by which HMGB1 induces lysosomal destabilization. HMGB1 is an amphipathic protein and is able to bind multiple types of phospholipids, including phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol (Rouhiainen et al., 2007). As revealed by single-molecule imaging, Alexa 488-labeled HMGB1 rapidly accumulated in the cell membrane of macrophages (Figure 3E). This finding prompted us to test whether HMGB1 could directly permeabilize phospholipid bilayers. Cell membrane permeabilization leads to the formation of an inward current that can be measured using whole-cell patch-clamp techniques (Moyes et al., 2016). HMGB1 exposure rapidly induced an inward current across cell membranes (Figure 3F). The threshold concentration of HMGB1 to induce inward current was eight times lower in an acidic environment (pH = 5.0) than at neutral pH (Figure 3F). The exposure to LPS, albumin, LBP, or IL-26, an amphipathic protein that binds LPS, failed to induce an inward current (Figure 3F). LPS did not alter the ability of HMGB1 to disrupt the membrane (Figure 3F). Truncated HMGB1 containing A-box retained the ability to permeabilize membrane (Figure S3H). To confirm that HMGB1 could directly permeabilize phospholipid bilayers, we conducted a liposome-leakage assay and found that HMGB1 induced liposome leakage in a time-dependent manner (Figure 3G). The capacity of HMGB1 to induce liposome leakage was markedly increased under acidic conditions (pH = 5.0) in a manner similar to that of the human immunodeficiency virus-1 TAT protein (Figure 3G). Together, these findings suggest that HMGB1 destabilizes lysosomal membranes leading to leakage of LPS into the cytosol, which culminates in the activation of caspase-11.
Hepatocyte-Released HMGB1 Is Important for Caspase-11-Dependent Pyroptosis and Lethality in Endotoxemia
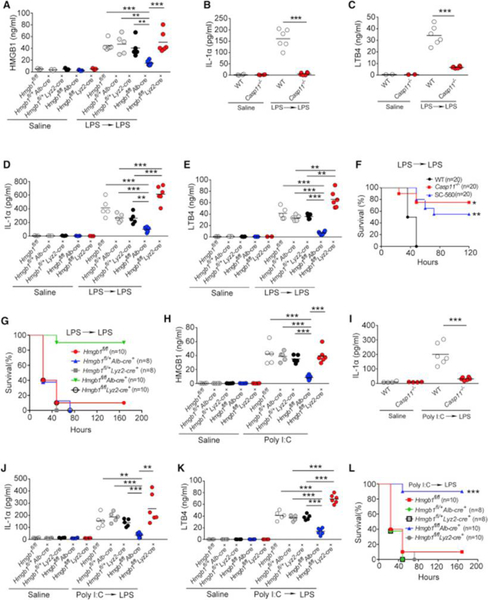
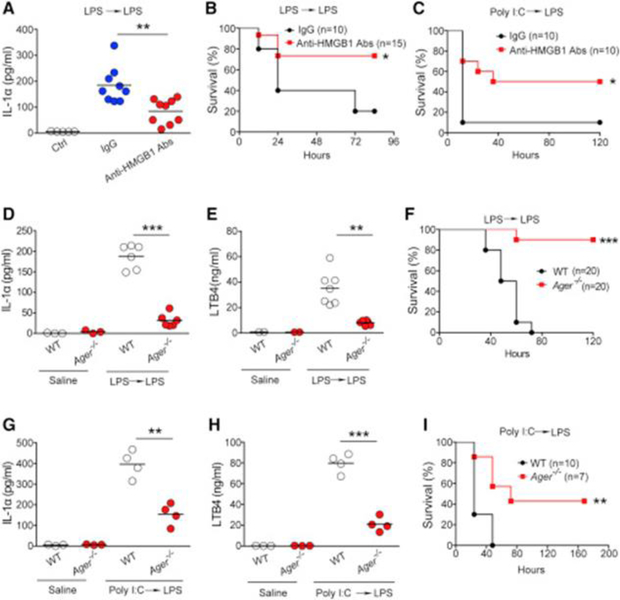
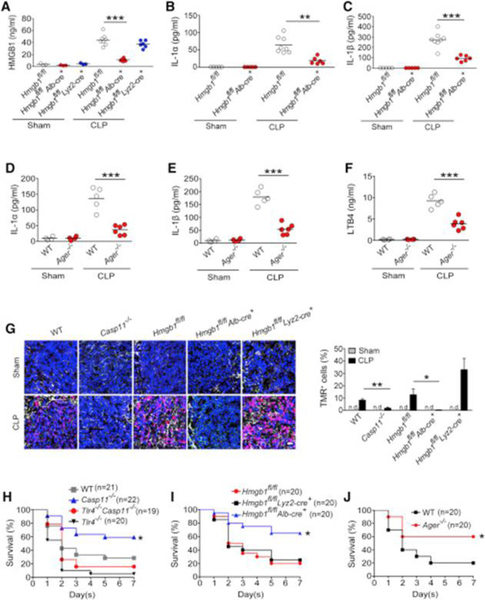
Extracellular HMGB1 at the concentration of 0.2 μg/mL enabled LPS to induce caspase-11-dependent pyroptosis in vitro (Figure 1E). As serum HMGB1 concentrations in endotoxemia can exceed 0.25 μg/mL (Lamkanfi et al., 2010), we next investigated whether HMGB1 contributes to the caspase-11-dependent pyroptosis in endotoxemia. Because both myeloid cells and hepatocytes have been shown to actively and passively release HMGB1 during inflammation or hypoxia (Lu et al., 2012, Scaffidi et al., 2002, Tsung et al., 2007), we generated mice with selective Hmgb1deletion in either myeloid cells (Hmgb1f/fLyz2-cre+) or hepatocytes (Hmgb1f/fAlb-cre+). Though global deletion of Hmgb1 leads to early postnatal death (Calogero et al., 1999), these mice displayed no detectable defects under physiological conditions. We found that hepatocytes, but not myeloid cells, were the major source of the increased circulating HMGB1 concentrations during endotoxemia (Figure 4A). Global deletion of Casp11 or hepatocyte selective deletion of Hmgb1markedly reduced the systemic release of IL-1α, IL-1β, and LTB4 in endotoxemia (Figures 4B–4E and S4A–S4B). Consistent with previous findings (Hagar et al., 2013), genetic deletion of Casp11 or pharmacological inhibition of COX-1 led to improved host survival during lethal endotoxemia (Figure 4F). Importantly, deletion of Hmgb1 in hepatocytes, which prevents the accumulation of HMGB1 and LTB4 in the circulation, also significantly improved survival in lethal endotoxemia (Figure 4G). HMGB1 deficiency in myeloid cells has been shown to increase cell death including pyroptosis (Yanai et al., 2013). Although myeloid-specific deletion of Hmgb1 did not reduce circulating concentrations of HMGB1 during endotoxemia, consistent with the previous observations (Yanai et al., 2013), we did observe significantly increased serum concentrations of the pyroptotic markers IL-1α and LTB4 with myeloid-specific Hmgb1 deletion (Figures 4D and 4E). Similar observations were made in poly(I:C)-primed and LPS-challenged mice (Figures 4H–4L). Thus, hepatocytes are the major source of HMGB1 released systemically in endotoxemia that mediates pyroptosis.
Figure 4. Hepatocyte-Released HMGB1 Is Critical for Caspase-11-Dependent Pyroptosis and Lethality in Endotoxemia.
(A–E) Plasma HMGB1, IL-1α, and LTB4 concentrations from mice of indicated genotypes injected with low dose of LPS (400 μg/kg) for 6 hr and then challenged with high dose of LPS (10 mg/kg) for 4 hr.
(F and G) Kaplan-Meier survival plots for mice with indicated genotypes injected with low dose of LPS (400 μg/kg) for 6 hr and then challenged with high dose of LPS (10 mg/kg) with or without SC-560 (5 mg/kg).
(H) Plasma HMGB1 concentrations from mice of indicated genotypes injected with poly(I:C) (10 mg/kg) for 6 hr.
(I–K) Plasma IL-1α or LTB4 concentrations from mice of indicated genotypes injected with poly(I:C) (10 mg/kg) for 6 hr and then challenged with LPS (10 mg/kg) for 4 hr.
(L) Kaplan-Meier survival plots for mice with indicated genotypes injected with poly(I:C) (10 mg/kg) for 6 hr and then challenged with LPS (10 mg/kg).
Circles represent individual mice.∗p < 0.05;∗∗p < 0.01;∗∗∗p < 0.001; NS: not significant (Student’s t test and log-rank test for survival). See also Figure S4.
Neutralizing Extracellular HMGB1 or Inhibition of HMGB1-LPS Binding Prevents Caspase-11-Dependent Pyroptosis and Death in Endotoxemia
To further confirm that extracellular HMGB1 is critical for pyroptosis and death in endotoxemia, mice were injected with monoclonal HMGB1 neutralizing antibodies or isotype control IgG. Neutralizing extracellular HMGB1 significantly reduced the release of IL-1α and IL-1β as well as lethality in endotoxemia as compared to the IgG controls (Figures 5A, 5B, and S4C). Similar observations were made in poly(I:C)-primed and LPS-challenged mice (Figure 5C). Next, we determined whether LPS-HMGB1 binding is critical for pyroptosis in vivo. Mice were injected with HPep1, a peptide known to specifically block the HMGB1-LPS binding (Youn et al., 2011). Administration of HPep1 significantly attenuated the release of IL-1α and IL-1β in endotoxemia as compared to that of control peptide (Figure S4D). Moreover, intra-peritoneal injection of LPS-RS significantly promoted survival during lethal endotoxemia (Figure S4E). Further, administration of HMGB1 alone, even at high doses, was not lethal in mice (Figure S4F), supporting the notion that HMGB1 requires binding to LPS to initiate lethal responses. Collectively, these results demonstrate that neutralizing extracellular HMGB1 or inhibition of HMGB1-LPS binding prevents pyroptosis and lethality during endotoxemia.
Figure 5. Neutralizing Extracellular HMGB1 or Deletion of Ager Prevents Pyroptosis and Lethality in Endotoxemia.
(A) Plasma IL-1α concentrations from WT mice injected with low-dose LPS (400 μg/kg) for 6 hr and then subjected to administration of high-dose LPS (10 mg/kg) with monoclonal HMGB1 neutralizing antibody (2G7, 150 μg per mouse) or isotype control IgG (15 μg per mouse) for 4 hr.
(B and C) Kaplan-Meier survival plots for WT mice injected with low-dose LPS (400 μg/kg) (B) or poly(I:C) (10 mg/kg) (C) for 6 hr and then subjected to administration of a high-dose LPS (10 mg/kg) with monoclonal HMGB1 neutralizing antibodies (2G7, 150 μg per mouse) or isotype control IgG (150 μg per mouse).
(D and E) Plasma IL-1α or LTB4 concentrations from WT or Ager−/− mice injected with low-dose LPS (400 μg/kg) for 6 hr and then challenged with high-dose LPS (10 mg/kg) for 4 hr.
(F) Kaplan-Meier survival plots for WT or Ager−/− mice injected with a low dose of LPS (400 μg/kg) for 6 hr and then challenged with a high dose of LPS (10 mg/kg).
(G and H) Plasma IL-1α or LTB4 concentrations from WT or Ager−/− mice injected with poly(I:C) (10 mg/kg) for 6 hr and then challenged with LPS (10 mg/kg) for 4 hr.
(I) Kaplan-Meier survival plots for WT or Ager−/− mice injected with poly(I:C) (10 mg/kg) for 6 hr and then challenged with LPS (10 mg/kg).
Circles represent individual mice.∗p < 0.05;∗∗p < 0.01;∗∗∗p < 0.001; NS: not significant (Student’s t test and log-rank test for survival). See also Figure S4.
RAGE Is Required for Pyroptosis and Lethality in Endotoxemia
Both endothelial cells and macrophages express functional RAGE (Wautier et al., 1996, Hofmann et al., 1999). As RAGE is essential for HMGB1+LPS-induced caspase-11 activation in both macrophages and endothelial cells in vitro (Figures 2E–2H), we reasoned that RAGE might be critical for caspase-11-dependent pyroptosis and lethality in endotoxemia. Ager deficiency significantly inhibited IL-1α, LTB4, and IL-1β release during endotoxemia (Figures 5D, 5E, and S4G). Deletion of Ageror administration of soluble RAGE protected mice from lethal endotoxemia (Figures 5F and S4H). Furthermore, Ager deletion significantly reduced IL-1α and LTB4 release and improved survival in mice treated with poly(I:C) followed by LPS (Figures 5G–5I). Similar to caspase-11-deficient mice (Kayagaki et al., 2013), RAGE-deficient mice were less protected after poly(I:C)+LPS challenge than after LPS+LPS challenge (Figures 5I). Together with other data, these observations support the conclusion that hepatocyte-released HMGB1 drives caspase-11 activation in RAGE-expressing cells leading to pyroptosis and death in endotoxemia.
RAGE and Hepatocyte-Released HMGB1 Are Important for Caspase-11-Dependent Pyroptosis and Lethality in Bacterial Sepsis
Experiments were then conducted to determine whether hepatocyte-released HMGB1 and RAGE are critical for caspase-11-dependent lethality in bacterial sepsis. Using cecum ligation and puncture (CLP), a clinically relevant murine model of Gram-negative polymicrobial sepsis, we observed that hepatocytes, but not myeloid cells, were the major source of circulating HMGB1 during bacterial sepsis (Figure 6A). Hepatocyte-specific deletion of Hmgb1 or global deletion of Casp11significantly reduced the release of IL-1α and IL-1β following CLP (Figures 6B, 6C, and S4I). Ager deletion also significantly reduced the release of IL-1α, IL-1β, and LTB4 in this model of intra-abdominal sepsis (Figures 6D–6F). The spleen but not the kidneys highly expressed caspase-11 in sepsis (Figure S4J). Accordingly, bacterial sepsis caused considerable caspase-11-dependent immune cell death in spleen (Figure 6G). Hepatocyte-specific deletion of Hmgb1 or global deletion of Ager markedly attenuated sepsis-induced cell death in the spleen (Figures 6G and S4K). In agreement with previous work (Yanai et al., 2013), deletion of HMGB1 in myeloid cells significantly enhanced cell death in the spleen (Figure 6G). Similar findings were obtained by analyzing peritoneal immune cells using flow cytometry (Figure S4L). Global deletion of Casp11 or Ager or hepatocyte-specific deletion of Hmgb1 significantly improved survival after CLP (Figures 6H–6J). In contrast, deletion of Hmgb1 in myeloid cells failed to improve survival (Figure 6I). Global simultaneous deletion of both Tlr4 and Casp11 resulted in lethality after CLP in between that seen in Tlr4−/− or WT mice (Figure 6H). This could be explained by previous findings showing that TLR4 is essential for bacterial clearance and survival in bacterial sepsis (Deng et al., 2013). These data support the conclusion that hepatocyte-released HMGB1 and RAGE are required for caspase-11-dependent pyroptosis and lethality in bacterial sepsis.
Figure 6. Caspase-11, Hepatocyte-Released HMGB1, and RAGE Are Critical for Pyroptosis and Lethality in Bacterial Sepsis.
(A–F) Plasma HMGB1, IL-1α, IL-1β, and LTB4 concentrations from mice of indicated genotypes were subjected to either cecum ligation and puncture (CLP) or sham operation.
(G) Cell death in the spleens of indicated transgenic mouse strains was assessed by TUNEL staining. Shown in the right panel is the percentage of TMR-positive cells. Scale bar: 20 μm.
(H–J) Kaplan Meier survival curves for the indicated transgenic mouse strains subjected to CLP.
Circles represent individual mice.*p < 0.05; **p < 0.01; ***p < 0.001; NS: not significant (Student’s t test and log-rank test for survival). See also Figure S4.
LPS Stimulates HMGB1 Release from Hepatocytes in a TLR4- and Caspase-11-Dependent Manner
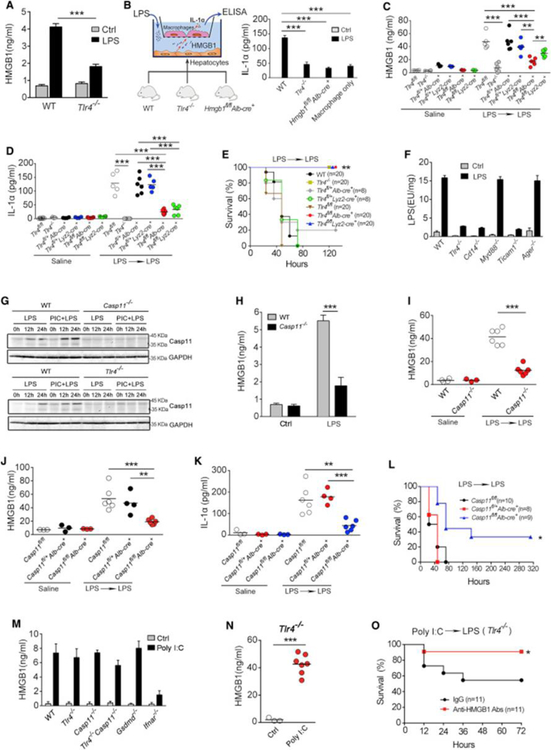
Next, we investigated the mechanisms by which hepatocytes release HMGB1 in response to LPS. Hepatocytes express TLR4 that senses extracellular LPS (Scott and Billiar, 2008, Deng et al., 2013). Upon LPS exposure, cultured hepatocytes utilized TLR4 to trigger HMGB1 release, which promoted caspase-11-dependent IL-1α release from co-cultured macrophages (Figures 7A, 7B, and S5A–5C). Selective deletion of Tlr4 in hepatocytes significantly reduced the release of HMGB1, IL-1α, and IL-1β as well as lethality in endotoxemia (Figures 7C–7E and S5D). Similar observations were made in bacterial sepsis (Figures S5E–S5G). We also confirmed that TLR4 in myeloid cells was important for IL-1α release and lethality in endotoxemia using specific Tlr4−/− mice (Figures 7D and 7E).
Figure 7. Hepatocytes Release HMGB1 in Response to LPS or Poly(I:C) by Distinct Mechanisms.
(A) HMGB1 release from isolated primary WT or Tlr4−/− hepatocytes stimulated by LPS (0.1 μg/mL) for 16 hr.
(B) Primary hepatocytes isolated from mice of indicated genotypes co-cultured with WT mouse peritoneal macrophages are shown in the left panel. IL-1α released from macrophages after stimulation of LPS (0.1 μg/mL) is shown in the right panel.
(C and D) Plasma HMGB1 and IL-1α concentrations from mice of indicated genotypes injected with a low-dose LPS (400 μg/kg) for 6 hr and then challenged with high-dose LPS (10 mg/kg) for 4 hr.
(E) Kaplan-Meier survival plots for mice with indicated genotypes injected with low-dose LPS (400 μg/kg) for 6 hr and then challenged with high-dose LPS (10 mg/kg).
(F) LPS activity assay of the cytosolic fraction from LPS (0.1 μg/mL)-stimulated primary hepatocytes from indicated transgenic mouse strains.
(G) Primary hepatocytes isolated from mice of indicated genotypes were stimulated by LPS alone (100 ng/mL) or LPS+poly(I:C) (PIC) (10 μg/mL) for indicated time. Then the expression of Caspase-11 and GAPDH were determined by immunoblot.
(H) HMGB1 release from isolated primary WT or Casp11−/− hepatocytes stimulated by LPS (0.1 μg/mL) for 16 hr.
(I–K) Plasma HMGB1 or IL-1α concentrations from mice of indicated genotypes injected with low-dose LPS (400 μg/kg) for 6 hr and then challenged with LPS (10 mg/kg) for 4 hr.
(L) Kaplan-Meier survival plots for mice with indicated genotypes injected with low-dose LPS (400 μg/kg) for 6 hr and then challenged with LPS (10 mg/kg).
(M) HMGB1 release from isolated WT, Tlr4−/−, Casp11−/−, Tlr4−/−Casp11−/−, Gsdmd−/−, or Ifnar1−/− hepatocytes stimulated by poly(I:C) (10 μg/mL) for 24 hr.
(N) Plasma HMGB1 concentrations for Tlr4−/− mice injected with poly(I:C) (10 mg/kg) for 6 hr.
(O) Kaplan-Meier survival plots for Tlr4−/− mice injected with poly(I:C) (10 mg/kg) for 6 hr and then challenged with LPS (10 mg/kg) with monoclonal HMGB1 neutralizing antibodies (2G7, 150 μg per mouse) or isotype control IgG (150 μg per mouse).
*p < 0.05; **p < 0.01; ***p < 0.001 (Student’s t test and log-rank test for survival). Circles represent individual mice. See also Figures S5, S6, and S7.
In line with our previous findings (Scott and Billiar, 2008, Deng et al., 2013), hepatocytes utilize TLR4-CD14-Trif signaling, rather than RAGE or HMGB1, to promote the uptake of LPS into the cytosol (Figures 7F and S6A–S6E). This led us to hypothesize that an intracellular LPS-sensing pathway, such as caspase-11, could be involved in HMGB1 release by hepatocytes in endotoxemia. We confirmed that hepatocytes express caspase-11 in response to LPS (Figure 7G). Using hepatocytes isolated from transgenic mice, we found that LPS-induced HMGB1 release from living hepatocytes required both caspase-11 and GSDMD (Figures 7H and S6F–S6J). Furthermore, live cell imaging confirmed the extracellular release of GFP-labeled HMGB1 from living hepatocytes exposed to LPS (Figure S6K).
To understand the relative importance of caspase-11 in myeloid cell versus non-myeloid cells in endotoxin lethality, we generated bone marrow chimeras wherein Casp11 deletion is restricted to hematopoieticor non-hematopoietic cells using WT (CD45.1+) and global caspase-11 (CD45.2+) strains as donors and recipients (Figures S7A–S7C). Deletion of Casp11 only in hematopoietic cells significantly inhibited lethality in endotoxemia (Figure S7A), confirming that myeloid cell caspase-11 contributes to LPS-induced lethality. However, deletion of Casp11 only in non-hematopoietic compartments reduced LPS lethality to an even greater extent and to the same rate of that observed in global caspase-11-deficient mice (Figure S7B). These observations are consistent with a recent study showing that genetic deletion of Casp11 in endothelial cells reduces lethality by more than 50% in endotoxemia (Cheng et al., 2017), but also could reflect the importance of hepatocyte caspase-11 to HMGB1 release to the lethal LPS response. To more directly test this possibility, we generated mice with selective Casp11 deletion in hepatocytes (Casp11f/fAlb-cre+) and confirmed that hepatocyte caspase-11 was required for systemic release of HMGB1 release and increased in systemic markers of pyroptosis in endotoxemia and bacterial sepsis. Accordingly, selective deletion of Casp11 in hepatocytes also prevented LPS-induced lethality (Figures 7I–7L and S7D–S7F).
Poly(I:C) Stimulates HMGB1 Release from Hepatocytes Independent of TLR4 and Caspase-11
Similar to LPS, poly(I:C) priming triggered HMGB1 release from hepatocytes in vivo (Figure 4H) and is known to dramatically promote LPS lethality in a caspase-11-dependent manner (Hagar et al., 2013, Kayagaki et al., 2013). We next investigated whether poly(I:C) utilizes the same signaling pathway to induce hepatocyte HMGB1 release as LPS. WT, Tlr4−/−, Casp11−/−, Tlr4−/−Casp11−/−, and Gsdmd−/− hepatocytes released comparable amounts of HMGB1 in response to poly(I:C) (Figure 7M), indicating that poly(I:C) induces hepatocyte HMGB1 release independent of TLR4 and caspase-11. Poly(I:C) is known to stimulate robust type 1 interferon (IFN) production in hepatocytes (Zhang et al., 2016). As it is known that type 1 IFN signaling mediates HMGB1 release in macrophages (Lu et al., 2014, Kim et al., 2009), we next tested whether type 1 IFN signaling is required for poly(I:C)-induced hepatocyte HMGB1 release. Deletion of Ifnar1−/−, the receptor for type 1 IFN, markedly decreased poly(I:C)-induced HMGB1 release from hepatocytes (Figure 7M).
In line with the in vitro findings (Figure 7M), administration of poly(I:C) markedly elevated plasma HMGB1 concentrations in Tlr4−/− mice (Figure 7N). Consistent with previous reports (Hagar et al., 2013, Kayagaki et al., 2013), priming with poly(I:C) enabled LPS to bypass the requirement for TLR4 to cause host death. The role of HMGB1 in the lethality induced by poly(I:C) was confirmed using neutralizing antibodies (Figures 7O). As hepatocyte-released HMGB1 and RAGE are critical for LPS lethality in poly(I:C)-primed mice (Figures 4I–4L and 5G–5I), these data suggest that poly(I:C) priming promotes caspase-11 activation and LPS lethality, at least in part, through hepatocyte-released HMGB1 and RAGE.
Discussion
Taken together, our results identify HMGB1 as a critical mediator that drives caspase-11 activation during endoxemia and bacterial sepsis. Hepatocytes can sense circulating pathogen-associated molecular patterns (PAMPs) such as LPS or poly(I:C) and subsequently release HMGB1 into the blood stream. Hepatocyte-released HMGB1 delivers extracellular LPS into the cytosol of RAGE-expressing cells and this is critical for caspase-11-dependent lethality in endotoxemia and bacterial sepsis. Important steps in this process are RAGE-mediated internalization of HMGB1-LPS complexes followed by the destabilization of lysosomesby HMGB1 for the delivery of LPS to cytosolic caspase-11. Both endothelial cells and myeloid cells express RAGE and caspase-11 (Hofmann et al., 1999, Liliensiek et al., 2004). Although endothelial cell caspase-11 plays a major role in endotoxemia-induced lethality (Cheng et al., 2017), caspase-11 in myeloid cells, such as macrophages, also contributes to the lethality in sepsis. These findings unravel how the host initiates lethal immune responses to LPS at the level of the whole organism and provide mechanistic insight into how HMGB1 delivers extracellular LPS into the cytosol at the molecular level.
HMGB1 physically binds to extracellular LPS and subsequently utilizes a RAGE-dependent internalization process to deliver LPS into acidic lysosomes, where HMGB1 directly permeabilizes the phospholipid bilayer of the lysosomes. Using whole cell patch-clamp analysis and a liposome leakage assay, we observed that the capacity of HMGB1 to permeabilize the phospholipid bilayer was enhanced at more acidic pH. This finding not only elucidates the importance of pH in regulating the capacity of HMGB1 to induce rupture of lysosomes from inside the organelle, but also could explain why HMGB1 in the cytosol or the nucleus does not disrupt cytoplasmic or nuclear membranes under the physiological conditions. By inducing lysosomal rupture, HMGB1 can also activate NLRP3inflammasome independent of caspase-11 in TLR agonist-primed macrophages. A recent study shows that HMGB1 alone triggers ASC-dependent and caspase-11-independent pyroptosis (Xu et al., 2014). In our study, however, HMGB1 alone failed to induce pyroptosis. The discrepancy between the two studies is likely due to the distinct redox status of HMGB1 recombinant proteins. Oxidation of HMGB1 results in the formation of intramolecular Cysteine 23-Cysteine 45 bonds, which enables HMGB1 to signal through TLR4-MD2 and function as a priming signal for the NLRP3 inflammasome activation (Frank et al., 2016, Yang et al., 2015, Hornung et al., 2008). As most circulating HMGB1 in sepsis is fully reduced and unable to activate TLR4 (Valdés-Ferrer et al., 2013), we used reduced HMGB1 proteins in this study and did not observe pyroptosis with HMGB1 alone.
We also found that myeloid immune cells are not the major source of circulating HMGB1 during endotoxemia (Yanai et al., 2013) and established that hepatocytes are the dominant source of systemic HMGB1 release in bacterial sepsis. During the course of studying how hepatocytes release HMGB1 in response to LPS, we found that hepatocyte caspase-11 mediates LPS-induced HMGB1 release from hepatocytes and that LPS delivery to the cytosol of hepatocytes is enhanced by TLR4 signaling. We had previously shown that hepatocytes utilize a unique TLR4 complex involving β2-integrin as well as CD14 to promote LPS uptake (Scott and Billiar., 2008). Internalized LPS delivered to caspase-11 in the cytosol led to HMGB1 release from hepatocytes. Living hepatocytes could actively release HMGB1 in response to LPS in a process that required both caspase-11 and GSDMD. In agreement with our findings, a recent study shows that GSDMD mediates IL-1 release from living macrophages (Evavold et al., 2018). However, poly(I:C)-induced hepatocyte HMGB1 release depends on type 1 IFN signaling, rather than TLR4, caspase-11, or GSDMD. These observations are in an agreement with previous findings that type 1 IFN or type 2 IFN stimulates HMGB1 release in the absence of LPS (Lu et al., 2014, Kim et al., 2009, Rendon-Mitchell et al., 2003). Though we previously reported that type 1 IFN induces hyperacetylation of the nuclear location sequences in HMGB1 leading to the translocation of HMGB1 from the nucleus to the cytosol (Lu et al., 2014), how cytosolic HMGB1 is subsequently released into the extracellular space remains unknown.
In the current study, we utilized a “double-hit” endotoxemia model and confirmed that priming with a low dose of LPS or poly(I:C) enhanced LPS lethality in a caspase-11-dependent manner (Hagar et al., 2013, Kayagaki et al., 2013). In this model and in a model of bacterial sepsis, we found that hepatocyte-released HMGB1 and RAGE are critical for caspase-11-dependent lethality. However, hepatocyte Hmgb1 deletion fails to protect the mice from a single high dose of LPS (Huebener et al., 2015) and even may enhance inflammasome activation (Yanai et al., 2013). Extremely high concentrations of extracellular LPS alone induce pyroptosis in vitroand bypass the need for HMGB1 to activate caspase-11. Taken together, our observations provide a mechanistic basis for the lethal effects of HMGB1 in endotoxemia and sepsis. Because HMGB1 release and caspase-11 upregulation evolve over several hours after the onset of sepsis, both molecules could be targeted to prevent lethality in sepsis.
Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HMGB1 monoclonal antibody (2G7) for in vivo exps and ELISPOT | Prepared in K.J.T. Lab | Qin et al., 2006 |
| Anti-caspase-11 antibody | Sigma | C1354; RRID: AB_258736 |
| Anti-IL-1α antibody | Abcam | ab109555 |
| Anti-caspase-1 antibody | Abcam | ab179515 |
| Anti-HMGB1 antibody | Abcam | ab79823; RRID: AB_1603373 |
| Anti-IL-1 βantibody | R&D system | AF-401-NA; RRID: AB_416684 |
| Anti-LAMP1 antibody (clone 1D4B) | eBioscience | 14–1071-85; RRID: AB_65753 |
| Sodium Potassium ATPase Alpha 1 Antibody | Novus Biological | NB300–146; RRID: AB_2060981 |
| Anti-Rab7 antibody | Cell Signaling Technologies | 9367S; RRID: AB_1904103 |
| Anti-EEA1 antibody | Cell Signaling Technologies | 3288S; RRID: AB_2096811 |
| Anti-Capase-8 antibody | Cell Signaling Technologies | 4927S; RRID: AB_10694563 |
| Anti-cleaved Caspase-8 antibody | Cell Signaling Technologies | 8592S; RRID: AB_10891784 |
| Anti-Caspase-4 antibody | MBL international | M029–3 |
| Anti-Gasdermin D-antibody | Santa cruz | Sc393581 |
| Anti-EEA1 antibody | Proteintech | 22266–1-AP |
| Anti-LAMP1 antibody | Proteintech | 21997–1-AP |
| Anti-E.coli LPS antibodies | Abcam | Ab35654; RRID: AB_732222 |
| Anti-Mouse CD144 (VE-Cadherin) APC | eBioscience | 85–17-1441–80 |
| APC | ||
| Anti-Mouse CD309 (FLK1) PE | eBioscience | 85–12-5821–81 |
| Anti-Mouse CD31 (PECAM-1) Biotin | eBioscience | 85–13-0311–85; RRID: AB_466421 |
| Anti-CD11b-FITC | eBioscience | 11–0112-41; RRID: AB_11042156 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ultrapure LPS (E. coli 0111:B4) | InvivoGen | tlrl-3pelps |
| LPS-RS | InvivoGen | tlrl-prslps |
| LPS-EB Biotin | InvivoGen | tlrl-3blps |
| Pam3CSK4 | InvivoGen | tlrl-pms |
| Poly(I:C) LMW | InvivoGen | tlrl-picw-250 |
| MCC950 (CP-456773) | Selleck | S7809 |
| Dynasore | Sigama | D7693 |
| Cholera Toxin B Subunit (Choleragenoid) from Vibrio cholerae | List Biological Laboratories, INC. | 103B |
| Lipofectamine 3000 Transfection Reagent | ThermoFisher Scientific | L3000015 |
| Recombinant HMGB1 protein | Prepared in K.J.T. Lab | Wang et al., 1999 |
| Mouse Albumin protein | Abcam | ab183228 |
| Mouse immunoglobin IgG protein | Abcam | ab198772 |
| Recombinant Mouse LBP Protein | RD systems | 6635-LP-025/CF |
| Box A from HMGB1 | TECAN | REHM013 |
| Fully reduced HMGB1 | TECAN | REHM114 |
| RAGE Antagonist Peptide | MERK Millipore | 553031 |
| Hpep1 peptide | Chinese Peptide Company. | Youn et al., 2011 |
| Recombinant Mouse M-CSF Protein | R&D System Inc. | 416-ML-050 |
| Critical Commercial Assays | ||
| In Situ Cell Death Detection Kit, TMR red | ThermoFisher Scientific | L3000015 |
| Mouse IL-6 DuoSet ELISA | R&D System Inc. | DY406 |
| Mouse TNF-α DuoSet ELISA | R&D System Inc. | DY410 |
| Mouse IL-1 alpha DuoSet ELISA | R&D System Inc. | DY400 |
| Mouse IL-1 beta DuoSet ELISA | R&D System Inc. | DY401 |
| HMGB1 ELISA | TECAN | ST51011 |
| EasySep Release Mouse Biotin Positive Selection Kit | STEMCELL Technologies | 17655 |
| Experimental Models: Cell Lines | ||
| MEF cells | HMGBiotech, Inc | N/A |
| Mouse Hepatocytes | Prepared in T.R.B. Lab | Described in current manuscript |
| Mouse Macrophages | Prepared in B.L. Lab | Described in current manuscript |
| Mouse Macrophages | Prepared in B.L. Lab | Described in current manuscript |
| Mouse Endothelial cells | Prepared in B.L. Lab | Described in current manuscript |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mice | Jackson Laboratories | 000664 |
| Casp11−/−mice | University of Pittsburgh Animal Housing | Described in current manuscript |
| Nlrp3−/− mice | Genentech Inc | Mariathasan et al., 2006 |
| Asc−/− mice | Genentech Inc | Mariathasan et al., 2006 |
| Gsdmd−/− mice | University of Pittsburgh Animal Housing | He et al., 2015 |
| Ager−/− mice | University of Pittsburgh Animal Housing | Wendt et al., 2003 |
| Tlr4−/− mice | University of Pittsburgh Animal Housing | Deng et al., 2013 |
| B6.129P2-Lyz2tm1(cre)Ifo/J | Jackson Laboratories | 004781 |
| B6.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J | Jackson Laboratories | 003574 |
| Tlr4fl/flmice | University of Pittsburgh Animal Housing | Deng et al., 2013 |
| Casp11fl/flmice | University of Pittsburgh Animal Housing | Described in current manuscript |
| Hmgb1fl/flmice | University of Pittsburgh Animal Housing | Described in current manuscript |
| Oligonucleotides | ||
| 5′-ACT TTC TCT CTT CTC ACT −3′, 5′-TGT CTA ACT ATA TTG AAA TGT G-3′ | Invitrogen | Casp11-specific primers |
| TCTACACTATAGTCCAGACCC | Shi et al., 2014 | CASP4-specific siRNA |
| GTCTGGACTATAGTGTAGATG | Shi et al., 2014 | CASP4-specific siRNA |
| CGTACGCGGAATACTTCGA | Shi et al., 2014 | Control siRNA |
| Software and Algorithms | ||
| Graphpad Prism 5 software | Graphpad Prism 5 software | N/A |
| Adobe Illustrator CC 2015 | Adobe | N/A |
| NIS Elements software | Nikon Instruments | N/A |
| Microsoft Excel | Microsoft | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Ben Lu (xybenlu@csu.edu.cn).
Experimental Model and Subject Details
Mouse studies
Experimental protocols were approved by the Institutional Animal Care and Use Committees of the University of Pittsburgh and Central South University. We used moribundity as the endpoint for our survival study following the Animal Research Advisory Committee Guidelines from National Institutes of Health. Briefly, mice were monitor twice daily by personnel experienced in recognizing signs of moribundity. Mice were euthanized with CO2, when they became moribund.
Generation of Casp11−/− mice
Casp11−/−mice mice were prepared by bringing the mutated Casp11 allelethat spontaneously occurred in SV129 mouse (Kayagaki et al., 2011) to C57BL/6 background. Five nucleotides deletion at the exon 7 splicing site resulted into a shorter mRNA due to the alternative splicing skipping exon 7. Premature termination due to reading frameshift generated undetectable caspase-11 protein in the deficient mice. The F1 mouse was backcrossed with C57BL/6 mouse for 9 generations before experiments were carried out. The mutated alleles were monitored and tracked in the progenies with a set of PCR genotyping primers designed specific for the mutated alleles. The sequences of the primers are, forward, 5′-ACT TTC TCT CTT CTC ACT −3′, reverse, 5′-TGT CTA ACT ATA TTG AAA TGT G-3′. In the current study, we used WT littermates (C57BL/6J background) as the controls for the Casp11−/− mice.
Cell-type specific Hmgb1−/− or Tlr4−/− or Casp11−/− mice
Casp11fl/fl mice were generated by KOMP repository (www.komp.org). The Casp4 mouse strain used for this research project was created from ES cell clone EPD0208_5_H03, obtained from the KOMP Repository (www.komp.org) and generated by the Wellcome Trust Sanger Institute (WTSI). Targeting vectors used were generated by the Wellcome Trust Sanger Institute and the Children’s Hospital Oakland Research Institute as part of the Knockout Mouse Project (3U01HG004080). This strain has the Knockout First, Reporter-tagged insertion with conditional Casp4(Promoter Driven Cassette) allele, tm1a. The first sliced allele is exposed to the Flp construct (by in vivo Flp breeding) to remove the trapping cassette and render the allele fully conditional. The mouse strain used for this research project, C57BL/6-Tg(CAG-Flpo)1Afst-mucd, identification number 036512-UCD, was obtained from the Mutant Mouse Reginonal Resource Center, a NIH funded strain repository, and was donated by the MMRRC at UC Davis. The original transgenic was donated by Dr. Konstantinos Anastassiadis from Technische Universitaet Dresden.
For the generation of Hmgb1fl/fl mice, the Hmgb1fl/fl allele was created by inserting loxP sites within intron 1 and intron 2, flanking exon 2 of Hmgb1.
Tlr4fl/fl mice were generated as previously described (Deng et al., 2013). In brief, the Tlr4fl/fl allele was created by inserting loxP sites within intron 1 and intron 2, flanking exon 2 of Tlr4.
Casp11fl/fl, Hmgb1fl/fl or Tlr4fl/fl mice were interbred with heterozygous stud males to generate desired genotype (Casp11fl/+Alb-cre; Hmgb1fl/+Alb-cre; Hmgb1fl/+Lyz2-cre; Tlr4fl/+Alb-cre and Tlr4fl/+Lyz2-cre). Transgenic miceused for experiments were confirmed to be desired genotype via standard genotyping techniques.
Chimeric mice
10 to 12-week-old recipient B6.SJL mice (CD45.1) or Casp11−/− mice (CD45.2) received a total 11 Gy irradiation from an X-ray source irradiator delivered in 2 split doses with a 4-hour interval. 5 × 106 total bone marrow cells (BM cells) from donor Casp11−/− mice or B6.SJL mice were injected intravenously into the irradiated recipients. Four weeks later, peripheral blood mononuclear cells were collected from the recipients and stained with anti-CD45.1 and anti-CD45.2 antibodies to analyze engraftment by flow cytometry.
Endotoxemia model
Male mice that were 25 to 30 g in weight were injected intraperitoneally with low dose of LPS (400μg/kg) or Poly I:C (10mg/kg) with or without SC-560 or soluble RAGE (sRAGE) or anti-HMGB1 antibody (2G7, 150μg per mouse) or HMGB1 peptide1 (200μg per mouse) or their control isotypeIgG (150μg per mouse) or control peptides (200μg per mouse) for 6h and then challenged with high dose of LPS (10mg/kg).
CLP bacterial sepsis model
Sepsis was induced by CLP. Male mice that were 25 to 30 g in weight were used. The skin was disinfected with a 2% iodine tincture. Laparotomy was performed under 2% isoflurance (Piramal Critical Care) with oxygen. For the sub lethal model, 50% of the cecum was ligated and punctured twice with a 22-gauge needle. Saline (1 mL) was given subcutaneously for resuscitation immediately after operation. Mice were sacrificed at 18 hours after CLP. For the lethal model, 75% of the cecum was ligated and punctured twice with an 18-gauge needle. Mice were monitor twice daily by personnel experienced in recognizing signs of a moribund state. Mice were euthanized with CO2, when they became moribund or at observation endpoint (7 days).
Macrophage preparation and stimulation
Mouse peritoneal macrophages were isolated and cultured as described previously (Lu et al., 2012). Briefly, mice (7–12 wk old) were injected with 3 mL of sterile 3% thioglycollate broth intraperitoneally to elicit peritoneal macrophages. Cells were collected by lavage of the peritoneal cavity with 5 mL of RPMI medium 1640 (GIBCO) 72 h later. After washing, cells were resuspended in RPMI medium 1640 supplemented with 10% fetal bovine serum and antibiotics (GIBCO). Peritoneal macrophages (106 cells per well) plated in 12-well plates were stimulated with ultra-pure LPS and HMGB1 which were pre-mixed at room temperature for 20 min. In some experiments, Cell lysates and supernatants were collected 16h later for immune-blot, ELISA and LDH release. Primary bone marrow derived macrophages (BMDMs) were cultured and differentiated in vitro as described previously (Lu et al., 2012). Briefly, bone marrow were collected and incubated with collagenase at 37°C for 1 hour and then filtered through a 70-μm cell strainer. Erythrocytes were lysed with lysis solution. Bone marrow cells were differentiated in DMEM supplement with 10% fetal bovine serum and 20% M-CSF for 6 days.
Mouse lung endothelial cell preparation and stimulation
Mouse lung endothelial cells were isolated as previously described (Cheng et al., 2017). In brief, blood-free mouse lungs were minced, digested with collagenase at 37°C for 45 min, triturated, and centrifuged at 1,000 g. Cell suspensions were incubated with Anti-Mouse CD31(PECAM-1) Biotin antibody, after which ECs were magnetically sorted by EasySep Release Mouse Biotin Positive Selection Kit. Isolated ECs were plated on fibronectin-coated T-25 flasks and cultured with DME containing endothelial growth supplement. Cells were trypsinized and characterized by FACS analysis using anti–VE-cadherin, anti-VEGFR2, and anti-CD31 antibodies, which are known EC surface markers.
Mouse hepatocyte preparation and stimulation
Cells were isolated from mice through an in situ collagenases (type VI, Sigma) perfusion technique, modified as described previously (Deng et al., 2013, Scott and Billiar, 2008). Hepatocyte purity exceeded 99% by flow cytometric assay. Cell viability was typically > 90% by trypan blue exclusion. Hepatocytes (150,000 cells/mL) were cultured on gelatin-coated culture plates or coverslips coated with collagen I (BD PharMingen) in Williams medium E with 10% calf serum, 15 mM HEPES, 1 μM insulin, 2 mM L-glutamine, penicillin (100 U/mL), and streptomycin(100 U/mL). Hepatocytes were allowed to attach to plates overnight, and the culture media was replaced with fresh media before the cells were treated for experiments. After treatment, 1 mL of culture media was collected and centrifuged at 400 g for 5 min. The supernatant was removed to a new tube and stored at −80°C for further analysis.
Method Details
RNAi Silencing
For siRNA Silencing of CASP4, THP1 were cultured in 12-well plates at 3 × 105 cells/well at the time of transfection. siRNA transfection was performed using the Lipofectamine RNAi MAX Transfection Reagent by following the manufacturer’s instructions. 72 h after transfection, cells were stimulated with LPS alone or LPS+HMGB1.The siRNA target sequences are TCTACACTATAGTCCAGACCC (CASP4–1), GTCTGGACTATAGTGTAGATG (CASP4–2) and CGTACGCGGAATACTTCGA (control), which were described previously (Shi et al., 2014). The silencing efficiency was examined by western-blotusing the corresponding antibodies.
Necrotic cell lysate preparation
Wild-type and Hmgb1−/− fibroblasts (MEF) were purchased from HMGBiotech, Inc. Cells were cultured in DMEM medium supplemented with 10% FBS, 2-mecaptoethanol (5 × 10−5 M), 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 Mm HEPES. Necrotic cells were prepared by ten cycles of freezing and thawing. Necrosis was verified by microscopic evaluation showing cell fragments but no intact cells. The ratio of necrotic cells to macrophages in Figure 1F is 1:1. Briefly, 108 MEF cells were freezed and thawed 10 times in 1 mL serum-free RPMI 1640 medium, and then subjected to high-speed centrifugation. The supernatant was collected for macrophage stimulation. 106 macrophages were stimulated with 10ìl of such necrotic cell supernatant in the presence or the absence of LPS (1 μg/ml).
Isolation of cytosol fraction and LPS activity assay
Subcellular fractionation of mouse peritoneal macrophages was conducted by a digitonin-based fractionation method as described previously with modifications (Vanaja et al., 2016). Briefly, 5 × 106 cells were stimulated with LPS (100ng/mL) and HMGB1 (400ng/mL). After 2 h of treatment, the cells were washed with sterile cold PBS 4 times. Cells were subsequently treated with 300 μl of 0.005% digitonin extraction buffer for 10–15 min on ice and the supernatant containing cytosol was collected. The residual cell fraction containing cell membranes, organelles and nuclei were collected in 300 μl of 0.1% CHAPS buffer. BCA assay was used for protein quantification and LPS activity assay for LPS quantification. Additionally the fractions were immunoblotted for Na+-K+ ATPase, Rab7, LAMP1, and beta-Actin to confirm the purity of cytosol fraction.
Proximity-ligation assay
A Proximity Ligation Assay kit (Sigma) was used to study the interaction between LPS and HMGB1 or caspase11 protein in mouse peritoneal macrophages, which is a unique method developed to visualize subcellular localization and protein-protein interactions in situ. Macrophages were cultured and stimulated on a six-well object glass in RPMI medium 1640. For LPS and HMGB1 interaction, cells were treated with or without LPS (5μg/mL) plus HMGB1 (10μg/mL) or LPS (5μg/mL) alone for 2h. To assess LPS and caspase11 interaction, cells were primed with 100ng/mL LPS for 4h before this treatment. After fixation with 4% formaldehyde and permeabilization with Triton, cells were incubated over night with primary antibody pair of different species directed to LPS (mouse monoclonal 2D7/1), HMGB1 (rabbit monoclonal EPR3507) or caspase11 (rat monoclonal 17D9). In situ PLA was performed according to manufacturer’s instructions. Briefly, after incubation with primary antibodies, the cells were incubated with a combination of corresponding PLA probes, secondary antibodies conjugated to oligonucleotides (mouse MINUS and rabbit PLUS for LPS and HMGB1 interaction, mouse PLUS and rat MINUS for LPS and caspase11 interaction). Subsequently, ligasewas added forming circular DNA strands when PLA probes were bound in close proximity, along with polymerase and oligonucleotides to allow rolling circle amplification. Fluorescently labeled probes complementary to sequence to the rolling circle amplification product was hybridized to the rolling circle amplification product. Thus, each individual pair of proteins generated a spot that could be visualized using fluorescent microscopy. Images were taken using a Leica confocal laser scanning microscope and quantified using Image-J software. In some experiments, cells were incubated with antibodies recognized mouse EEA1 and LAMP1 (Proteintech) before PLA procedure to stain the lysosomes or endosomes, respectively.
Immunofluorescence microscopy
For immunofluorescence staining of spleen and gut tissue, the whole animal was perfused with PBS and fixed in 2% paraformaldehyde. Tissue was then placed in 2% paraformaldehyde for 2 h and then switched to 30% sucrose in distilled water solution for 12 h. Tissue sections (5 μm) were incubated with 2% bovine serum albumin (BSA) in PBS for 1 h, followed by five washes with PBS containing 0.5% BSA (PBB). The samples were then incubated overnight with primary antibodies. TMR red-labeled In Situ Cell Death Detection Reagent kit (Roche) was used to detect cell death according to manufacturer’s instructions. Samples were washed with PBS prior to being coverslipped using Gelvatol (23 g polyvinyl alcohol 2000, 50mL glycerol, 0.1% sodium azide to 100 mL PBS). Imaging conditions were maintained at identical settings within each antibody-labeling experiment with original gating performed using the negative control. Imaging was performed using a Nikon A1 confocal microscope (purchased with 1S10OD019973–01 awarded to Dr. Simon C. Watkins). Quantification was performed using NIS Elements Software (Nikon).
ELISA and LDH Assay
Serum and cell culture supernatant samples were analyzed using IL-1α, IL-1β, TNF-α, IL-6 and HMGB1 ELISA kits. Cell death was assessed by LDH Cytotoxicity Assay kit.
Immuno-blot
Proteins from cell-free supernatants were extracted by methanol-chloroform precipitation and cell extracts. Samples were separated by 12% SDS-PAGE and transferred onto PVDF membranes (Millipore). Antibodies to mouse caspase-11 were used at 1:500 dilution, IL-1α was used at 1:1000 dilution; Antibodies to mouse gasdermin D was used at 1:500; HMGB1 was used at 1:2500 dilution. Antibodies to Na-K-ATP, Lamp1, and Rab7 were used at 1:1000 dilution. Blots were normalized to β-actin expression.
Flow cytometry
For evaluation of lysosomal rupture, cells were incubated with 1 μg/mL acridine orange for 15 min, washed three times and subsequently stimulated as indicated. Lysosomal rupture can be assessed by loss of emission at 600–650 nm using flow cytometry as described previously (Hornung et al., 2008).
For measurement of immune cell death, cells were blocked for Fc receptors with anti-mouse CD16/32 (BD Bioscience) for 5 min and then were stained with fluorochrome-conjugated Ab for 30 min, at 4°C in dark. Cells isolated from PLF were incubated with cell surface markers for myeloid cells (CD11b) and subsequently stained with red-labeled TMR (in situ cell death detection reagent) (sigma). Data were acquired with a BD FACS LSR Fortessa flow cytometer (BD Bioscience) and analyzed with FlowJo analytical software (TreeStar). Each experiment was repeated three times.
Imaging of HMGB1 molecules
Single particle fluorescence imaging experiments were performed on an inverted optical microscope (Ti-U, Nikon, Japan). The mercury lamp was replaced with a solid-state diode laser (532 nm, CNI laser Changchun, China). The expended laser line was reflected with a dichroic mirror and focused onto the back port of an oilimmersion objective (NA 1.49, 100 ×, Nikon, Japan). The emitted fluorescence from the sample was then filtered with a band pass filter (605/55, Semrock, U.S.A.). In order to reduce the background noise from the sample and to image only the cell membrane, the incidence angle of the laser line was slightly adjusted to achieve a highly inclined thin illumination mode. The fluorescence image was captured by an EMCCD camera (Ultra 897, Andor, UK). The exposure time was set to 20 ms. Under this condition the data acquisition rate can reach 32 Hz for dynamic tracking. The diffusion trajectories of HMGB1 single particles on cell membrane were analyzed with ImageJ (http://rsb.info.nih.gov/ij/). The detailed procedures for single particle trajectory analysis can be found in the previous work. With this imaging setup (the pixel size of the camera is 16 μ m × 16 μ m, that is in the image plane each pixel represents an area of 160 nm × 160 nm), the temporal localization accuracy (with S/N better than 10) is comparable to the results in our previous work (better than 10 nm).
Whole-cell Patch-clamp Recording
Inward currents were recorded using an EPC-10 patch-clamp amplifier (HEKA, Germany) under whole-cell patch-clamp conditions at room temperature (20–25°C) as described previously (Moyes et al., 2016). Briefly, fire-polished electrodes (0.8–1.5MΩ) were fabricated from borosilicate glass tubing (VWR micropipettes; VWR Co., West Chester, PA, USA) using atwo-stage vertical microelectrode puller (PC-10; Narishige, Tokyo, Japan). For K+ current recording in HEK293T cells, pipette solution contained the following:140 mMKCl,2.5 mM MgCl2,10 mM HEPES, and 11 mM EGTA (pH7.2),the external bath solution contained the following: 150, 30 mM KCl, 5 mM CaCl2, 4 mM MgCl2, 0.3 mM TTX, 10 mM HEPES, and 10 mM D-glucose, adjusted to pH 7.4 or 5.0. The P4 protocol was used to subtract linear capacitive and leakage currents. Experimental data were acquired and analyzed on a Pentium IV computer using the Pulse program (version 8.31; HEKA).
Liposome leakage assay
Liposomes were prepared by a filming-rehydration method. Soy bean phosphatidylcholine (Lipoid GmbH, Ludwigshafen, Germany) was dissolved in chloroform, and then subjected to a rotary evaporator (RE-2000A, Yarong Biochem, Shanghai, China) to remove the solvent in flask, thus leading to the lipid film formation. The neutral-pH hydration buffer was composed of 100 mMKCl, 10 mMTris, and 100 mM MES. The fluorescent dye calcein (J&K Chem, Shanghai, China) was dissolved in the hydration buffer to make a saturated solution. The liposomes encapsulating calcein was prepared by adding the saturated solution of calcein to the flask with lipid film, followed by treatment in ultrasonic water bath. The thus-formed liposomes were extruded through 0.1 μm nucleopore polycarbonate membranes by using Avati Mini-Extruder (Avanti Polar Lipids, Inc. Alabaster, USA). The free calcein was removed by gel filtration on a Sephadex G-50 column (GE Healthcare, USA), and the liposomes were collected and adjusted to a lipid concentration of 0.8mg/mL before use. The liposomes were incubated with the peptide and bovine serum albumin (BSA, control) to investigate the interaction between the peptide and the lipid bilayer. The liposome leakage was measured by monitoring the fluorescence intensity at preset time points with excitation wavelength of 490 nm and emission wavelength 520 nm using a Multimode Plate Reader (Perkin Elmer, Waltham, USA). At the experimental endpoint, 0.1% Triton X-100 was added to disrupt the liposomes.
Mass-spectrometric analysis
Male C57BL/6 mice that were 25 to 30 g in weight were injected intraperitoneally with 10 mg/kg Biotin labeled LPS or PBS and sacrificed 2 hours after injection. 2ml of PBS was injected into peritoneal lavage and then filtered with 100 kDa Amicon Ultra-centrifugal filter (Merck Millipore). The filtered peritoneal lavage fluid (PLF) was incubated with 200μl Streptavidin-Agrose beads (Thermo Fisher Scientific) overnight at 4°C. Protein complexes were incubated at 95°C for 5minutes after sufficient washing, and then and were subjected to immuno-blot analysis and SDS-PAGE for Mass-spectrometric analysis: Excised gel bands were cut into approximately 1 mm3 pieces. The gel pieces were subjected to in-gel trypsin digestion, and the peptides extracted were sequenced on a Q Exactive HF liquid chromatography-mass spectrometer (Thermo Scientific). Peptide sequences (and hence protein identity) were determined by matching the acquired fragmentation pattern with protein databases using MaxQuant (Version1.5.2.8). Data were searched with a 20 ppm precursor ion tolerance window and a 50 mmu fragment iontolerance window against the UniProt database (Musmusculus FASTA database updated on April 4, 2015). For each peptide, up to two missed-cleavages are allowed. Cysteine carboxamidomethylation was specified as a static modification, oxidation of methionine residue and the Acetyl (protein-N) were allowed as variable modifications for each sample set. Reverse decoy databases were included for all searches to estimate false discovery rates (FDR), and the data was filtered using a 1% FDR.
Surface plasmon resonance
Analysis of LPS binding to HMGB1, A-box or B-box was conducted using a BIAcore4000 instrument (BIAcore). In brief, HBS running buffer was used for sample dilution and analysis (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20 (pH 7.4)). The Pre-Functionalized CM Dextran Sensor chip was activated with equal amounts of 0.4 M N-ethyl-N’-[3-diethylamino-propyl]-carbodiimide and 0.1 MN-hydroxysuccinimide. HMGB1 boxes A and B proteins at a concentration of 100 μg/mL were immobilized in 10 mM sodium acetate buffer (pH 4.0) followed by 1 M ethanolamine-hydrochloride (pH 8.0) to deactivate excess N-hydroxysuccinimide-esters. To evaluate binding, LPS was diluted in HBS buffer and passed over the sensor chip at a flow rate of 10μl/min. An activated and blocked flow-cell without immobilized ligand was used to evaluate nonspecific binding. For all samples, response curves were also recorded on control surfaces. Results were calculated after subtraction of the control values using the BI Data Analysis 2.8.4 software.
ELISPOT assays
Multiscreen 96-well high-throughput screening plates were pretreated with 70% ethanol, washed with sterile water, and coated with 20 ug/mL mouse mAb 2G7 at 4°C overnight in a moist chamber. The plates were blocked with complete medium for 2 h. Primary hepatocytes isolated from mice of indicated genotypes co-cultured with WT mouse peritoneal macrophages, and both were stimulated with 1μg/mL LPS (E. coli 0111:B4, InvivoGen) at 37°C with 5% CO2 for 24 h. Cells and medium were decanted from plates, and the plates were washed for three times with washing buffer. monoclonal rabbit anti-HMGB1 antibodies (abcam ab79823) 2μg/mL were added and incubated overnight at 4°C in a moist chamber. Plates were washed three times with washing buffer and incubated with 1μg/mL biotinylated donkey anti-rabbit antibody (Jackson ImmunoResearch Lab) at room temperature for 3 h in a moist chamber. Plates were washed three times with washing buffer, followed by two times washed with PBS alone, and further incubated with Streptavidin-alkaline phosphatase(Mabtech AB, Stockholm, Sweden) for 2 h at room temperature in a moist chamber, following by washing three times. Wells were developed for 10~60 min with substrate solution containing AEC (Sigma) and H2O2(Sigma-Aldrich), the reaction was stopped by washing six times with H2O. After drying, the plates were read using CTL analyzer (CTL S5 micro, Cellular Technology Ltd.). Cell viability was assessed at every experimental set-up and determined to be 90%–100% using Trypan blue (Merck, Darmstadt, Germany) exclusion.
Live cell imaging
Primary hepatocytes at a density of 2 × 106/mL in William E media with 10% calf serum were transfected with plasmids encoding HMGB1-EGFP for 24 hr. After LPS (1μg/mL) stimulation for 3h, time-lapse images were collected with a Nikon swept filed slit scanning confocal microscope in 3D stacks over time with a 1.49 NA 60 × objective lens (Nikon) using Nikon NIS-Elements software. Images of the EGFP-HMGB1 were acquired with a 488 nm laser with a 100 ms exposure at multiple positions in the cell every 2 min for 2h.
Quantification and Statistical Analysis
All data were analyzed using GraphPad Prism software (version 5.01). Data were analyzed using by Student’s t test were used for comparison between two groups or one-way ANOVA followed by post hoc Bonferroni test for multiple comparisons. For measurements of bacterial CFU, groups were compared using nonparametric Mann Whitney U statistics. Survival data were analyzed using the log-rank test. A p value < 0.05 was considered statistically significant for all experiments. All values are presented as the mean ± SD.
Supplementary Material
Acknowledgments
The authors thank R. Zhou, A. Bierhaus, and J. Hai for sharing key mouse strains (Nlrp3−/−, Ager–/–, Asc–/–, and Gsdmd–/– mice). We thank P. Loughran and H. Liao for their excellent technical support. We thank F. Liu, R. Meng, L. Gu, D. Wang, Y. Lu, and Q. Xue for managing mouse colonies and research assistance. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was supported by National key scientific project 2015CB910700(B.L.), National Natural Science Foundation of China (Nos. 81422027 and 81470345(B.L.), No. 81400149 (Y.T.), and No. 81571879 (T.R.B.), Innovation-driven scientific project of CSU (B.L.), Outstanding young investigator fund of Hunan province (B.L.), and NIH grants RO1GM50441 (T.R.B.), R01GM063075 (H.W.), No. R21AG052912 (Q.W.), No. R01 GM102146 (M.J.S.), and No. R01GM115366 (D.T.).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Supplemental Information
References
- Aachoui, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, Tan MH, Cotter PA, Vance RE, Aderem A, Miao EA. Caspase-11 protects against bacteria that escape the vacuole. Science, 339 (2013), pp. 975–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu. Rev. Immunol, 29 (2011), pp. 139–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus DC, van der Poll T. Severe sepsis and septic shock. N. Engl. J. Med, 369 (2013), pp. 840–851 [DOI] [PubMed] [Google Scholar]
- Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet, 22 (1999), pp. 276–280 [DOI] [PubMed] [Google Scholar]
- Cheng KT, Xiong S, Ye Z, Hong Z, Di A, Tsang KM, Gao X, An S, Mittal M, Vogel SM, Miao EA, Rehman J, Malik AB. Caspase-11-mediated endothelial pyroptosis underlies endotoxemia-induced lung injury. J. Clin. Invest, 127 (2017), pp. 4124–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll RC, Robertson AA, Chae JJ, Higgins SC, Muñoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med, 21 (2015), pp. 248–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Scott MJ, Loughran P, Gibson G, Sodhi C, Watkins S, Hackam D, Billiar TR. Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis. J. Immunol, 190 (2013), pp. 5152–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity, 48 (2018), pp. 35–44.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MG, Weber MD, Fonken LK, Hershman SA, Watkins LR, Maier SF. The redox state of the alarmin HMGB1 is a pivotal factor in neuroinflammatory and microglial priming: A role for the NLRP3 inflammasome. Brain Behav. Immun, 55 (2016), pp. 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science, 341 (2013), pp. 1250–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res, 25 (2015), pp. 1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell, 97 (1999), pp. 889–901 [DOI] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol, 9 (2008), pp. 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebener P, Pradere JP, Hernandez C, Gwak GY, Caviglia JM, Mu X, Loike JD, Jenkins RE, Antoine DJ, Schwabe RF. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J. Clin. Invest, 125 (2015), pp. 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature, 479 (2011), pp. 117–121 [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszyński A, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science, 341 (2013), pp. 1246–1249 [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signaling. Nature, 526 (2015), pp. 666–671 [DOI] [PubMed] [Google Scholar]
- Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity, 29 (2008), pp. 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim SJ, Lee IS, Lee MS, Uematsu S, Akira S, Oh KI. Bacterial endotoxin induces the release of high mobility group box 1 via the IFN-beta signaling pathway. J. Immunol, 182 (2009), pp. 2458–2466 [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Sarkar A, Vande Walle L, Vitari AC, Amer AO, Wewers MD, Tracey KJ, Kanneganti TD, Dixit VM. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J. Immunol, 185 (2010), pp. 4385–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liliensiek B, Weigand MA, Bierhaus A, Nicklas W, Kasper M, Hofer S, Plachky J, Gröne HJ, Kurschus FC, Schmidt AM, et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J. Clin. Invest, 113 (2004), pp. 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Nakamura T, Inouye K, Li J, Tang Y, Lundbäck P, Valdes-Ferrer SI, Olofsson PS, Kalb T, Roth J, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature, 488 (2012), pp. 670–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Antoine DJ, Kwan K, Lundbäck P, Wähämaa H, Schierbeck H, Robinson M, Van Zoelen MA, Yang H, Li J, et al. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc. Natl. Acad. Sci. USA, 111 (2014), pp. 3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature, 440 (2006), pp. 228–232 [DOI] [PubMed] [Google Scholar]
- Meunier E, Dick MS, Dreier RF, Schürmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature, 509 (2014), pp. 366–370 [DOI] [PubMed] [Google Scholar]
- Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature, 532 (2016), pp. 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Wang H, Yuan R, Li H, Ochani M, Ochani K, Rosas-Ballina M, Czura CJ, Huston JM, Miller E, et al. Role of HMGB1 in apoptosis-mediated sepsis lethality. J. Exp. Med, 203 (2006), pp. 1637–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon-Mitchell B, Ochani M, Li J, Han J, Wang H, Yang H, Susarla S, Czura C, Mitchell RA, Chen G, et al. IFN-gamma induces high mobility group box 1 protein release partly through a TNF-dependent mechanism. J. Immunol, 170 (2003), pp. 3890–3897 [DOI] [PubMed] [Google Scholar]
- Rittirsch D, Flierl MA, Nadeau BA, Day DE, Huber-Lang M, Mackay CR, Zetoune FS, Gerard NP, Cianflone K, Köhl J, et al. Functional roles for C5a receptors in sepsis. Nat. Med, 14 (2008), pp. 551–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco C, Piccinni P, KellumRationale J of extracorporeal removal of endotoxin in sepsis: theory, timing and technique. Contrib. Nephrol, 167 (2010), pp. 25–34 [DOI] [PubMed] [Google Scholar]
- Rouhiainen A, Tumova S, Valmu L, Kalkkinen N, Rauvala H. Pivotal advance: analysis of proinflammatory activity of highly purified eukaryotic recombinant HMGB1 (amphoterin). J. Leukoc. Biol, 81 (2007), pp. 49–58 [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature, 418 (2002), pp. 191–195 [DOI] [PubMed] [Google Scholar]
- Scott MJ, Billiar TR. Beta2-integrin-induced p38 MAPK activation is a key mediator in the CD14/TLR4/MD2-dependent uptake of lipopolysaccharide by hepatocytes. J. Biol. Chem, 283 (2008), pp. 29433–29446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature, 514 (2014), pp. 187–192 [DOI] [PubMed] [Google Scholar]
- Thompson DB, Villaseñor R, Dorr BM, Zerial M, Liu DR. Cellular uptake mechanisms and endosomal trafficking of supercharged proteins. Chem. Biol, 19 (2012), pp. 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsung A, Klune JR, Zhang X, Jeyabalan G, Cao Z, Peng X, Stolz DB, Geller DA, Rosengart MR, Billiar TR. HMGB1 release induced by liver ischemia involves Toll-like receptor 4 dependent reactive oxygen species production and calcium-mediated signaling. J. Exp. Med, 204 (2007), pp. 2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Ferrer SI, Rosas Ballina M, Olofsson PS, Lu B, Dancho ME, Ochani M, Li JH, Scheinerman JA, Katz DA, Levine YA, et al. HMGB1 mediates splenomegaly and expansion of splenic CD11b+ Ly-6C(high) inflammatory monocytes in murine sepsis survivors. J. Intern. Med, 274 (2013), pp. 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam VAK. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell, 165 (2016), pp. 1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell, 92 (1998), pp. 501–509 [DOI] [PubMed] [Google Scholar]
- Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science, 285 (1999), pp. 248–251 [DOI] [PubMed] [Google Scholar]
- Wang H, Liao H, Ochani M, Justiniani M, Lin X, Yang L, Al-Abed Y, Wang H, Metz C, Miller EJ, et al. Cholinergic agonists inhibit HMGB1 release and improve survival in experimental sepsis. Nat. Med, 10 (2004), pp. 1216–1221 [DOI] [PubMed] [Google Scholar]
- Wautier JL, Zoukourian C, Chappey O, Wautier MP, Guillausseau PJ, Cao R, Hori O, Stern D, Schmidt AM. Receptor-mediated endothelial cell dysfunction in diabetic vasculopathy. Soluble receptor for advanced glycation end products blocks hyperpermeability in diabetic rats. J. Clin. Invest, 97 (1996), pp. 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt TM, Tanji N, Guo J, Kislinger TR, Qu W, Lu Y, Bucciarelli LG, Rong LL, Moser B, Markowitz GS, et al. RAGE drives the development of glomerulosclerosis and implicates podocyte activation in the pathogenesis of diabetic nephropathy. Am. J. Pathol, 162 (2003), pp. 1123–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Jiang Y, Wang J, Shi X, Liu Q, Liu Z, Li Y, Scott MJ, Xiao G, Li S, et al. Macrophage endocytosis of high-mobility group box 1 triggers pyroptosis. Cell Death Differ, 21 (2014), pp. 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Matsuda A, An J, Koshiba R, Nishio J, Negishi H, Ikushima H, Onoe T, Ohdan H, Yoshida N, Taniguchi T. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc. Natl. Acad. Sci. USA, 110 (2013), pp. 20699–20704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Ju Z, Ragab AA, Lundbäck P, Long W, Valdes-Ferrer SI, He M, Pribis JP, Li J, et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J. Exp. Med, 212 (2015), pp. 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JH, Oh YJ, Kim ES, Choi JE, Shin JS. High mobility group box 1 protein binding to lipopolysaccharide facilitates transfer of lipopolysaccharide to CD14 and enhances lipopolysaccharide-mediated TNF-alpha production in human monocytes. J. Immunol, 180 (2008), pp. 5067–5074 [DOI] [PubMed] [Google Scholar]
- Youn JH, Kwak MS, Wu J, Kim ES, Ji Y, Min HJ, Yoo JH, Choi JE, Cho HS, ShinIdentification JS of lipopolysaccharide-binding peptide regions within HMGB1 and their effects on subclinical endotoxemia in a mouse model. Eur. J. Immunol, 41 (2011), pp. 2753–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xiang W, Wang G, Yan Z, Zhu Z, Guo Z, Sengupta R, Chen AF, Loughran PA, Lu B, et al. Interferon β (IFN-β) production during the double-stranded RNA (dsRNA) response in hepatocytes involves coordinated and feedforward signaling through Toll-like Receptor 3 (TLR3), RNA-dependent protein kinase (PKR), inducible nitric oxide synthase (iNOS), and Src protein. J. Biol. Chem, 291 (2016), pp. 15093–15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.