Abstract
To explore novel genetic loci for diabetic nephropathy, we performed genome-wide association studies (GWAS) for diabetic nephropathy in Japanese patients with type 2 diabetes. We analyzed the association of 5,768,242 single nucleotide polymorphisms (SNPs) in Japanese patients with type 2 diabetes, 2,380 nephropathy cases and 5,234 controls. We further performed GWAS for diabetic nephropathy using independent Japanese patients with type 2 diabetes, 429 cases and 358 controls and the results of these two GWAS were combined with an inverse variance meta-analysis (stage-1), followed by a de novo genotyping for the candidate SNP loci (p < 1.0 × 10−4) in an independent case-control study (Stage-2; 1,213 cases and 1,298 controls). After integrating stage-1 and stage-2 data, we identified one SNP locus, significantly associated with diabetic nephropathy; rs56094641 in FTO, P = 7.74 × 10−10. We further examined the association of rs56094641 with diabetic nephropathy in independent Japanese patients with type 2 diabetes (902 cases and 1,221 controls), and found that the association of this locus with diabetic nephropathy remained significant after integrating all association data (P = 7.62 × 10−10). We have identified FTO locus as a novel locus for conferring susceptibility to diabetic nephropathy in Japanese patients with type 2 diabetes.
Introduction
Diabetic nephropathy is a leading cause of end-stage renal disease (ESRD), and its prevalence is progressively increasing according to the increase of number of patients with diabetes mellitus [1,2]. Prolonged persistent hyperglycemia is a principal cause of diabetic microvascular complications, but it has been shown that only ~30% of all patients with diabetes develop overt nephropathy, whereas most patients develop diabetic retinopathy 20–30 years after the onset of diabetes [3]. In addition, familial clustering of diabetic nephropathy has been observed in both type 1 and type 2 diabetes [4–6], implying that genetic factors are involved in the development and/or progression of diabetic nephropathy. Identification of disease susceptibility loci for many common diseases such as type 2 diabetes, has been achieved by the introduction of genome-wide association studies (GWAS). Worldwide efforts to identify susceptibility loci for diabetic nephropathy, however, have not yet met with success so much. Several susceptibility loci to diabetic nephropathy or its related traits which showed their lowest p-values close to or with a genome-wide significance level have been unveiled; rs2268388 in ACACB [7], rs7583877 in AFF3 locus, rs17709344 in RGMA-MCTP2 locus [8], rs4972593 in Sp3-CDCA7 locus [9], rs1801239 in CUBN locus [10], rs161740 in EPO locus [11]. However, the results have not been conclusive, and thus most susceptibility loci for diabetic nephropathy remain to be un-identified, suggesting heterogeneity of the disease or contribution of non-genetic factors, which have not been taken into account, may produce inconsistent results among the studies.
In this study, to identify novel susceptibility loci to diabetic nephropathy, we performed a GWAS meta-analysis for diabetic nephropathy using existing Japanese GWAS data for patients with type 2 diabetes.
Materials and methods
Study subjects
Discovery stage (Stage-1)
We selected 7,641 individuals having type 2 diabetes registered in Biobank Japan, and divided these patients into two groups (stage-1, set-1); 1) 2,380 nephropathy cases, defined as patients with overt albuminuria or under renal replacement therapy and 2) 5,234 controls with normoalbuminuria and with diabetes duration of 5 years or longer or with diabetic retinopathy. We also used independent patients with type 2 diabetes who were extracted from previously reported GWAS data [12], and performed GWAS for diabetic nephropathy (stage-1 set 2, cases, n = 429, controls, n = 358). There was no overlap between set-1 and set-2. Diabetes was diagnosed according to the World Health Organization (WHO) criteria [13], and those who were diagnosed as type 1 diabetes, mitochondrial diseases or maturity-onset diabetes of the young were excluded.
Validation analysis (Stage-2)
We examined independent 2,511 patients with Japanese patients with type 2 diabetes, 1,213 diabetic nephropathy cases and 1,298 controls, from the BioBank Japan that were not included in the discovery stage.
Clinical characteristics of participants for Stage-1 (set-1, set-2) and Stage-2 are shown in Table 1. Genomic DNA was extracted from peripheral leukocytes using the standard procedure. All individuals provided written informed consent to participate in this study.
Table 1. Clinical characteristics of participants.
| Stage1 | Stage2 | |||||
|---|---|---|---|---|---|---|
| Set1 | Set2 | |||||
| case | control | case | control | case | control | |
| n | 2,380 | 5,234 | 429 | 358 | 1,213 | 1,298 |
| M/F | 1,611/769 | 3,162/2,072 | 304/125 | 226/131** | 833/380 | 759/539 |
| Men% | 67.6 | 60.4 | 70.9 | 63.1 | 68.6 | 58.5 |
| age | 65.9 ± 10.5 | 66.3 ± 9.6 | 67.2 ± 10.0 | 66.9 ± 9.2 | 63.9 ± 11.0 | 63.9 ± 10.5 |
| BMI | 23.9 ± 4.0* | 23.6 ± 3.6 | 23.5 ± 4.0 | 23.8 ± 3.8 | 24.1 ± 3.9 | 23.6 ± 3.8 |
| Diabetes duration | 13.1 ± 9.8* | 14.4 ± 8.5 | 13.4 ± 10.2 | 13.2 ± 10.0 | 12.0 ± 9.5 | 10.5 ± 8.8 |
* P < 0.01 vs. control
** information of sex for one participant is not available
Genotyping, quality control and imputation in the discovery stage
Set-1 individuals in stage 1 were genotyped using the Human Omni Express Exome Bead Chip. There were 628,670 autosomal SNPs that passed quality control, a call rate ≥ 0.99, Hardy-Weinberg equilibrium test P ≥ 1 × 10−6 in controls and minor allele frequency (MAF) ≥ 0.01. Set-2 samples were genotyped using the Illumina Human 610K SNP array, and 504,984 autosomal SNPs passed the quality control described above and used for further analysis. For sample quality control, we evaluated cryptic relatedness for each sample using an identity-by-state method and removed samples that exhibited second-degree or closer relatedness. We performed principal component analysis to select individuals belong to the major Japanese cluster (Hondo cluster, S1 Fig) as reported previously [14], and data for 7,614 individuals (2,380 diabetic nephropathy cases and 5,234 controls) in set-1 and 787 individuals (429 diabetic nephropathy cases and 358 controls) in set-2 were used in subsequent analyses. We performed genotype imputation with MACH and Minimac [15,16] using linkage disequilibrium data in the 1000 Genomes Project (phased JPT, CHB and Han Chinese South data n = 275, March 2012) as reference populations. To evaluate the potential effect of population stratification, we used a quantile-quantile (qq) plot of the observed P-values.
De novo genotyping
We genotyped 1,213 individuals with diabetic nephropathy and 1,298 controls (Stage-2) registered as type 2 diabetes in BioBank Japan, who were not included in a discovery stage using a multiplex PCR-invader assay [17] for SNPs with p values < 10−4 in the meta-analysis. We also genotyped additional 2,123 Japanese patients with type 2 diabetes (902 diabetic nephropathy cases and 1,221 controls), who visited out-patient clinics of Tokai university hospital, Shiga university of medical science hospital, Juntendo university hospital, Kawasaki medical school hospital, Tokyo women’s medical university hospital, Iwate Medical University, Toride Kyodo Hospital, Kawai Clinic, Osaka City General Hospital, Chiba Tokusyukai Hospital or Osaka Rosai Hospital.
Genotyping success rates < 95% or concordance rates < 99.9% were excluded from the analyses.
Statistical analysis
The association between each SNP and diabetic nephropathy was assessed using the logistic regression test with an additive model with or without adjusting for age, sex, and log-transformed body mass index (BMI) using Mach2dat. We combined data from the each GWAS, validation and replication studies using METAL [18] as an inverse variance method. Heterogeneity in effect sizes among the studies was evaluated with a Cochran’s Q test. Regional association plots were generated using LocusZoom [19].
Ethics approval
The protocol of this study conformed to the provisions of the Declaration of Helsinki and was approved by the ethical committees at the RIKEN Yokohama Institute, Tokai university hospital, Shiga university of medical science, Juntendo university, Kawasaki medical school, Tokyo women’s medical university and Iwate Medical University, and the institutional review boards at Toride Kyodo Hospital, Osaka City General Hospital, Chiba Tokusyukai Hospital and Osaka Rosai Hospital.
Results
A meta-analysis of GWAS for diabetic nephropathy in the Japanese patients with type 2 diabetes
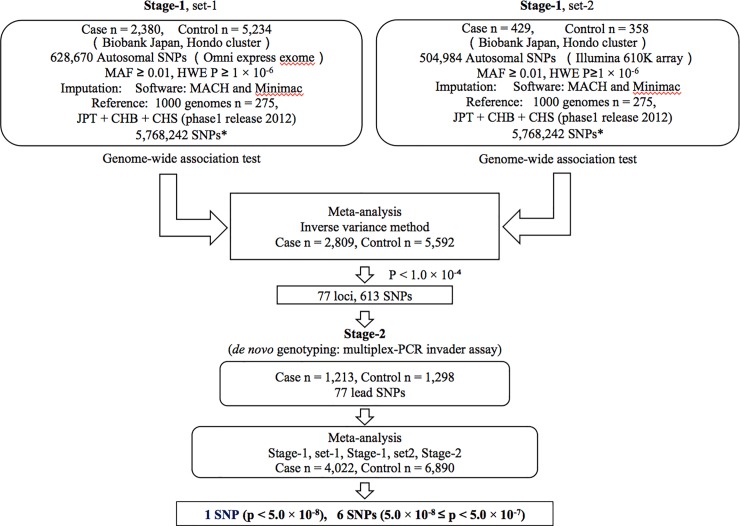
We obtained genotype data for 7,521,074 SNPs by imputation, and among them, 5,768,242 SNPs those passed quality control (r2 > 0.7) in both studies (Stage-1 set-1 and set-2) were evaluated in this meta-analysis (Fig 1).
Fig 1. Outline of this study.
* r2 > 0.7 in both studies, SNP; single nucleotide polymorphism, MAF; minor allele frequency, HWE P; Hardy-Weinberg Equilibrium test P, RSQ, r square.
There was no genomic inflation in the qq plots in the both studies (S2 Fig). We selected 77 loci associated with diabetic nephropathy in the meta-analysis (P < 1 × 10−4, S1 Table), and the associations of the lead SNPs from the 77 loci with DN were examined in an independent case-control study (Stage-2, 1,213 cases and 1,298 controls). In stage-2 analyses, we successfully obtained the genotype data for 65 loci by the multiplex-PCR invader assay. After combining all association data (Stage-1 set-1, set-2 and Stage-2) using the inverse-variance fixed-effects meta-analysis, we found that one SNP locus, rs56094641 in FTO at chromosome (Chr) 16, 16q12.2, showed a genome-wide significant association with diabetic nephropathy (P = 7.74 × 10−10, odds ratio (OR) = 1.23, 95% confidence interval (CI) 1.15−1.31, Table 2 and S2 Table). The association of the FTO locus with overt nephropathy was not affected by an adjustment for age, sex and BMI (rs9936385, r2 = 1 with rs56094641, S3 Table). We additionally identified suggestive evidence for the associations of six SNP loci with diabetic nephropathy (5.0 × 10−8 < P < 5.0 × 10−7, Table 2 and S2 Table): rs895157 in PRCD (17q25.1, P = 7.70 × 10−8, OR = 1.28, 95% CI 1.17−1.41), rs10144968 near RAD51B (14q24.1–24.2, P = 1.22 × 10−7, OR = 1.35, 95% CI 1.21−1.51), rs13306536 in LRP8 (1p32.3, P = 2.70 × 10−7, OR = 1.33, 95% CI 1.19−1.48), rs7544082 near TRABD2B (1p33, P = 3.08 × 10−7, OR = 1.17, 95% CI 1.10−1.24), rs11101179 in CHAT (10q11.2–21.1, P = 3.85 × 10−7, OR = 1.19, 95% CI 1.11−1.28), rs710375 near CCNH (5q14-15, P = 3.96 × 10−7, OR = 1.21, 95% CI 1.12−1.30). Regional plots for these 7 loci are shown in S3 Fig.
Table 2. Association of 7 SNP loci with diabetic nephropathy in Japanese patients with type 2 diabetes.
| SNP ID Gene Chromosome |
Alleles | study | RAF | OR (95%CI) | P | Phet |
|---|---|---|---|---|---|---|
| rs56094641 | G/A | Stage-1, set-1 | 0.247/0.211 | 1.22 (1.13–1.32) | 1.19 × 10−6 | |
| FTO | Stage-1, set-2 | 0.236/0.207 | 1.19 (0.93–1.52) | 0.164 | ||
| Ch16 | Stage-2 | 0.252/0.209 | 1.27 (1.11–1.46) | 3.76 × 10−4 | ||
| Combined | 1.23 (1.15–1.31) | 7.74 × 10−10 | 0.85 | |||
| rs895157 | C/A | Stage-1, set-1 | 0.130/0.108 | 1.28 (1.14–1.43) | 2.85 × 10−5 | |
| PRCD | Stage-1, set-2 | 0.126/0.113 | 1.14 (0.83–1.58) | 0.422 | ||
| Ch17 | Stage-2 | 0.140/0.109 | 1.35 (1.13–1.60) | 7.02 × 10−4 | ||
| Combined | 1.28 (1.17–1.41) | 7.70 × 10−8 | 0.67 | |||
| rs10144968 | G/T | Stage-1, set-1 | 0.078/0.062 | 1.27 (1.11–1.46) | 3.75 × 10−4 | |
| RAD51B | Stage-1, set-2 | 0.083/0.060 | 1.41 (0.95–2.10) | 8.48 × 10−2 | ||
| Stage-2 | 0.078/0.051 | 1.58 (1.25–1.99) | 1.15 × 10−4 | |||
| Combined | 1.35 (1.21–1.51) | 1.22 × 10−7 | 0.28 | |||
| rs13306536 | T/C | Stage-1, set-1 | 0.089/0.068 | 1.27 (1.11–1.46) | 3.75 × 10−4 | |
| LRP8 | Stage-1, set-2 | 0.068/0.075 | 0.87 (0.58–1.30) | 0.496 | ||
| Ch1 | Stage-2 | 0.076/0.058 | 1.35 (1.08–1.69) | 1.15 × 10−4 | ||
| Combined | 1.33 (1.19–1.48) | 2.70 × 10−7 | 0.1 | |||
| rs7544082 | A/C | Stage-1, set-1 | 0.525/0.492 | 1.18 (1.09–1.27) | 3.22 × 10−5 | |
| TRABD2B | Stage-1, set-2 | 0.550/0.514 | 1.15 (0.77–1.72) | 0.164 | ||
| Ch1 | Stage-2 | 0.552/0.514 | 1.16 (1.04–1.31) | 8.70 × 10−3 | ||
| Combined | 1.17 (1.10–1.24) | 3.08 × 10−7 | 0.97 | |||
| rs11101179 | C/T | Stage-1, set-1 | 0.231/0.197 | 1.22 (1.12–1.33) | 2.23 × 10−6 | |
| CHAT | Stage-1, set-2 | 0.205/0.205 | 1.00 (0.77–1.31) | 0.977 | ||
| Ch10 | Stage-2 | 0.226/0.198 | 1.17 (1.02–1.34) | 2.10 × 10−2 | ||
| Combined | 1.19 (1.11–1.28) | 3.85 × 10−7 | 0.37 | |||
| rs710375 | C/T | Stage-1, set-1 | 0.201/0.169 | 1.24 (1.14–1.36) | 2.23 × 10−6 | |
| CCNH-TMEM161B | Stage-1, set-2 | 0.197/0.154 | 1.38 (1.04–1.77) | 0.977 | ||
| Ch5 | Stage-2 | 0.187/0.176 | 1.08 (0.93–1.24) | 2.10 × 10−2 | ||
| Combined | 1.21 (1.12–1.30) | 3.96 × 10−7 | 0.16 |
Replication study
We examined the association of rs56094641 in FTO with overt nephropathy in 2,123 Japanese patients with type 2 diabetes (cases n = 902, controls n = 1,221). As shown in Table 3, rs56094641 in FTO showed the same direction of effect with the original finding in the discovery stage. The association of rs56094641 in FTO with overt nephropathy was still genome-wide significant level after integration of all association data (P = 7.62 × 10−10, OR = 1.21, 95% CI 1.14−1.29, Table 3) though the association was not statistically significant in the replication study alone (P = 0.258, OR = 1.11, 95% CI 0.93−1.32, Table 3).
Table 3. Replication studies for 7 loci associated with diabetic nephropathy.
| SNP ID gene |
Alleles | Ethnicity | RAF | OR (95%CI) | P | Phet |
|---|---|---|---|---|---|---|
| rs56094641 | G/A | Japanese | Discovery | 1.23 (1.15–1.31) | 7.74 × 10−10 | |
| FTO | Replication | 1.11 (0.93–1.32) | 0.258 | |||
| Combined | 1.21 (1.14–1.29) | 7.62 × 10−10 | 0.67 | |||
| Europeans | 1.06 (1.19–0.95) | 0.300 | ||||
| rs895157 | C/A | Japanese | Discovery | 1.28 (1.17–1.41) | 7.70 × 10−8 | |
| PRCD | Replication | 0.96 (0.77–1.21) | 0.748 | |||
| Combined | 1.23 (1.13–1.34) | 1.15 × 10−6 | 0.11 | |||
| Europeans | 0.92 (0.80–1.06) | 0.195 | ||||
| rs10144968 | G/T | Japanese | Discovery | 1.35 (1.21–1.51) | 1.22 × 10−7 | |
| RAD51B | Replication | 1.04 (0.76–1.41) | 0.808 | |||
| Combined | 1.31 (1.18–1.45) | 4.23 × 10−7 | 0.17 | |||
| Europeans | 0.91 (0.80–1.03) | 0.289 | ||||
| rs13306536 | T/C | Japanese | Discovery | 1.33 (1.19–1.48) | 2.70 × 10−7 | |
| LRP8 | Replication | 0.98 (0.72–1.34) | 0.914 | |||
| Combined | 1.30 (1.16–1.42) | 1.38 × 10−6 | 0.05 | |||
| Europeans | N/A | N/A | ||||
| rs7544082 | A/C | Japanese | Discovery | 1.17 (1.10–1.24) | 3.08 × 10−7 | |
| TRABD2B | Replication | 0.99 (0.86–1.15) | 0.946 | |||
| Combined | 1.16 (1.04–1.31) | 2.58 × 10−6 | 0.24 | |||
| Europeans | N/A | N/A | ||||
| rs11101179 | C/T | Japanese | Discovery | 1.19 (1.11–1.28) | 3.85 × 10−7 | |
| CHAT | Replication | 0.92 (0.77–1.10) | 0.365 | |||
| Combined | 1.15 (1.08–1.23) | 9.36 × 10−6 | 0.03 | |||
| Europeans | N/A | N/A | ||||
| rs710375 | C/T | Japanese | Discovery | 1.21 (1.12–1.30) | 3.96 × 10−7 | |
| CCNH | Replication | 0.85 (0.70–1.03) | 9.86 × 10−2 | |||
| Combined | 1.16 (1.08–1.24) | 3.02 × 10−5 | 0.002 | |||
| Europeans | 1.12 (0.999–1.12) | 3.58 × 10−2 |
N/A, data is not available
By in silico replication using SUMMIT consortium data for type 2 diabetes, the association of FTO variants with diabetic nephropathy was not replicated in European patients with type 2 diabetes (Table 3).
Evaluation of previously reported loci
We reflected our results (meta-analysis of Stage-1 set-1 and set-2) on previously- reported susceptibility loci to overt nephropathy. There was no overlap between the above seven SNP loci and 21 loci from previously-reported GWAS for susceptibility to overt nephropathy (S4 Table). None of the SNPs in the original reports showed significant association with overt nephropathy in the discovery stage (P > 2.38 x 10−3 = 0.05/21). Regional lead SNPs within the ELMO1, RGMA-MCTP2, ERBB4 and SP3-CDCA7 in the discovery stage attained the threshold of the correction of multiple testing error.
Discussion
From a result of GWAS meta-analysis for diabetic nephropathy followed by validation studies in Japanese patients with type 2 diabetes, which comprising in 4,022 cases and 6,890 controls, we identified significant association between rs56094641 in FTO and susceptibility to diabetic nephropathy in Japanese patients with type 2 diabetes.
FTO locus has been repeatedly reported to be associated with obesity and/or adiposity [20], and, in this study, we have shown that the risk allele for obesity was significantly associated with susceptibility to diabetic nephropathy in Japanese patients with type 2 diabetes.
Smemo et al. mentioned that obesity-associated noncoding sequences within FTO are functionally connected, at megabase distances, with the homeobox gene IRX3 [21]. IRX3 is a member of the Iroquois homeobox gene family and plays a role in an early step of neural development [22]. Members of this family including IRX3 and IRX5 appear to play multiple roles during pattern formation of vertebrate embryos [23]. However, there is no evidence showing functional link between IRX3/IRX5 and diabetic nephropathy.
Obesity has been recognized as an important risk factor for chronic kidney disease (CKD) [24,25]. The incidence of obesity has actualized an increase the number of patients with obesity-related glomerulopathy and proteinuria [26]. Furthermore, obesity has been reported to be an independent risk factor for ESRD by a large historical cohort study of 177,570 adults after adjustment for multiple epidemiologic and clinical conditions including diabetes [27] Moreover, a FTO variant, rs17817449, which are in absolute linkage disequilibrium to rs56094641, was shown to be associated with ESRD [28], suggesting FTO variants confer susceptibility to CKD/ESRD through the mechanisms mediated by obesity/adipocity.
In European patients with type 1 or type 2 diabetes, a genetic risk score constructed from confirmed obesity related SNP loci was associated with diabetic nephropathy [29–31], although FTO variant alone did not have significant effect on any of renal phenotype. Therefore, our report is the first to show a robust association of FTO variant with diabetic nephropathy. In addition, adjustment for BMI did not influence the association of the SNP with diabetic nephropathy in this study, suggesting that the SNP gets involved in susceptibility to diabetic nephropathy through the mechanism other than increasing BMI. Values of BMI used in this study, however, were obtained after making diagnosis of type 2 diabetes and some therapeutic interventions might affect the BMI values used in this study. Therefore, we might underestimate the effects of BMI on susceptibility to diabetic nephropathy.
We also obtained six SNP loci, which have shown borderline associations with overt nephropathy at the discovery stage (Table 1: 5 × 10−8 < combined P ≤ 5 × 10−7), but all candidate SNPs did not show significant effects in the replication stage in an independent Japanese case-control study (Table 3). In this study, we could not replicate almost all of variants identified by meta-analysis of stage 1 and stage 2 in the additional dataset with a comparable sample size with stage 2 GWAS. Although genome-wide SNPs data are not available for individuals in the replication stage, all samples for the replication stage were collected in the Japan main-island, and it has been shown individuals living in Japan main-island are genetically homogeneous [14]; therefore, we think there is no evidence for population heterogeneity between the discovery stage and the replication stage.
We further performed a GWAS meta-analysis for diabetic nephropathy including patients with microalbuminuria as cases, and found that rs56094641 in FTO was significantly associated with diabetic nephropathy also in this analysis (P = 2.40 × 10−9, OR = 1.18, 95% CI 1.15−1.31, S5 and S6 Tables). However, the association was not stronger than that in the analysis using overt nephropathy as cases, suggesting the FTO variant was associated with advanced stages of diabetic nephropathy.
Our study has some limitations. Firstly, the detail clinical information, i.e. blood pressure, Hemoglobin A1c, lipid profiles, prescription of antihypertensive treatments, was not available in many participants in Stage-1 samples. It means that we cannot exclude the possibility that the association of the SNP with diabetic nephropathy is mediated by some risk factors except age, sex and BMI. Second, by in silico replication using SUMMIT consortium data for type 2 diabetes, the association of FTO variants with diabetic nephropathy was not replicated in European patients with type 2 diabetes (Table 3, S7 Table), and the association of FTO variants with diabetic nephropathy showed a genome-wide significant association only in a joint analysis for the discovery stage; therefore, these observations may not be in line with standards for GWA studies; further study, such as a large-scaled longitudinal study, is required to elucidate the association of FTO locus with diabetic nephropathy.
Conclusions
We performed GWAS for diabetic nephropathy in Japanese patients with type 2 diabetes. One SNP locus, FTO locus, showed a significant association with susceptibility to diabetic nephropathy, and the association attained a genome-wide significant level. Further studies are required to confirm the association of this locus with diabetic nephropathy.
Supporting information
(PDF)
A: Stage-1, set-1, B: Stage-1, set-2.
(PDF)
Results of stage-1 GWAS meta-analysis are shown. Red, diamond-shaped plots indicate the most significant variants in each locus after combining stage 1 and stage 2 data. r2, linkage disequilibrium coefficient; chr., chromosome.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
Acknowledgments
The authors thank all participating doctors, staff, and patients from the collaborating institutes for providing DNA samples. We also thank Mr. Takashi Morizono, Drs. Michiaki Kubo, Momoko, Horikoshi, RIKEN Center for Integrative Medical Sciences for their support of data managements, and technical staffs of the Laboratory for Endocrinology, Metabolism and Kidney diseases, RIKEN Center for Integrative Medical Sciences for performing de novo genotyping.
Members of the SUMMIT consortium.
Michael Mark, Markus Albertini, Carine Boustany, Alexander Ehlgen, Martin Gerl, Jochen Huber, Corinna Schölch, Heike Zimdahl-Gelling at Boehringer-Ingelheim, Ingelheim, Germany
Leif Groop, Elisabet Agardh, Emma Ahlqvist, Tord Ajanki, Nibal Al Maghrabi, Peter Almgren, Jan Apelqvist, Eva Bengtsson, Lisa Berglund, Harry Björckbacka, Ulrika Blom-Nilsson, Mattias Borell, Agneta Burström, Corrado Cilio, Magnus Cinthio, Karl Dreja, Pontus Dunér, Daniel Engelbertsen, Joao Fadista, Maria Gomez, Isabel Goncalves, Bo Hedblad, Anna Hultgårdh, Martin E. Johansson, Cecilia Kennbäck, Jasmina Kravic, Claes Ladenvall, Åke Lernmark, Eero Lindholm, Charlotte Ling, Holger Luthman, Olle Melander, Malin Neptin, Jan Nilsson, Peter Nilsson, Tobias Nilsson, Gunilla Nordin Fredriksson, Marju Orho-Melander, Emilia Ottoson-Laakso, Annie Persson, Margaretha Persson, Mats-Åke Persson, Jacqueline Postma, Elisabeth Pranter, Sara Rattik, Gunnar Sterner, Lilian Tindberg, Maria Wigren, Anna Zetterqvist, Mikael Åkerlund, Gerd Östling at Lund University Clinical Research Centre, Malmö, Sweden.
Timo Kanninen, Anni Ahonen-Bishopp, Anita Eliasson, Timo Herrala, Päivi Tikka-Kleemola at Biocomputing Platforms, Espoo, Finland.
Anders Hamsten, Christer Betsholtz, Ami Björkholm, Ulf de Faire, Fariba Foroogh, Guillem Genové, Karl Gertow, Bruna Gigante, Bing He, Karin Leander, Olga McLeod, Maria Nastase-Mannila, Jaako Patrakka, Angela Silveira, Rona Strawbridge, Karl Tryggvason, Max Vikström, John Öhrvik, Anne-May Österholm at Karolinska Institute, Stockholm, Sweden.
Barbara Thorand, Christian Gieger, Harald Grallert, Tonia Ludwig, Barbara Nitz, Andrea Schneider, Rui Wang-Sattler, Astrid Zierer at Helmholtz Centre, Munich, Germany.
Giuseppe Remuzzi, Ariela Benigni, Maria Domenica Lesti, Marina Noris, Norberto Perico, Annalisa Perna, Rossella Piras, Piero Ruggenenti, Erica Rurali at Mario Negri Institute for Pharmacological Research, Bergamo, Italy.
David Dunger, Ludo Chassin, Neil Dalton, John Deanfield, Jane Horsford, Clare Rice, James Rudd, Neil Walker, Karen Whitehead, Max Wong at University of Cambridge, UK.
Helen Colhoun, Fiona Adams, Tahira Akbar, Jill Belch, Harshal Deshmukh, Fiona Dove, Angela Ellingford, Bassam Farran, Mike Ferguson, Gary Henderson, Graeme Houston, Faisel Khan, Graham Leese, Yiyuan Liu, Shona Livingstone, Helen Looker, Margaret McCann, Andrew Morris, David Newton, Colin Palmer, Ewan Pearson, Gillian Reekie, Natalie Smith at University of Dundee, Scotland.
Angela Shore, Kuni Aizawa, Claire Ball, Nick Bellenger, Francesco Casanova, Tim Frayling, Phil Gates, Kim Gooding, Andrew Hatttersley, Roland Ling, David Mawson, Robin Shandas, David Strain, Clare Thorn at Peninsula Medical School, Exeter, UK.
Ulf Smith, Ann Hammarstedt, Hans Häring, Oluf Pedersen, Georgio Sesti at University of Gothenburg, Sweden.
Per-Henrik Groop, Emma Fagerholm, Carol Forsblom, Valma Harjutsalo, Maikki Parkkonen, Niina Sandholm, Nina Tolonen, Iiro Toppila, Erkka Valo at Folkhälsan, Helsinki, Finland.
Veikko Salomaa, Aki Havulinna, Kati Kristiansson, Pia Okamo, Tomi Peltola, Markus Perola, Arto Pietilä, Samuli Ripatti, Marketta Taimi at The National Institute for Health and Welfare, Helsinki, Finland.
Seppo Ylä-Herttuala, Mohan Babu, Marike Dijkstra, Erika Gurzeler, Jenni Huusko, Ivana Kholová, Markku Laakso, Mari Merentie, Marja Poikolainen at University of Eastern Finland, Kuopio, Finland.
Mark McCarthy, Chris Groves, Thorhildur Juliusdottir, Fredrik Karpe, Vasiliki Lagou, Andrew Morris, Will Rayner, Neil Robertson, Natalie van Zuydam at University of Oxford, UK.
Claudio Cobelli, Barbara Di Camillo, Francesca Finotello, Francesco Sambo, Gianna Toffolo, Emanuele Trifoglio at University of Padova, Italy.
Riccardo Bellazzi, Nicola Barbarini, Mauro Bucalo, Christiana Larizza, Paolo Magni, Alberto Malovini, Simone Marini, Francesca Mulas, Silvana Quaglini, Lucia Sacchi, Francesca Vitali at University of Pavia, Italy.
Ele Ferrannini, Beatrice Boldrini, Michaela Kozakova, Andrea Mari, Carmela Morizzo, Lucrecia Mota, Andrea Natali, Carlo Palombo, Elena Venturi, Mark Walker at University of Pisa, Italy.
Carlo Patrono, Francesca Pagliaccia, Bianca Rocca at Catholic University of Rome, Italy.
Pirjo Nuutila, Johanna Haukkala, Juhani Knuuti, Anne Roivainen, Antti Saraste at University of Turku, Finland.
Paul McKeague, Norma Brown, Marco Colombo at University of Edinburgh, Scotland.
Birgit Steckel-Hamann, Krister Bokvist, Sudha Shankar, Melissa Thomas at Eli Lilly.
Li-ming Gan, Suvi Heinonen, Ann-Cathrine Jönsson-Rylander, Remi Momo, Volker Schnecke, Robert Unwin, Anna Walentinsson, Carl Whatling at AstraZeneca.
Everson Nogoceke, Gonzalo Durán Pacheco, Ivan Formentini, Thomas Schindler at Roche.
Piero Tortoli, Luca Bassi, Enrico Boni, Alessandro Dallai, Francesco Guidi, Matteo Lenge, Riccardo Matera, Alessandro Ramalli, Stefano Ricci, Jacopo Viti at University of Florence, Italy
Bernd Jablonka, Dan Crowther, Johan Gassenhuber, Sibylle Hess, Thomas Hübschle, Hans-Paul Juretschke, Hartmut Rütten, Thorsten Sadowski, Paulus Wohlfart at Sanofi-aventis.
Julia Brosnan, Valerie Clerin, Eric Fauman, Craig Hyde, Anders Malarstig, Nick Pullen, Mera Tilley, Theresa Tuthill, Ciara Vangjeli, Daniel Ziemek at Pfizer.
Lead authors of the SUMMIT consortium are MM (mark.mccarthy@drl.ox.ac.uk) and LG (leif.groop@med.lu.se).
Data Availability
All summary statistics data are available in the National Bioscience Database Center website at (http://humandbs.biosciencedbc.jp/en/.) The associated Accession number(s)/URL for our data is as follows: hum0014.v12.T2DMwN.v1 and URL: https://humandbs.biosciencedbc.jp/hum0014-v12.
Funding Statement
This work was partly supported by a grant from the Ministry of Education, Culture, Science and Technology, Japan (to SM), and from the Japan Agency of Medical Research and Development (17km0405202h0802) (TK, TY, SM). The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) for the Innovative Medicine Initiative under grant agreement n° 115006, the SUMMIT Consortium.
References
- 1.Saran R, Robinson B, Abbott KC, Agodoa LYC, Bhave N, Bragg-Gresham J, et al. US Renal Data System 2017 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am J Kidney Dis. 2018;71: S1–S672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama H, Kawai K, Kobayashi M; Japan Diabetes Clinical Data Management Study Group. Microalbuminuria is common in Japanese type 2 diabetic patients: a nationwide survey from the Japan Diabetes Clinical Data Management Study Group (JDDM 10). Diabetes Care. 2007; 30: 989–992. 10.2337/dc06-1859 [DOI] [PubMed] [Google Scholar]
- 3.Krolewski AS, Warram JH, Rand LI, Kahn CR. Epidemiologic approach to the etiology of type I diabetes mellitus and its complications. New Engl J Med. 1987; 317: 1390–1398. 10.1056/NEJM198711263172206 [DOI] [PubMed] [Google Scholar]
- 4.Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia. 1996; 39: 940–945. [DOI] [PubMed] [Google Scholar]
- 5.Pettitt DJ, Saad MF, Bennett PH, Nelson RG, Knowler WC. Familial predisposition to renal disease in two generation of Pima Indians with type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia. 1990; 33: 438–443. [DOI] [PubMed] [Google Scholar]
- 6.Fava S, Azzopardi J, Hattersley AT, Watkins PJ. Increased prevalence of proteinuria in diabetic sibs of proteinuric type 2 diabetic subjects. Am J Kidney Dis. 2000; 35: 708–712. [DOI] [PubMed] [Google Scholar]
- 7.Maeda S, Kobayashi MA, Araki S, Babazono T, Freedman BI, Bostrom MA, et al. A single nucleotide polymorphism within the acetyl-coenzyme A carboxylase beta gene in associated with proteinuria in patients with type 2 diabetes. PLoS Genet. 2010; 6: e1000842 10.1371/journal.pgen.1000842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012; 8: e1002921 10.1371/journal.pgen.1002921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandholm N, McKnight AJ, Salem RM, Brennan EP, Forsblom C, Harjutsalo V, et al. Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J Am Soc Nephrol. 2013; 24: 1537–1543. 10.1681/ASN.2012111122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böger CA, Chen MH, Tin A, Olden M, Köttgen A, de Boer IH, et al. CUBN is a gene locus for albuminuria. J Am Soc Nephrol. 2011; 22: 555–570. 10.1681/ASN.2010060598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci USA. 2008;105: 6998–7003. 10.1073/pnas.0800454105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara K, Fujita H, Johnson TA, Yamauchi T, Yasuda K, Horikoshi M, et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet. 2014; 23: 239–246. 10.1093/hmg/ddt399 [DOI] [PubMed] [Google Scholar]
- 13.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998; 15: 539–553. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, Kubo M, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008; 83: 445–456. 10.1016/j.ajhg.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010; 34: 816–834. 10.1002/gepi.20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie B1, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012; 44: 955–959. 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y. A high-throughput SNP typing system for genome-wide association studies. J Hum Genet. 2001; 46: 471–477. 10.1007/s100380170047 [DOI] [PubMed] [Google Scholar]
- 18.Willer CJ1, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genome wide association scan. Bioinformatics. 2010; 26: 2190–2191. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruim RJ1, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010; 26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014; 10: 51–61. 10.1038/nrendo.2013.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, et al. Obesity–associated variants within FTO form long-range functional connections with IRX3. Nature. 2014; 507: 371–375. 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellefroid EJ1, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 1998; 17: 191–203. 10.1093/emboj/17.1.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis MT, Ross S, Strickland PA, Snyder CJ, Daniel CW. Regulated expression patterns of IRX-2, an Iroquois-class homeobox gene, in the human breast. Cell Tissue Res. 1999; 296: 549–554. [DOI] [PubMed] [Google Scholar]
- 24.Stenvinkel P, Zoccali C, Ikizler TA. Obesity in CKD—what should nephrologists know? J Am Soc Nephrol. 2013; 24: 1727–1736. 10.1681/ASN.2013040330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickman C., Kramer H. Obesity and kidney disease: potential mechanisms. Semin Nephrol. 2013; 33: 14–22. 10.1016/j.semnephrol.2012.12.006 [DOI] [PubMed] [Google Scholar]
- 26.D’Agati VD, Chagnac A, de Vries AP, Levi M, Porrini E, Herman-Edelstein M, et al. Obesity-related glomerulopathy: clinical and pathologic characteristics and pathogenesis. Nat Rev Nephrol. 2016; 12: 453–471. 10.1038/nrneph.2016.75 [DOI] [PubMed] [Google Scholar]
- 27.Hsu CY, McCulloch CE, Iribarren C, Darbinian J, Go AS. Body mass index and risk factor for end-stage renal disease. Ann Intern Med. 2006; 144: 21–28. [DOI] [PubMed] [Google Scholar]
- 28.Hubacek JA, Viklicky O, Dlouha D, Bloudickova S, Kubinova R, Peasey A, et al. The FTO gene polymorphism is associated with end-stage renal disease: two large independent case-control studies in a general population. Nephrol Dial Transplant. 2012; 27: 1030–1035. 10.1093/ndt/gfr418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandholm N, Van Zuydam N, Ahlqvist E, Juliusdottir T, Deshmukh HA, Rayner NW, et al. The Genetic Landscape of Renal Complications in Type 1 Diabetes. J Am Soc Nephrol. 2017; 28: 557–574. 10.1681/ASN.2016020231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todd JN, Dahlström EH, Salem RM, Sandholm N, Forsblom C, McKnight AJ, et al. Genetic Evidence for a Causal Role of Obesity in Diabetic Kidney Disease. Diabetes. 2015; 64: 4238–4246. 10.2337/db15-0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Zuydam NR, Ahlqvist E, Sandholm N, Deshmukh H, Rayner NW, Abdalla M, et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects With Type 2 Diabetes. Diabetes. 2018; 67:1414–1427. 10.2337/db17-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A: Stage-1, set-1, B: Stage-1, set-2.
(PDF)
Results of stage-1 GWAS meta-analysis are shown. Red, diamond-shaped plots indicate the most significant variants in each locus after combining stage 1 and stage 2 data. r2, linkage disequilibrium coefficient; chr., chromosome.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
Data Availability Statement
All summary statistics data are available in the National Bioscience Database Center website at (http://humandbs.biosciencedbc.jp/en/.) The associated Accession number(s)/URL for our data is as follows: hum0014.v12.T2DMwN.v1 and URL: https://humandbs.biosciencedbc.jp/hum0014-v12.