ABSTRACT
The matricellular protein SPARC (secreted proteome acidic and rich in cysteine) is known to bind collagens and regulate fibrillogenesis. Cleavage of SPARC at a single peptide bond, increases the affinity for collagens up to 20-fold. To investigate if this specific cleavage has pathological relevance in fibrotic disorders, we developed a competitive ELISA targeting the generated neo-epitope on the released fragment and quantified it in serum from patients with lung cancer, idiopathic pulmonary fibrosis (IPF), chronic obstructive pulmonary disease (COPD) and healthy subjects. Furthermore, the ability of SPARC to protect fibrillar collagens from proteolytic degradation was investigated in vitro, potentially adding a new collagen chaperone function to SPARC. The fragment was significantly elevated in lung cancer patients when compared to healthy subjects measured in a discovery cohort (p = 0.0005) and a validation cohort (p < 0.0001). No significant difference was observed for IPF and COPD patients compared to healthy subjects. When recombinant SPARC was incubated with type I or type III collagen and matrix metalloproteinase-9, collagen degradation was completely inhibited. Together, these data suggest that cleavage of SPARC at a specific site, which modulates collagen binding, is a physiological mechanism increased during pathogenesis of lung cancer. Furthermore, inhibition of fibrillar collagen degradation by SPARC adds a new chaperone function to SPARC which may play additional roles in the contribution to increased collagen deposition leading to a pro-fibrotic and tumorigenic environment.
Keywords: SPARC, lung cancer, serum biomarker, fibrillar collagens, chaperone, the extracellular matrix, collagen deposition
Introduction
Fibrosis is a part of the pathology and/or an end-point in many diseases such as cancer, liver cirrhosis and fibrotic lung disorders. Fibrosis is characterized by an increased deposition of extracellular matrix (ECM), including collagens, which interferes with normal tissue function leading to organ failure. There is a persuasive amount of data showing that ‘secreted proteome acidic and rich in cysteine’ (SPARC), also referred to as osteonectin or basement membrane protein 40 (BM-40) is an important factor for fibrogenesis,1-5 and SPARC expression has been shown to be upregulated in fibrosis and cancer.6–9 SPARC is a 32-kDa matricellular protein known to regulate ECM assembly and deposition, growth factor signaling and interactions between cells and their surrounding ECM.10,11 The expression of SPARC is increased in epithelial/endothelial cells with a high ECM turnover, during abnormal tissue growth associated with neoplasia and during tissue injury and inflammation, highlighting the importance of SPARC in tissue remodeling.12-14
The SPARC protein is divided into three different structural and functional modules. Studies have shown that these modules contains bioactive peptides with different biological functions (Figure 1).15,16,17 For example, small synthetic peptides with sequences derived from module II (follistatin-like domain) are able to regulate proliferation of endothelial cells, stimulate fibroblast proliferation and promote angiogenesis. Module III (extracellular calcium binding domain) contains collagen binding sites and peptide domains that are able to induce MMP production, stimulate angiogenesis and inhibit endothelial cell proliferation. These data suggest that the activity of SPARC is modulated upon cleavage leading to unmasking of domains with biological functions that are distinct from those observed for the native protein. SPARC binds multiple ECM proteins in a calcium-dependent manner within module III, with collagens being the best characterized binding partners. It has been suggested that SPARC acts as an extracellular chaperone due to its many chaperone-like properties. Several studies have shown that SPARC binds different collagens (collagen type I, II, III, IV and V) in the ECM and is important for correct collagen deposition and assembly.18-24 The cleavage of a single peptide bond by metalloproteinases (MMP’s) increases the affinity for collagens up to 20-fold25,26,(Figure 1). Cleavage of SPARC at this specific site has been detected in mouse tissues, suggesting a physiological mechanism of modulating collagen binding.
Figure 1.
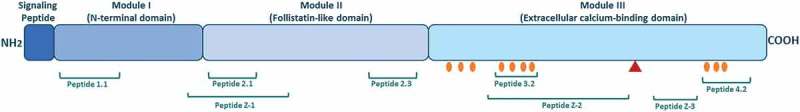
The structure of SPARC and bioactive peptides.
The SPARC protein is divided into three different modules containing bioactive peptides. Peptide 1.1 inhibits spreading of endothelial cells and fibroblasts and potentiates MMP-2 activation. Peptide 2.1 inhibits proliferation of endothelial cells but stimulates proliferation of fibroblasts. Peptide 2.3 stimulates endothelial cell proliferation and angiogenesis. Peptide 3.2 induces MMP production. Peptide 4.2 inhibits cell spreading of endothelial cells and fibroblasts, but stimulates endothelial cell migration. Peptide Z-1 has biphasic effect on endothelial cell proliferation and stimulates vascular growth. Peptides Z-2 and Z-3 inhibit endothelial cell proliferation, but stimulate their migration. Collagen binding sites are shown with orange circles. The red triangle represents the cleavage site associated with increased collagen affinity.
Even though SPARC is considered of importance in collagen processing, oncology and fibrosis, the exact pathological function of the different subparts of the molecule remains to be understood. In the present study, we investigated if MMP-cleavage of SPARC at a specific site known to be involved in increased collagen affinity, has pathological relevance in fibrotic disorders. We developed and validated a competitive enzyme-linked immunosorbent assay (ELISA) quantifying this specific fragment in the circulation. Additionally, we examined if binding of SPARC to fibrillar collagens (type I and III collagen) interfered with their degradation by MMP-9 and MMP-13, proteases known to play important roles in tumor progression.27,28
Results
Specificity of the SPARC-M ELISA assay
The target sequence, LLARDFEKNY, was blasted for homology to other human secreted extracellular matrix proteins using NPS@: Network Protein Sequence Analysis with the UniprotKB/Swiss-prot database. The target sequence was found to be unique to human SPARC when compared to other secreted ECM proteins. Allowing one amino acid mismatch, four secreted extracellular matrix proteins, Von Willebrand factor, glucagon, SPARC-like protein 1 and ADAMTS15, were identified with mismatches at the 6th, 2nd, 3rd and 6th position, respectively (Table 1). There was no reactivity against the sequence of these four peptides (Figure 2A) suggesting high specificity of the antibody for the target sequence. The specificity of the competitive SPARC-M ELISA was further evaluated by analyzing the reactivity towards the calibrator peptide, a non-sense peptide, an elongated peptide, a truncated peptide and using a non-sense biotinylated coating peptide. All peptide sequences are shown in Table 1 and results are shown in Figure 2B. The antibody only reacted with the calibrator peptide and the calibrator peptide clearly inhibited the signal in a dose-dependent manner compared to the other peptides. No detectable signal was observed when using the non-sense biotinylated coating peptide.
Table 1.
Synthetic peptides used for development and validation of the SPARC-M ELISA assay.
| Peptide name | Amino acid sequence |
|---|---|
| Calibrator peptide | LLARDFEKNY |
| Immunogenic peptide | LLARDFEKNY-GGC-KLH |
| Biotinylated coating peptide | LLARDFEKNY-K-biotin |
| Elongated peptide | ELLARDFEKNY |
| Truncated peptide | LARDFEKNY |
| Non-sense calibrator peptide | VPKDLPPDTT |
| Non-sense coating peptide | VPKDLPPDTT-biotin |
| Von Willebrand factor (VWF) | LLARDCQDHS |
| Glucagon (GCG) | LAARDFINWL |
| SPARC-like protein 1 (SLP1) | LLLRDFKKNY |
| ADAMTS15 (A15) | LLARDQCNLH |
Figure 2.
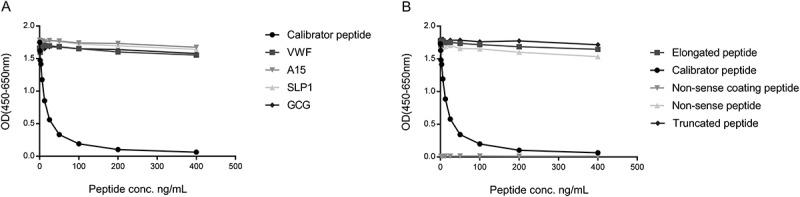
Specificity of the SPARC-M monoclonal antibody.
Monoclonal antibody reactivity towards (A) the calibrator peptide (LLARDFEKNY), the elongated peptide (ELLARDFEKNY), the truncated peptide (LARDFEKNY) a non-sense peptide (VPKDLPPDTT) and a non-sense coating peptide (VPKDLPPDTT-biotin) and (B) Von Willebrand factor (VWF), ADAMTS15 (A15), SPARC-like protein 1 (SLP1) and glucagon (GCG), was tested for in the competitive SPARC-M ELISA. Signals are shown as optical density (OD) at 450 nm (subtracted the background at 650 nm) as a function of peptide concentration.
Together, these data suggest that the selected antibody exhibits high neo-epitope specificity.
Degradation of SPARC by MMP-8 and MMP-13
To further evaluate the specificity of the antibody and to investigate which proteases generate SPARC-M, different gelatinases (MMP-2 and MMP-9) and collagenases (MMP-8 and MMP-13) were incubated with recombinant full-length SPARC. As shown in Figure 3, the collagenases were able to generate the fragment, with MMP-13 giving the highest level of SPARC-M. In contrast, no SPARC-M was detected without the collagenases or when incubated with MMP-9. MMP-2 was able to generate a small amount of SPARC-M as compared to the collagenases.
Figure 3.
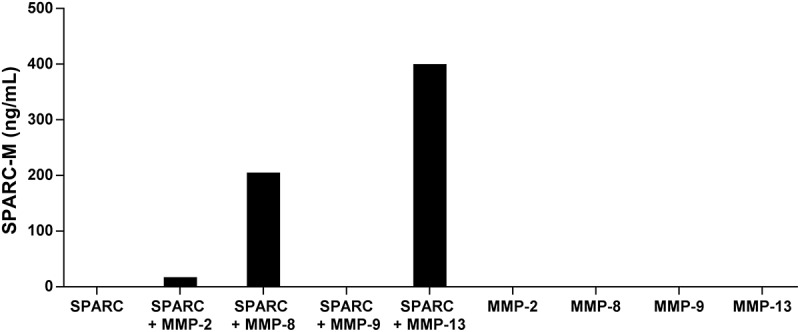
Cleavage of SPARC by MMP-8 and MMP-13.
SPARC was incubated with different MMP’s and SPARC-M levels were measured after 24 hours. Data were normalized by subtracting the background measured in buffer alone. The graph below is representative of two experiments.
These results indicate that the antibody is specific for the cleavage site and that collagenases compared to gelatinases have a higher preference for SPARC at this specific site.
Technical evaluation of the SPARC-M ELISA
The technical performance of the SPARC-M ELISA was further evaluated according to inter – and intra-assay variation, linearity, lower limit of detection, upper limit of detection, analyte stability (freeze/thaw and storage) and interference. The results from the different validation steps and SPARC-M performance are summarized in Table 3. The measuring range (LLOD to ULOD) of the assay was determined to 2.7–300.7 ng/mL. The intra- and inter-assay variation was 6% and 10%, respectively. The acceptance criterion was below 10% for the intra-assay variation and below 15% for the inter-assay variation and therefore acceptable. To obtain linearity, human serum needed to be diluted 1:4. The mean dilution recovery for human serum was 96% calculated with 1:4 pre-diluted samples as references. The analyte stability was analyzed according to freeze/thaw cycles and storage stability at 4°C and 20°C with an acceptance criterion of the recovery within 100% ± 20%. The analyte recovery in serum was 92% after 4 freeze/thaw cycles. After storage at 4°C for 48 hours the recovery was 84%. Analyte stability was also tested at 20°C for 4, 24 and 48 hours. The recovery after 4 hours was 88%. However after 24 hours the analyte could not be recovered within the acceptance range (50% recovery). These data indicate that the analyte in serum is stable at 4°C up to 48 hours, however upon analysis serum samples should not be stored above 20°C for more than four hours. No interference was detected from either low or high contents of biotin, lipids or hemoglobin with recoveries ranging from 80–98%. The acceptance criterion was a recovery within 100% ± 20%.
Table 3.
Technical validation data of the SPARC-M ELISA assay.
| Tecnical validation step | SPARC-M performance |
|---|---|
| Detection range (LLOD-ULOD) | 2.7–300.7 ng/mL |
| Intra-assay variation | 6% |
| Inter-assay variation | 10% |
| Dilution of serum samples | 1:4 |
| Dilution recovery (1:4 pre-dilution) | 96% (77–102%) |
| Freeze/thaw recovery (4 cycles) | 92% (86–103%) |
| Analyte stability up to 48 h, 4°C and 4 h, 20°C | 88% (84–96%) |
| Interference Lipids, low/high | 96%/97% |
| Interference Biotin, low/high | 96%/98% |
| Interference Hemoglobin, low/high | 96%/80% |
Percentages are reported as mean with range shown in brackets
Clinical evaluation of SPARC-M
To investigate whether SPARC-M had clinical disease relevance and biomarker potential, SPARC-M was measured in patients with different fibrotic lung disorders and healthy controls. The discovery cohort (cohort 1) consisted of patients with lung cancer, IPF, COPD and healthy controls (Table 2). As shown in Figure 4A, SPARC-M was significantly elevated in lung cancer patients compared to healthy controls (p = 0.0005) and COPD patients (p = 0.0003). IPF patients also had an increased level of SPARC-M compared to healthy controls although not significant (p = 0.66). To validate the findings in lung cancer patients, SPARC-M was measured in a validation cohort (cohort 2) including 40 lung cancer patients and 20 healthy controls (Table 2). A significant increase in SPARC-M in lung cancer patients as compared to healthy controls was observed in this cohort as well (p < 0.0001) (Figure 4B).
Table 2.
Clinical sample overview and patients demographics.
| Cohort | Samples | No. of subjects | Mean age (range) | Gender, % females | Mean BMI (range) | Tumor stage I | Tumor stage II | Tumor stage III | Tumor stage IV |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Lung cancer patients | 8 | 61 (47–77) | 13 | - | - | - | - | - |
| 1 | IPF patients | 7 | 73 (55–81) | 57 | - | - | - | - | - |
| 1 | COPD patients | 8 | 75 (69–82) | 50 | - | - | - | - | - |
| 1 | Healthy controls | 6 | 55 (44–65) | 83 | - | - | - | - | - |
| 2 | Lung cancer patients | 40 | 62 (55–66) | 50 | 25 (16–35) | 10 | 10 | 10 | 10 |
| 2 | Healthy controls | 20 | 62 (60–65) | 50 | 26 (22–32) | - | - | - | - |
Figure 4.
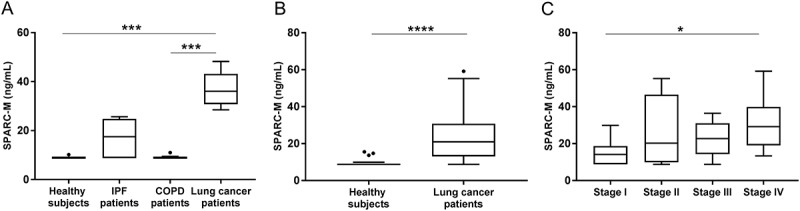
Serum SPARC-M levels in patients with fibrotic disorders and healthy controls.
(A) Cohort 1: Serum SPARC-M was assessed in healthy controls (n = 6), IPF patients (n = 7), COPD patients (n = 8) and lung cancer patients (n = 8). Groups were compared using Kruskal-Wallis adjusted for Dunn’s multiple comparisons test. (B) Cohort 2: Serum SPARC-M was assessed in healthy controls (n = 20) and lung cancer patients (n = 40). Groups were compared using unpaired, two-tailed Mann-Whitney test. (C) Lung cancer patients (from cohort 2) were stratified according to their cancer stage (stage I-IV, n = 10 in each group). Data were compared using Kruskal-Wallis adjusted for Dunn’s multiple comparisons test. All Data are shown as Tukey box plots. Significance level: *: p < 0.05, ***: p < 0.001, ****: p < 0.0001.
The area under the receiver operating characteristics (AUROC) was used to evaluate the discriminative power of SPARC-M in relation to lung cancer patients and healthy controls (cohort 2). SPARC-M was able to discriminate between patients and healthy controls with an AUROC of 0.87 (95%CI: 0.78–0.96).
To examine if the level of SPARC-M was different in patients with metastasis (high tumor burden) compared to patients with localized tumors, patients from cohort 2 were stratified according to their tumor stage (stage I-IV). A significantly higher level of SPARC-M was found in metastatic patients (stage IV) compared to stage I patients (Figure 4C). Moreover, the discriminative accuracy increased with tumor stage with an AUC of 0.71 for stage I, an AUC of 0.87 for stage II, an AUC of 0.91 for stage III and an AUC of 0.99 for stage IV.
Together, these data demonstrate that the investigated cleavage site, which modulates collagen binding and measured by SPARC-M, is a physiological mechanism that is increased during progression and invasion of lung cancer.
Inhibition of fibrillar collagen degradation by SPARC
To investigate if the binding of SPARC to collagens interfered with and inhibited fibrillar collagen degradation, type I collagen or type III collagen was incubated together with MMP-9 alone or together with MMP-9 and SPARC. The degradation of collagens was measured by ELISAs measuring type I collagen degradation by MMP-9 and MMP-13 (C1M) and type III collagen degradation by MMP-9 (C3M). As shown in Figure 5, MMP-9, degraded both collagens in a time-dependent manner illustrated by an increase in C1M (Figure 5A) and C3M (Figure 5B) concentration. The addition of SPARC to collagen completely inhibited both type I and type III collagen degradation by MMP-9.
Figure 5.
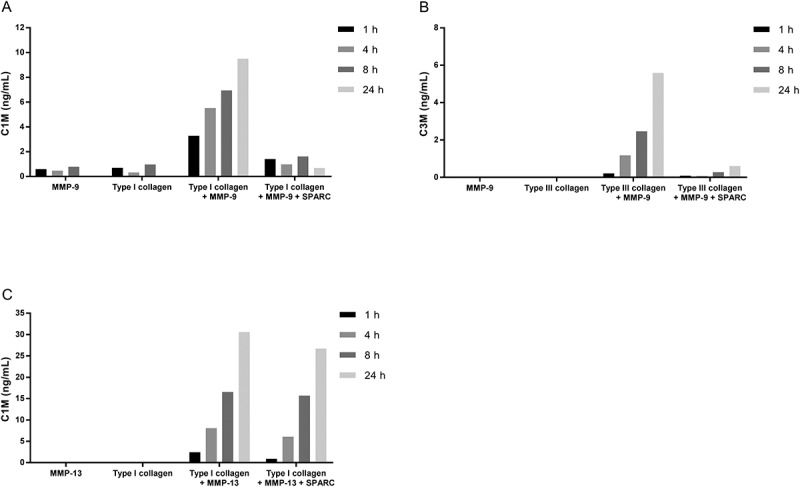
SPARC inhibits fibrillar collagen degradation by MMP-9.
(A) Type I collagen or (B) type III collagen was incubated with MMP-9 alone or together with MMP-9 and SPARC. (C) Type I collagen was incubated with MMP-13 alone or together with MMP-13 and SPARC. The solutions incubated at 37°C for 1 h, 4 h, 8 h and 24 h. The reaction was stopped by adding 1 µM EDTA to the solutions. Collagen degradation was measured with ELISAs targeting MMP-9 and MMP-13 degraded type I collagen, C1M (A)(C) and MMP-9 degraded type III collagen, C3M (B). MMP-buffer with either MMP’s or collagen alone were included as negative controls. Data were normalized by subtracting the background measured in buffer alone. The graphs below are representative of two experiments.
To examine if SPARC also had a protective function in collagenase-mediated degradation of collagens, type I collagen and MMP-13 was incubated with or without SPARC and degradation was measured by C1M. Interestingly, no change in type I collagen degradation was observed by the addition of SPARC (Figure 5C).
These data suggest a new chaperone function of SPARC, i.e. protecting fibrillar collagens from degradation by gelatinases but not by collagenases.
Discussion
The present study validates a new serum biomarker reflecting increased collagen binding by SPARC and demonstrates a new collagen chaperone function of SPARC. The main findings of this study were: 1) the investigated fragment, SPARC-M, was detectable in serum and significantly elevated in lung cancer patients compared to healthy controls, 2) the SPARC-M ELISA was technically robust and specific towards a MMP-degraded fragment of SPARC and 3) SPARC was able to inhibit MMP-9-mediated degradation of fibrillar collagens. To our knowledge, this is the first biological validation of this specific fragment in human serum and the first study to show that SPARC acts by preventing collagen degradation.
Studies have shown that the collagen binding function of SPARC can be modulated by extracellular proteolytic processing.25,26,29 We found that MMP-8 and MMP-13 had preference for the investigated cleavage site compared to MMP-2 and MMP-9. The fact that some MMPs show preference for this site over others, suggest a way for the stroma to regulate collagen binding to SPARC and thereby fibril formation. Previous studies using SDS-gel electrophoresis are in concordance with our findings, showing that MMP-13 is able to cleave SPARC at this site.25,29 However, Sasaki et al.25 demonstrated cleavage by MMP-9 and MMP-2, although to a lesser extent than MMP-13. As shown in Figure 3, MMP-2 is able to generate a small amount of the fragment whereas MMP-9 is negative. The discrepancy between our data and the data presented by Sasaki et al. with MMP-9 might be due to different detection methods (ELISA vs. SDS-gel electrophoresis) and warrants further investigations.
The investigated cleavage site of SPARC has been shown to be present in mouse tissue quantified by immunohistochemistry using polyclonal antibodies against the cleavage site,26 however this is the first time this cleavage is demonstrated in humans. SPARC-M was significantly elevated in patients with lung cancer compared to healthy controls. An increase of SPARC-M was also observed in IPF patients, although it was found not to be significantly elevated. We hypothesize that the SPARC-M fragment is released to the circulation upon MMP-cleavage and here represents a surrogate measure of the bioactive part of SPARC which is retained within the matrix, and have increased collagen affinity. Interestingly, SPARC itself has been shown to increase the expression of MMP’s in fibroblasts 30–32 causing a positive feedback loop with MMP-cleavage of SPARC which may, if uncontrolled, be involved in the pathology of ECM remodeling diseases with increased collagen deposition, such as lung cancer and IPF. The fact that patients with IPF and lung cancer, and not COPD, had elevated levels of SPARC-M, supports this hypothesis. In accordance with our findings, several studies have shown an increased expression of SPARC in cancer and fibrosis. 6-9 As SPARC-M was elevated in stage IV patients and the discriminative power increased with tumor stage support that this cleavage is in fact a pathological mechanism in lung cancer that increases with tumor burden. These results indicate a prognostic value of SPARC-M, although further studies are needed to evaluate this.
The limitations of the present clinical studies are the relatively small population sizes and limited clinical information. However, as we could confirm the findings in two independent cohorts, increases their validity. Larger longitudinal studies are needed to validate the potential of SPARC-M as a biomarker in fibrotic lung diseases.
This study also demonstrates a new collagen chaperone function of SPARC. In general, the chaperone function of SPARC has been linked to its ability to inhibit thermal aggregation of alcohol dehydrogenase in a concentration-dependent manner33 and its importance for correct collagen deposition and assembly.18-24 Here, we show that SPARC is able to interfere with the degradation of fibrillar collagens by MMP-9 but not MMP-13. These findings may indicate that SPARC plays a chaperone role in maintaining a collagen structure that does not enable gelatinolytic (MMP-9) processing, but collagenolytic (MMP-13) processing. How this translates to physiological conditions remains to be established.
The observed collagen chaperone function of SPARC could be involved in the pathogenesis of fibrotic disorders by contributing to increased collagen deposition. We hypothesize that stress, such as malignant transformation or tissue injury, causes activation of fibroblast and increased SPARC expression which induces MMP expression resulting in a positive feed-back mechanism with cleavage of SPARC by MMP’s. Cleavage at this specific site will enhance binding of SPARC to collagens, preventing collagen degradation by MMP’s. This will result in increased collagen deposition and thereby play a role in fibrogenesis and tumorigenesis.
In summary, we have shown that SPARC is able to inhibit degradation of fibrillar collagens and that cleavage of SPARC at a specific site, known to modulate collagen binding, is a pathological mechanism in lung cancer. Whether this is a cause or consequence of lung cancer needs further investigation.
Materials and methods
Development of SPARC-M (SPARC degraded by mmp’s) ELISA
Selection of peptides
The selection of target peptide for ELISA development was based on the following cleavage site (↓) on SPARC previously identified by Edman degradation and published by Sasaki et al.:25 211HPVE ↓ LLARDFEKNYNMYIFP230.
To generate an antibody specific for the N-terminal of the cleavage fragment, a sequence of 10 amino acids adjacent to the site was chosen as the target: ↓215LLARDFEKNY224. The sequence was blasted for homology to other human secreted extracellular matrix proteins using NPS@: Network Protein Sequence Analysis with the UniprotKB/Swiss-prot database 34.
Synthetic peptides used for monoclonal antibody production and validation of the ELISA were purchased from Genscript and shown in Table 1. The target sequence was used as the calibrator peptide (LLARDFEKNY). A biotinylated peptide (LLARDFEKNY-K-biotin) was included as a coating peptide with addition of a lysine residue to the C-terminal end to ensure biotin linking. The specificity of the antibody was tested by including an elongated calibrator peptide with an additional amino acid added to the N-terminal of the target peptide sequence (ELLARDFEKNY), a truncated calibrator peptide with a removal of the first N-terminal amino acid (LARDFEKNY) as well as a non-sense calibrator peptide (VPKDLPPDTT) and a non-sense biotinylated coating peptide (VPKDLPPDTT-biotin) in the assay validation. To screen for any potential cross-reactivity to other ECM proteins and further test the antibody specificity, four peptides (derived from Von Willebrand factor, glucagon, SPARC-like protein 1 and ADAMTS15) with one amino acid mismatch compared to the first six amino acids in the target sequence were also included (Table 1). The immunogenic peptide (LLARDFEKNY-GGC-KLH) was generated by covalently cross-linking the standard peptide to Keyhole Limpet Hemocyanin (KLH) carrier protein using Succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate, SMCC (Thermo Scientific, cat.no. 22336). Glycine and cysteine residues were added at the C-terminal end to ensure right linking of the carrier protein.
Monoclonal antibody production
Six week old Balb/C mice were immunized by subcutaneous injection of 200 µL emulsified antigen containing 100 µg immunogenic peptide (LLARDFEKNY-GGC-KLH) mixed with Stimune Immunogenic Adjuvant (Thermo fisher, cat.no. 7925000). Consecutive immunizations were performed at 2-week intervals until stable sera titer levels were reached. The mouse with the highest titer rested for four weeks and was then boosted with 100 µg immunogenic peptide in 100 µL 0.9% NaCl solution intravenously. Hybridoma cells were produced by fusing spleen cells with SP2/0 myeloma cells as previously described.35 The resultant hybridoma cells were then cultured in 96-well microtiter plates and standard limited dilution was used to secure monoclonal growth.
Clone characterization
The reactivity of the monoclonal antibody from different clones was evaluated by displacement using human serum samples and the calibrator peptide (LLARDFEKNY) in a preliminary ELISA using 10 ng/mL biotinylated coating peptide on streptavidin-coated microtiter plates (Roche, cat. no. 11940279) and the supernatant from the antibody producing monoclonal hybridoma cells. The clone with the best reactivity towards the calibrator peptide was purified using protein-G-columns according to the manufacturer’s instructions (GE Healthcare Life Sciences, cat. no. 17–0404-01).
SPARC-M ELISA protocol
Optimal incubation buffer, -time and -temperature, as well as the optimal concentrations of antibody and coating peptide were determined and the finalized SPARC-M competitive ELISA protocol was as follows:
A 96-well streptavidin-coated microtiter plate was coated with 1.1 ng/mL biotinylated coating peptide dissolved in assay buffer (50 mM Tris-BTB, 4 g/L NaCl, pH 8.0) and incubated for 30 min. at 20°C with shaking (300 rpm) in darkness shaking. Twenty µL calibrator peptide or pre-diluted serum (1:4) were added to appropriate wells, followed by the addition of 100 µL monoclonal antibody dissolved in assay buffer to a concentration of 14 ng/mL per well and incubated 1 hour at 20°C in darkness with shaking (300 rpm). One hundred µL of goat anti-mouse horse-radish peroxidase (POD)-conjugated IgG antibody (Thermo Scientific, cat. no. 31437) diluted 1:6000 in assay buffer was added to each well and incubated 1 hour at 20°C in darkness with shaking. All incubation steps were followed by five washes in washing buffer (20 mM Tris, 50 mM NaCl, pH 7.2). Finally, 100 µL tetramethylbenzidine (TMB) (Kem-En-Tec Diagnostics, cat. no. 438OH) was added to each well and the plate was incubated for 15 minutes at 20°C in darkness with shaking. The enzymatic reaction was stopped by adding 0.18 M H2SO4 and absorbance was measured at 450 nm with 650 nm as reference. A calibration curve was plotted using a 4-parameter logistic curve fit. Data were analyzed using the SoftMax Pro v.6.3 software.
Technical evaluation of the SPARC-M ELISA
To evaluate the technical performance of the SPARC-M ELISA, the following validation tests were carried out: Inter- and intra-assay variation, linearity, lower limit of detection, upper limit of detection, analyte stability (freeze/thaw and storage) and interference.
The inter- and intra-assay variation was determined by ten independent runs on different days using seven quality control samples covering the detection range, with each run consisting of double-determinations of the samples. The seven quality control samples consisted of: two human serum samples and five samples with standard peptide in buffer. Intra-assay variation was calculated as the mean coefficient of variance (CV%) within plates and the inter-assay variation was calculated as the mean CV% between the ten individual runs analyzed on different days. To assess linearity of the assay, two-fold dilutions of human serum samples were performed and dilution linearity was calculated as a percentage of recovery of the un-diluted sample. The lower limit of detection (LLOD) was determined from 21 measurements using assay buffer as sample and was calculated as the mean + three standard deviations. The upper limit of detection (ULOD) was determined from ten independent runs of the highest standard peptide concentration and was calculated as the mean back-calibration calculation + three standard deviations. Analyte stability was first determined by the effect of repeated freeze/thaw of serum samples by measuring the SPARC-M level in three human serum samples in four freeze/thaw cycles. The freeze/thaw recovery was calculated with the first cycle as reference. Second, analyte stability in relation to storage was determined by a 48 hour study performed at 4°C or 20°C. The SPARC-M level in three human serum samples was measured after 0 h, 4 h, 24 h and 48 h of storage, and recovery was calculated with samples stored at −20°C as reference. Interference was determined by adding a low/high content of hemoglobin (0.155/0.310 mM), lipemia/lipids (4.83/10.98 mM) and biotin (30/90 ng/mL) to a serum sample of known concentration. Recovery percentage was calculated with the serum sample as reference.
Cleavage of SPARC in vitro
Recombinant human SPARC (PeproTech, cat. no. 120–36) was reconstituted to a final concentration of 1000 ug/mL in MMP-buffer (50 mM Tris-HCl, 150 mM NaCl, 10 mM CaCl2, 10uM ZnCl, 0.05%Brij35, pH 7.5). MMP-2, MMP-8, MMP-9 and MMP-13 (Giotto, cat. no. G04MP02C, G04MP08C, G04MP09C, G04MP13C) were added 1:10 (1 µg MMP and 10 µg SPARC). Digestion of carboxymethylated transferrin (a natural substrate of MMP’s) was included as a positive control. The solutions incubated at 37°C for 24 h. The reaction was stopped by adding 1 µM EDTA to the solutions. MMP-buffer added the different proteases alone were included as negative controls. Samples were stored at −80°C until analysis. The activity of the proteases was confirmed by silverstaining according to the manufacturer’s instructions (SilverXpress®, Invitrogen, cat. no. LC6100) and coomassie blue (data not shown).
Clinical validation of SPARC-M
Patient serum samples were obtained from the commercial vendor ProteoGenex. The discovery cohort (cohort 1) consisted of patients with lung cancer, idiopathic pulmonary fibrosis (IPF), chronic obstructive pulmonary disease (COPD) and healthy colonoscopy-negative controls with no symptomatic or chronic disease (Table 2). The validation cohort (cohort 2) included 40 patients with different stages of lung cancer, and 20 age- and gender-matched healthy colonoscopy-negative controls with no symptomatic or chronic disease (Table 2). Appropriate Institutional Review Board/Independent Ethical Committee approved sample collection and all subjects filed informed consent.
Effect of SPARC on fibrillar collagen degradation
Recombinant human SPARC (PeproTech, cat. no. 120–36) was reconstituted to a final concentration of 1000 ug/mL in MMP-buffer. Natural human type I collagen (Abcam, cat. no. ab7533) and type III collagen (Abcam, cat. no. ab7535) was dialyzed for 2 days to remove the acetic acid, against MMP buffer using Slide-A-Lyzer™ Dialysis Cassettes, 3.5 K MWCO, 0.5 mL (Thermofisher, cat. no. 66333) according to the manufacturer’s instructions. The collagens were either incubated with MMP-9 (Giotto, Firenze, cat. no. G04MP09C) alone (MMP:collagen ratio of 1:17) or together with MMP-9 and SPARC (collagen:SPARC molar ratio of 1:10). In addition type I collagen was also incubated with MMP-13 (Giotto, cat. no. G04MP13C) with or without SPARC. The solutions incubated at 37°C for 1 h, 4 h, 8 h and 24 h. The reaction was stopped by adding 1 µM EDTA. MMP-buffer with either collagen or MMP’s alone were included as negative controls. Digestion of carboxymethylated transferrin (a natural substrate of MMP’s) was included as a positive control and this reaction was stopped after 24 h. Samples were stored at −80°C until analysis. MMP-9 and −13 mediated degradation of type I collagen was investigated by an ELISA measuring type I collagen degradation (C1M) (Nordic Bioscience) and type III collagen was investigated by an ELISA measuring MMP-9 mediated degradation of type III collagen (C3M) (Nordic Bioscience). The C1M analyte has previously been shown to be generated by MMP-9 and MMP-13, and the C3M analyte by MMP-9, and the assays do not react to non-cleaved collagen36,37. The activity of the MMP’s was confirmed by Coomassie blue staining (data not shown).
Statistical analysis
The level of SPARC-M in serum samples was compared using unpaired, two-tailed Mann-Whitney test or Kruskal-Wallis adjusted for Dunn’s multiple comparisons test. Patients in cohort 2 were stratified according to their tumor stage and the level of SPARC-M in each group was compared using Kruskal-Wallis adjusted for Dunn’s multiple comparisons test. The discriminative power was investigated by the area under the receiver operating characteristics (AUROC) comparing patients with lung cancer and healthy controls. Graph design and statistical analyses were performed using GraphPad Prism version 7 (GraphPad Software, Inc.).
Funding Statement
This work was supported by The Danish Research Foundation (Den Danske Forskningsfond) under Grant number [20130067];
Acknowledgments
We acknowledge the Danish Research Foundation (“Den Danske Forskningsfond”).
Disclosure statement
T. Manon-Jensen, S. Sun, D.J. Leeming, M.A. Karsdal and N. Willumsen are employed at Nordic Bioscience A/S which is a company involved in discovery and development of biochemical biomarkers. M.A. Karsdal owns stocks at Nordic Bioscience. S.N. Kehlet and S. Brix reports no conflict of interest.
References
- 1.Sangaletti S, Tripodo C, Cappetti B, Casalini P, Chiodoni C, Piconese S, Santangelo A, Parenza M, Arioli I, Miotti S, et al. 2011. SPARC oppositely regulates inflammation and fibrosis in bleomycin-induced lung damage. Am J Pathol. 179:3000–3010. doi: 10.1016/j.ajpath.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strandjord TP, Madtes DK, Weiss DJ, Sage EH.. Collagen accumulation is decreased in SPARC-null mice with bleomycin-induced pulmonary fibrosis. Am J Physiol. 277;1999:L628–35. [DOI] [PubMed] [Google Scholar]
- 3.Wang J-C, Lai S, Guo X, Zhang X, De Crombrugghe B, Sonnylal S, Arnett FC, Zhou X. 2010. Attenuation of fibrosis in vitro and in vivo with SPARC siRNA. Arthritis Res Ther. 12:1–9. doi: 10.1186/ar2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pichler RH, Hugo C, Shankland SJ, Reed MJ, Bassuk JA, Andoh TF, Lombardi DM, Schwartz SM, Bennett WM, Alpers CE, et al. 1996. SPARC is expressed in renal interstitial fibrosis and in renal vascular injury. Kidney Int. 50:1978–1989. doi: 10.1038/ki.1996.520. [DOI] [PubMed] [Google Scholar]
- 5.Camino AM, Atorrasagasti C, Maccio D, Prada F, Salvatierra E, Rizzo M, Alaniz L, Aquino JB, Podhajcer OL, Silva M, et al. 2008. Adenovirus-mediated inhibition of SPARC attenuates liver fibrosis in rats. J Gene Med. 10:993–1004. doi: 10.1002/jgm.v10:9. [DOI] [PubMed] [Google Scholar]
- 6.Neuzillet C, Tijeras-Raballand A, Cros J, Faivre S, Hammel P, Raymond E Stromal expression of SPARC in pancreatic adenocarcinoma. Cancer and Metastasis Rev. 2013;32:585–602. doi: 10.1007/s10555-012-9414-4. [DOI] [PubMed] [Google Scholar]
- 7.Wong SLI, Sukkar MB. 2017. The SPARC protein: an overview of its role in lung cancer and pulmonary fibrosis and its potential role in chronic airways disease. Br J Pharmacol. 174:3–14. doi: 10.1111/bph.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frizell E, Liu SL, Abraham A, Ozaki I, Eghbali M, Sage EH, Zern MA Expression of SPARC in normal and fibrotic livers. Hepatology. 1995;21:847–854. [PubMed] [Google Scholar]
- 9.Kuhn C, Mason RJ. Immunolocalization of SPARC, tenascin, and thrombospondin in pulmonary fibrosis. Am J Pathol. 147;1995:1759–1769. [PMC free article] [PubMed] [Google Scholar]
- 10.Lane TF, Sage EH. 1994. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 8:163–173. doi: 10.1096/fasebj.8.2.8119487. [DOI] [PubMed] [Google Scholar]
- 11.Bradshaw AD. 2009. The role of SPARC in extracellular matrix assembly. J Cell Commun Signal. 3:239–246. doi: 10.1007/s12079-009-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix. Matrix Biol. 19;2000:569–580. [DOI] [PubMed] [Google Scholar]
- 13.Chiodoni C, Colombo MP, Sangaletti S. 2010. Matricellular proteins: from homeostasis to inflammation, cancer, and metastasis. Cancer and Metastasis Rev. 29:295–307. doi: 10.1007/s10555-010-9221-8. [DOI] [PubMed] [Google Scholar]
- 14.Yan Q, Sage EH. 1999. SPARC, a Matricellular Glycoprotein with Important Biological Functions. J Histochem Cytochem. 47:1495–1505. doi: 10.1177/002215549904701201. [DOI] [PubMed] [Google Scholar]
- 15.Ribeiro N, Sousa SR, Brekken RA, Monteiro FJ. 2014. Role of sparc in bone remodeling and cancer-related bone metastasis. J Cell Biochem. 115:17–26. doi: 10.1002/jcb.v115.1. [DOI] [PubMed] [Google Scholar]
- 16.Tai IT, Tang MJ. 2008. SPARC in cancer biology: its role in cancer progression and potential for therapy. Drug Resist Updat. 11:231–246. doi: 10.1016/j.drup.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Chlenski A, Cohn SL. 2010. Modulation of matrix remodeling by SPARC in neoplastic progression. Semin Cell Dev Biol. 21:55–65. doi: 10.1016/j.semcdb.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Rosset EM, Bradshaw AD. 2016. SPARC/osteonectin in mineralized tissue. Matrix Biol. 52–54:78–87. doi: 10.1016/j.matbio.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradshaw AD, Baicu CF, Rentz TJ, Van Laer AO, Boggs J, Lacy JM, Zile MR. 2009. Pressure overload-induced alterations in fibrillar collagen content and myocardial diastolic function: role of secreted protein acidic and rich in cysteine (SPARC) in post-synthetic procollagen processing. Circulation. 119:269–280. doi: 10.1161/CIRCULATIONAHA.108.773424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradshaw AD, Puolakkainen P, Wight TN, Helene Sage E, Dasgupta J, Davidson JM. 2003. SPARC-null mice display abnormalities in the dermis characterized by decreased collagen fibril diameter and reduced tensile strength. J Invest Dermatol. 120:949–955. doi: 10.1046/j.1523-1747.2003.12241.x. [DOI] [PubMed] [Google Scholar]
- 21.Delany AM, Amling M, Priemel M, Howe C, Baron R, Canalis E. 2000. Osteopenia and decreased bone formation in osteonectin-deficient mice. J Clin Invest. 105:915–923. doi: 10.1172/JCI7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentz TJ, Poobalarahi F, Bornstein P, Sage EH, Bradshaw AD. 2007. SPARC regulates processing of procollagen I and collagen fibrillogenesis in dermal fibroblasts. J Biol Chem. 282:22062–22071. doi: 10.1074/jbc.M700167200. [DOI] [PubMed] [Google Scholar]
- 23.Trombetta-eSilva J. 2012. The Function of SPARC as a Mediator of Fibrosis. Open Rheumatol J. 6:146–155. doi: 10.2174/1874312901206010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trombetta JM, Bradshaw AD, Johnson RH. 2010. SPARC/Osteonectin Functions to Maintain Homeostasis of the Collagenous Extracellular Matrix in the Periodontal Ligament. J Histochem Cytochem. 58:871–879. doi: 10.1369/jhc.2009.954354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki T, Göhring W, Mann K, Maurer P, Hohenester E, Knäuper V, Murphy G, Timpl R. 1997. Limited cleavage of extracellular matrix protein BM-40 by matrix metalloproteinases increases its affinity for collagens. J Biol Chem. 272:9237–9243. doi: 10.1074/jbc.272.14.9237. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki T, Miosge N, Timpl R. 1999. Immunochemical and tissue analysis of protease generated neoepitopes of BM-40 (osteonectin, SPARC) which are correlated to a higher affinity binding to collagens. Matrix Biol. 18:499–508. doi: 10.1016/S0945-053X(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 27.Brinckerhoff CE, Rutter JL, Benbow U. Interstitial collagenases as markers of tumor progression. Clin Cancer Res. 6;2000:4823–4830. [PubMed] [Google Scholar]
- 28.Orlichenko LS, Radisky DC. 2008. Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clin Exp Metastasis. 25:593–600. doi: 10.1007/s10585-008-9143-9. [DOI] [PubMed] [Google Scholar]
- 29.Maurer P, Göhring W, Sasaki T, Mann K, Timpl R, Nischt R Recombinant and tissue-derived mouse BM-40 bind to several collagen types and have increased affinities after proteolytic activation. Cell Mol Life Sci. 1997;53:478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilles C, Bassuk JA, Pulyaeva H, Sage EH, Foidart JM, Thompson EW SPARC/osteonectin induces matrix metalloproteinase 2 activation in human breast cancer cell lines. Cancer Res. 1998;58:5529–5536. [PubMed] [Google Scholar]
- 31.Jacob K, Webber M, Benayahu D, Kleinman HK. Osteonectin promotes prostate cancer cell migration and invasion: A possible mechanism for metastasis to bone. Cancer Res. 59;1999:4453–4457. [PubMed] [Google Scholar]
- 32.Tremble PM, Lane TF, Sage EH, Werb Z. 1993. SPARC, a secreted protein associated with morphogenesis and tissue remodeling, induces expression of metalloproteinases in fibroblasts through a novel extracellular matrix-dependent pathway. J Cell Biol. 121:1433–1444. doi: 10.1083/jcb.121.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emerson RO, Sage EH, Ghosh JG, Clark JI. 2006. Chaperone-like activity revealed in the matricellular protein SPARC. J Cell Biochem. 98:701–705. doi: 10.1002/(ISSN)1097-4644. [DOI] [PubMed] [Google Scholar]
- 34.Combet C, Blanchet C, Geourjon C, Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem Sci. 25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 35.Gefter ML, Margulies DH, Scharff MD. 1977. A simple method for polyethylene glycol-promoted hybridization of mouse myeloma cells. Somatic Cell Genet. 3:231–236. doi: 10.1007/BF01551818. [DOI] [PubMed] [Google Scholar]
- 36.Barascuk N, Veidal SS, Larsen L, Larsen DV, Larsen MR, Wang J, Zheng Q, Xing R, Cao Y, Rasmussen LM, et al. 2010. A novel assay for extracellular matrix remodeling associated with liver fibrosis: an enzyme-linked immunosorbent assay (ELISA) for a MMP-9 proteolytically revealed neo-epitope of type III collagen. Clin Biochem. 43:899–904. doi: 10.1016/j.clinbiochem.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Leeming D, He Y, Veidal SS, Nguyen Q, Larsen DV, Koizumi M, Segovia-Silvestre T, Zhang C, Zheng Q, Sun S, et al. 2011. A novel marker for assessment of liver matrix remodeling: an enzyme-linked immunosorbent assay (ELISA) detecting a MMP generated type I collagen neo-epitope (C1M). Biomarkers. 16:616–628. doi: 10.3109/1354750X.2011.620628. [DOI] [PubMed] [Google Scholar]


