Abstract
Polycystic ovary syndrome (PCOS) is a disorder characterized by hyperandrogenism, ovulatory dysfunction and polycystic ovarian morphology. Affected women frequently have metabolic disturbances including insulin resistance and dysregulation of glucose homeostasis. PCOS is diagnosed with two different sets of diagnostic criteria, resulting in a phenotypic spectrum of PCOS cases. The genetic similarities between cases diagnosed based on the two criteria have been largely unknown. Previous studies in Chinese and European subjects have identified 16 loci associated with risk of PCOS. We report a fixed-effect, inverse-weighted-variance meta-analysis from 10,074 PCOS cases and 103,164 controls of European ancestry and characterisation of PCOS related traits. We identified 3 novel loci (near PLGRKT, ZBTB16 and MAPRE1), and provide replication of 11 previously reported loci. Only one locus differed significantly in its association by diagnostic criteria; otherwise the genetic architecture was similar between PCOS diagnosed by self-report and PCOS diagnosed by NIH or non-NIH Rotterdam criteria across common variants at 13 loci. Identified variants were associated with hyperandrogenism, gonadotropin regulation and testosterone levels in affected women. Linkage disequilibrium score regression analysis revealed genetic correlations with obesity, fasting insulin, type 2 diabetes, lipid levels and coronary artery disease, indicating shared genetic architecture between metabolic traits and PCOS. Mendelian randomization analyses suggested variants associated with body mass index, fasting insulin, menopause timing, depression and male-pattern balding play a causal role in PCOS. The data thus demonstrate 3 novel loci associated with PCOS and similar genetic architecture for all diagnostic criteria. The data also provide the first genetic evidence for a male phenotype for PCOS and a causal link to depression, a previously hypothesized comorbid disease. Thus, the genetics provide a comprehensive view of PCOS that encompasses multiple diagnostic criteria, gender, reproductive potential and mental health.
Author summary
We performed an international meta-analysis of genome-wide association studies combining over 10,000,000 genetic markers in more than 10,000 European women with polycystic ovary syndrome (PCOS) and 100,000 controls. We found three new risk variants associated with PCOS. Our data demonstrate that the genetic architecture does not differ based on the diagnostic criteria used for PCOS. We also demonstrate a genetic pathway shared with male pattern baldness, representing the first evidence for shared disease biology in men, and shared genetics with depression, previously postulated based only on observational studies.
Introduction
Polycystic ovary syndrome (PCOS) is the most common endocrine disorder in reproductive aged women, with a complex pattern of inheritance [1–5]. Two different diagnostic criteria based on expert opinion have been utilized: The National Institutes of Health (NIH) criteria require hyperandrogenism (HA) and ovulatory dysfunction (OD) [6] while the Rotterdam criteria include the presence of polycystic ovarian morphology (PCOM) and requires at least two of three traits to be present, resulting in four phenotypes (S1 Fig) [6,7]. PCOS by NIH criteria has a prevalence of ~7% in reproductive age women worldwide [8]; the use of the broader Rotterdam criteria increases this to 15–20% across different populations [9–11].
PCOS is commonly associated with insulin resistance, pancreatic beta cell dysfunction, obesity and type 2 diabetes (T2D). These metabolic abnormalities are most pronounced in women with the NIH phenotype [12]. In addition, the odds for moderate or severe depression and anxiety disorders are higher in women with PCOS [13]. However, the mechanisms behind the association between the reproductive, metabolic and psychiatric features of the syndrome remain largely unknown.
Genome-wide association studies (GWAS) in women of Han Chinese and European ancestry have reproducibly identified 16 loci [14–17]. The observed susceptibility loci in PCOS appeared to be shared between NIH criteria and self-reported diagnosis [17], which is particularly intriguing. Genetic analyses of causality (by Mendelian Randomization analysis) among women of European ancestry with self-reported PCOS suggested that body mass index (BMI), insulin resistance, age at menopause and sex hormone binding globulin contribute to disease pathogenesis [17].
We performed the largest GWAS meta-analysis of PCOS to date, in 10,074 cases and 103,164 controls of European ancestry diagnosed with PCOS according to the NIH (2,540 cases and 15,020 controls) or Rotterdam criteria (2,669 cases and 17,035 controls), or by self-reported diagnosis (5,184 cases and 82,759 controls) (Tables 1 and S1). We investigated whether there were differences in the genetic architecture across the diagnostic criteria, and whether there were distinctive susceptibility loci associated with the cardinal features of PCOS; HA, OD and PCOM. Further, we explored the genetic architecture with a range of phenotypes related to the biology of PCOS, including male-pattern balding [18–21].
Table 1. Characteristics of PCOS cases and controls from each cohort included in the meta-analysis.
| Cohort | Subject Type | Number | Age (years) | BMI (kg/m2) | PCOS Definition | HA(1) n(%) | OD n(%) | PCOM n(%) |
|---|---|---|---|---|---|---|---|---|
| Rotterdam | Cases** | 1184 | 28.8 (4.8) | 26.1 (6.3) | NIH (41%) & Rotterdam (100%)(2) | 439 (37.0) | 946 (79.8) | 661 (55.8) |
| Controls | 5799 | 60.5 (7.9) | 27.6 (4.7) | Population Based Rotterdam Study | NA | NA | NA | |
| UK (London/ Oxford) | Cases** | 670 | 32.1 (6.8) | 28.2 (7.9) | NIH (33%) & Rotterdam (100%)(2) | 455 (67.9) | 537 (80.1) | 383 (57.2) |
| Controls | 1379 | 45 (0)§ | 26.8 (5.5) | 1958 British Birth Cohort | NA | NA | NA | |
| EGCUT | Cases** | 157 | 30.7 (8.2) | 26.2 (6.7) | Rotterdam(2) | NA | NA | NA |
| Controls | 2807 | 31.5 (7.3) | 23.1 (5.5) | Population Based | NA | NA | NA | |
| deCODE | Cases** | 658 | 41.3 (8.7) | 30.1 (7.8) | NIH (56%) & Rotterdam (100%)(2) | 644 (97.9) | 380 (57.7) | 507 (77.1) |
| Controls | 6774 | 49.0 (9.9) | 25.1 (4.9) | Population Based | NA | NA | NA | |
| Chicago | Cases* | 984 | 28.6 (5.5) | 35.9 (8.5) | NIH | 984 (100) | 984 (100) | NA |
| Controls | 2963 | 46.8 (15.2) | 27.0 (7.4) | Population Based NUgene | NA | NA | NA | |
| Boston | Cases* | 485 | 28.4 (6.7) | 30.8 (8.7) | NIH | 485 (100) | 485 (100) | 441 (90.9) |
| Controls | 407 | 27.2 (6.5) | 23.8 (4.1) | Screened controls(3) | 0 | 0 | 177 (43.4) | |
| 23andMe | Cases*** | 5,184 | 45.1 (13.6) | 29.2 (8.2) | Self report (defined by questionnaire) | NA | NA | NA |
| Controls | 82,759 | 51.1 (15.7) | 26.1 (6.1) | No PCOS by self report (defined by questionnaire) | NA | NA | NA |
(1) Clinical or Biochemical.
(2) Rotterdam diagnostic criteria include the NIH criteria. All subjects from the indicated cohorts were used in the Rotterdam analysis.
(3) Controls were screened for regular menses and no hyperandrogenism.
* PCOS diagnosis was based on NIH criteria,
** Rotterdam criteria, or
*** self report.
Results are reported as mean (SD) or a number (%).
Abbreviations: BMI: body mass index, NA: not available, HA: hyperandrogenism, OD: ovulatory dysfunction (<10 menses per year), PCOM: polycystic ovarian morphology.
§All subjects are from the British Birth Cohort (born in 1958).
Results
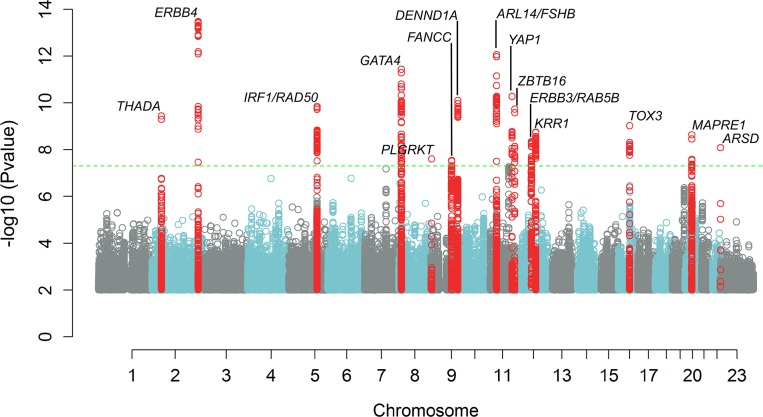
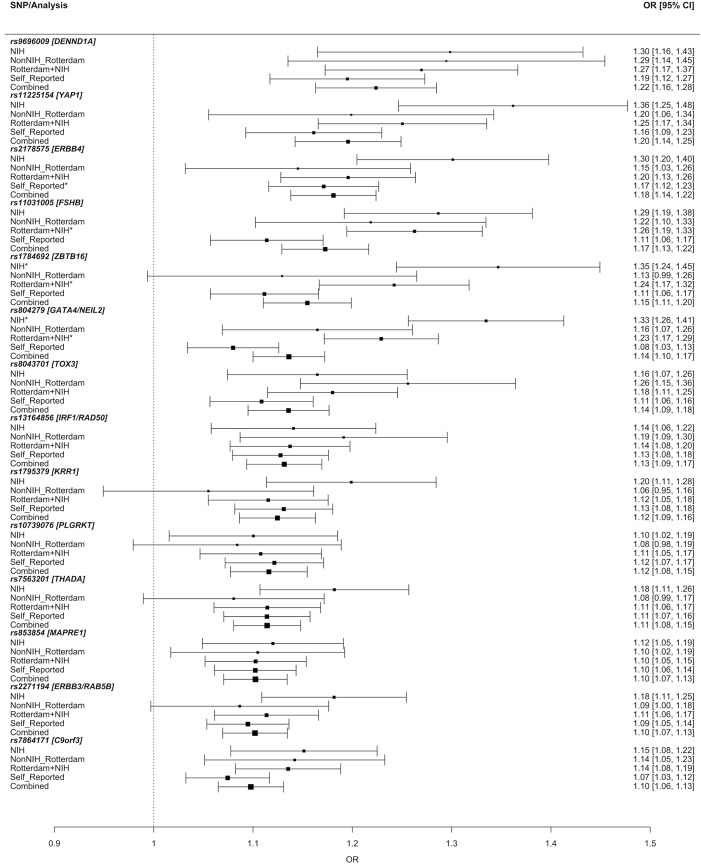
We identified 14 genetic susceptibility loci associated with PCOS, adjusting for age, at the genome-wide significance level (P < 5.0 x 10−8) bringing the total number of PCOS associated loci to nineteen (Tables 2 and S2 and Fig 1). Three of these loci were novel associations (near PLGRKT, ZBTB16 and MAPRE1, respectively; shown in bold in Table 2). Six of the 11 reported associations were previously observed in Han Chinese PCOS women [14,15]. Eight loci have been reported in European PCOS cohorts [16,17]. Obesity is commonly associated with PCOS and in most of the cohorts, cases were heavier than controls (Table 1). However, adjusting for both age and BMI did not identify any novel loci; and the 14 loci remained genome-wide significant. All variants demonstrated the same direction of effect across all phenotypes including NIH, non-NIH Rotterdam, and self-report (Fig 2 and S2 Table). Only one SNP near GATA4/NEIL2 showed significant evidence of heterogeneity across the different diagnostic groups (rs804279, Het P = 2.6x10-5; Fig 2 and S3 Table). For this SNP, the largest effect was seen in NIH cases and the smallest in self-reported cases. Credible set analysis, which prioritises variants in a given locus with regards to being potentially causal, was able to reduce the plausible interval for the causal variant(s) at many loci (S4 Table). Of note, 95% of the signal at the THADA locus came from two SNPs. Examination of previously published genome-wide significant loci from Han Chinese PCOS [14,15] demonstrated that index variants from the THADA, FSHR, C9orf3, YAP1 and RAB5B loci were significantly associated with PCOS after Bonferroni correction for multiple testing in our European ancestry subjects (S5 Table).
Table 2. The 14 genome-wide significant variants associated with PCOS in the meta-analysis.
| Chr:Position1 | rsID | Alleles2 | EAF3 | Beta | Odds Ratio (95% CI)4 | Std. Error | Nearest Gene | P-value | Effective N5 | Ref6 |
|---|---|---|---|---|---|---|---|---|---|---|
| 2:43561780 | rs7563201 | A/[G] | 0.4507 | -0.1081 | 0.90 (0.87–0.93) | 0.0172 | THADA | 3.678e-10 | 17192 | |
| 2:213391766 | rs2178575 | G/[A] | 0.1512 | 0.1663 | 1.18 (1.13–1.23) | 0.0219 | ERBB4 | 3.344e-14 | 17192 | 17 |
| 5:131813204 | rs13164856 | [T]/C | 0.7291 | 0.1235 | 1.13 (1.09–1.18) | 0.0193 | IRF1/RAD50 | 1.453e-10 | 17192 | 17 |
| 8:11623889 | rs804279 | A/[T] | 0.2616 | 0.1276 | 1.14 (1.10–1.18) | 0.0184 | GATA4/NEIL2 | 3.761e-12 | 16895 | 16 |
| 9:5440589 | rs10739076 | C/[A] | 0.3078 | 0.1097 | 1.12 (1.07–1.16) | 0.0197 | PLGRKT | 2.510e-08 | 17192 | |
| 9:97723266 | rs7864171 | G/[A] | 0.4284 | -0.0933 | 0.91 (0.88–0.94) | 0.0168 | FANCC | 2.946e-08 | 17192 | 16 |
| 9:126619233 | rs9696009 | G/[A] | 0.0679 | 0.202 | 1.22 (1.15–1.30) | 0.0311 | DENND1A | 7.958e-11 | 17192 | |
| 11:30226356 | rs11031005 | [T]/C | 0.8537 | -0.1593 | 0.85 (0.82–0.89) | 0.0223 | ARL14EP/FSHB | 8.664e-13 | 17192 | 16,17 |
| 11:102043240 | rs11225154 | G/[A] | 0.0941 | 0.1787 | 1.20 (1.13–1.26) | 0.0272 | YAP1 | 5.438e-11 | 17192 | 17 |
| 11:113949232 | rs1784692 | [A]/G | 0.8237 | 0.1438 | 1.15 (1.10–1.14) | 0.0226 | ZBTB16 | 1.876e-10 | 17192 | |
| 12:56477694 | rs2271194 | A/[T] | 0.416 | 0.0971 | 1.10 (1.07–1.14) | 0.0166 | ERBB3/RAB5B | 4.568e-09 | 17192 | 17 |
| 12:75941042 | rs1795379 | C/[T] | 0.2398 | -0.1174 | 0.89 (0.86–0.92 | 0.0195 | KRR1 | 1.808e-09 | 17192 | 17 |
| 16:52375777 | rs8043701 | [A]/T | 0.815 | -0.1273 | 0.88 (0.85–0.92) | 0.0208 | TOX3 | 9.610e-10 | 17192 | |
| 20:31420757 | rs853854 | A/[T] | .4989 | -.0975 | 0.91 (0.88–0.94) | 0.0163 | MAPRE1 | 2.358e-09 | 17192 |
1Chr—Chromosome:Position (bp) in hg19;
2Alleles are shown as Major/Minor by allele frequency in 1000G EUR cohort, with the effect allele shown within [];
3Effect allele frequency;
495% Confidence Interval of the Odds Ratio;
5Effective N—effective sample size;
6Ref = Reference.
Loci previously identified in GWAS studies of European ancestry are referenced. Novel associations with PCOS not previously reported are shown in bold. EAF = Effect Allele Frequency.
Fig 1. Manhattan plot showing results of meta-analysis for PCOS status, adjusting for age.
The inverse log10 of the p value (-log10(p)) is plotted on the Y axis. The green dashed line designates the minimum p value for genome-wide significance (<5.0 x 10−8). Genome wide significant loci are denoted with a label showing the nearest gene to the index SNP at each locus. SNPs with p values ≤1.0x10-2 are not depicted.
Fig 2. Odds ratio of polycystic ovary syndrome (PCOS) as a function of diagnostic criteria applied.
The Y-axis specifies the diagnostic criteria and the X-axis indicates the odds ratio (OR) and 95% confidence intervals (CI) for PCOS (black circle and horizontal error bars). Data derived as follows: NIH = groups recruiting only NIH diagnostic criteria; NonNIH_Rotterdam = Rotterdam diagnostic criteria excluding the subset fulfilling NIH diagnostic criteria; Rotterdam+NIH = all groups except self-reported; self-reported = 23andMe; and combined = all groups. Specific OR’s [95% CI, 5% CI] are indicated on the right. rs804279 in the GATA4/NEIL2 locus demonstrates significant heterogeneity (Het P = 2.6x10-5). The * indicates statistically significant association for PCOS and the variant in that specific stratum.
We assessed the association of the PCOS susceptibility variants identified in the GWAS meta-analysis with the PCOS related traits: HA, OD, PCOM, testosterone, FSH and LH levels, and ovarian volume in PCOS cases (Tables 3 and S6 and S2 Fig). We found four variants associated with HA, eight variants associated with PCOM and nine variants associated with OD. Of the eight loci associated with PCOM, seven were also associated with OD. Three of the four loci associated with HA were also associated with OD and PCOM. Two additional loci were associated with OD alone, one of which was the locus near FSHB (S6 Table). This locus was also associated with LH and FSH levels. There was a single PCOS locus near IRF1/RAD50 associated with testosterone levels (S6 Table). We repeated this analysis with susceptibility variants reported previously in Han Chinese PCOS cohorts [14,15]. In this analysis, there was one association with HA (near DENND1A), three with PCOM and three with OD (S2 Fig and S5 Table). A limitation of these analyses is the variable sample size across the phenotypes analysed. Additionally, the known referral bias for the more severely affected NIH phenotype (patients having both OD and HA) may result in more PCOS diagnoses than the other criteria [22], and may have contributed to the number of associations between the identified PCOS risk loci and these phenotypes.
Table 3. Association of PCOS GWAS meta-analysis susceptibility variants and PCOS related traits.
| Chr:Position | rsID | Gene | Ref. allele | Other allele | Hyperandrogenism | PCOM | OD | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | Beta | P-value | Beta | P-value | Beta | P-value | |||||
| 2:213391766 | rs2178575 | ERBB4* | G | A | 0.83 | -0.126 | 4.3E-03 | -0.24 | 1.4E-05 | -0.23 | 1.2E-11 |
| 2:43561780 | rs7563201 | THADA*† | G | A | 0.56 | 0.061 | 8.0E-02 | 0.16 | 3.7E-04 | 0.08 | 1.5E-03 |
| 5:131813204 | rs13164856 | IRF1/RAD50* | T | C | 0.73 | 0.092 | 1.8E-02 | 0.16 | 1.4E-03 | 0.08 | 5.6E-03 |
| 8:11623889 | rs804279 | GATA4/NEIL2* | A | T | 0.27 | 0.126 | 8.7E-04 | 0.22 | 1.5E-06 | 0.16 | 9.9E-09 |
| 9:126619233 | rs9696009 | DENND1A† | G | A | 0.94 | -0.330 | 2.9E-07 | -0.32 | 4.0E-05 | -0.36 | 4.4E-15 |
| 9:5440589 | rs10739076 | PLGRKT | A | C | 0.30 | 0.026 | 5.3E-01 | 0.10 | 5.9E-02 | 0.00 | 8.9E-01 |
| 9:97723266 | rs7864171 | C9orf3*† | G | A | 0.60 | 0.124 | 3.8E-04 | 0.19 | 1.3E-05 | 0.10 | 2.3E-04 |
| 11:30226356 | rs11031005 | ARL14EP/FSHB* | T | C | 0.85 | -0.079 | 8.2E-02 | -0.18 | 1.3E-03 | -0.13 | 2.8E-04 |
| 11:102043240 | rs11225154 | YAP1*† | G | A | 0.91 | -0.144 | 1.4E-02 | -0.24 | 3.5E-04 | -0.23 | 5.7E-08 |
| 11:113949232 | rs1784692 | ZBTB16 | T | C | 0.85 | 0.146 | 4.6E-03 | 0.30 | 2.8E-06 | 0.21 | 6.6E-09 |
| 12:75941042 | rs1795379 | KRR1* | T | C | 0.24 | -0.104 | 8.0E-02 | -0.16 | 1.5E-03 | -0.11 | 1.8E-04 |
| 12:56477694 | rs2271194 | ERBB3/RAB5B† | A | T | 0.42 | 0.126 | 2.7E-04 | 0.17 | 7.9E-05 | 0.13 | 1.4E-06 |
| 16:52375777 | rs8043701 | TOX3† | A | T | 0.82 | -0.166 | 1.4E-04 | -0.17 | 1.5E-03 | -0.08 | 9.2E-03 |
| 20:31420757 | rs853854 | MAPRE1 | T | A | 0.50 | 0.111 | 9.8E-04 | 0.10 | 2.1E-02 | 0.05 | 3.8E-02 |
Significant associations are highlighted in bold. Variant previously reported as a PCOS risk variant in
*European or
†Han Chinese populations.
In the analyses looking at the weighted genetic risk score in the Rotterdam cohort, we observed an increase in the risk for PCOS (S3 Fig). Compared to individuals in the third quintile (reference group), individuals in the top 5th quintile of risk score have an OR of 1.9 (1.4–2.5; 95% CI) for PCOS based on NIH criteria and an OR of 2.1 (1.7–2.5; 95% CI) for Rotterdam criteria based PCOS. Of the associations, only the effect estimate for the Rotterdam criteria was significant, possibly due to the smaller size available with cases diagnosed according to the NIH criteria. When looking at the area under the ROC curves at SNPs with different P-value thresholds, we found a maximum AUC of 0.54 using SNPs with a P-value < 5x10-6 for both diagnostic criteria. While this is significantly better than chance, it is unlikely that a risk score generated from the variants discovered to date would represent a clinically relevant tool.
LD score regression analysis revealed genetic correlations with childhood obesity, fasting insulin, T2D, HDL, menarche timing, triglyceride levels, cardiovascular diseases and depression (Table 4) suggesting that there is shared genetic architecture and biology between these phenotypes and PCOS. There were no genetic correlations with menopause timing or male pattern balding. Mendelian randomization suggested that there was a causal role for BMI, fasting insulin and depression pathways (Table 5). Interestingly, while there was no genetic correlation detected for male pattern balding or menopause timing with PCOS, the Mendelian randomization analyses were significant. The difference in the genetic correlation compared to the Mendelian randomization result suggests that there may be a small number of key biological process that are common between the phenotypes, and that the common genetic causal variants are limited only to the variants shared by the subset of key biological processes. The importance of BMI pathways on reproductive phenotypes was further demonstrated by the attenuation of significance of Mendelian randomization analysis for age-at-menarche when BMI-associated variants were excluded from the analysis.
Table 4. LD Score regression results using the LDSC method.
| Phenotype | Genetic Correlation | SE | Z | P-value |
|---|---|---|---|---|
| Body mass index | 0.34 | 0.039 | 8.60 | 8.21×10−18 |
| Childhood obesity | 0.34 | 0.066 | 5.17 | 2.40×10−7 |
| Fasting insulin levels | 0.44 | 0.087 | 5.01 | 5.33×10−7 |
| Type 2 diabetes | 0.31 | 0.068 | 4.47 | 7.84×10−6 |
| High-density lipoprotein levels | -0.23 | 0.059 | -3.96 | 7.40×10−5 |
| Menarche | -0.16 | 0.042 | -3.76 | 1.71×10−4 |
| Triglyceride levels | 0.19 | 0.052 | 3.61 | 3.05×10−4 |
| Coronary artery disease | 0.23 | 0.069 | 3.32 | 8.86×10−4 |
| Depression | 0.205 | 0.0582 | 3.5203 | 0.0004 |
| Menopause | -0.014 | 0.0183 | -0.762 | 0.4461 |
| Male pattern balding | 0.0149 | 0.0168 | 0.8861 | 0.3756 |
Table 5. Mendelian randomization using an inverse weighted variant method.
| Potential Risk factor | IVW method1 | MR-EGGER intercept p-value2 | ||
|---|---|---|---|---|
| Beta | SE | P-value | ||
| Body mass index | 0.72 | 0.072 | 1.56 x 10−23 | 0.13 |
| Fasting insulin levels* | 0.03 | 0.007 | 1.73 x 10−5 | 0.06 |
| Male pattern balding | 0.05 | 0.017 | 0.0034 | 0.93 |
| Menopause | 0.1 | 0.022 | 1.31 x 10−5 | 0.39 |
| Depression | 0.77 | 0.213 | 0.00029 | 0.64 |
*Loci used were initially reported in an analysis of fasting insulin adjusted for BMI.
1IVW = inverse weighted variant,
2Mendelian Randomization (MR)-Egger intercept p values were not significant. Therefore, MR-Egger results are not presented.
Discussion
We found 14 independent loci significantly associated with the risk for PCOS, including three novel loci. The 11 previously reported loci implicated neuroendocrine and metabolic pathways that may contribute to PCOS (1.1 Note in S1 Data). Two of the novel loci contain potential endocrine related candidate genes. The locus harbouring rs10739076 contains several interesting candidate genes; PLGRKT, a plasminogen receptor and several genes in the insulin superfamily; INSL6, INSL4 and RLN1, RLN2 which are endocrine hormones secreted by the ovary and testis and are suspected to impact follicle growth and ovulation [23]. ZBTB16 (also known as PLZF) has been marked as an androgen-responsive gene with anti-proliferative activity in prostate cancer cells [24]. PLZF activates GATA4 gene transcription and mediates cardiac hypertrophic signalling from the angiotensin II receptor 2 [25]. Furthermore, PLZF is upregulated during adipocyte differentiation in vitro [26] and is involved in control of early stages of spermatogenesis [27] and endometrial stromal cell decidualization [28]. The third novel locus harbours a metabolic candidate gene; MAPRE1 (interacts with the low-density lipoprotein receptor related protein 1 (LRP1), which controls adipogenesis [29] and may additionally mediate ovarian angiogenesis and follicle development [30] (1.2 Note in S1 Data). Thus, all the new loci contain genes plausibly linked to both the metabolic and reproductive features of PCOS.
We found that there was no significant difference in the association with case status for the majority of the PCOS-susceptibility loci by diagnostic criteria. All susceptibility variants demonstrated the same direction of effect for the NIH phenotype, non-NIH Rotterdam phenotype and self-report, with only one variant demonstrating significant heterogeneity among the groups. It is of considerable interest that the cohort of research participants from the personal genetics company 23andMe, Inc., identified by self-report, had similar risks to the other cohorts where the diagnosis was clinically confirmed. Our findings suggest that the genetic architecture of these PCOS definitions does not differ for common susceptibility variants. Only one locus, GATA4/NEIL2 (rs804279), was significantly different across diagnostic criteria: most strongly associated in NIH compared to the Rotterdam phenotype and self-reported cases. Deletion of GATA4 results in abnormal responses to exogenous gonadotropins and impaired fertility in mice [31]. The locus also encompasses the promoter region of FDFT1, the first enzyme in the cholesterol biosynthesis pathway [32], which is the substrate for testosterone synthesis, and is associated with non-alcoholic fatty liver disease [33]. The major difference between the NIH phenotype and the additional Rotterdam phenotypes is metabolic risk; the NIH phenotype is associated with more severe insulin resistance [34]. rs804279 does not show association with any of the metabolic phenotypes in the T2D diabetes knowledge portal {Type 2 Diabetes Knowledge Portal. type2diabetesgenetics.org. 2015 Feb 1; http://www.type2diabetesgenetics.org/variantInfo/variantInfo/rs804279} so it may represent a PCOS-specific susceptibility locus.
The significant association of PCOS GWAS meta-analysis susceptibility variants with the cardinal PCOS related traits OD, HA and PCOM further strengthened the hypothesis that specific variants may confer risk for PCOS through distinct mechanisms. Three variants at the C9orf3, DENND1A, and RAB5B were associated with all PCOS related traits. The findings were consistent with the Han Chinese DENND1A variant association with HA, as suggested previously [35]. Thus, these loci, along with GATA4/NEIL2 (as discussed above) may help identify pathways that link specific PCOS related traits with greater metabolic risk. In contrast, the variants at the ERBB4, YAP1, and ZBTB16 loci were strongly associated with OD and PCOM, and therefore, might be more important for links to menstrual cycle regularity and fertility. In addition, the FSHB variant was associated with the levels of FSH and LH [16,17], suggesting that it may act by affecting gonadotropin levels. This variant maps 2kb upstream from open chromatin (identified by DNase-Seq) and an enhancer (identified by peaks for both H3K27Ac and H3K4me1) in a lymphoblastoid cell line from ENCODE, indicating a potential role for a regulatory element ~25kb upstream from the FSHB promoter. Furthermore, the association between the IRF1/RAD50 variant and testosterone levels may indicate a regulatory role in testosterone production.
Of note, results of the follow-up analysis show a high level of shared biology between PCOS and a range of metabolic outcomes consistent with the previous findings [17]. In particular, there is genetic evidence for increased BMI as a risk factor for PCOS. There is also genetic evidence that fasting insulin might be an independent risk factor. This study also confirmed a causal association with the pathways that underlie menopause [17], suggesting that PCOS has shared aetiology with both classic metabolic and reproductive phenotypes. Furthermore, there was an apparent effect of depression-associated variants on the likelihood of PCOS, suggesting a role for psychological factors on hormonally related diseases. However, the links between PCOS and depression might be complicated by pathways that are also related to BMI, as BMI pathways are causal in both PCOS and depression [36]. In addition, male-pattern balding-associated variants showed strong effects on PCOS, suggesting that this might be a male manifestation of PCOS pathways, as has been previously suggested [18,20,21,37]. This observation may reflect the biology of hair follicle sensitivity to androgens, seen in androgenetic alopecia, a well-recognised feature of HA and PCOS [38,39]. The Mendelian randomization results for male-pattern balding and menopause are significant despite non-significant genetic correlation results, suggesting that the shared aetiology may be specific to only a few key pathways.
In conclusion, the genetic underpinnings of PCOS implicate neuroendocrine, metabolic and reproductive pathways in the pathogenesis of disease. Although specific phenotype stratified analyses are needed, genetic findings were consistent across the diagnostic criteria for all but one susceptibility locus, suggesting a common genetic architecture underlying the different phenotypes. There was genetic evidence for shared biologic pathways between PCOS and a number of metabolic disorders, menopause, depression and male-pattern balding, a putative male phenotype. Our findings demonstrate the extensive power of genetic and genomic approaches to elucidate the pathophysiology of PCOS.
Methods
Ethics statement
All research involving human participants has been approved by the authors' Institutional Review Board (IRB) or an equivalent committee, and all clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all participants. The Boston cohort was approved by the Partners IRB (# 2002P001924) and the University of Utah IRB (IRB_00076659). The deCODE cohort was approved by the National Bioethics Committee of Iceland (VSN 03–007), which was conducted in agreement with conditions issued by the Data Protection Authority of Iceland. Personal identities of the participants’ data and biological samples were encrypted by a third-party system (Identity Protection System), approved and monitored by the Data Protection Authority. The UK cohort was approved by the Parkside Health Authority (Now—NHS Health Research Authority, NRES Committee—West London & GTAC, UK, London, UK) under EC2359 "The Molecular Genetics of Polycystic Ovaries." The Rotterdam PCOS cohort, the COLA study, was approved by institutional review board (Medical Ethics Committee) of the Erasmus Medical Center (04–263). Controls from the Rotterdam Study were approved by the Medical Ethics Committee of the Erasmus MC (registration number MEC 02.1015) and by the Dutch Ministry of Health, Welfare and Sport (Population Screening Act WBO, license number 1071272-159521-PG). The Rotterdam Study Personal Registration Data collection is filed with the Erasmus MC Data Protection Officer under registration number EMC1712001. The Rotterdam Study has been entered into the Netherlands National Trial Register (NTR; www.trialregister.nl) and into the WHO International Clinical Trials Registry Platform (ICTRP; www.who.int/ictrp/network/primary/en/) under shared catalogue number NTR6831. The Chicago PCOS cohort was approved by the Northwestern IRB (#STU00008096). The control subjects from the NUgene study were approved by the Northwestern IRB (# STU00010003). The Estonia cohort was approved by the Research Ethics Committee of the University of Tartu approved the study (198T-18). The Twins UK study was approved by the St Thomas' Hospital Research Ethics Committee (EC04/015). The Nurses' Health Study (NHS I and II) was approved by the Partners Human Research Committee (#1999-P-011114).
Subjects
The meta-analysis included 10,074 cases and 103,164 controls from seven cohorts of European descent. For the analysis of PCOS related traits three additional cohorts, the Northern Finnish Birth Cohort (NFB66) [40], Twins UK [41] and the Nurses’ Health Study (NHS) [42] were included. Cases were diagnosed with PCOS based on NIH or Rotterdam Criteria or by self-report. The NIH criteria require the presence of both OD and clinical and/or biochemical HA for a diagnosis of PCOS [6]. The Rotterdam criteria require two out of three features 1) OD defined by oligo- or amenorrhea (chronic menstrual cycle interval >35 days in all cohorts), 2) clinical and/or biochemical hyperandrogenism (HA) and/or 3) PCOM for a diagnosis of PCOS [7]. Non-NIH Rotterdam was defined by OD and PCOM or clinical and/or biochemical hyperandrogenism (HA) and PCOM. Self-reported female cases from research participants in the 23andMe, Inc. (Mountain View, CA, USA) cohort either responded “yes” to the question “Have you ever been diagnosed with polycystic ovary syndrome?” or indicated a diagnosis of PCOS when asked about fertility (“Have you ever been diagnosed with PCOS?” or “What was your diagnosis? Please check all that apply.” Answer = PCOS), hair loss in men or women (“Have you been diagnosed with any of the following? Please check all that apply.” Answer = PCOS) or research question (“Have you ever been diagnosed with PCOS?”) [17]. 23andMe controls were female, only.
HA was defined as hirsutism and quantified by the Ferriman-Gallwey (FG) score. The FG score assesses terminal hair growth in a male pattern in females, and a score above the upper limit of normal controls (>8) is considered hirsutism [43]. Hyperandrogenemia was defined as testosterone, androstenedione or DHEAS greater than the 95% confidence limits in control subjects in the individual population. OD was defined as cycle interval <21 or >35 days [44]. PCOM was defined as 12 or more follicles of 2–9 mm in at least one ovary or an ovarian volume >10 mL [7]. The quantitative PCOS traits included levels of total testosterone (T), follicle-stimulating hormone (FSH), and luteinizing hormone (LH) and ovarian volume (S1 Table). An overview of the cohorts, diagnostic criteria and number of subjects included in each subphenotype or trait analysis are summarized in Tables 1 and S1.
Data collection and quality control
Each study provided summary results of genetic per-variant estimates produced in either case-control or trait association analyses. Adjustment for principle components was performed at the study level. The collected files underwent quality control (QC) by two independent analysts using the EasyQC pipeline [45]. Variants were excluded based on minor allele frequency (MAF) < 1%, imputation quality (R2) < 0.3 or info < 0.4 for MACH and IMPUTE2 respectively [46,47]. Per-cohort QC results from EasyQC are shown (S7 Table), and allele frequency spectrum for each cohort, and the combined cohort after meta-analysis is shown (S4 Fig).
Meta-analysis of PCOS status and PCOS related traits
The per-variant estimates collected from the summary statistics of contributing studies were meta-analysed using a fixed-effect, inverse-weighted-variance meta-analysis that employed either GWAMA [48] or METAL [49]. In addition to the overall meta-analysis, we performed meta-analyses for studies with available data for the separate PCOS diagnostic criteria: NIH, non-NIH Rotterdam [7] and self-report [17], as well as for the PCOS related traits of HA, OD and PCOM. The meta-analysis of PCOS status was performed using two models; (1) age-adjusted, (2) age and BMI-adjusted, given the high prevalence of obesity in affected women that resulted in cases being significantly heavier than controls in most cohorts (Table 1).
We removed any variants that were not present in more than 50% of the effective sample size prior to combining with 23andMe as this was the largest cohort in the meta-analysis, providing approximately 51% of the PCOS cases and 80% of controls. We also removed any variants only present in one study. The meta-analysis of PCOS related traits was performed adjusting for age and BMI. Identified variants were annotated for insight into their biological function using ANNOVAR [50] to assign refGene gene information, SIFT score [51], PolyPhen2 scores [52], CADD scores [53], GERP scores [54] and SiPhy log odds [55].
Comparison of PCOS diagnostic criteria
In order to compare different PCOS diagnostic criteria [(1) NIH, (2) non-NIH Rotterdam and (3) self-reported] included in the PCOS meta-analysis, an additional meta-analysis was performed to test for heterogeneity across these independent PCOS case groups. These three PCOS case groups were combined in an inverse variance weighted fixed meta-analysis and the heterogeneity statistics (Cochran’s Q and I2) were obtained using GWAMA [48]. Any variant with a statistically significant Cochran’s Q p-value (P<0.05/14 = 0.0036 corrected for multiple testing) and I2>70% were considered exhibiting heterogeneity across the PCOS case groups. Further analysis of the heterogeneity included comparison of the 95% confidence intervals for the direction of effect and overlaps.
Identifying associations between PCOS Loci and PCOS related traits
In order to understand biology relevant to identified PCOS susceptibility, we assessed the association between index SNPs at each genome-wide-significant locus and the PCOS related traits HA, OD, PCOM as well as the quantitative traits testosterone, LH and FSH levels and ovarian volume. The threshold for significance in this analysis was p<4.5×10−4 (Bonferroni correction [0.05/(14 independent loci x 8 traits)].
Identifying shared risk loci between European ancestry and Han Chinese PCOS
In order to identify shared risk loci between the previously reported GWAS in Han Chinese PCOS cases and our European ancestry cohort, 13 independent signals (represented by 15 SNPs) at 11 genome-wide significant loci reported by Chen et al. [14] and Shi et al. [15] were investigated for association in our meta-analyses of PCOS and PCOS related traits. The adjusted P-value for this analysis was <0.00048 (Bonferroni correction [0.05/(13 independent signals x 8 traits)]).
Biologic function of genes in associated loci
Information on the biological function of the nearest gene (or genes, if variants were equidistant from more than one coding transcript and annotated as such by ANNOVAR [49] to the index SNP of each identified risk locus) was collected by performing a search of the Entrez Gene Database [56], and collecting the co-ordinates of the gene (genome build 37; hg19) as well as the cytogenetic location and the summary of the gene function. In addition to the EntrezGene Database queries, the gene symbol was used as a search term in the PubMed database [57], either alone or combined with the additional search term “PCOS” to identify relevant published literature in order to obtain information on putative biological function and involvement in the pathogenesis of PCOS (summarized in 1.1 Note in S1 Data).
Weighted genetic risk score and prediction
One potential use of genetic risk scores is prediction of disease. The ability of genetic risk scores calculated from loci discovered in analysis of the different diagnostic criteria to discriminate cases from alternative criteria was measured. We constructed a weighted genetic risk score based on a meta-analysis excluding the Rotterdam Study subjects. The weighted genetic risk score was divided into quintiles and tested for association with PCOS in the Rotterdam cohort. The middle quintile was used as the reference and the odds for having PCOS based on both Rotterdam and NIH criteria was then calculated.
Additionally, the 23andMe results were used to select independent SNPs with cut-offs of p<5×10−4 to p<5×10−8. The Rotterdam cohort was then used to calculate risk scores and the area-under-the curve (AUC) for both NIH and Rotterdam diagnostic criteria. Analyses were performed using PLINK v1.9 and SPSS v21 (IBM Corp, Armonk, NY) [58].
Linkage disequilibrium (LD) score regression
To assess the level of shared etiology between PCOS and related traits, we performed genetic correlation analysis using LD-score regression [59]. Publicly available genome-wide summary statistics for body mass index (BMI) [60], childhood obesity [61], fasting insulin levels (adjusted for BMI) [62], type 2 diabetes [63], high-density lipoprotein (HDL) levels [64], menarche timing [65], triglyceride levels [64], coronary artery disease [66], depression [36], menopause [17] and male pattern balding [67] were used to estimate the genome-wide genetic correlation with PCOS. The adjusted P-value for this analysis was p<0.0045 after a Bonferroni correction (0.05/11 traits).
Mendelian randomization
Phenotypes of interest, both where there was evidence of shared genetic architecture and where there was previous evidence for genetic links, were assessed using Mendelian randomization methods. Mendelian randomization differs from LD score regression in that one phenotype is analysed as a potential causal factor for another. Mendelian randomization was performed using both inverse weighted variance and Egger’s regression methods [68], with inverse weighted methods being more powerful, but Egger’s methods being resistant to directional pleiotropy (where there are a set of SNPs that appear to have an alternative pathway of effect). We report here the results of the IVW methods as none of the analysis suggested that the MR-EGGERs results were more appropriate given that none of the EGGERs intercepts were significant (Table 5). In addition to the phenotypes implicated by the LD-score regression measures, male pattern balding has a strong biological rationale and was therefore included. The genetic score for childhood obesity substantially overlaps with the score for adult BMI (such that the INSIDE violation—where the effect of SNPs on a confounding factor scales with that on the trait of interest—of Mendelian randomization would likely occur [69], so only a score for BMI was used, with the proviso that this represents BMI across the whole of the life course after very early infancy. The SNPs for depression were drawn from the results of a more recent analysis, for which there was not, at time of analysis, publicly available genome-wide data.
Credible sets
We defined a locus as mapping within 500kb of the lead SNP. For each locus, we first calculated the posterior probability, πCj, that the jth variant is driving the association, given by:
where the summation is over all retained variants in the locus. In this expression, Λj is the approximate Bayes’ factor [70] for the jth variant, given by
where βj and Vj denote the estimated allelic effect (log-OR) and corresponding variance from the meta-analysis. The parameter ω denotes the prior variance in allelic effects, taken here to be 0.04 [70]. The 99% credible set [71] for each signal was then constructed by: (i) ranking all variants according to their Bayes’ factor, Λj; and (ii) including ranked variants until their cumulative posterior probability of driving the association attained or exceeded 0.99.
Supporting information
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the research participants and employees of 23andMe for contributing to this study. EGCUT Computations were performed in the High Performance Computing Center, University of Tartu.
¶ 23andMe Research Team (23andMe, Inc., Mountain View, California, United States of America): Michelle Agee, Babak Alipanahi, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, David A. Hinds, Karen E. Huber, Aaron Kleinman, Nadia Kenref. Litterman, Matthew H. McIntyre, Joanna L. Mountain, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, J. Fah Sathirapongsasuti, Olga V. Sazonova, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Vladimir Vacic, Catherine H. Wilson.
Data Availability
Summary statistic GWAS meta-analysis results for the combined dataset excluding 23andMe are available at https://doi.org/10.17863/CAM.27720. The most significant 10,000 SNPs for the meta-analysis including 23andMe are available at https://doi.org/10.17863/CAM.27720.
Funding Statement
This work has been supported by MRC grant MC_U106179472 (FD, KO, JRBP), Samuel Oschin Comprehensive Cancer Institute Developmental Funds, Center for Bioinformatics and Functional Genomics and Department of Biomedical Sciences Developmental Funds (MRJ), NCI P30CA177558 (CH), NCI UM1CA186107 (PK), European Regional Development Fund (Project No. 2014-2020.4.01.15-0012) and the European Union’s Horizon 2020 research and innovation program under grant agreements No 692065 (TL, RM, AS) and 692145 (RM), NICHD R01HD065029 (RS), Estonian Ministry of Education and Research (grant IUT34-16 to TL), NICHD R01HD057450 (MU), NICHD P50HD044405 (AD), NICHD R01HD057223 (AD), R01HD085227 (MGH, AD), deCode Genetics (GT, UT, KS, US), Raine Medical Research Foundation Priming Grant (BHM), SCGOPHCG RAC 2015-16/034 (SGW, BGAS), 2016-17/018 (BGAS), NIHR BRC, Wellcome Trust, MRC (TDS), Eris M. Field Chair in Diabetes Research (MOG), NIDDK P30 DK063491 (MOG), NIDDK U01DK094431, U01DK048381 (DE), NICHD U10HD38992 (RL), Estonian Ministry of Education and Research (grant IUT34-16), Enterprise Estonia (grant EU48695); the EU-FP7 Marie Curie Industry-Academia Partnerships and Pathways (IAPP, grant SARM, EU324509 to AS), Wellcome (090532, 098381, 203141); European Commission (ENGAGE: HEALTH-F4-2007-201413 to MIM), MRC G0802782, MR/M012638/1 (SF), Li Ka Shing Foundation, WT-SSI/John Fell Funds, NIHR Biomedical Research Centre, Oxford, Widenlife and NICHD 5P50HD028138-27 (CML), NICHD R01HD065029, ADA 1-10-CT-57, Harvard Clinical and Translational Science Center, from the National Center for Research Resources 1UL1 RR025758 (CKW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vink J.M., Sadrzadeh S., Lambalk C.B. & Boomsma D.I. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab 91, 2100–4 (2006). 10.1210/jc.2005-1494 [DOI] [PubMed] [Google Scholar]
- 2.Kahsar-Miller M.D., Nixon C., Boots L.R., Go R.C. & Azziz R. Prevalence of polycystic ovary syndrome (PCOS) in first-degree relatives of patients with PCOS. Fertil Steril 75, 53–8 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Legro R.S., Driscoll D., Strauss J.F. 3rd, Fox J. & Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci U S A 95, 14956–60 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahanfar S., Eden J.A., Nguyen T., Wang X.L. & Wilcken D.E. A twin study of polycystic ovary syndrome and lipids. Gynecol Endocrinol 11, 111–7 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Jahanfar S., Eden J.A., Warren P., Seppala M. & Nguyen T.V. A twin study of polycystic ovary syndrome. Fertil Steril 63, 478–86 (1995). [PubMed] [Google Scholar]
- 6.Zawadzki J.K. & Dunaif A. Diagnostic criteria for polycystic ovary syndrome: toward a rational approach in Polycystic ovary syndrome (eds. Dunaif A., Givens J.R., Haseltine F. & Merriam G.R.) 377–84 (Blackwell Scientific Publications, Cambridge, 1992). [Google Scholar]
- 7.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 81, 19–25 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Knochenhauer E.S. et al. Prevalence of the polycystic ovary syndrome in unselected black and white women of the southeastern United States: a prospective study. J Clin Endocrinol Metab 83, 3078–82 (1998). 10.1210/jcem.83.9.5090 [DOI] [PubMed] [Google Scholar]
- 9.Tehrani F.R., Simbar M., Tohidi M., Hosseinpanah F. & Azizi F. The prevalence of polycystic ovary syndrome in a community sample of Iranian population: Iranian PCOS prevalence study. Reprod Biol Endocrinol 9, 39 (2011). 10.1186/1477-7827-9-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.March W.A. et al. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum Reprod 25, 544–51 (2010). 10.1093/humrep/dep399 [DOI] [PubMed] [Google Scholar]
- 11.Yildiz B.O., Bozdag G., Yapici Z., Esinler I. & Yarali H. Prevalence, phenotype and cardiometabolic risk of polycystic ovary syndrome under different diagnostic criteria. Hum Reprod 27, 3067–73 (2012). 10.1093/humrep/des232 [DOI] [PubMed] [Google Scholar]
- 12.Diamanti-Kandarakis E. & Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev 33, 981–1030 (2012). 10.1210/er.2011-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooney L.G., Lee I., Sammel M.D. & Dokras A. High prevalence of moderate and severe depressive and anxiety symptoms in polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod 32, 1075–1091 (2017). 10.1093/humrep/dex044 [DOI] [PubMed] [Google Scholar]
- 14.Chen Z.J. et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet 43, 55–9 (2011). 10.1038/ng.732 [DOI] [PubMed] [Google Scholar]
- 15.Shi Y. et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet 44, 1020–5 (2012). 10.1038/ng.2384 [DOI] [PubMed] [Google Scholar]
- 16.Hayes M.G. et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun 6, 7502 (2015). 10.1038/ncomms8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day F.R. et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun 6, 8464 (2015). 10.1038/ncomms9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carey A.H. et al. Evidence for a single gene effect causing polycystic ovaries and male pattern baldness. Clin Endocrinol (Oxf) 38, 653–8 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Fabre D. et al. Identification of patients with impaired hepatic drug metabolism using a limited sampling procedure for estimation of phenazone (antipyrine) pharmacokinetic parameters. Clin Pharmacokinet 24, 333–43 (1993). 10.2165/00003088-199324040-00006 [DOI] [PubMed] [Google Scholar]
- 20.Sanke S., Chander R., Jain A., Garg T. & Yadav P. A Comparison of the Hormonal Profile of Early Androgenetic Alopecia in Men With the Phenotypic Equivalent of Polycystic Ovarian Syndrome in Women. JAMA Dermatol 152, 986–91 (2016). 10.1001/jamadermatol.2016.1776 [DOI] [PubMed] [Google Scholar]
- 21.Govind A., Obhrai M.S. & Clayton R.N. Polycystic ovaries are inherited as an autosomal dominant trait: analysis of 29 polycystic ovary syndrome and 10 control families. J Clin Endocrinol Metab 84, 38–43 (1999). 10.1210/jcem.84.1.5382 [DOI] [PubMed] [Google Scholar]
- 22.Ezeh U., Yildiz B.O. & Azziz R. Referral bias in defining the phenotype and prevalence of obesity in polycystic ovary syndrome. J Clin Endocrinol Metab 98, E1088–96 (2013). 10.1210/jc.2013-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand-Ivell R. & Ivell R. Regulation of the reproductive cycle and early pregnancy by relaxin family peptides. Mol Cell Endocrinol 382, 472–9 (2014). 10.1016/j.mce.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 24.Jiang F. & Wang Z. Identification and characterization of PLZF as a prostatic androgen-responsive gene. Prostate 59, 426–35 (2004). 10.1002/pros.20000 [DOI] [PubMed] [Google Scholar]
- 25.Wang N. et al. Promyelocytic leukemia zinc finger protein activates GATA4 transcription and mediates cardiac hypertrophic signaling from angiotensin II receptor 2. PLoS One 7, e35632 (2012). 10.1371/journal.pone.0035632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambele M.A., Dessels C., Durandt C. & Pepper M.S. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res 16, 725–34 (2016). 10.1016/j.scr.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 27.Lovelace D.L. et al. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development 143, 1893–906 (2016). 10.1242/dev.132761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kommagani R. et al. The Promyelocytic Leukemia Zinc Finger Transcription Factor Is Critical for Human Endometrial Stromal Cell Decidualization. PLoS Genet 12, e1005937 (2016). 10.1371/journal.pgen.1005937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masson O. et al. LRP1 receptor controls adipogenesis and is up-regulated in human and mouse obese adipose tissue. PLoS One 4, e7422 (2009). 10.1371/journal.pone.0007422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenaway J. et al. Thrombospondin-1 inhibits VEGF levels in the ovary directly by binding and internalization via the low density lipoprotein receptor-related protein-1 (LRP-1). J Cell Physiol 210, 807–18 (2007). 10.1002/jcp.20904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efimenko E. et al. The transcription factor GATA4 is required for follicular development and normal ovarian function. Dev Biol 381, 144–58 (2013). 10.1016/j.ydbio.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Do R., Kiss R.S., Gaudet D. & Engert J.C. Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway. Clin Genet 75, 19–29 (2009). 10.1111/j.1399-0004.2008.01099.x [DOI] [PubMed] [Google Scholar]
- 33.Chalasani N. et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology 139, 1567–76, 1576 e1-6 (2010). 10.1053/j.gastro.2010.07.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fauser B.C. et al. Consensus on women's health aspects of polycystic ovary syndrome (PCOS): the Amsterdam ESHRE/ASRM-Sponsored 3rd PCOS Consensus Workshop Group. Fertil Steril 97, 28-38.e25 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Welt C.K. et al. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab 97, E1342–7 (2012). 10.1210/jc.2011-3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wray N. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681 (2018). 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norman R.J., Masters S. & Hague W. Hyperinsulinemia is common in family members of women with polycystic ovary syndrome. Fertil Steril 66, 942–7 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Cela E. et al. Prevalence of polycystic ovaries in women with androgenic alopecia. Eur J Endocrinol 149, 439–42 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Quinn M. et al. Prevalence of androgenic alopecia in patients with polycystic ovary syndrome and characterization of associated clinical and biochemical features. Fertil Steril 101, 1129–34 (2014). 10.1016/j.fertnstert.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 40.Pinola P. et al. Menstrual disorders in adolescence: a marker for hyperandrogenaemia and increased metabolic risks in later life? Finnish general population-based birth cohort study. Hum Reprod 27, 3279–86 (2012). 10.1093/humrep/des309 [DOI] [PubMed] [Google Scholar]
- 41.Spector T.D. & Williams F.M. The UK Adult Twin Registry (TwinsUK). Twin Res Hum Genet 9, 899–906 (2006). 10.1375/183242706779462462 [DOI] [PubMed] [Google Scholar]
- 42.Lindstrom S. et al. A comprehensive survey of genetic variation in 20,691 subjects from four large cohorts. PLoS One 12, e0173997 (2017). 10.1371/journal.pone.0173997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferriman D. & Gallwey J.D. Clinical assessment of body hair growth in women. J Clin Endocrinol Metab 21, 1440–7 (1961). 10.1210/jcem-21-11-1440 [DOI] [PubMed] [Google Scholar]
- 44.Solomon C.G. et al. Long or highly irregular menstrual cycles as a marker for risk of type 2 diabetes mellitus. JAMA 286, 2421–6 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Winkler T.W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9, 1192–212 (2014). 10.1038/nprot.2014.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howie B.N., Donnelly P. & Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529 (2009). 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott L.J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–5 (2007). 10.1126/science.1142382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magi R. & Morris A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 11, 288 (2010). 10.1186/1471-2105-11-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willer C.J., Li Y. & Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders C.T. & Baker D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J Mol Biol 322, 891–901 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Adzhubei I.A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–9 (2010). 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kircher M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–5 (2014). 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davydov E.V. et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6, e1001025 (2010). 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garber M. et al. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25, i54–62 (2009). 10.1093/bioinformatics/btp190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown G.R. et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res 43, D36–42 (2015). 10.1093/nar/gku1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coordinators N.R. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res 45, D12–D17 (2017). 10.1093/nar/gkw1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.International Schizophrenia C. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–52 (2009). 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bulik-Sullivan B.K. et al. LD An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–41 (2015). 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Locke A.E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015). 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Felix J.F. et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet 25, 389–403 (2016). 10.1093/hmg/ddv472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manning A.K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44, 659–69 (2012). 10.1038/ng.2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris A.P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44, 981–90 (2012). 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willer C.J. et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 45, 1274–1283 (2013). 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perry J.R. et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 514, 92–97 (2014). 10.1038/nature13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nikpay M. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47, 1121–1130 (2015). 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagenaars S.P. et al. Genetic prediction of male pattern baldness. PLoS Genet 13, e1006594 (2017). 10.1371/journal.pgen.1006594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yavorska O.O. & Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 46, 1734–1739 (2017). 10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bowden J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol 45, 1961–1974 (2016). 10.1093/ije/dyw220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 81, 208–27 (2007). 10.1086/519024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wellcome Trust Case Control, C. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet 44, 1294–301 (2012). 10.1038/ng.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Summary statistic GWAS meta-analysis results for the combined dataset excluding 23andMe are available at https://doi.org/10.17863/CAM.27720. The most significant 10,000 SNPs for the meta-analysis including 23andMe are available at https://doi.org/10.17863/CAM.27720.