Abstract
We used a modified Walker–Mason scald burn rat model to demonstrate that Pseudomonas aeruginosa, a common opportunistic pathogen in the burn ward and notable biofilm former, establishes biofilms within deep partial-thickness burn wounds in rats.
Deep partial-thickness burn wounds, ~10% of the TBSA, were created in anesthetized male Sprague-Dawley rats (350–450 g; n = 84). Immediately post-burn, 100 µl of P. aeruginosa in phosphate-buffered saline at 1 × 103, 1 × 104, or 1 × 105 cells/wound was spread over the burn surface . At 1, 3, 7, and 11 days post-burn, animals were euthanized and blood and tissue were collected for complete blood counts, colony-forming unit (CFU) counts, biofilm gene expression, histology, scanning electron microscopy (SEM), and myeloperoxidase activity in the burn eschar.
P. aeruginosa developed robust biofilm wound infections, plateauing at ~1 × 109 CFU/g burn tissue within 7 days regardless of inoculum size. Expression of Pseudomonas alginate genes and other virulence factors in the infected wound indicated formation of mature P. aeruginosa biofilm within the burn eschar. Compared to un-inoculated wounds, P. aeruginosa infection caused both local and systemic immune responses demonstrated by changes in systemic neutrophil counts, histology, and myeloperoxidase activity within the burn wound. Additionally, SEM showed P. aeruginosa enmeshed within an extracellular matrix on the burn surface as well as penetrating 500–600 µm deep into the eschar.
P. aeruginosa establishes biofilms within deep partial-thickness burn wounds and invades deep into the burned tissue. This new in vivo biofilm infection model is valuable for testing novel anti-biofilm agents to advance burn care.
Pseudomonas aeruginosa is a gram-negative opportunistic pathogen known for its propensity to form biofilms. Biofilms are extracellular matrices composed of bacteria-derived exopolysaccharides, extracellular DNA, proteins, and bacterial cells themselves.1–6 Similarly, Pseudomonas biofilms contain several forms of exopolysaccharides (alginate, Psl, and Pel), extracellular DNA, proteins, and lipids. A key attribute afforded by biofilm lifestyle is the inherent drug resistance imparted to bacterial cells within the matrix. Bacterial cells dwelling within the biofilm require nearly 1000× the antimicrobial concentration needed to kill their planktonic, single-cell living, counterparts.7–9 The same phenomena holds true for their ability to resist attack and phagocytosis by a host’s immune system.10–12 The mechanisms of this resistance are partly attributed to the exopolysaccharides that physically block, sequester, or inactivate/degrade antibiotics before interacting with bacterial cells13–16; additionally, cells dwelling deep within the biofilm may be sessile or non-growing, and thereby evade the mechanisms of action of many antimicrobial agents.17–20 Not surprisingly, bacterial biofilms are often associated with many long-lasting infections such as cystic fibrosis,21–24 chronic otitis media,25–27 chronic skin wounds (pressure, venous, and diabetic foot ulcers),28–31 and dental plaques.32–34
During the initial hours and days post-burn, gram-positive staphylococci, as members of normal skin flora, colonize the wound surface because they often survive the thermal injury.35 Often within a week following a burn, secondary colonization of the burn wound surface by other bacterial and fungal species occurs, including P. aeruginosa. Contamination of the burn wound with P. aeruginosa may lead to an invasive burn wound infection and potentially fatal sepsis.36,37 This was the basis for development of the original Walker–Mason rat scald model at the U.S. Army Institute of Surgical Research (USAISR) in the 1960s to evaluate treatment modalities to reduce mortality associated with Pseudomonas burn wound infection. Upon infection, of either partial- or full-thickness burn wounds in rats, with clinical burn wound isolates of P. aeruginosa, severe invasive burn wound infection developed resulting in animal death.38–43 This recapitulated the clinical observations of P. aeruginosa burn infection. Directly out of this model, two treatments—mafenide acetate (Sulfamylon®) and silver sulfadiazine (Silvadene®)—were examined and showed near complete protection of the animal from infection with high inoculums, 106–108 colony-forming unit (CFU)/wound, of the pathogen.39,44–46 Additionally, the role of early excision was noted for improving survival of a large TBSA burn with concomitant P. aeruginosa infection.47 This formed the basis of current clinical practice for treatment of severe burn wounds at the USAISR burn center. These treatments, while immensely effective at preventing invasive P. aeruginosa burn wound infection, primarily target planktonic Pseudomonas bacteria and are severely limited by their ease of application and require a great number of person-hours to perform on a daily basis.45 In addition, delaying treatment by only 48–72 hours after the burn decreased the effectiveness of the antimicrobial treatments.39 We hypothesized that P. aeruginosa, left untreated, forms biofilms within the burn eschar, thereby resulting in both exacerbated wound progression and systemic inflammatory effects.
Here, we describe the development of P. aeruginosa biofilms within partial-thickness burn wounds using a modified Walker–Mason scald model.48 We determined that the time required for creating partial- and full-thickness burn wounds in 350–450 g Sprague-Dawley rats was 3 and 6 seconds at 99–100°C, respectively. We used the same clinical P. aeruginosa isolate as the original Walker–Mason scald burn model, strain 12-4-4(59). This strain was isolated at the Brooke Army Medical Center (Joint Base San Antonio [JBSA]-Fort Sam Houston, TX) from a female burn patient with partial- and full-thickness burns comprising 70% TBSA.38,49P. aeruginosa formed robust wound infections regardless of inoculation level. In the wound tissue, several biofilm-associated genes were significantly upregulated. Scanning electron microscopy (SEM) revealed the presence of Pseudomonas communities embedded within matrix material and the invasive nature of the pathogen to penetrate the burn eschar. To our knowledge, this is the first report of a biofilm-based infection in partial-thickness burn wounds using a rat model.
METHODS
Partial-Thickness Infection Scald Model
Animal Ethics Statement.
The Institutional Animal Care and Use Committee (IACUC) at the USAISR (JBSA-Fort Sam Houston, TX) approved this study (animal protocol numbers A-16-020 and A-16-035) on January 22, 2016 and May 31, 2016, respectively. Research was conducted in compliance with the Animal Welfare Act, the implementing Animal Welfare Regulations, and the principles of the Guide for the Care and Use of Laboratory Animals, National Research Council. The facility’s IACUC approved all research conducted in this study. The facility where this research was conducted is fully accredited by AAALAC International.
Deep partial-thickness burns were created on the dorsum of 84 male Sprague-Dawley rats weighing 350–450 g. The burn area equaled ~10% of the TBSA as determined by Meeh’s formula, , where A is the TBSA in cm2, k is the Meeh constant (9.46 for Sprague-Dawley rats), and W is body weight in grams.50
One day prior to scalding, rats were anesthetized with 2.5–4% isoflurane (Forane, Baxter Healthcare Corporation, Deerfield, IL) and the dorsum shaved and depilated with Nair (Church & Dwight Co., Inc., Ewing, NJ). Buprenorphine SR LAB (1.2 mg/kg; Zoopharm Pharmacy) was administered subcutaneously for proactive pain management. Rats were housed individually following shaving. On the day of the burn, rats were anesthetized with 2.5% isoflurane for 15 minutes. Once anesthetized, eye lube (Artificial Tears Ointment, Akorn, Inc., Lake Forrest, IL) was applied and initial vital signs (ie, blood oxygen content and pulse rate) were measured using a pulse oximeter (2500A Vet Pulse Oximeter, Nonin Medical, Inc., Plymouth, MN).
To induce the burn injury, anesthetized rats were positioned in an insulated polyvinyl chloride mold (Figure 1A) with a 9 × 5.3 cm opening allowing exposure of ~10% of the TBSA to near-boiling water. Upon correct positioning in the template (Figure 1B), anesthetic surgical plane was verified via toe pinch and the rats were locked in place with Velcro straps. The entire mold was lowered into the near-boiling water. Exposure to 99°C water for 3 seconds resulted in partial-thickness burn injury, while exposure for 6 seconds resulted in full-thickness burn injury (see “Results”). A Precision CIR 35 circulating water bath (Thermo Scientific, Waltham, MA) set at 99°C water temperature was used to induce the scald burns.
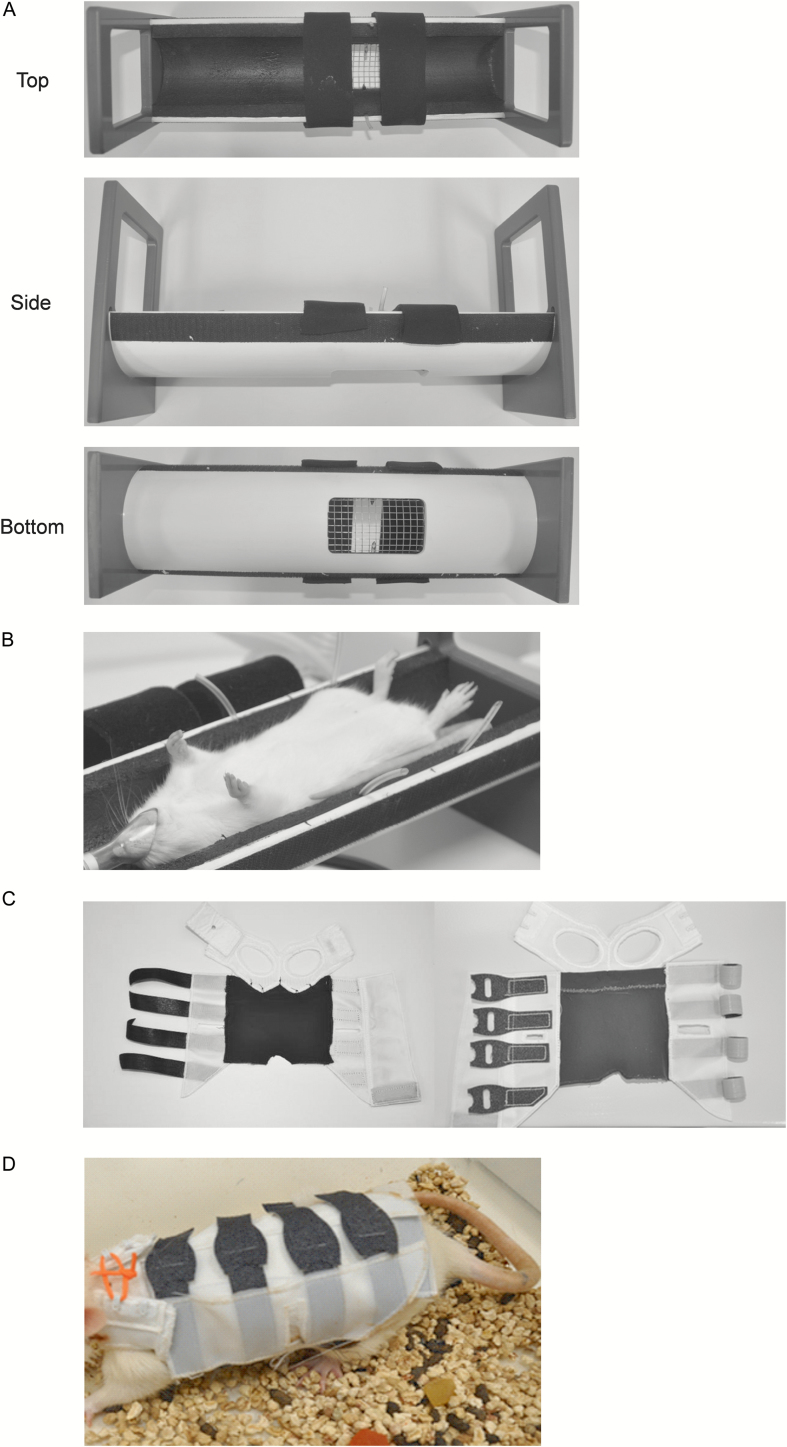
Figure 1.
A. Side and top views of the burn template designed to expose ~10% of the TBSA to near-boiling water. Note two Velcro straps hold the rat in place and 2-mm-diameter tubing on either side of the burn window allows escape of trapped air as the burn mold is lowered into the water bath. B. Demonstration of a rat positioned correctly in the burn template. C. Top (left panel) and bottom (right panel) views of the composite rat jacket. The jacket comprised three pieces: a front leg strap, in which the front legs are inserted, sutured to the piece that wraps around the ventral and dorsal surfaces and a Velcro tab that holds the front strap closed when placed on the rat. D. Demonstration of a rat wearing the jacket, which remained on the animals and was well tolerated for up to 11 days.
Immediately after the burn, excess water was blotted from the skin using damp papers towels (room temperature). Warmed (37°C) lactated Ringer’s solution (LRS) (Baxter Healthcare Corporation) was used to resuscitate the rats after the burn based on Parkland’s formula: Volume resuscitation (ml) = 4 × Body weight (kg) × Percent burn of TBSA. On average, the rats were resuscitated with 16 ml of LRS, split into four equal intraperitoneal injections over 24 hours following the burn. The burn injury was imaged using a Nikon D90 with an attached Nikkor AF-S lens (18–105 mm, 1:3.5–5.6 G) immediately following the burn and on each of the experimental end dates.
The burn surface was inoculated with P. aeruginosa strain 12-4-4(59). P. aeruginosa was grown in Trypticase Soy Broth (TSB; Becton, Dickinson and Co., Sparks, MD) overnight at 37°C and subcultured the next morning into fresh TSB to reach the mid-logarithm (log) growth phase. Bacteria cells in the mid-log growth phase were centrifuged at 3000g for 15 minutes at 4°C and re-suspended in 1× phosphate-buffered saline (PBS) (Sigma, St. Louis, MO) and adjusted to different inoculum sizes based on optical density at the wavelength of 600 nm. Bacterial cells (in 100 µl PBS) of different inoculum sizes (103, 104, or 105 CFU/wound) were spread over the burn area using a sterile pipette tip. Sterile PBS was used as a control (un-inoculated). The total number of animals included in the study was 84, with 12 serving as controls and 24 in each P. aeruginosa inoculation group. The experiment was repeated three times. In each experimental repeat (n = 28), the number of animals in each inoculation group was set to two per day and the number of controls was set to one per day for each of the experimental end points, post-operative day (POD) 1, 3, 7, and 11. In addition to the primary study, a baseline data set (n = 30) was generated using animals exposed to the exact same experimental conditions minus the burn and surface inoculation of P. aeruginosa (sham burn group). An additional 36 rats were used to optimize the scald times for partial- and full-thickness burn wounds in Sprague-Dawley rats.
The wounds were covered with a non-adherent dressing (N-Terface Wound Contact Layer, Winfield Laboratories, Inc., Richardson, TX). Tegaderm™ Film (3M Health Care, St. Paul, MN) was placed over the non-adherent dressing and sealed to the skin surrounding the burn with NOTAPE professional silicone bonding adhesive (Vapon, Inc., Fairfield, NJ). After bandaging, rats were placed into a composite rat jacket (developed in house by co-authors L.Q. and P.C.; Figure 1C and D) to prevent disruption of the Tegaderm Film and self-inflicted damage to the underlying burn wound. The rats were recovered in a ThermoCare Portable Animal Intensive Care Unit (Daisy Products LLC, Paso Robles, CA). Daily weights and pain assessments were made throughout the experiment to determine if additional pain relief or fluid resuscitation was required.
End-Point Procedures
On each euthanasia day, rats were anesthetized with 100 mg/kg Ketamine HCl (Zetamine, MWI Veterinary Supply Co., Boise, ID) and 10 mg/kg Xylazine (Akorn Animal Health, Inc., Lake Forest, IL). Additionally, 4% isoflurane was delivered via nosecone to ensure depth of surgical anesthetic plane, as needed. Blood was obtained via cardiac puncture followed immediately by direct intra-cardiac injection of Fatal-Plus® (Vortech Pharmaceuticals, Ltd., Dearborn, MI). Euthanasia was confirmed by lack of cardiac movement, pulse, and breathing as specified by the AVMA Guidelines for the Euthanasia of Animals: 2013 Edition.51
After confirming euthanasia, the Tegaderm dressings were removed and the wounds were imaged. The entire burn area was excised from the dorsum and three separate histologic cross-sections were taken from the cranial, middle, and caudal parts of the burn. Additionally, wound tissue was isolated using a 7-mm biopsy punch for: quantitative bacteriology, bacterial mRNA expression levels, myeloperoxidase (MPO) activity, and SEM of the burn wound surface. Spleens, liver, and kidney samples were also taken at each experimental end point, flash frozen in Liquid Nitrogen, and stored at −80°C for future analysis. After collecting the cardiac blood samples in EDTA-coated blood collection tubes (BD Vacutainer 367841) and analyzing on an Abbott CELL-DYN® 3700 Blood Count Analyzer (Abbott Laboratories, Abbott Park, IL), systemic neutrophil counts were determined.
Bacterial Load Quantification
Four random 7-mm biopsy punches from the burn wound were placed in pre-weighed MagNA Lyser Green Beads tubes (Roche Diagnostics GmbH, Mannheim, Germany), weighed to determine the tissue weight, and homogenized with 1 ml PBS using a FastPrep®-24 Tissue Homogenizer (MP Biomedicals, LLC, Santa Ana, CA). The samples were serial diluted with PBS and plated on Trypticase soy agar containing 5% sheep’s blood (Becton, Dickinson and Co.) as well as P. aeruginosa isolation agar (Hardy Diagnostics, Santa Maria, CA) using a WASP 2 Spiral Plater (Microbiology International, Frederick, MD). Viable CFUs were enumerated using a ProtoCOL 3 Colony Counter (Microbiology International) and plotted as log10 (CFU/g wound tissue) ± SEM.
Fifty microliters from each of the four biopsy homogenates per animals were pooled for analysis by PCR using P. aeruginosa-specific and universal primers. Bacterial DNA was isolated using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. For quantification of P. aeruginosa tissue load by qPCR, primers and probe were designed using the outer membrane lipoprotein gene oprL sequence.52 For the quantification of total bacterial load in each wound, the sequences of the universal primers and probe set used were based on the amplification of 16S rDNA.53 Primers and probes, sequences listed in Table 1, were synthesized by Applied Biosystems (Carlsbad, CA). The specificity of the primers and probe set for P. aeruginosa was confirmed by lack of amplification when genomic DNA from other bacteria was used as the template in the PCR reactions. Genomic P. aeruginosa DNA from the mid-log growth phase culture was isolated to establish a standard curve for bacterial quantification of wound samples. The concentration of isolated genomic DNA was determined using a Quant-iT ds DNA BR Assay Kit (Invitrogen, Carlsbad, CA) and confirmed by gel electrophoresis.
Table 1.
List of PCR primers for 16s ribosomal target regions and qRT–PCR
| Gene name | Primer sequence 5ʹ to 3ʹ |
|---|---|
| P. aeruginosa oprL – forward | 5ʹ-ATGGAAATGCTGAAATTCGGC-3ʹ |
| P. aeruginosa oprL – reverse | 5ʹ-CTTCTTCAGCTCGACGCGACG-3ʹ |
| P. aeruginosa oprL – probe | (6FAM)-5ʹ-TGCGATCACCACCTTCTACTTCGAGT-3ʹ-MGBNFQ |
| Total 16S rDNA – forward | 5ʹ-TCCTACGGGAGGCAGCAGT-3ʹ |
| Total 16S rDNA – reverse | 5ʹ-GGACTACCAGGGTATCTAATCCTGTT-3ʹ |
| Total 16S rDNA – probe | (6FAM)-5ʹ-CGTATTACCGCGGCTGCTGGCAC-3ʹ-MGBNFQ |
| FabD – forward | 5ʹ-GGTCCAGAATGGTCCTGAAGAG-3ʹ |
| FabD – reverse | 5ʹ-CGATCGAAACCGTAAGGATGGC-3ʹ |
| PvdS – forward | 5ʹ-GATAACCGTACGATCCTGGTGAAG-3ʹ |
| Pvds – reverse | 5ʹ-GACGATCTGGAACAGGTAGCTGAG-3ʹ |
| Alg8 – forward | 5ʹ-CAAGGATTTCATCCTGCTTATCGG-3ʹ |
| Alg8 – reverse | 5ʹ-GAAACTGGTGACCATCAGGAACA-3ʹ |
| AlgE – forward | 5ʹ-GACAACCTCGACAAGACCTATACC-3ʹ |
| AlgE – reverse | 5ʹ-CTCGAACGATAGTTGTAGGCATCG-3ʹ |
All real-time PCR reactions were performed with a StepOne Plus Real-Time PCR System (Applied Biosystems) using optical grade 96-well plates. All samples were run in triplicate. Each PCR reaction was performed in a total volume of 20 µl of TaqMan Gene Expression Master Mix (Applied Biosystems) containing 100 nM of each of the forward and reverse primers, TaqMan MGB probe, and 5 µl of the sample template. The reaction conditions for DNA amplification were 95°C for 5 minutes, 40 cycles (95°C for 15 seconds followed by 60°C for 1 minute). StepOne software provided by Applied Biosystems was used to analyze the data. The amount of P. aeruginosa (or total) genomic DNA in each sample was converted into genome copy number and normalized by the weight of the wound sample.
Histology
Three tissue cross-sections (5–10 mm) from each wound were fixed in 10% buffered formalin in PBS (Fisher Diagnostics, Kalamazoo, MI) for at least 48 hours and processed for routine embedding in paraffin wax. Thin 4–5-µm sections from each sample were stained with hematoxylin–eosin (H&E) and terminal deoxnucleotidyl transferase dUTP nick-end labeling (TUNEL) for evaluation of tissue damage and burn depth. Penetration of P. aeruginosa into the eschar was visualized by SEM and peptide nucleic acid (PNA)-fluorescent in situ hybridization (FISH).
TUNEL Staining
To detect apoptotic cells in the burn tissue, histologic cross-sections were stained using a Click-iT® Plus TUNEL Assay kit (Life Technologies, Carlsbad, CA) following the manufacturer’s directions. Briefly, tissue sections on slides were deparaffinized with xylene and re-hydrated with an ethanol series before fixation in 4% paraformaldehyde for 15 minutes. The tissue was permeabilized with Proteinase K for 15 minutes and additionally fixed with 4% paraformaldehyde for 5 minutes. The tissue was incubated in TdT Reaction Buffer for 10 minutes before addition of the TdT Reaction Mixture for 60 minutes at 37°C. Tissue sections were then washed with 3% bovine serum albumin (BSA) in 0.1% Triton X-100 for 5 minutes, rinsed with PBS, and incubated with Click-iT Plus TUNEL reaction cocktail for 30 minutes at 37°C in the dark before washing with 3% BSA in PBS for 5 minutes. The samples received a final rinse in PBS before being cover slipped using ProLong Gold Antifade Mountant with 4ʹ,6-diamidino-2-phenylindole (DAPI; Life Technologies). Fluorescent images were acquired using a ×10 objective and Ex/Em wavelengths of 590/615 nm for Alexa Fluor 594 to show damaged DNA strands and 358/461 nm for DAPI as a general nuclei stain on a Leica Aperio Versa 200 (Leica Biosystems, Inc., Buffalo Grove, IL) slide scanner.
Histopathology Assessment
The semi-quantitative analyses were based on the following scale: 0 is normal skin; 1 is epithelial necrosis within the epidermis, but the basement membrane remains intact; no dermal changes; 2 is epithelial ulceration, thinning, or regeneration; denatured collagen in top half of dermis; hair follicles and sebaceous glands in top half of dermis are necrotic (+/−); inflammation (+/−); 3 is epithelial ulceration, thinning, or regeneration; denatured collagen in top and bottom half of dermis; inflammation from dermis extends into muscle layer; degeneration, necrosis, and/or regeneration of superficial individual myocytes in muscle layer; hair follicles and sebaceous glands in top half and hair follicles in bottom half are necrotic (+/−); hemorrhage, congestion, and edema in mid to deep dermis (+/−); inflammation (+/−); 4 is epithelial ulceration, thinning, or regeneration; denatured collagen in top and bottom half of dermis; hair follicles and sebaceous glands in top half of dermis and hair follicles of bottom half of dermis are necrotic; hemorrhage, congestion, and edema in deep dermis present; degeneration, necrosis, and/or regeneration of entire muscle layer present; inflammation present.
Peptide Nucleic Acid-Fluorescent in situ Hybridization
Histology sections of un-inoculated and P. aeruginosa-infected burn wound tissue on PODs 7 and 11 were probed with the AdvanDx Escherichia coli/P. aeruginosa PNA-FISH Culture Identification Kit KT007 (OpGen, Inc., Gaithersburg, MD) to confirm the penetration of P. aeruginosa into the burn eschar. Briefly, 4-µm-thick sections were deparaffinized using xylene. AvanDx fixative solution was applied to cover the tissue section on the slide and incubated for 20 minutes on a 55°C slide warmer. One drop of the PNA-FISH probe was hybridized to each tissue section using a coverslip for 45 minutes on the slide warmer. The coverslip was removed by allowing the slide to soak in heated wash buffer, and the tissue section was rinsed in 55°C wash buffer (included in the kit) for 45 minutes. After washing, the samples were cover slipped using the included mounting medium and edges were sealed with Permount. Tissue sections were evaluated using the Texas Red filter (596/615 nm) with a ×10 objective on a Leica Aperio Versa 200 slide scanner.
Scanning electron microscopy
For observation of burn wound surface-associated P. aeruginosa biofilms, biopsy tissues were fixed by 2.5% phosphate-buffered glutaraldehyde for 24 hours at 4°C. The fixed samples were dehydrated in a graded series of cold ethanol/water (increasing from 10, 30, 50, 70, 80, 90, 95% to 100% ethanol) for 10 minutes each. The samples were dehydrated using a critical point dryer (EM CPD300, Leica Biosystems, Inc.) and coated with carbon and gold/palladium using a Leica ACE600 Coater.
To determine P. aeruginosa tissue penetration, paraffin-embedded histologic sections on microscopy slides were deparaffinized with 100% xylene and allowed to air dry before being coated with carbon and gold/palladium. All samples were observed with a Sigma VP40 field emission scanning electron microscope (Carl Zeiss, Inc., Germany) in high vacuum mode at 2 kV.
RNA Extraction and Quantitative RT–PCR for P. aeruginosa Biofilm Genes
Wound tissue used to quantify biofilm gene expression were placed immediately in tubes pre-filled with 1 ml of RNA stabilization mix (3 volumes of RNAprotect Bacteria Reagent [Qiagen, Germantown, MD] with 1 volume of PBS) and MagNA Lyser Green beads. Tissues were homogenized for 90 seconds at 5000 rpm in a FastPrep®-24 Tissue Homogenizer. Bacterial RNA was isolated with TRIzol (Life Technologies) and purified using RNEasy Mini-Kit (Qiagen). Genomic DNA was removed by treatment with DNAse I (Ambion, ThermoFisher) and the absence of DNA contamination was confirmed using a control without reverse transcriptase and demonstrating a Ct value 10 cycles higher than the reverse transcribed samples. The RNA was quantified (A260/A280, NanoDrop, Thermo Scientific). The purified RNA was reverse transcribed to cDNA using the iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA). The RNA was isolated and pooled from four 7-mm biopsy samples harvested from each animal. Each gene transcript-level determination was from RNA pooled from 16 to 20 infected wounds. Quantitative real-time PCRs were performed using the SYBR green master mix (Bio-Rad) with specified primers (Table 1) and analysis by the StepOne System (Applied Biosystems) with relative changes using fabD housekeeping gene, and fold difference with 2−ΔΔCt method. Unpaired Student’s t test and p<.05 were implemented.
MPO Assay
Tissue samples were freeze fractured under Liquid Nitrogen using a Bessman Tissue Pulverizer (Spectrum, Inc., Rancho Dominguez, CA) and collected in pre-weighed Eppendorf tubes prior to homogenization. Samples were homogenized in accordance with the Fluoro MPO Myeloperoxidase Detection Kit (Cell Technology, Inc., Mountain View, CA). Briefly, samples were processed using an IKA T10 basic Ultra Turrax tissue homogenizer (IKA Works, Inc., Wilmington, NC), centrifuged at 12,000g for 20 minutes at 4°C. The tissue pellets were re-suspended in MPO solubilization buffer, reprocessed with the tissue homogenizer, sonicated with a Sonic Dismembrator Model 100 (Fisher Scientific, Kalamazoo, MI) for 5 seconds, and subjected to two freeze-thaw cycles before isolating and storing the supernatant at −80°C. Fifty microliters of MPO enzyme standard (from the kit) or sample were mixed with an equal volume of MPO reaction cocktail and incubated for 60 minutes at room temperature in black-opaque, clear-bottom, microtiter plates before measuring fluorescence (530/590 nm). Relative fluorescence was converted to MPO activity using the standard curve generated from the MPO standard included in the kit.
Statistical Analysis
GraphPad Prism 7.03 (GraphPad Software, Inc., San Diego, CA) was used to plot and analyze the data. Results were compared using two-way analysis of variance with Tukey’s multiple comparisons test to determine significant differences between experimental groups. Data were plotted as the mean ± SEM.
RESULTS
Partial-Thickness Rat Scald Burn Model Development
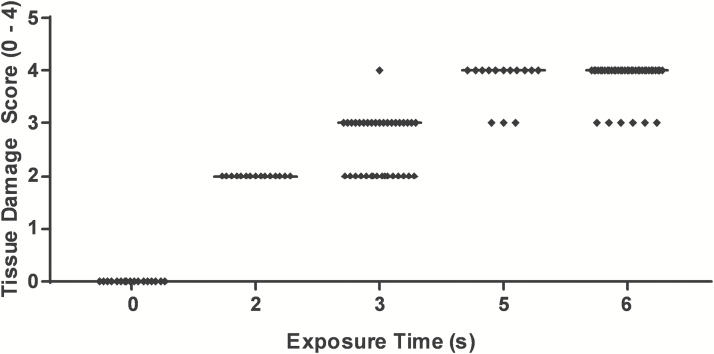
Figure 2 shows a scatter plot of the pathologists’ assessment of the tissue damage with regards to exposure time to near-boiling water. The samples evaluated by the pathologists included sham burned skin (n = 6), 2 seconds (n = 5), 3 seconds (n = 12), 5 seconds (n = 5), and 6 seconds (n = 14) burned skin. Supplementary Figure 1 shows representative photomicrographs of the H&E-stained tissue sections exposed to near-boiling water for varying times (0–6 seconds). Normal skin (sham burn) is shown in Supplementary Figure 1A–C. A 2-second exposure time resulted primarily in superficial to partial-thickness burn injury (Supplementary Figure 1D and E). Increasing exposure time to 3 seconds resulted in consistent deep partial-thickness burn wounds (Supplementary Figure 1F and G). Exposure to near-boiling water for 6 seconds resulted in a consistent full-thickness burn wounds, without damaging the underlying skeletal muscle (Supplementary Figure 1H and I). An additional study with three rats, lasting through POD 21, confirmed the full-thickness burn exposure time of 6 seconds by the lack of hair regrowth within the burn region.
Figure 2.
Pathologist assessment of tissue damage score (1–4) versus skin exposure time to near-boiling (99°C) water for 0 second (sham burn, n = 6), 2 seconds (n = 6), 3 seconds (n = 12), 5 seconds (n = 5), and 6 seconds (n = 14).
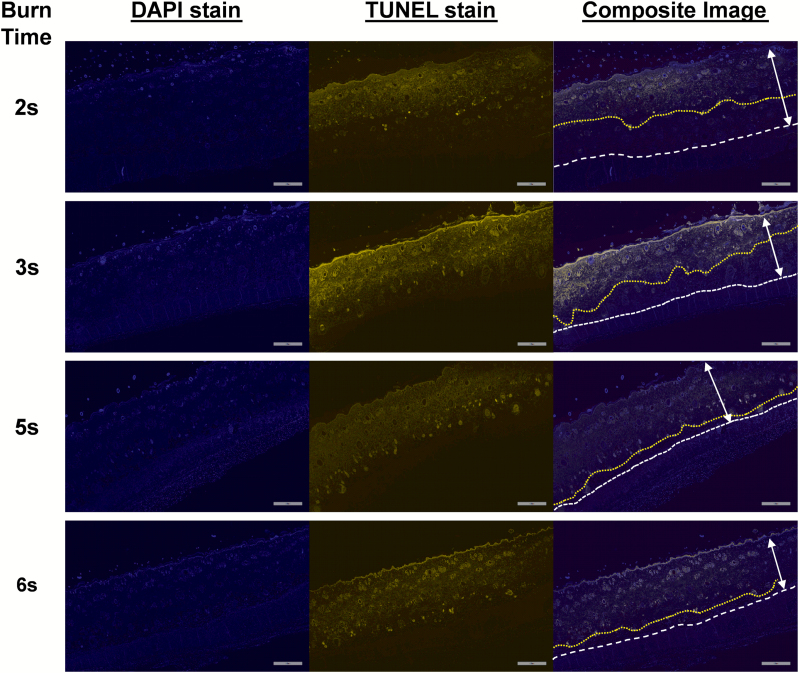
In addition to standard H&E staining, tissue sections were stained using the Click-iT Plus TUNEL Assay kit to visualize damaged DNA in necrotic cells. The damaged DNA was labeled with an Alexa Fluor 594 dye and DAPI used as a nuclear counterstain. Figure 3 shows representative tissue sections from 2-, 3-, 5-, and 6-second exposure to near-boiling water. Increasing the burn time resulted in greater DNA damage deeper in the skin, as indicated by TUNEL labeling.
Figure 3.
Representative histological sections of burn tissue from 2-, 3-, 5-, and 6-second scald times stained with TUNEL to visualize the depth of the burn by the fluorescently tagged damaged DNA strands. The two-headed arrow shows the distance from the epidermidis to the panniculus carnosus. The dashed white line shows the interface of the dermis and panniculus carnosus. The yellow dotted line indicates the deepest location of the TUNEL labeling visible in the dermis. The scale bar is 800 µm. DAPI, 4ʹ,6-diamidino-2-phenylindole; TUNEL, terminal deoxnucleotidyl transferase dUTP nick-end labeling.
P. aeruginosa Burn Infection
We used the newly modified Walker–Mason scald burn model to characterize the kinetics of a sublethal P. aeruginosa infection in partial-thickness burn injury. Overall, the rats tolerated the burn and infection well with limited weight loss. Figure 4 shows representative gross images of the burn wound designated by inoculum group and POD. Little visual difference was observed on POD 1 between any of the groups. By POD 3, the burned area was clearly delineated from the adjacent skin. Additionally, a majority of the P. aeruginosa-inoculated wounds began to show exudate on the wound’s surface in comparison to the un-inoculated group. During the tissue harvest on POD 7, all the P. aeruginosa-inoculated wounds showed maceration of the eschar surface and edema in the tissue underneath. By POD 11, the infection and massive host response to the wound infection caused separation of the burn eschar from the underlying muscle tissue in several of the 1 × 104 and 1 × 105 CFU/inoculum wound groups. Comparatively, the 1 × 103 CFU/inoculum showed signs of infection, but tissue separation was not observed. The un-inoculated burn wounds appeared much healthier with little to no tissue separation and edema.
Figure 4.
Representative images of partial-thickness burn wounds inoculated with or without P. aeruginosa over the course of 11 days post-burn. P. aeruginosa-infected burns showed presence of wound exudate by post-operative day (POD) 3 compared to little or none seen in the controls. By POD 7, wound discoloration and necrosis due to P. aeruginosa infection were clearly seen in the inoculated samples.
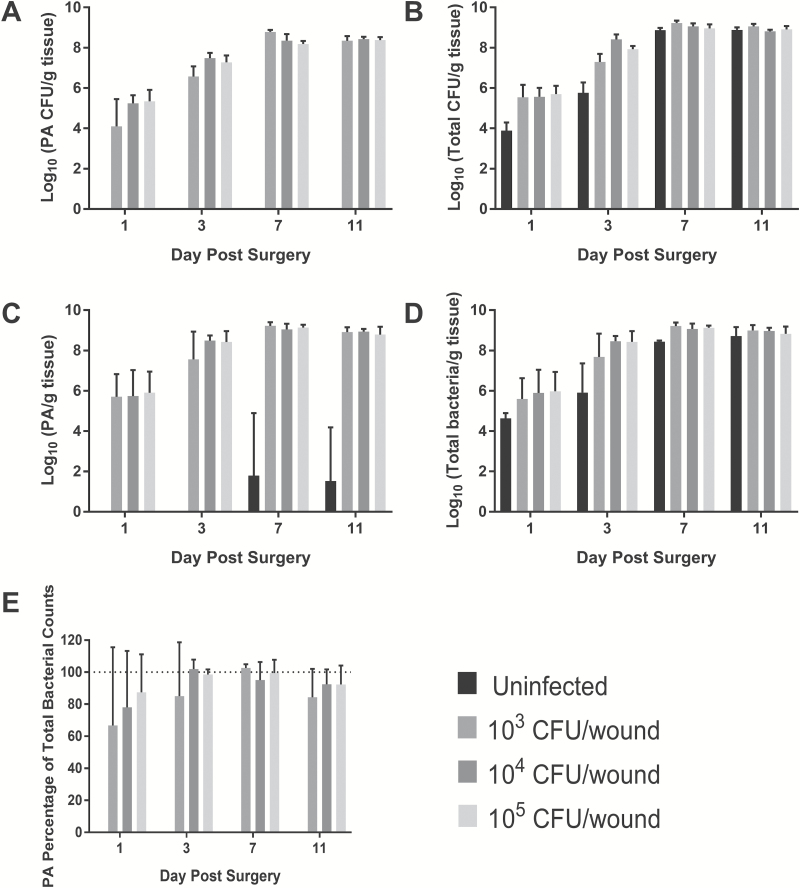
P. aeruginosa CFU counts, as plated on P. aeruginosa isolation agar, are shown in Figure 5A and was not recovered from any control wound. A clear dose effect in regard to inoculum level was observed at PODs 1 and 3, but by PODs 7 and 11, all P. aeruginosa CFU counts were equivalent at approximately 1 × 108 CFU/g burn tissue. When the wound homogenates were plated on non-selective nutrient blood agar, bacterial growth of Pseudomonas and non-Pseudomonas could be observed in every group as shown in Figure 5B. The total CFU counts in the P. aeruginosa-inoculated burn wounds mirrored the results seen from the selective agar. However, the un-inoculated burn wounds showed as high of a bacterial load as the others, albeit slightly less at PODs 1 and 3. Many of the microorganisms contaminating the un-inoculated burn wounds appeared to be part of the normal skin flora consisting of many gram-positive species (ie, Staphylococcus sciuri) and also included at least one Proteus species as determined using a GEN III OmniLog® Combo System (Biolog, Inc. Hayward, CA) (data not shown).
Figure 5.
Bacterial counts recovered from infected partial-thickness burn tissue over 11 days post-burn. P. aeruginosa (A) and total CFU counts (B) recovered as determined by serial dilution and plating on either P. aeruginosa isolation agar or Trypticase soy agar containing 5% sheep's blood, respectively. C. Total P. aeruginosa (live and dead) counts as determined by P. aeruginosa-specific primers. D. Total CFU counts as determined by general gram-negative primers. E. Percentage of total gram-negative bacterial load in the burn tissue that is P. aeruginosa. As time post-infection increased, P. aeruginosa accounted for a greater number of total bacterial cells isolated from wound tissue. P. aeruginosa eventually dominated as the primary wound pathogen. Burns without P. aeruginosa inoculation also became colonized with bacterial cells to the same level as the inoculated groups, but had greater diversity of gram-negative and positive cells. PA, P. aeruginosa.
PCR amplification of bacterial DNA using P. aeruginosa-specific and universal primers mirrored the CFU counts as shown in Figure 5C and D, respectively. Unfortunately, 2 of 12 un-inoculated biopsy punch samples were contaminated during the processing of the tissue, which led to positive PCR results in the P. aeruginosa-specific primer data set on PODs 7 and 11 for the un-inoculated samples. P. aeruginosa dominated the partial-thickness burn wound as the primary microorganism by POD 3, comprising 80–100% of the bacterial species within the wound as shown in Figure 5E. No significant differences in P. aeruginosa percentage of total wound bacterial cells, comprising >95% of the total, were observed between bacterial inoculation groups on PODs 7 and 11.
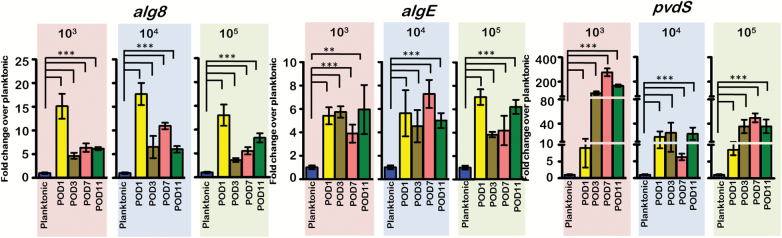
Expression of P. aeruginosa biofilm matrix components, alg8 and algE, and iron acquisition gene, pvdS, over the 11 days following the burn is shown in Figure 6. Regardless of infection level, alginate biosynthesis gene alg8 was upregulated 10–15× more than the planktonic inoculum on POD 1. After POD 3, the expression dropped, but still remained 5–10× higher than the inoculum. Conversely, algE was consistently upregulated 4–6× above the level of the planktonic cells on all PODs. Expression of pvdS was significantly upregulated compared to the planktonic inoculum at all-time points. The lowest inoculum had pvdS expression levels reaching 200× the planktonic cells between PODs 3 and 11. The pvdS expression levels of the 104 and 105 CFU/wound inoculums were upregulated ~40× more than the inoculum.
Figure 6.
Expression of biofilm matrix (alg8 and algE) and iron acquisition (pvdS) genes. Alg8, algE, and pvdS were significantly elevated in the P. aeruginosa-infected burn tissue as compared to the planktonic inoculum suggesting formation of mature P. aeruginosa biofilms within the burn wound. (***p < .001; **p < .01, unpaired Student’s t test). POD, post-operative day.
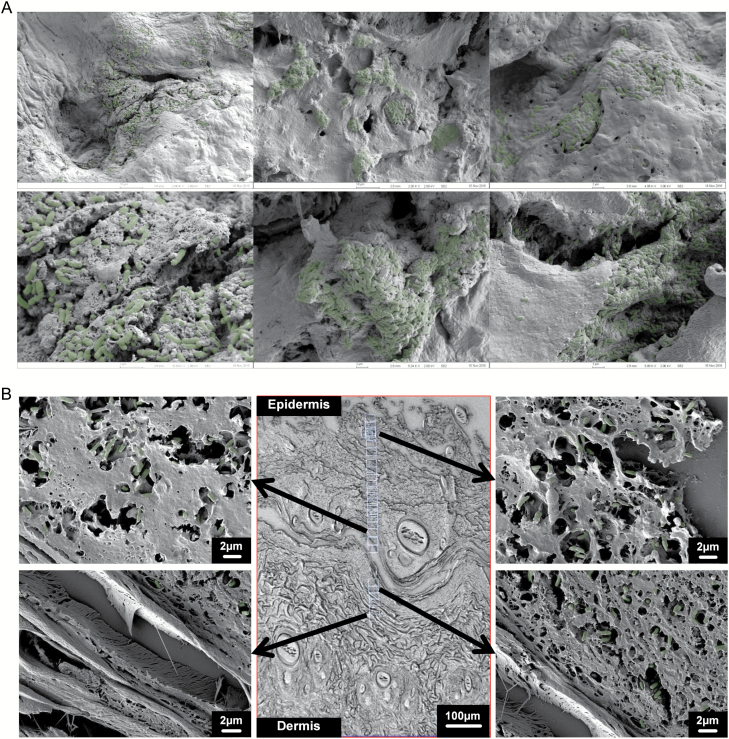
Surface imaging of the P. aeruginosa-infected burn eschar revealed communities of bacterial cells buried in a layer of extracellular debris as seen in Figure 7A. Examination of wound cross-sections revealed penetration of P. aeruginosa deep (500–600 µm) into the eschar layer of the burn wound, perhaps through at least the zone of coagulation and possibly zone of stasis as seen in Figure 7B. Penetration into the eschar by P. aeruginosa was confirmed using PNA-FISH with probes specific for P. aeruginosa as shown in Supplementary Figure 2.
Figure 7.
Scanning electron microscopy (SEM) of P. aeruginosa-infected burn tissue. A. Representative SEM images of the surface partial-thickness burn wounds infected with P. aeruginosa. Bacterial cells are pseudo-colored green for easier viewing. B. Representative SEM images of histological cross-sections of partial-thickness burn wound infected with P. aeruginosa. The bacterial cells penetrate deep (500–600 µm) into the eschar.
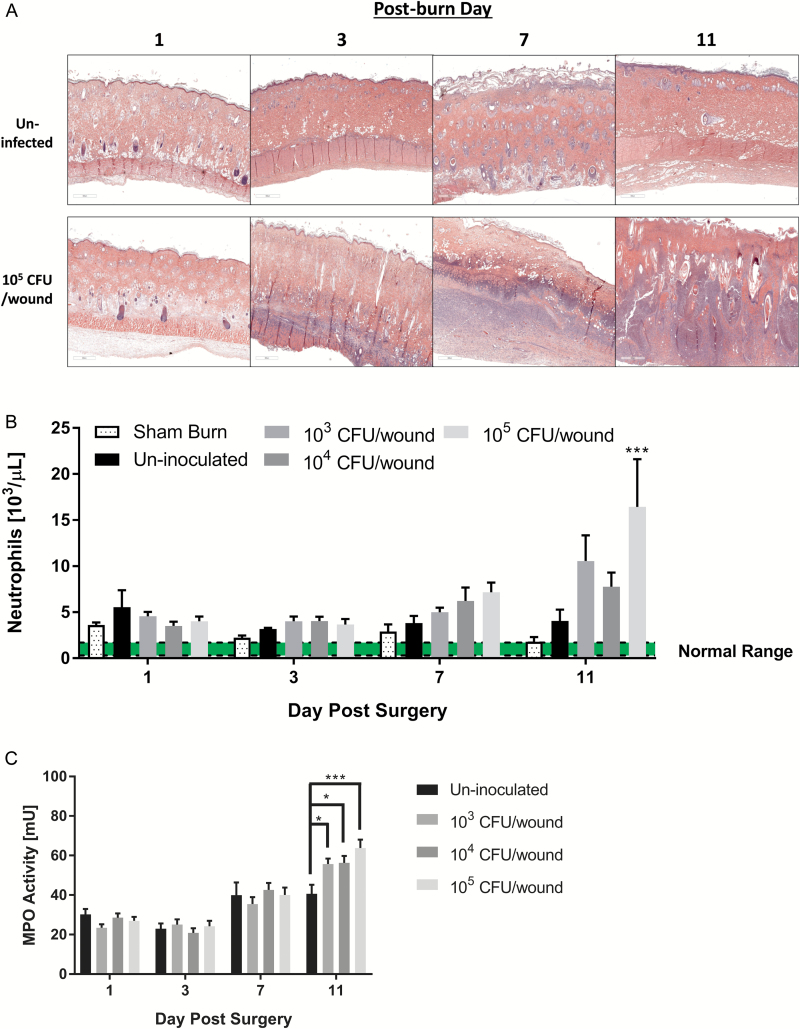
We stained histological sections with H&E to visualize the tissue damage from the burn and resulting immune response to the burn and infection. Figure 8A shows representative micrographs of un-inoculated and 105 CFU/wound groups over 11 days. Compared to controls, a massive immune response was seen in the P. aeruginosa-infected burn by POD 3 and later time points. Systemic neutrophil counts shown in Figure 8B correlated with the increased influx of inflammatory cells observed in the tissue sections. As shown in Figure 8C, significantly increased MPO activity, a commonly used marker of neutrophil activity, was detected in P. aeruginosa-infected wound tissue compared to controls by POD 11.
Figure 8.
Histological examination of P. aeruginosa-infected burn tissue. A. Representative hematoxylin–eosin stain tissue sections from un-inoculated and P. aeruginosa (105 CFU/wound) inoculated partial-thickness burn wounds over 11 days following the burn. Compared to the un-inoculated, a large number of inflammatory cells (purple features) infiltrated into the burn eschar in the P. aeruginosa-infected wounds. B. Systemic neutrophil cell counts, prior to euthanasia obtained via cardiac puncture. Neutrophil counts showed increasing trends with P. aeruginosa burn surface inoculation with significant increases detected on post-operative day (POD) 11 between the highest inoculum level and un-inoculated animals (***p < .005, two-way analysis of variance [ANOVA]). C. Myeloperoxidase activity of P. aeruginosa-infected deep partial-thickness burn tissue compared to controls. Significant increases in myeloperoxidase activity were detected on POD 11 in the P. aeruginosa-infected animals, (*p < .05; ***p < .0005, two-way ANOVA).
DISCUSSION
P. aeruginosa is one of the greatest microbiological barriers to healing in burn care. Previous research, conducted at the USAISR, focused exclusively on improving survival upon infection of full-thickness burn wounds with P. aeruginosa.38,54–56 Results of this work led to the current “gold standard” prophylaxis treatment for infection control in burn injuries, namely mafenide acetate and silver sulfadiazine.39,45,46 As such, the clinical mortality associated with invasive P. aeruginosa burn wound infection rapidly declined.36 However, contamination of the burn wound with P. aeruginosa or other opportunistic pathogens may still occur in circumstances where treatment is delayed, such as during evacuation from the battlefield and transport to higher echelons of medical care.45,57,58 The literature shows that delaying application of mafenide acetate cream by only 48–72 hours after full-thickness burn and infection with P. aeruginosa in rats resulted in 12–40% mortality; and delaying treatment up to 96 hours resulted in 100% mortality within 9 days.39 This suggests that resistance to the antimicrobial treatment, specifically after a 48–96-hour treatment delay, could be a deciding factor in burn infection-related mortality. Because Pseudomonas readily forms biofilms on both biotic and abiotic surfaces within hours, it is conceivable that Pseudomonas could form biofilms within the burn eschar. These biofilms may potentially withstand the debridement process and act as a nidus for sustained wound infection and graft failure.41 While, Pseudomonas biofilms are commonly found in chronic non-healing skin wounds,28,31,59 biofilm formation by P. aeruginosa has not been documented in the clinically accepted rat scald burn model.
To test our hypothesis that P. aeruginosa forms biofilms in partial-thickness burn wounds, we used the Walker–Mason rat scald burn model, originally developed at USAISR. When this model was originally developed, the end-point analysis focused on survival. Specifically, 1 × 106–1 × 108P. aeruginosa cells were flooded over the burn wound and time until death with or without antimicrobial treatment measured. Early work also showed that P. aeruginosa invaded full-thickness 35-cm2 burn wounds within 9 hours of surface inoculation and obtained a growth plateau of 1 × 108 CFU/g of burn tissue within 8 days post-infection.60
Instead of survival, we characterized the kinetics of a sublethal P. aeruginosa infection that resulted in biofilm formation within the burn wound. Regardless of inoculum size, robust infections developed plateauing at approximately 1 × 108 CFU/g of burn tissue, thereby recapitulating the bacterial load observed previously, albeit with higher inoculums. Visual appearance of the wound consistently replicated clinical observations of P. aeruginosa burn infections showing areas of necrosis, edema, and maceration.43,55,61,62 While the non-inoculated burns became contaminated, presumably with the native microflora, the P. aeruginosa-inoculated wounds primarily contained only the pathogen of interest, accounting for 80–100% of the total bacterial load recovered. Similar observations of P. aeruginosa’s dominance of microbial communities have been previously reported in a number of mixed-species infections both in vitro and in vivo.63–67 This phenomenon only helps underscore the importance of effective burn care to prevent or limit the growth of P. aeruginosa.
Our hypothesis that P. aeruginosa forms biofilms within partial-thickness burn wounds was confirmed via qRT–PCR. Important P. aeruginosa biofilm genes alg8, algE, and pvdS were highly upregulated in bacterial cells obtained from burn tissue. Both alg8, a glycosyl transferase, and algE, an export protein, associated with alginate biosynthesis are hallmarks of mature P. aeruginosa biofilms. Alginate along with other polysaccharides such as psl, pel, and DNA comprise structural matrix components of mature P. aeruginosa biofilms.30,68–70 Likewise, pyoverdine synthesis gene pvdS, which sequesters iron, was also highly upregulated in bacterial cells recovered from burn tissue. Iron is a critical nutrient for P. aeruginosa biofilm formation, both in vitro and in vivo.71–73 Pyoverdine, an essential virulence factor, liberates bound iron from the local microenvironment (eg, hemoglobin), thereby making it available to bacterial cells.74,75 These significant increases in gene expression of markers associated with P. aeruginosa virulence and biofilm formation confirmed that the bacterial cells were establishing biofilms within the wound.
Visualization of dense bacterial growth enmeshed within thick extracellular matrices covering the surface of the wound by SEM also helped confirm the biofilm nature of the wound infection. Whether the matrix material was derived solely from host or bacterial cells, or a mixture of both, has yet to be determined. Presumptively, the material is a combination of both wound exudate (eg, albumin, denatured collagen, etc.) and P. aeruginosa-derived polysaccharides such as alginate and extracellular nucleic acids. The morphological appearance of the bacterial cells on the wound bed closely resembles bacterial biofilms found within chronic skin wounds reported by other research groups.31 Additionally, P. aeruginosa penetrated into the burn eschar to at least the layer of undamaged dermal collagen (500–600 µm). In the cross-sections of the infected burn wounds, P. aeruginosa was closely associated with the denatured collagen fibers, caused by the thermal injury, and in some cases forming what appeared to be micro-colonies, another classical feature of P. aeruginosa biofilm formation. Visualization of P. aeruginosa with PNA-FISH showed dense localization surrounding the hair follicles. This reconfirms earlier work in full-thickness burn wounds regarding penetration of the P. aeruginosa into the burn wound via hair shafts and localizing around the hair follicles.60 These hallmarks of Pseudomonas biofilm formation coupled with their location deep beneath the surface of the burn may explain, in part, the decreased effectiveness of mafenide acetate cream upon a 48-hour treatment delay. This deep localization of P. aeruginosa within the eschar may also exacerbate the progression of partial-thickness burn wounds to full-thickness tissue damage. P. aeruginosa expresses several proteases including collagenase, which may contribute to its ability to invade burned skin.76 Furthermore, the biofilm infection induced a noticeable inflammatory response characterized by both the systemic increase in circulating neutrophils and elevated activity of MPO in the local P. aeruginosa-infected burn skin suggesting that the inflammatory cells observed in the H&E-stained tissue sections were largely neutrophils. Additional work is ongoing to characterize the host response to P. aeruginosa biofilm formation in the context of deep partial-thickness burn wounds.
In this study, we characterized the kinetics of a sublethal infection of P. aeruginosa in deep partial-thickness burn wounds using Sprague-Dawley rats. Our results confirmed the development of a robust infection upon application of at least 1000 CFU of P. aeruginosa over the 10% TBSA burn area. The infection was well tolerated in this model with no premature deaths due to the underlying burn wound infection or invasive P. aeruginosa infection. We observed formation of P. aeruginosa biofilms within the burn eschar using both SEM and RT–qPCR. This new in vivo burn wound biofilm infection model in Sprague-Dawley rats allows for testing novel anti-biofilm treatments in a clinically relevant and accepted scald burn animal model.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge all members of the USAISR DCTRTRD for their help and support in the production of this manuscript. The authors would also like to acknowledge the advice provided by Dr. Ravi Shankar, PhD. Additionally, the authors acknowledge the support of the USAISR Research Support Division for their assistance with animal handling and laboratory support in obtaining systemic neutrophil counts.
The study was funded, in part, by the Naval Medical Research Center’s Advanced Medical Development Program (MIPR N3239815MHX040) and the Combat Casualty Care Research Directorate, U.S. Army Medical Research and Materiel Command (USAMRMC). This research was also supported, in part, by an appointment to the Postgraduate Research Participation Program at the U.S. Army Institute of Surgical Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRMC.
REFERENCES
- 1. Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol 2005;13:20–6. [DOI] [PubMed] [Google Scholar]
- 2. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol 2010;8:623–33. [DOI] [PubMed] [Google Scholar]
- 3. López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol 2010;2:a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Limoli DH, Jones CJ, Wozniak DJ. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr 2015; 3:MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirkpatrick CL, Viollier PH. Cell dispersal in biofilms: an extracellular DNA masks nature’s strongest glue. Mol Microbiol 2010;77:801–4. [DOI] [PubMed] [Google Scholar]
- 6. Dominiak DM, Nielsen JL, Nielsen PH. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol 2011;13:710–21. [DOI] [PubMed] [Google Scholar]
- 7. Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol 2005;13:34–40. [DOI] [PubMed] [Google Scholar]
- 8. Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2011;55:4469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. González JF, Alberts H, Lee J, Doolittle L, Gunn JS. Biofilm formation protects Salmonella from the antibiotic ciprofloxacin in vitro and in vivo in the mouse model of chronic carriage. Sci Rep 2018;8:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol 2010;59:253–68. [DOI] [PubMed] [Google Scholar]
- 11. Jensen PO Givskov M, Bjarnsholt T, Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol 2010;59:292–305. [DOI] [PubMed] [Google Scholar]
- 12. Jones CJ, Wozniak DJ. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. MBio 2017;8:e00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thurnheer T, Gmür R, Shapiro S, Guggenheim B. Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl Environ Microbiol 2003;69:1702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z, Nadezhina E, Wilkinson KJ. Quantifying diffusion in a biofilm of Streptococcus mutans. Antimicrob Agents Chemother 2011;55:1075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bryers JD, Drummond F. Local macromolecule diffusion coefficients in structurally non-uniform bacterial biofilms using fluorescence recovery after photobleaching (FRAP). Biotechnol Bioeng 1998;60:462–73. [PubMed] [Google Scholar]
- 16. Hornemann JA, Lysova AA, Codd SL et al. Biopolymer and water dynamics in microbial biofilm extracellular polymeric substance. Biomacromolecules 2008;9:2322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Absalon C, Van Dellen K, Watnick PI. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog 2011;7:e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sendi P, Frei R, Maurer TB, Trampuz A, Zimmerli W, Graber P. Escherichia coli variants in periprosthetic joint infection: diagnostic challenges with sessile bacteria and sonication. J Clin Microbiol 2010;48:1720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trafny EA. Susceptibility of adherent organisms from Pseudomonas aeruginosa and Staphylococcus aureus strains isolated from burn wounds to antimicrobial agents. Int J Antimicrob Agents 1998;10:223–8. [DOI] [PubMed] [Google Scholar]
- 20. Vlassova N, Han A, Zenilman JM, James G, Lazarus GS. New horizons for cutaneous microbiology: the role of biofilms in dermatological disease. Br J Dermatol 2011;165:751–9. [DOI] [PubMed] [Google Scholar]
- 21. Maurice NM, Bedi B, Sadikot RT. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol 2018;58:428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maunders E, Welch M. Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol Lett 2017;364:fnx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rada B. Neutrophil extracellular trap release driven by bacterial motility: relevance to cystic fibrosis lung disease. Commun Integr Biol 2017;10:e1296610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christophersen LJ, Trøstrup H, Malling Damlund DS et al. Bead-size directed distribution of Pseudomonas aeruginosa results in distinct inflammatory response in a mouse model of chronic lung infection. Clin Exp Immunol 2012;170:222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Galli J, Calò L, Giuliani M et al. Biofilm’s role in chronic cholesteatomatous otitis media: a pilot study. Otolaryngol Head Neck Surg 2016;154:914–6. [DOI] [PubMed] [Google Scholar]
- 26. Wessman M, Bjarnsholt T, Eickhardt-Sørensen SR, Johansen HK, Homøe P. Mucosal biofilm detection in chronic otitis media: a study of middle ear biopsies from Greenlandic patients. Eur Arch Otorhinolaryngol 2015;272:1079–85. [DOI] [PubMed] [Google Scholar]
- 27. Kaya E, Dag I, Incesulu A, Gurbuz MK, Acar M, Birdane L. Investigation of the presence of biofilms in chronic suppurative otitis media, nonsuppurative otitis media, and chronic otitis media with cholesteatoma by scanning electron microscopy. ScientificWorldJournal 2013;2013:638715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 2003;112:1466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dass CL, Walsh MF, Seo S, Shiratsuchi H, Craig DH, Basson MD. Irrigant divalent cation concentrations influence bacterial adhesion. J Surg Res 2009;156:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 2006;188:8213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. James GA, Swogger E, Wolcott R et al. Biofilms in chronic wounds. Wound Repair Regen 2008;16:37–44. [DOI] [PubMed] [Google Scholar]
- 32. Leung KP, Crowe TD, Abercrombie JJ et al. Control of oral biofilm formation by an antimicrobial decapeptide. J Dent Res 2005;84:1172–7. [DOI] [PubMed] [Google Scholar]
- 33. Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 2017;15:740–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z, Shen Y, Haapasalo M. Antibiofilm peptides against oral biofilms. J Oral Microbiol 2017;9:1327308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev 2006;19:403–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McManus AT, Mason AD Jr., McManus WF, Pruitt BA Jr. A decade of reduced gram-negative infections and mortality associated with improved isolation of burned patients. Arch Surg 1994;129:1306–9. [DOI] [PubMed] [Google Scholar]
- 37. McManus AT, Mason AD Jr., McManus WF, Pruitt BA Jr. Twenty-five year review of Pseudomonas aeruginosa bacteremia in a burn center. Eur J Clin Microbiol 1985;4:219–23. [DOI] [PubMed] [Google Scholar]
- 38. Walker HL, Mason AD Jr., Raulston GL. Surface infection with Pseudomonas aeruginosa. Ann Surg 1964;160:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindberg RB, Moncrief JA, Mason AD Jr. Control of experimental and clinical burn wounds sepsis by topical application of sulfamylon compounds. Ann N Y Acad Sci 1968;150:950–60. [DOI] [PubMed] [Google Scholar]
- 40. Yurt RW, McManus AT, Mason AD Jr., Pruitt BA Jr. Increased susceptibility to infection related to extent of burn injury. Arch Surg 1984;119:183–8. [DOI] [PubMed] [Google Scholar]
- 41. Chu CS, McManus AT, Mason AD, Pruitt BA Jr. Topical silver treatment after escharectomy of infected full thickness burn wounds in rats. J Trauma 2005;58:1040–6. [DOI] [PubMed] [Google Scholar]
- 42. McManus AT, Moody EE, Mason AD. Bacterial motility: a component in experimental Pseudomonas aeruginosa burn wound sepsis. Burns 1980;6:235–9. [Google Scholar]
- 43. Rabin ER, Graber CD, Vogel EH Jr., Finkelstein RA, Tumbusch WA. Fatal Pseudomonas infection in burned patients. A clinical, bacteriologic and anatomic study. N Engl J Med 1961;265:1225–31. [DOI] [PubMed] [Google Scholar]
- 44. Chu CS, McManus AT, Pruitt BA Jr., Mason AD Jr. Therapeutic effects of silver nylon dressings with weak direct current on Pseudomonas aeruginosa-infected burn wounds. J Trauma 1988;28:1488–92. [DOI] [PubMed] [Google Scholar]
- 45. Kauvar DS, Acheson E, Reeder J, Roll K, Baer DG. Comparison of battlefield-expedient topical antimicrobial agents for the prevention of burn wound sepsis in a rat model. J Burn Care Rehabil 2005;26:357–61. [DOI] [PubMed] [Google Scholar]
- 46. McManus AT, McLeod CG Jr., Mason AD Jr. Experimental Proteus mirabilis burn surface infection. Arch Surg 1982;117:187–91. [DOI] [PubMed] [Google Scholar]
- 47. Levine NS, Salisbury RE, Mason AD Jr. The effect of early surgical excision and homografting on survival of burned rats and of intraperitoneally-infected burned rats. Plast Reconstr Surg 1975;56:423–9. [DOI] [PubMed] [Google Scholar]
- 48. Walker HL, Mason AD Jr. A standard animal burn. J Trauma 1968;8:1049–51. [DOI] [PubMed] [Google Scholar]
- 49. Karna SLR, Chen T, Chen P, Peacock TJ, Abercrombie JJ, Leung KP. Genome sequence of a virulent Pseudomonas aeruginosa strain, 12-4-4(59), isolated from the blood culture of a burn patient. Genome Announc 2016;4:e00079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gilpin DA. Calculation of a new Meeh constant and experimental determination of burn size. Burns 1996;22:607–11. [DOI] [PubMed] [Google Scholar]
- 51. Leary S, Underwood W, Anthony R et al. AVMA guidelines for the euthanasia of animals: 2013 edition. Introduction and General Comments. Schaumburg (IL): American Veterinary Medical Association; 2013; p16. [Google Scholar]
- 52. Pirnay JP, De Vos D, Duinslaeger L et al. Quantitation of Pseudomonas aeruginosa in wound biopsy samples: from bacterial culture to rapid ‘real-time’ polymerase chain reaction. Crit Care 2000;4:255–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002;148:257–66. [DOI] [PubMed] [Google Scholar]
- 54. Walker HL, McLeod CG Jr., Leppla SH, Mason AD Jr. Evaluation of Pseudomonas aeruginosa toxin A in experimental rat burn wound sepsis. Infect Immun 1979;25:828–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Order SE, Mason AD Jr., Walker HL, Lindberg RB, Switzer WE, Moncrief JA. Vascular destructive effects of thermal injury and its relationship to burn wound sepsis. J Trauma 1965;5:62–71. [DOI] [PubMed] [Google Scholar]
- 56. Lieberman MM, Walker HL, Ayala E, Chapa I. Active and passive immunization with Pseudomonas aeruginosa ribosomal vaccines and antisera in the burned rat model. J Surg Res 1986;40:138–144. [DOI] [PubMed] [Google Scholar]
- 57. Chan RK, Siller-Jackson A, Verrett AJ, Wu J, Hale RG. Ten years of war: a characterization of craniomaxillofacial injuries incurred during operations Enduring Freedom and Iraqi Freedom. J Trauma Acute Care Surg 2012;73(6 Suppl. 5):S453–8. [DOI] [PubMed] [Google Scholar]
- 58. Akers KS, Mende K, Cheatle KA et al. Biofilms and persistent wound infections in United States military trauma patients: a case-control analysis. BMC Infect Dis 2014;14:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kennedy P, Brammah S, Wills E. Burns, biofilm and a new appraisal of burn wound sepsis. Burns 2010;36:49–56. [DOI] [PubMed] [Google Scholar]
- 60. Teplitz C, Davis D, Mason AD Jr., Moncrief JA. Pseudomonas burn wound sepsis. I pathogenesis of experimental Pseudomonas burn wound sepsis. J Surg Res 1964;4:200–16. [DOI] [PubMed] [Google Scholar]
- 61. Foley FD, Greenawald KA, Nash G, Pruitt BA Jr. Pathology of Pseudomonas infection. Tex Med 1969;65:36–9. [PubMed] [Google Scholar]
- 62. Pruitt BA Jr., McManus AT. Opportunistic infections in severely burned patients. Am J Med 1984;76:146–54. [DOI] [PubMed] [Google Scholar]
- 63. Seth AK, Geringer MR, Hong SJ, Leung KP, Galiano RD, Mustoe TA. Comparative analysis of single-species and polybacterial wound biofilms using a quantitative, in vivo, rabbit ear model. PLoS One 2012;7:e42897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Miller CL, Van Laar TA, Chen T, et al. Global transcriptome responses including small RNAs during mixed-species interactions with methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa. Microbiology Open 2017;6:e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thet NT, Wallace L, Wibaux A, Boote N, Jenkins ATA. Development of a mixed-species biofilm model and its virulence implications in device related infections. J Biomed Mater Res B Appl Biomater 2018;00B:000–000. [DOI] [PubMed] [Google Scholar]
- 66. Chew SC, Kundukad B, Seviour T et al. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. MBio 2014;5:e01536–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Malic S, Hill KE, Playle R, Thomas DW, Williams DW. In vitro interaction of chronic wound bacteria in biofilms. J Wound Care 2011;20:569–70, 572, 574. [DOI] [PubMed] [Google Scholar]
- 68. Colvin KM, Irie Y, Tart CS et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 2012;14:1913–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ghafoor A, Hay ID, Rehm BH. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 2011;77:5238–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 2007;10:644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tettmann B, Niewerth C, Kirschhöfer F et al. Enzyme-mediated quenching of the Pseudomonas quinolone signal (PQS) promotes biofilm formation of Pseudomonas aeruginosa by increasing iron availability. Front Microbiol 2016;7:1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wiens JR, Vasil AI, Schurr MJ, Vasil ML. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. MBio 2014;5:e01010–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Llamas MA, Imperi F, Visca P, Lamont IL. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev 2014;38:569–97. [DOI] [PubMed] [Google Scholar]
- 74. Vasil ML, Ochsner UA. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol 1999;34:399–413. [DOI] [PubMed] [Google Scholar]
- 75. Kruczek C, Wachtel M, Alabady MS, Payton PR, Colmer-Hamood JA, Hamood AN. Serum albumin alters the expression of iron-controlled genes in Pseudomonas aeruginosa. Microbiology 2012;158: 353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wellisch G, Cohen E, Cahane Z, Horowitz J. Simple method for collagenase determination in 38 Pseudomonas aeruginosa strains. J Clin Microbiol 1984;20:1020–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.