Abstract
Drug resistance in Mycobacterium leprae is assumed to be due to genetic alterations in the drug targets and reduced cell wall permeability. However, as observed in Mycobacterium tuberculosis, drug resistance may also result from the overactivity of efflux systems, which is mostly unexplored. In this perspective, we discuss known efflux pumps involved in M. tuberculosis drug resistance and virulence and investigate similar regions in the genome of M. leprae. In silico analysis reveals that the major M. tuberculosis efflux pumps known to be associated with drug resistance and virulence have been retained during the reductive evolutionary process that M. leprae underwent, e.g., RND superfamily, the ABC transporter BacA, and the MFS P55. However, some are absent (DinF, MATE) while others are derepressed (Mmr, SMR) in M. leprae reflecting the specific environment where M. leprae may live. The occurrence of several multidrug resistance efflux transporters shared between M. leprae and M. tuberculosis reveals potential implications in drug resistance and virulence. The conservation of the described efflux systems in M. leprae upon genome reduction indicates that these systems are potentially required for its intracellular survival and lifestyle. They potentially are involved in M. leprae drug resistance, which could hamper leprosy treatment success. Studying M. leprae efflux pumps as new drug targets is useful for future leprosy therapeutics, enhancing the global efforts to eradicate endemic leprosy, and prevent the emergence of drug resistance in afflicted countries.
Keywords: antimicrobial resistance, efflux pumps, leprosy, mycobacteria, tuberculosis, virulence
Introduction
Leprosy and tuberculosis are public health threatening infectious diseases with similar problems of ongoing human-to-human transmission, inherent drug resistance to several antimicrobial agents, propensity to develop resistance to antimycobacterial drugs, and virulence (Singh et al., 2016; Dheda et al., 2017). Whilst there is extensive knowledge about the mechanisms of M. tuberculosis drug resistance, less is known about the mechanisms by which M. leprae develops drug resistance. M. leprae is an obligate intracellular pathogen and one of the few known microorganisms that still cannot be cultured in vitro which have been hindering the study of the mechanisms of drug resistance by biochemical and functional studies. The overexpression of multidrug (MDR) efflux pump genes is a common mechanism of antimicrobial resistance in M. tuberculosis (da Silva et al., 2016; Machado et al., 2017). Likewise, efflux pumps certainly contribute to drug resistance in M. leprae, which is mostly unexplored.
Efflux pumps are one of the most widespread resistance determinants in bacteria. Usually, they are chromosomally encoded and are greatly conserved at both gene and protein level across bacterial species. More than 50 putative efflux pumps have been associated with the transport of several drugs in M. tuberculosis (De Rossi et al., 2006; Louw et al., 2009; Kapopoulou et al., 2011; Black et al., 2014; da Silva et al., 2016). Although they are mostly known due to their role in the efflux of antimicrobials, efflux pumps are mainly involved in physiological processes such as cell-to-cell communication, bacterial virulence, cellular homeostasis, detoxification of intracellular metabolites, and intracellular signal trafficking (De Rossi et al., 2006; Martinez et al., 2009; Viveiros et al., 2012; Black et al., 2014; da Silva et al., 2016; Li et al., 2016; Sandhu and Akhter, 2018). Recently, it was shown that the loss of the efflux pump AcrAB in Salmonella enterica serovar Typhymurium reduces virulence leading to the accumulation of noxious molecules inside the bacteria reducing the bacterial factors required for infection (Wang-Kan et al., 2017). From a biological point of view, drug resistance and virulence are required for pathogen survival. In normal conditions, the expression of these systems is tightly downregulated by specific transcriptional regulators (Grkovic et al., 2001) and their overexpression is achieved only in the presence of specific stressors capable of binding to the transcriptional regulators. The induction of efflux systems in the presence of inducers such as antimicrobials or host factors during infection promote a low-level resistance phenotype that allows the bacteria to survive during prolonged periods in the presence of drugs contributing for the development and stabilization of resistant phenotypes (Machado et al., 2012; Schmalstieg et al., 2012).
In this perspective, we compared in silico M. tuberculosis efflux pumps involved in drug resistance and virulence with those of M. leprae investigating their possible involvement in antimicrobial resistance and virulence in M. leprae.
Reductive Evolution
The genome of the non-pathogenic Mycobacterium smegmatis mc2155 has 7 Mb in size; the genomes of the pathogenic M. tuberculosis and M. leprae are much smaller in length and for M. leprae this is even more dramatic with almost 2000 genes lost in comparison with M. tuberculosis. Compared to non-pathogenic mycobacteria, M. tuberculosis and M. leprae evolved by extensive reductive evolution suggesting that pathogenic mycobacteria evolved toward pathogenicity by the loss of genetic material as the result of niche adaptation. Contrary to M. tuberculosis, M. leprae is an obligate intracellular pathogen. Adaptation to a permanent pathogenic lifestyle in constant association with the host led to gene loss toward a minimal gene set, such as those coding for metabolism and respiration, needed for a successful obligate intracellular parasitism (Moran, 2002; Scollard et al., 2006) and limited capacity to survive extracellularly (Cole et al., 2001; Eiglmeier et al., 2001). Of interest is the fact that the G+C content is lower in M. leprae pseudogenes (56.5%) than in its active ORFs (60.1%) (Cole et al., 2001). Changes in G+C content during the path of evolution may confer an advantage in response to environmental changes (Mann and Chen, 2010). Free-living organisms have an average G+C content higher than obligatory pathogens and symbionts. The shift toward lower G+C contents and smaller genomes in obligate pathogenic mycobacteria seems to occur in response to environmental adaptation where they encounter low selective pressure (Mann and Chen, 2010). In this context some genes became inactivated, as they are not required in these highly specialized niches meeting the theoretical principles of Morris, Lenski, and Zinser's Black Queen Hypothesis for the symbiotic reductive genome evolution of microorganisms (Morris et al., 2012), now applied to a bacterium and his long-lasting and almost exclusive host—the human being. In this case, M. leprae relies on his host functions to live efficiently, losing burdensome genes for functions it does not have to perform for itself (Morris et al., 2012). Nevertheless, it is not clear why M. leprae maintains such high number of pseudogenes in the genome. It has been hypothesized that the maintenance of pseudogenes is due to the slow-growth rate (McLeod et al., 2004) or lack of recombination (Bolotin and Hershberg, 2015). Additionally, pseudogene maintenance may allow the bacteria to revert back and forward from a non-functional protein to a functional one (Bolotin and Hershberg, 2015). If true, this may explain why some genes are loss and others are maintained as pseudogenes in M. leprae genome.
In this evolutionary context, where an obligate intracellular pathogen evolved to become dependent on his host, the central question of this work is the impact of M. leprae genome downsizing on antimicrobial resistance. This can be viewed as the time when the host decides that he no longer wants to maintain this intimate relation and starts antibiotic treatment with the assistance of his clinician and the health system.
Drug Resistance and Efflux Systems in M. leprae
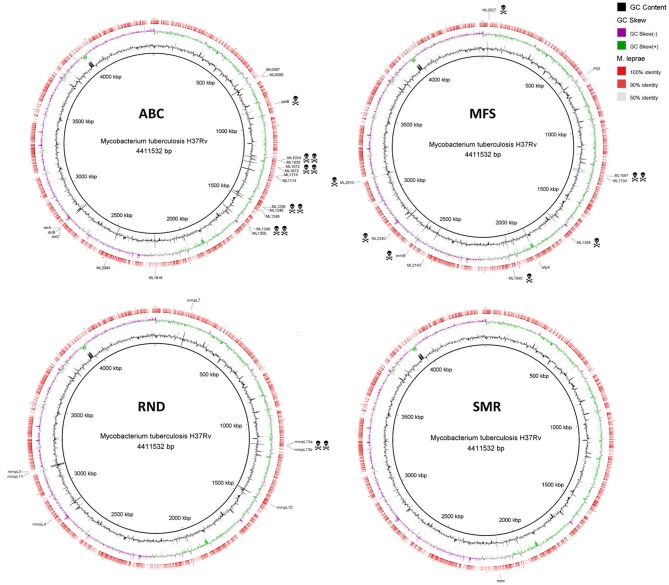
Several mycobacterial drug efflux pumps have been described in M. tuberculosis (Table 1). Comparative analysis of M. leprae genome shows the presence of approximately half of these transporters while several others are inactivated or absent, probably lost as consequence of reductive evolution. In silico, M. tuberculosis H37Rv genome encodes 267 putative transporters, of which 129 belong to the ATP-binding cassette (ABC) superfamily, 31 to the major facilitator superfamily (MFS), 14 to the resistance nodulation and cell division (RND) superfamily, 1 to the small multidrug resistance (SMR) family, and 1 to the multidrug and toxic compound extrusion (MATE) family. M. leprae genome encodes for 114 transporters, of which 62 corresponds to ABC transporters, 6 MFS, 5 RND, and 1 SMR (Elbourne et al., 2017). Those that have been associated with drug resistance in M. tuberculosis are discussed below. Alignment visualization of the M. tuberculosis and the M. leprae whole genome sequences, with the predicted CDS regions of the efflux transporters of M. leprae highlighted is shown in Figure 1.
Table 1.
Putative drug membrane transporters encoded by M. tuberculosis H37Rv and its orthologous in M. leprae TN.
| Efflux pump family | Gene | Gene locus tag* | Identity (%)** | M. tuberculosis | ||
|---|---|---|---|---|---|---|
| M. tuberculosis | M. leprae | Antimicrobial substrates | Main references | |||
| ABC | ||||||
| “One gene” | bacA | Rv1819c | ML2084 | 75 | RIF, INH, BL, CHL, TET, VAN, MAC, NOV, AGs, AP | Danilchanka et al., 2008; Domenech et al., 2009; Gupta et al., 2010a; Kapopoulou et al., 2011; Li et al., 2015 |
| Rv0194 | Rv0194 | Absent | - | BL, CHL, STR, TET, VAN, MAC, NOV, EMB, EtBr | Danilchanka et al., 2008; Kapopoulou et al., 2011; Garima et al., 2015 | |
| pstB | Rv0933 | ML0741c | - | FQs, INH, RIF, EMB | Banerjee et al., 1996, 2000; Braibant et al., 1996; Gupta et al., 2006; Srivastava et al., 2010; Kapopoulou et al., 2011; Brandis and Hughes, 2013; Lu et al., 2014 | |
| Rv1473 | Rv1473 | ML1816c | 88 | MAC | Kapopoulou et al., 2011 | |
| Rv2477c | Rv2477c | ML1248 | 92 | MAC, FQs | Gupta et al., 2010a; Kapopoulou et al., 2011 | |
| “Two-genes” | Rv1218c- Rv1217c | Rv1218c- Rv1217c |
ML1073c- ML1072c |
- | BL, NOV, BP, PD, PR, BSP, PA, INH, RIF | Balganesh et al., 2010; Kapopoulou et al., 2011; Dinesh et al., 2013; Wang et al., 2013 |
| Rv1273c- | Rv1273c- | ML1114c- | 78 | Unknown | Kapopoulou et al., 2011 | |
| Rv1272c | Rv1272c | ML1113c | 75 | |||
| Rv1668c- Rv1667c | Rv1668c- Rv1667c |
ML1240c- ML1239c |
- | MAC | Kapopoulou et al., 2011 | |
| Rv1687c- Rv1686c | Rv1687c- Rv1686c |
ML1350c- ML1349c |
- | MAC | Kapopoulou et al., 2011 | |
| “Three-genes” | Rv1458c- | Rv1458c- | ML0590c- | 88 | RIF, INH, STR, EMB | Hao et al., 2011; Kapopoulou et al., 2011; Caleffi-Ferracioli et al., 2016 |
| Rv1457c- | Rv1457c- | ML0589c- | 83 | |||
| Rv1456c | Rv1456c | ML0587c | 83 | |||
| Rv2688c- | Rv2688c- | Absent | - | FQs | Pasca et al., 2004; Gupta et al., 2010a; Kapopoulou et al., 2011 | |
| Rv2687c- | Rv1687c- | ML1035 | - | |||
| Rv2686c | Rv1686c | ML1034 | - | |||
| drrA- | Rv2936- | ML2352c- | 85 | TET, EMB, MAC, AGs, CHL, RIF, EtBr, NOR, PUR, BCECF, DAU, DOX | Choudhuri et al., 2002; Kapopoulou et al., 2011; Pang et al., 2013; Li et al., 2015 | |
| drrB- | Rv2937- | ML2351c- | 64 | |||
| drrC | Rv2938 | ML2350c | 79 | |||
| MFS | ||||||
| Rv0037c | Rv0037c | ML0027c | - | Unknown | Kapopoulou et al., 2011 | |
| Rv0191 | Rv0191 | ML2610 | - | RIF | Kapopoulou et al., 2011; Li et al., 2015 | |
| emrB | Rv0783c | ML2224 | - | Multiple drugs | De Rossi et al., 2002; Gupta et al., 2010a; Kapopoulou et al., 2011; Brandis and Hughes, 2013; Li et al., 2015 | |
| Rv0842 | Rv0842 | Absent | - | RIF | Kapopoulou et al., 2011; Li et al., 2015 | |
| Rv0849 | Rv0849 | Absent | - | BL, INH, RIF | Kapopoulou et al., 2011; Balganesh et al., 2012 | |
| Rv0876c | Rv0876c | ML2143 | 81 | Unknown | Kapopoulou et al., 2011 | |
| Rv1250 | Rv1250 | ML1097 | - | INH | Kapopoulou et al., 2011; Garima et al., 2015; Li et al., 2015 | |
| Rv1258c | Rv1258c | ML1104c | - | TET, FQs, RIF, CFZ, INH, EMB, ERY, EtBr, SPE | Ainsa et al., 1998; Siddiqi et al., 2004; Gupta et al., 2006; Ramón-García et al., 2006, 2012; Jiang et al., 2008; Kapopoulou et al., 2011; Balganesh et al., 2012; Machado et al., 2012, 2017 | |
| p55 | Rv1410c | ML0556c | 82 | TET, AGs, RIF, INH, CFZ | da Silva et al., 2001; Jiang et al., 2008; Ramón-García et al., 2009; Bianco et al., 2011a,b; Kapopoulou et al., 2011; Machado et al., 2012, 2017; Li et al., 2015 | |
| Rv1634 | Rv1634 | ML1388 | - | FQs; SKI | De Rossi et al., 2002; Kapopoulou et al., 2011; Harris et al., 2014 | |
| Rv1672c | Rv1672c | Absent | - | Unknown | Kapopoulou et al., 2011 | |
| Rv1877 | Rv1877 | Absent | - | RIF, EtBr, ACR, ERY, KAN, TET | De Rossi et al., 2002; Li et al., 2004; Kapopoulou et al., 2011; Louw et al., 2011 | |
| Rv2265 | Rv2265 | Absent | - | Unknown | Kapopoulou et al., 2011 | |
| stp | Rv2333c | Absent | - | SPE, TET, RIF | Ramón-García et al., 2007; Kapopoulou et al., 2011; Li et al., 2015 | |
| Rv2456c | Rv2456c | Absent | - | Unknown | Kapopoulou et al., 2011 | |
| Rv2459 | Rv2459 | Absent | - | INH, EMB, RIF, EtBr | De Rossi et al., 2002; Gupta et al., 2010b; Kapopoulou et al., 2011; Machado et al., 2012; Li et al., 2015 | |
| efpA | Rv2846c | ML1562c | 81 | INH, RIF, EtBr, ACR, ERY, FQs | Doran et al., 1997; Wilson et al., 1999; Li et al., 2004, 2015; Gupta et al., 2010a; Kapopoulou et al., 2011; Machado et al., 2012, 2017 | |
| Rv2994 | Rv2994 | ML1690 | - | STR, RIF | Gupta et al., 2010a; Kapopoulou et al., 2011; Louw et al., 2011 | |
| Rv3239c | Rv3239c | Absent | - | Unknown | Kapopoulou et al., 2011 | |
| Rv3728 | Rv3728 | ML2340 | - | RIF | Gupta et al., 2010a; Kapopoulou et al., 2011 | |
| RND | ||||||
| mmpS1-mmpL1 | Rv0403c- Rv0402c |
Absent | - | Unknown | Kapopoulou et al., 2011 | |
| mmpS2-mmpL2 | Rv0506- Rv0507 |
Absent | - | Unknown | Kapopoulou et al., 2011 | |
| mmpL3 | Rv0206c | ML2620c | 76 | SQ109, BM212, AU, IA | Kapopoulou et al., 2011; La Rosa et al., 2012; Tahlan et al., 2012; Li et al., 2014 | |
| mmpS4- | Rv0451c- | ML2377 | 75 | CMB, MB, RIF | Kapopoulou et al., 2011; de Knegt et al., 2013; Wells et al., 2013 | |
| mmpL4 | Rv0450c | ML2378 | 79 | |||
| mmpS5-mmpL5 | Rv0677c- Rv0676c |
Absent | - | AZ, BDQ, CFZ, TET | Milano et al., 2009; Kapopoulou et al., 2011; Hartkoorn et al., 2014 | |
| mmpL6 | Rv1557 | Absent | - | Unknown | Kapopoulou et al., 2011 | |
| mmpL7 | Rv2942 | ML0137c | 69 | INH | Choudhuri et al., 1999; Domenech et al., 2005; Kapopoulou et al., 2011; Machado et al., 2012 Pasca et al., 2005 | |
| mmpL8 | Rv3823c | Absent | - | SQ109 | Domenech et al., 2004; Kapopoulou et al., 2011; Li et al., 2014 | |
| mmpL9 | Rv2339 | Absent | - | SQ109 | Kapopoulou et al., 2011; Li et al., 2014 | |
| mmpL10 | Rv1183 | ML1231 | 71 | Unknown | Kapopoulou et al., 2011 | |
| mmpL11 | Rv0202c | ML2617c | 73 | Unknown | Kapopoulou et al., 2011 | |
| mmpL12 | Rv1522c | Absent | - | Unknown | Kapopoulou et al., 2011 | |
| mmpL13a | Rv1145 | ML0971 | - | Unknown | Kapopoulou et al., 2011 | |
| mmpL13b | Rv1146 | ML0972 | - | Unknown | Kapopoulou et al., 2011 | |
| mmpS3 | Rv2198c | ML0877 | 68 | - | Kapopoulou et al., 2011 | |
| SMR | ||||||
| mmr | Rv3065 | ML1756 | 79 | ACR, EtBr, INH, MAC, FQs, TPP, PY | De Rossi et al., 1998; Kapopoulou et al., 2011; Balganesh et al., 2012; Machado et al., 2012; Rodrigues et al., 2013 | |
| MATE | ||||||
| dinF | Rv2836c | Absent | - | AGs, Phleo, sulpha drugs, CPC | Kapopoulou et al., 2011; Mishra and Daniels, 2013 | |
CDS, coding DNA sequence;
determined at protein level; pseudogenes are underlined. ABC, ATP-binding cassette; ACR, acriflavine; AGs, aminoglycosides; AP, antimicrobial peptides; AU, adamantyl ureas; AZ, azoles; BCECF, 2',7'-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein; BDQ, bedaquiline; BL, β-lactams; BP, biarylpiperazines; BSP, bisanilinopyrimidines; CFZ, clofazimine; CHL, chloramphenicol; CMB, carboxymycobactins; CPC, cetylpyridinium chloride; DAU, daunorubicin; DOX, doxorubicin; EMB, ethambutol; ERY, erythromycin; EtBr, ethidium bromide; FQs, fluoroquinolones; IA, indoleamides; INH, isoniazid; KAN, kanamycin; MAC, macrolides; MATE, multidrug and toxic compound extrusion; MB, mycobactins; MFS, major facilitator superfamily; NOR, norfloxacin; NOV, novobiocin; PA, pyrazolones; PD, pyridines; Phleo, phleomycin; PR, pyrroles; PUR, puromycin; PY, pyronin Y; RIF, rifampicin; RND, resistance nodulation division; SKI, imidazoline SKI-356313; SMR, small multidrug resistance; SPE, spectinomycin; STR, streptomycin; TET, tetracycline; TPP, tetraphenylphosphonium; VAN, vancomycin.
Figure 1.
Alignment of M. leprae TN predicted CDS regions of efflux transporters to those of M. tuberculosis H37Rv. Efflux transporters identified in M. tuberculosis are highlighted outside the ring in black. The death heads denote pseudogenes. See Table 1 for locus tag. The circular genomic comparison was generated using BRIG software (Alikhan et al., 2011). CDS, coding DNA sequence.
ABC Transporters
ABC transporters can be divided in those encoded by “one,” “two,” and “three genes” (Table 1). Among those coded by “one gene” in M. leprae is ML2084, homologue of BacA that is involved in virulence of M. tuberculosis (Domenech et al., 2009) and in the active transport of drugs across the membrane (Table 1). Absent from M. leprae genome is the transporter Rv0194, which was the first to be associated with β-lactam transport in M. tuberculosis-important taking into account since β-lactams can be useful in M. leprae chemotherapy (Danilchanka et al., 2008; Garima et al., 2015). The phosphate-specific ABC transporter (PstB) is known to be operative in M. tuberculosis during phosphate limiting conditions during infection (Banerjee et al., 2000). The pst operon encodes pseudogenes in M. leprae. As PstB is non-functional in M. leprae and active in M. tuberculosis this may indicate that M. leprae encounters stable phosphate content within the host, making the presence of PstB unnecessary. Besides its role on phosphate uptake, PstB is also associated to the extrusion of antibiotics (Table 1). Rv1473 and Rv2477c encode putative macrolide transporters with functional orthologues in M. leprae genome. To our knowledge, there is no evidence for M. leprae isolates resistant to clarithromycin so far, stressing the therapeutic usefulness of this antibiotic against M. leprae.
ABC transporters encoded by “two genes” are Rv1218c-Rv1217c, Rv1273c-Rv1272c, Rv1668c-Rv1667c, and Rv1687c-Rv1686c. Although nothing is known about their expression in M. tuberculosis, the orthologous genes of Rv1668c-Rv1667c and Rv1687c-Rv1686c in M. leprae, ML1240c-ML1239c, and ML1350c-ML1349c, respectively, are non-functional. The Rv1218c-Rv1217c efflux pump has been associated with M. tuberculosis resistance to a wide variety of chemical classes of compounds (Table 1). Orthologues of Rv1273c-Rv1272c can be found in M. leprae genome but because there are no studies about their role on M. tuberculosis drug resistance, nothing can be anticipated for M. leprae.
ABC transporters encoded by “three genes” are the operons Rv1458c-Rv1457c-Rv1456c, DrrABC and Rv2688c-Rv2687c-Rv2686c associated with the extrusion of several drugs in M. tuberculosis (Table 1). M. leprae orthologue of Rv1458c-Rv1457c-Rv1456c is ML0590c-ML0589c-ML0587c, which encodes a functional efflux transporter. In relation to Rv2688c-Rv2687c-Rv2686c, M. leprae chromosome has non-functional orthologues of two components, ML1034-ML1035 (positive strand), while the orthologue of Rv2688c is absent. The genes Rv2686c, Rv2687c, and Rv2688c are co-transcribed. Rv2686c and Rv2687c proteins possess six transmembrane segments, whereas Rv2688c has a nucleotide-binding domain and is likely involved in ATP hydrolysis. In consequence, Rv2688c probably coordinates the functionality of the whole operon. In this case, it is not surprising that the absence of the Rv2688c orthologue in M. leprae renders non-functional the other genes within this operon, contributing to the increased susceptibility of M. leprae to fluoroquinolones.
MFS Transporters
M. leprae genome possess 11 MFS drug transporters, whereas 20 can be detected in M. tuberculosis (Table 1). Among the nine transporters absent from M. leprae genome, nothing is known about their role in M. tuberculosis for five of them, while for the remaining four it has been described an association with resistance to several drugs (Table 1). Concerning the 11 MFS efflux transporters present in M. leprae, eight are non-functional of which some were found to be upregulated in M. tuberculosis in response to antibiotics (Table 1). Among these is noted that Rv1258c, also known as Tap-like efflux pump, is a pseudogene (ML1104c) in M. leprae presenting 58.4% similarity at nucleotide level with Rv1258c. Rv1258c is associated with reduced susceptibility to several drugs, namely to rifampicin and clofazimine (Table 1) and has an essential role in physiology, growth, and cell morphology (Ramón-García et al., 2012). These findings emphasize the important role of the Rv1258c efflux pump in the oxidative stress response, cell wall assembly and growth, intrinsic drug resistance (Ramón-García et al., 2012) and macrophage tolerance (Adams et al., 2011). Since M. leprae orthologue is non-functional, it is unlikely that Rv1258c play any role in M. leprae intrinsic drug resistance and virulence. This can be one more genomic trait of M. leprae that contributes to its notable susceptibility to rifampicin and clofazimine in vivo.
M. leprae chromosome encodes only three functional MFS efflux pumps associated with drug transport in M. tuberculosis (Table 1). Of these, the M. tuberculosis P55 efflux pump (Rv1410c), orthologue of M. leprae ML0556c, is one of the most relevant and well-studied efflux pumps of M. tuberculosis and has been associated with the resistance to several drugs (Table 1) and virulence. P55 forms an operon with LprG (Rv1411c), a conserved lipoprotein, which is required for in vivo growth of M. tuberculosis (Bigi et al., 2004; Farrow and Rubin, 2008), virulence (Bianco et al., 2011a) and accurate cell-wall assembly (Bigi et al., 2004; Bianco et al., 2011b). M. leprae encodes both proteins presenting high similarity with those of M. tuberculosis. P55 is also associated with cholesterol transport, carbon metabolism, and oxidative stress, which are of major importance for mycobacterial optimal survival and pathogenesis (Ramón-García et al., 2015). Contrary to that observed for Rv1258c (Tap-like efflux pump), the presence of P55 in M. leprae genome indicates a vital role of this transporter in M. leprae for which a significant contribution in providing intrinsic antibiotic resistance is plausible.
MATE Transporters
DinF (Rv2836c) is the only MATE transporter that M. tuberculosis genome encodes. The M. tuberculosis homologue in M. smegmatis (Mmp) is involved in the resistance to multiple drugs (Table 1). Importantly, DinF is absent from M. leprae genome which can be related with the Na+-dependent nature of the MATE transporters that may not exist in the environment where M. leprae resides. Youm and Saier (2012) also noted the absence of other NA+ transporters in M. leprae that are present in M. tuberculosis and suggested that these facilitators probably contribute to the maintenance of ion homeostasis and adaptation to several stress conditions.
SMR Transporters
M. tuberculosis genome harbors only one gene belonging to the SMR family, the mmr gene, orthologue of ML1756 of M. leprae. Mmr overexpression was showed to decrease susceptibility of M. smegmatis and M. tuberculosis to intercalating dyes, quaternary ammonium compounds and antibiotics (Table 1). Mmr is controlled by the TetR-like transcriptional repressor Rv3066 (Bolla et al., 2012) whose orthologue in M. leprae is ML1757. In both species, the transcriptional repressor is located immediately downstream of mmr or ML1756. However, while in M. tuberculosis the Rv3066 gene encodes a 202-aminoacidic protein, its orthologue in M. leprae is a pseudogene. This means that mmr transcription is no longer repressed in M. leprae. Nothing is known about M. leprae susceptibility to biocides and dyes thus the significance of mmr depression cannot be unveiled.
RND Transporters
M. tuberculosis genome contains 13 genes that encode MmpLs (Mycobacterial membrane protein, Large), and five auxiliary proteins, the MmpSs (Mycobacterial membrane protein, Small) (Table 1). The MmpLs efflux pumps are responsible for the transport of lipids, mainly mycolic acids, essential for mycobacterial survival and pathogenesis, and heme transport (Cox et al., 1999; Camacho et al., 2001; Converse et al., 2003; Domenech et al., 2005; Tullius et al., 2011; Grzegorzewicz et al., 2012; Tahlan et al., 2012; Rodríguez et al., 2013). The expression of M. tuberculosis MmpL proteins is controlled by a complex regulatory network that includes orthologues of TetR (Rv1816 and Rv3249c) and MarR (Rv0678) transcriptional regulators (Radhakrishnan et al., 2014; Delmar et al., 2015). The transcriptional regulator Rv0678 has no orthologue in M. leprae and Rv3249c and Rv1816, whose M. leprae counterparts are ML0770 and ML0933, are non-functional. This indicate that some of the M. leprae mmpL genes are out of regulation and are being constitutively expressed or MmpLs regulation in M. leprae involves a different regulatory network from that found in M. tuberculosis. Of the 13 MmpLs encoded in M. tuberculosis, only MmpL3, MmpL4, MmpL7, MmpL10, and MmpL11 are functional proteins in M. leprae, while the remaining are absent or non-functional (Table 1). Of these, only MmpL3 is essential for M. tuberculosis survival (Domenech et al., 2005). Moreover, MmpL3 has emerged as a novel therapeutic target in M. tuberculosis (Li et al., 2014). Due to high degree of similarity between M. leprae MmpL7, MmpL11, and MmpL3 and their orthologous in M. tuberculosis (Table 1), we can anticipate a similar function for both proteins in M. leprae.
MmpS5-MmpL5, one of the most important RND transporters of M. tuberculosis, is absent in the M. leprae genome. During the reductive evolutionary process that M. leprae experienced, the MmpS5-MmpL5 efflux transporter was eliminated probably to maintain only the pathways required for a strict intracellular lifestyle, typical of M. leprae. The overexpression of the MmpS5-MmpL5 efflux transporter was shown to be associated with resistance of M. tuberculosis to azoles (Milano et al., 2009) and bedaquiline and cross-resistance to clofazimine (Andries et al., 2014). The absence of the MmpS5-MmpL5 explains way clofazimine is so efficient against M. leprae. So far, very rare M. leprae strains were described with clofazimine resistance reinforcing the connection between MmpS5-MmpL5 and clofazimine resistance as well as its unique hypersusceptibility in M. leprae.
Conclusions and Future Prespectives
The occurrence of shared multidrug resistance efflux transporters between M. leprae and M. tuberculosis reveals implications for drug resistance and virulence. Multidrug resistance efflux pumps are ubiquitous in nature. Some efflux pumps exhibit a dual role in M. tuberculosis contributing to both drug resistance and virulence. Here, we have shown that the major M. tuberculosis efflux pumps that are associated with drug efflux and virulence have been retained during the reductive evolutionary process that M. leprae underwent. These efflux pumps are not only important for substrate transport across the inner membrane but are also responsible for drug resistance by extruding drugs from the periplasm to the outside of the cell. They may confer a selective advantage in hostile environments, therefore contributing to M. leprae pathogenicity and acquired drug resistance to therapy as seen in M. tubreculosis. It has been recently shown that resistance to effective multidrug therapy, especially in the high burden countries such as Brazil and India, is on rise, with noteworthy rates of resistance especially against rifampicin and dapsone (Cambau et al., 2018). Rifampicin resistance was found in new cases of leprosy that may relate to individual abuse of this antibiotic usage for treating other bacterial infections as it was also seen with ofloxacin resistance although an antibiotic not used for the first-line treatment of leprosy (Cambau et al., 2018). Future work should focus on efflux pumps, as those mentioned above, as new drug targets for new leprosy therapeutics. A comparative transcriptomic profile of these transporters may provide additional insights, since differences are expected in the efflux pump expression due to pathogen specificity as consequence of the obligate intracellular lifecycle of M. leprae. The modulation of these novel targets will enhance the eradication efforts of endemic leprosy and prevent emergence of drug resistance in afflicted countries. This comparative and perspective study identified these new targets using biological information gathered from M. tuberculosis and constitutes the first step for a more detailed computational studies to bring more mechanistic insights and biological analyses to be applied to M. leprae, susceptible and drug resistant clinical strains, similar to what have been done for M. tuberculosis (Sandhu and Akhter, 2016). The increase in the number of available sequenced genomes and structural data of these proteins together with the advances on experimental and computational biology will improve our knowledge on the relationship between M. leprae protein sequence, structure, dynamics and function (Li et al., 2017).
Author Contributions
DM and MV designed this conceptual study. DM performed the analyses. DM, EL, FM, EC and MV have made substantial contributions to the work and approved its final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. DM and MV work is partially supported by the Global Health and Tropical Medicine (GHTM) Research Center (Grant UID/Multi/04413/2013) from Fundação para a Ciência e a Tecnologia (FCT), Portugal and own funding. DM is supported by grant SFRH/BPD/100688/2014 from FCT, Portugal. EL, FM, and EC work on leprosy is supported by the 2016-2022 grant from Santé Publique France.
References
- Adams K. N., Takaki K., Connolly L. E., Wiedenhoft H., Winglee K., Humbert O., et al. (2011). Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53. 10.1016/j.cell.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsa J., Blokpoel M., Otal I., Young D., De Smet K., Martin C. (1998). Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 80, 5836–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan N -F., Petty N. K., Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K., Villellas C., Coeck N., Thys K., Gevers T., Vranckx L., et al. (2014). Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS ONE 9:e102135. 10.1371/journal.pone.0102135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balganesh M., Dinesh N., Sharma S., Kuruppath S., Nair A. V., Sharma U. (2012). Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 56, 2643–2651. 10.1128/AAC.06003-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balganesh M., Kuruppath S., Marcel N., Sharma S., Nair A., Sharma U. (2010). Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob. Agents Chemother. 54, 5167–5172. 10.1128/AAC.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. K., Bhatt K., Misra P., Chakraborti P. (2000). Involvement of a natural transport system in the process of efflux-mediated drug resistance in Mycobacterium smegmatis. Mol. Gen. Genet. 262, 949–956. 10.1007/PL00008663 [DOI] [PubMed] [Google Scholar]
- Banerjee S. K., Bhatt K., Rana S., Misra P., Chakraborti P. (1996). Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 226, 362–368. 10.1006/bbrc.1996.1362 [DOI] [PubMed] [Google Scholar]
- Bianco M. V., Blanco F., Forrellad M., Aguilar D., Campos E., Klepp L., et al. (2011a). Knockout mutation of p27-p55 operon severely reduces replication of Mycobacterium bovis in a macrophagic cell line and survival in a mouse model of infection. Virulence 2, 233–237. 10.4161/viru.2.3.15888 [DOI] [PubMed] [Google Scholar]
- Bianco M. V., Blanco F., Imperiale B., Forrellad M., Rocha R., Klepp L., et al. (2011b). Role of P27-P55 operon from Mycobacterium tuberculosis in the resistance to toxic compounds. BMC Infect. Dis. 11:195. 10.1186/1471-2334-11-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigi F., Gioffré A., Klepp L., de la Paz Santangelo M., Alito A., Caimi K., et al. (2004). The knockout of the lprG-Rv1410 operon produces strong attenuation of Mycobacterium tuberculosis. Microbes Infect. 6, 182–187. 10.1016/j.micinf.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Black P. A., Warren R., Louw G., van Helden P., Victor T., Kana B. (2014). Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2491–2503. 10.1128/AAC.02293-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolla J. R., Do S., Long F., Dai L., Su C., Lei H., et al. (2012). Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis. Nucleic Acids Res. 40, 9340–9355. 10.1093/nar/gks677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin E., Hershberg R. (2015). Gene loss dominates as a source of genetic variation within clonal pathogenic bacterial species. Genome Biol. Evol. 7, 2173–2187. 10.1093/gbe/evv135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braibant M., Lefevre P., de Wit L., Peirs P., Ooms J., Huygen K., et al. (1996). A Mycobacterium tuberculosis gene cluster encoding proteins of a phosphate transporter homologous to the Escherichia coli Pst system. Gene 176, 171–176. 10.1016/0378-1119(96)00242-9 [DOI] [PubMed] [Google Scholar]
- Brandis G., Hughes D. (2013). Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J. Antimicrob. Chemother. 68, 2493–2497. 10.1093/jac/dkt224 [DOI] [PubMed] [Google Scholar]
- Caleffi-Ferracioli K. R., Amaral R., Demitto F., Maltempe F., Canezin P., Scodro R., et al. (2016). Morphological changes and differentially expressed efflux pump genes in Mycobacterium tuberculosis exposed to a rifampicin and verapamil combination. Tuberculosis 97, 65–72. 10.1016/j.tube.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Camacho L., Constant P., Raynaud C., Laneelle M., Triccas J., Gicquel B., et al. (2001). Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276, 19845–19854. 10.1074/jbc.M100662200 [DOI] [PubMed] [Google Scholar]
- Cambau E., Saunderson P., Matsuoka M., Cole S., Kai M., Suffys P., et al. (2018). WHO surveillance network of antimicrobial resistance in leprosy. Antimicrobial resistance in leprosy: results of the first prospective open survey conducted by a WHO surveillance network for the period 2009-15. Clin. Microbiol. Infect. 24, 1305–1310. 10.1016/j.cmi.2018.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri B. S., Bhakta S., Barik R., Basu J., Kundu M., Chakrabarti P. (2002). Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J. 367, 279–285. 10.1042/bj20020615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri B. S., Sen S., Chakrabarti P. (1999). Isoniazid accumulation in Mycobacterium smegmatis is modulated by proton motive force-driven and ATP-dependent extrusion systems. Biochem. Biophys. Res. Commun. 256, 682–684. 10.1006/bbrc.1999.0357 [DOI] [PubMed] [Google Scholar]
- Cole S. T., Eiglmeier K., Parkhill J., James K., Thomson N., Wheeler P., et al. (2001). Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011. 10.1038/35059006 [DOI] [PubMed] [Google Scholar]
- Converse S. E., Mougous J., Leavell M., Leary J., Bertozzi C., Cox J. (2003). MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. U.S.A. 100, 6121–6126. 10.1073/pnas.1030024100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999). Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. [DOI] [PubMed] [Google Scholar]
- da Silva P., Bigi F., Santangelo M., Romano M., Martín C., Cataldi A., et al. (2001). Characterization of P55, a multidrug efflux pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 45, 800–804. 10.1128/AAC.45.3.800-804.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva P. E., Machado D., Ramos D., Couto I., von Groll A., Viveiros M. (2016). Efflux pumps in mycobacteria: antimicrobial resistance, physiological functions, and role in pathogenicity, in Efflux-Mediated Antimicrobial Resistance in Bacteria, 1st Edn eds Li X-Z., Elkins C. A., Zgurskaya H. I. (Cham: Springer International Publishing; ), 527–559. [Google Scholar]
- Danilchanka O., Mailaender C., Niederweis M. (2008). Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 52, 2503–2511. 10.1128/AAC.00298-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Knegt G. J., Bruning O., Marian T., de Jong M., van Belkum A., Endtz H. P., et al. (2013). Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberculosis 93, 96–101. 10.1016/j.tube.2012.10.013 [DOI] [PubMed] [Google Scholar]
- De Rossi E., Ainsa J., Ricardi G. (2006). Role of mycobacterial efflux transporters in drug resistance: and unresolved question. FEMS Microbiol. Rev. 30, 36–52. 10.1111/j.1574-6976.2005.00002.x [DOI] [PubMed] [Google Scholar]
- De Rossi E., Arrigo P., Bellinzoni M., da Silva P., Martin C., Ainsa J., et al. (2002). The multidrug transporters belonging to major facilitator superfamily in Mycobacterium tuberculosis. Mol. Med. 8, 714–724. 10.1007/BF03402035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E., Branzoni M., Cantoni R., Milano A., Riccardi G., Ciferri O. (1998). mmr, a Mycobacterium tuberculosis gene conferring resistance to small cationic dyes and inhibitors. J. Bacteriol. 180, 6068–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmar J., Chou T., Wright C., Licon M., Doh J., Radhakrishnan A., et al. (2015). Structural basis for the regulation of the MmpL transporters of Mycobacterium tuberculosis. J. Biol. Chem. 290, 28559–28574. 10.1074/jbc.M115.683797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K., Gumbo T., Maartens G., Dooley K. E., McNerney R., Murray M., et al. (2017). The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 5, 291–360. 10.1016/S2213-2600(17)30079-6 [DOI] [PubMed] [Google Scholar]
- Dinesh N., Sharma S., Balganesh M. (2013). Involvement of efflux pumps in the resistance to peptidoglycan synthesis inhibitors in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 1941–1943. 10.1128/AAC.01957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Kobayashi H., LeVier K., Walker G., Barry C. (2009). BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J. Bacteriol. 191, 477–485. 10.1128/JB.01132-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Reed M., Barry C. (2005). Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501. 10.1128/IAI.73.6.3492-3501.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Reed M., Dowd C., Manca C., Kaplan G., Barry C. (2004). The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279, 21257–21265. 10.1074/jbc.M400324200 [DOI] [PubMed] [Google Scholar]
- Doran J. L., Pang Y., Mdluli K., Moran A., Victor T., Stokes R., et al. (1997). Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin. Diagn. Lab. Immunol. 4, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiglmeier K., Parkhill J., Honore N., Garnier T., Tekaia F., Telenti A., et al. (2001). The decaying genome of Mycobacterium leprae. Lepr. Rev. 72, 387–398. [PubMed] [Google Scholar]
- Elbourne L. D., Tetu S., Hassan K., Paulsen I. (2017). TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324. 10.1093/nar/gkw1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow M. F., Rubin E. (2008). Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J. Bacteriol. 190, 1783–1791. 10.1128/JB.01046-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garima K., Pathak R., Tandon R., Rathor N., Sinha R., Bose M., et al. (2015). Differential expression of efflux pump genes of Mycobacterium tuberculosis in response to varied subinhibitory concentrations of antituberculosis agents. Tuberculosis 95, 155–161. 10.1016/j.tube.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Grkovic S., Brown M., Skurray R. (2001). Transcriptional regulation of multidrug efflux pumps in bacteria. Semin. Cell Dev. Biol. 12, 225–237. 10.1006/scdb.2000.0248 [DOI] [PubMed] [Google Scholar]
- Grzegorzewicz A. E., Pham H., Gundi V., Scherman M., North E., Hess T., et al. (2012). Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341. 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. K., Chauhan D., Srivastava K., Das R., Batra S., Mittal M., et al. (2006). Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J. Commun. Dis. 38, 246–254. [PubMed] [Google Scholar]
- Gupta A. K., Katoch V., Chauhan D., Sharma R., Singh M., Venkatesan K., et al. (2010a). Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb. Drug Resist. 16, 21–28. 10.1089/mdr.2009.0054 [DOI] [PubMed] [Google Scholar]
- Gupta A. K., Reddy V., Lavania M., Chauhan D., Venkatesan K., Sharma V., et al. (2010b). jefA (Rv2459), a drug efflux gene in Mycobacterium tuberculosis confers resistance to isoniazid and ethambutol. Indian J. Med. Res. 132, 176–188. [PubMed] [Google Scholar]
- Hao P., Shi-Liang Z., Ju L., Ya-Xin D., Biao H., Xu W., et al. (2011). The role of ABC efflux pump, Rv1456c-Rv1457c-Rv1458c, from Mycobacterium tuberculosis clinical isolates in China. Folia Microbiol. 56, 549–553. 10.1007/s12223-011-0080-7 [DOI] [PubMed] [Google Scholar]
- Harris K. K., Fay A., Yan G. H., Kunwar P., Socci N. D., Pottabathini N., et al. (2014). Novel imidazoline antimicrobial scaffold that inhibits DNA replication with activity against mycobacteria and drug resistant Gram-positive cocci. ACS Chem. Biol. 9, 2572–2583. 10.1021/cb500573z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkoorn R. C., Uplekar S., Cole S. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2979–2981. 10.1128/AAC.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Zhang W., Zhang Y., Gao F., Lu C., Zhang X., et al. (2008). Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microb. Drug Resist. 14, 7–11. 10.1089/mdr.2008.0772 [DOI] [PubMed] [Google Scholar]
- Kapopoulou A., Lew J., Cole S. (2011). The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis 91, 8–13. 10.1016/j.tube.2010.09.006 [DOI] [PubMed] [Google Scholar]
- La Rosa V., Poce G., Canseco J. O., Buroni S., Pasca M. R., Biava M., et al. (2012). MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324–331. 10.1128/AAC.05270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhang J., Guo Q., Wei J., Jiang Y., Zhao X., et al. (2015). Study of efflux pump gene expression in rifampicin-monoresistant Mycobacterium tuberculosis clinical isolates. J. Antibiot. 68, 431–435. 10.1038/ja.2015.9 [DOI] [PubMed] [Google Scholar]
- Li P., Gu Y., Li J., Xie L., Li X. Z., Xie J. (2017). Mycobacterium tuberculosis Major Facilitator superfamily transporters. J. Membrane Biol. 250, 573–585. 10.1007/s00232-017-9982-x [DOI] [PubMed] [Google Scholar]
- Li W., Upadhyay A., Fontes F., North E., Wang Y., Crans D., et al. (2014). Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 6413–6423. 10.1128/AAC.03229-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang L., Nikaido H. (2004). Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423. 10.1128/AAC.48.7.2415-2423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Z., Elkins C., Zgurskaya H. (2016). Efflux-Mediated Antimicrobial Resistance in Bacteria, 1st Edn. Cham: Springer International Publishing, 1–848. [Google Scholar]
- Louw G. E., Warren R., van Pittius N., Leon R., Jimenez A., Hernandez-Pando R., et al. (2011). Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am. J. Respir. Crit. Care Med. 184, 269–276. 10.1164/rccm.201011-1924OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw G. E., Warren R., van Pittius N., McEvoy C., van Helden P., Victor T. (2009). A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53, 3181–3189. 10.1128/AAC.01577-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Liu M., Wang Y., Pang Y., Zhao Z. (2014). Mechanisms of fluoroquinolone monoresistance in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 353, 40–48. 10.1111/1574-6968.12401 [DOI] [PubMed] [Google Scholar]
- Machado D., Coelho T., Perdigão J., Pereira C., Couto I., Portugal I., et al. (2017). Interplay between mutations and efflux in drug resistant clinical isolates of Mycobacterium tuberculosis. Front. Microbiol. 8:711. 10.3389/fmicb.2017.00711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Couto I, Perdigão J., Rodrigues L., Baptista P., Portugal I., et al. (2012). Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS ONE 7:e34538. 10.1371/journal.pone.0034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S., Chen Y-P. (2010). Bacterial genomic G+C composition-eliciting environmental adaptation. Gemonics 95, 7–15. 10.1016/j.ygeno.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Sánchez M., Martinez-Solano L., Hernandez A., Garmendia L., Fajardo A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449. 10.1111/j.1574-6976.2008.00157.x [DOI] [PubMed] [Google Scholar]
- McLeod M. P., Qin X., Karpathy S., Gioia J., Highlander S., Fox G., et al. (2004). Complete genome sequence of Rickettsia typhi and comparison with sequences of other Rickettsiae. J. Bacteriol. 186, 5842–5855. 10.1128/JB.186.17.5842-5855.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano A., Pasca M., Provvedi R., Lucarelli A., Manina G., Ribeiro A., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 89, 84–90. 10.1016/j.tube.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Mishra M., Daniels L. (2013). Characterization of the MSMEG_2631 gene (mmp) encoding a multidrug and toxic compound extrusion (MATE) family protein in Mycobacterium smegmatis and exploration of its polyspecific nature using biolog phenotype microarray. J. Bacteriol. 195, 1610–1621. 10.1128/JB.01724-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. (2002). Microbial minimalism: genome reduction in bacterial pathogens. Cell 108, 583–586. 10.1016/S0092-8674(02)00665-7 [DOI] [PubMed] [Google Scholar]
- Morris J. J., Lenski R., Zinser E. (2012). The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, e00036–e00012. 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Lu J., Wang Y., Song Y., Wang S., Zhao Y. (2013). Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 5, 893–900. 10.1128/AAC.01024-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca M. R., Guglierame P., Arcesi F., Bellinzoni M., De Rossi E., Riccardi G. (2004). Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 3175–3178. 10.1128/AAC.48.8.3175-3178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca M. R., Guglierame P., De Rossi E., Zara F., Riccardi G. (2005). mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 49, 4775–4777. 10.1128/AAC.49.11.4775-4777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A., Kumar N., Wright C. C., Chou T. H., Bolla J. R., Tringides M. L., et al. (2014). Crystal structure of the transcriptional regulator Rv0678 of Mycobacterium tuberculosis. J. Biol. Chem. 289, 16526–16540. 10.1074/jbc.M113.538959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-García S., Martín C., Aínsa J. A., De Rossi E. (2006). Characterization of tetracycline resistance mediated by the efflux pump Tap from Mycobacterium fortuitum. J. Antimicrob. Chemother. 57, 252–259. 10.1093/jac/dki436 [DOI] [PubMed] [Google Scholar]
- Ramón-García S., Martín C., De Rossi E., Aínsa J. A. (2007). Contribution of the Rv2333c efflux pump (the Stp protein) from Mycobacterium tuberculosis to intrinsic antibiotic resistance in Mycobacterium bovis BCG. J. Antimicrob. Chemother. 59, 544–547. 10.1093/jac/dkl510 [DOI] [PubMed] [Google Scholar]
- Ramón-García S., Martín C., Thompson C. J., Aínsa J. A. (2009). Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob. Agents Chemother. 53, 3675–3682. 10.1128/AAC.00550-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-García S., Mick V., Dainese E., Martin C., Thompson C., De Rossi E., et al. (2012). Functional and genetic characterization of the Tap efflux pump in Mycobacterium bovis BCG. Antimicrob. Agents Chemother. 56, 2074–2083. 10.1128/AAC.05946-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-García S., Stewart G., Hui Z., Mohn W., Thompson C. (2015). The mycobacterial P55 efflux pump is required for optimal growth on cholesterol. Virulence 6, 444–448. 10.1080/21505594.2015.1044195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L., Villellas C., Bailo R., Viveiros M., Ainsa J. (2013). Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 751–757. 10.1128/AAC.01482-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez J. E., Ramírez A., Salas L., Helguera-Repetto C., Gonzalez-y-Merchand J., Soto C., et al. (2013). Transcription of genes involved in sulfolipid and polyacyltrehalose biosynthesis of Mycobacterium tuberculosis in experimental latent tuberculosis infection. PLoS ONE 8:e58378. 10.1371/journal.pone.0058378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu P., Akhter Y. (2016). The drug binding sites and transport mechanism of the RND pumps from Mycobacterium tuberculosis: insights from molecular dynamics simulations. Arch. Biochem. Biophys. 592, 38–49. 10.1016/j.abb.2016.01.007 [DOI] [PubMed] [Google Scholar]
- Sandhu P., Akhter Y. (2018). Evolution of structural fitness and multifunctional aspects of mycobacterial RND family transporters. Arch. Microbiol. 200, 19–31. 10.1007/s00203-017-1434-6 [DOI] [PubMed] [Google Scholar]
- Schmalstieg A. M., Srivastava S., Belkaya S., Deshpande D., Meek C., Leff R., et al. (2012). The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob. Agents Chemother. 56, 4806–4815. 10.1128/AAC.05546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scollard D. M., Adams L., Gillis T., Krahenbuhl J., Truman R., Williams D. (2006). The continuing challenges of leprosy. Clin. Microbiol. Rev. 19, 338–381. 10.1128/CMR.19.2.338-381.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi N., Das R., Pathak N., Banerjee S., Ahmed N., Katoch V., et al. (2004). Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a Tap-like efflux pump. Infection 32, 109–111. 10.1007/s15010-004-3097-x [DOI] [PubMed] [Google Scholar]
- Singh I., Lavania M., Nigam A., Turankar R., Ahuja M., John A., et al. (2016). Symposium on emerging needs in leprosy research in the post elimination era: The Leprosy Mission Trust India. Lepr. Rev. 87, 132–143. [PubMed] [Google Scholar]
- Srivastava S., Musuka S., Sherman C., Meek C., Leff R., Gumbo T. (2010). Efflux-pump-derived multiple drug resistance to ethambutol monotherapy in Mycobacterium tuberculosis and the pharmacokinetics and pharmacodynamics of ethambutol. J. Infect. Dis. 201, 1225–1231. 10.1086/651377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K., Wilson R., Kastrinsky D., Arora K., Nair V., Fischer E., et al. (2012). SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 1797–1809. 10.1128/AAC.05708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius M. V., Harmston C. A., Owens C. P., Chim N., Morse R. P., McMath L. N. M., et al. (2011). Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108, 5051–5056. 10.1073/pnas.1009516108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros M., Martins M., Rodrigues L., Machado D., Couto I., Ainsa J., et al. (2012). Inhibitors of mycobacterial efflux pumps as potential boosters for anti-tubercular drugs. Expert Rev. Anti. Infect. Ther. 10, 983–998. 10.1586/eri.12.89 [DOI] [PubMed] [Google Scholar]
- Wang K., Pei H., Huang B., Zhu X., Zhang J., Zhou B., et al. (2013). The expression of ABC efflux pump, Rv1217c–Rv1218c, and its association with multidrug resistance of Mycobacterium tuberculosis in China. Curr. Microbiol. 66, 222–226. 10.1007/s00284-012-0215-3 [DOI] [PubMed] [Google Scholar]
- Wang-Kan X., Blair J., Chirullo B., Betts J., La Ragione R. M., Ivens A., et al. (2017). Lack of AcrB efflux function confers loss of virulence on Salmonella enterica serovar Typhimurium. mBio 8, e00968–e00917. 10.1128/mBio.00968-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. M., Jones C., Xi Z., Speer A., Danilchanka O., Doornbos K., et al. (2013). Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 9:e1003120. 10.1371/journal.ppat.1003120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M., DeRisi J., Kristensen H-H., Imboden P., Rane S., Brown P., et al. (1999). Exploring drug induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. U.S.A. 96, 12833–12838. 10.1073/pnas.96.22.12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm J., Saier M. (2012). Comparative analyses of transport proteins encoded within the genomes of Mycobacterium tuberculosis and Mycobacterium leprae. Biochim. Biophys. Acta Biomembranes. 1818, 776–797. 10.1016/j.bbamem.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]