Abstract
Deinococcus bacteria are famous for their extreme resistance to ionising radiation and other DNA damage- and oxidative stress-generating agents. More than a hundred genes have been reported to contribute to resistance to radiation, desiccation and/or oxidative stress in Deinococcus radiodurans. These encode proteins involved in DNA repair, oxidative stress defence, regulation and proteins of yet unknown function or with an extracytoplasmic location. Here, we analysed the conservation of radiation resistance-associated proteins in other radiation-resistant Deinococcus species. Strikingly, homologues of dozens of these proteins are absent in one or more Deinococcus species. For example, only a few Deinococcus-specific proteins and radiation resistance-associated regulatory proteins are present in each Deinococcus, notably the metallopeptidase/repressor pair IrrE/DdrO that controls the radiation/desiccation response regulon. Inversely, some Deinococcus species possess proteins that D. radiodurans lacks, including DNA repair proteins consisting of novel domain combinations, translesion polymerases, additional metalloregulators, redox-sensitive regulator SoxR and manganese-containing catalase. Moreover, the comparisons improved the characterisation of several proteins regarding important conserved residues, cellular location and possible protein–protein interactions. This comprehensive analysis indicates not only conservation but also large diversity in the molecular mechanisms involved in radiation resistance even within the Deinococcus genus.
Keywords: DNA repair, oxidative damage protection, regulatory proteins, stress response, metal homeostatis, biodiversity
The authors reviewed the mechanisms and factors involved in the extreme radiation and oxidative stress resistance in Deinococcus radiodurans in comparison with 10 other resistant Deinococcus species, and highlighted not only conserved pathways but also a large diversity of the repair, protection and regulation toolbox among the different deinococci.
INTRODUCTION
In 1956, scientists described a bacterium that was found as a contaminant in a can of ground meat. This bacterium had survived exposure to a high dose of ionising radiation (IR) that was supposed to sterilise the canned meat (Anderson et al.1956). Now known as Deinococcus radiodurans, this bacterial species is not only extremely tolerant to gamma radiation, but also to other DNA damage- and oxidative stress-generating conditions such as UV and desiccation (Battista 1997). Exposure to high doses of IR generates massive DNA damage, including hundreds of double-strand breaks, but D. radiodurans is able to reconstitute its genome completely within hours after irradiation. Therefore, D. radiodurans is a good model organism to study DNA repair, DNA damage and oxidative stress response, and radiation resistance.
Deinococcus radiodurans and other Deinococcus species show no loss of viability after exposure to IR doses up to 5 kGy. For comparion, a few hundred Gy will kill most known bacterial species, including Escherichia coli and Thermus thermophilus, and 5–10 Gy are lethal to most vertebrates, including humans (Daly 2012). Nevertheless, IR resistance is not unique to Deinococcus, and several organisms tolerating more than 1 kGy have been described, including not only bacteria (e.g. Chroococcidiopsis of the phylum Cyanobacteria) and archaea (e.g. Thermococcus gammatolerans), but also some small eukaryotes (e.g. tardigrades and bdelloid rotifers) (Cox and Battista 2005; Daly 2012). Of these IR-resistant species, D. radiodurans has been studied most extensively, which was accelerated after obtaining its genome sequence (White et al.1999) and by the development of techniques for its genetic manipulation. Characterisation of the mechanisms underlying IR resistance in Deinococcus is also useful to understand IR resistance, or sensitivity, in other organisms.
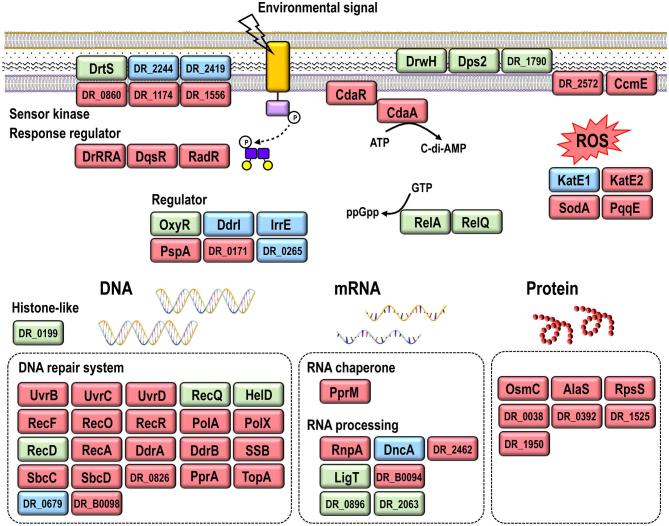
The various in vivo and in vitro approaches used in recent years to study D. radiodurans have indicated that its tolerance to radiation, desiccation and oxidative stress results from a combination of different physiological determinants and well-regulated molecular mechanisms (Fig. 1) (Cox and Battista 2005; Confalonieri and Sommer 2011; Slade and Radman 2011; Daly 2012; Agapov and Kulbachinskiy 2015; Timmins and Moe 2016). Compared to radiation-sensitive species such as E. coli, proteins in D. radiodurans and other radiation-resistant organisms are much better protected against oxidative damage (Daly et al.2007, 2010; Krisko and Radman 2010). Radiation and desiccation lead to generation of reactive oxygen species (ROS), but D. radiodurans has developed efficient enzymatic and non-enzymatic antioxidant systems to remove ROS and limit protein damage. Sufficient proteome protection is crucial for survival after irradiation because protein activity is required for essential processes including transcription, translation and DNA repair. Compared to IR-sensitive bacteria, the nucleoid of Deinococcus species appears more condensed, which may contribute to radiation resistance by limiting diffusion of DNA fragments (Levin-Zaidman et al.2003; Zimmerman and Battista 2005). Following exposure to IR or desiccation, the expression of many genes and proteins is induced in D. radiodurans, including DNA repair proteins and proteins of yet unknown function (Liu et al.2003; Tanaka et al.2004; Lu et al.2009; Basu and Apte 2012), and several regulator proteins involved in the radiation or oxidative stress response have been described (Agapov and Kulbachinskiy 2015).
Figure 1.
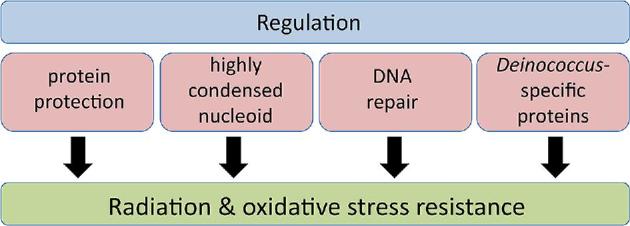
Extreme radiation and oxidative stress resistance in Deinococcus involves multiple factors and well-regulated mechanisms.
Deinococcus radiodurans was the first species of the genus Deinococcus that was isolated, and was also the first Deinococcus species for which the genome sequence was thoroughly analysed (White et al.1999; Makarova et al.2001). Deinococus bacteria are ubiquitous in nature and have been isolated from various environments and locations (e.g. hot and cold desert soil, air, high atmosphere, water). At present, more than 50 radiation-resistant Deinococcus species have been described, and for some of these a complete or draft genome sequence has been obtained. Here, we review the reported data about the mechanisms involved in radiation resistance, oxidative stress defence, DNA repair, and in their regulation in D. radiodurans. The conservation of the proteins involved in these processes was investigated in the 10 other radiation-resistant Deinococcus species for which a complete and assembled genome sequence was available. The 11 analysed Deinococcus species have been isolated from various locations worldwide (Table 1). This comparison showed a remarkable diversity of the radiation resistance-associated proteins among deinococci. Furthermore, sequence analysis improved the characterisation of several of these proteins. Throughout this article we discuss our findings regarding protein functions and resistance-associated mechanisms in the genus Deinococcus.
Table 1.
Information of complete genomes of Deinococcus species.
| Species | Identified in | Total genome size (Mb) | Replicons (sizes in kb) | Proteins | References |
|---|---|---|---|---|---|
| Deinococcus radiodurans (Drad) | Canned meat, USA | 3.28 | 4 (2649, 412, 177, 46) | 3167 | Anderson et al. (1956); Brooks and Murray (1981); White et al. (1999) |
| Deinococcus geothermalis (Dgeo) | Hot spring, Italy | 3.25 | 3 (2467, 574, 206) | 3003 | Ferreira et al. (1997); Makarova et al. (2007) |
| Deinococcus deserti (Ddes) | Sahara Desert sand, Morocco/Tunisia | 3.86 | 4 (2820, 325, 314, 396) | 3503 | de Groot et al. (2005, 2009) |
| Deinococcus maricopensis (Dmar) | Sonoran Desert soil, USA | 3.5 | 1 (3499) | 3242 | Rainey et al. (2005); Pukall et al. (2011) |
| Deinococcus gobiensis (Dgob) | Gobi Desert sand, China | 4.41 | 7 (3137, 433, 425, 232, 72, 55, 53) | 4140 | Yuan et al. (2009, 2012) |
| Deinococcus proteolyticus (Dpro) | Lama glama feces, Japan | 2.89 | 5 (2147, 315, 196, 132, 97) | 2645 | Kobatake, Tanabe and Hasegawa (1973); Brooks and Murray (1981); Copeland et al. (2012) |
| Deinococcus peraridilitoris (Dper) | Coastal desert soil, Chile | 4.51 | 3 (3882, 557, 75) | 4223 | Rainey et al. (2007) |
| Deinococcus swuensis (Dswu) | Mountain soil, South Korea | 3.53 | 1 (3531) | 3217 | Lee et al. (2013) |
| Deinococcus soli (Dsol) | Rice field soil, South Korea | 3.24 | 1 (3237) | 3055 | Cha et al. (2014); Joo et al. (2015) |
| Deinococcus actinosclerus (Dact) | Rocky hillside soil, South Korea | 3.26 | 1 (3264) | 3073 | Joo et al. (2016); Kim et al. (2016) |
| Deinococcus puniceus (Dpun) | Mountain soil, South Korea | 2.97 | 1 (2972) | 2681 | Lee et al. (2015) |
DEINOCOCCUS RADIODURANS MUTANTS AFFECTED IN RADIATION AND OXIDATIVE STRESS RESISTANCE
More than a hundred radiation- and/or oxidative stress-sensitive mutant strains of D. radiodurans have been described in numerous studies (Table S1, Supporting Information). A schematic overview of proteins required for radiation and oxidative stress resistance is shown in Fig. 2. Many of the mutants were obtained after deleting or disrupting a specific gene that was selected because of its expected or possible role in DNA repair, oxidative stress defence and regulation of radiation-resistance-associated genes, or because of its radiation-induced expression. Other mutants were obtained after chemical or transposon mutagenesis followed by screening for increased radiation sensitivity.
Figure 2.
Schematic overview of ionising radiation and oxidative stress resistance-associated proteins in D. radiodurans. Many D. radiodurans gene deletion or disruption mutants with more than 10-fold increased sensitivity compared to the wild-type strain have been described (Table S1, Supporting Information), and the corresponding proteins are indicated in the figure. Red box, ionising radiation sensitive; green box, oxidative stress sensitive; blue box, ionising radiation and oxidative stress sensitive.
Only a few mutant strains were found to be very sensitive to IR, showing a strong decrease in survival after exposure to relatively low doses (< 2 kGy) of IR (Table S1, Supporting Information). These strains are mutated for recA (locus tag DR_2340), recF (DR_1089), recO (DR_0819), recR (DR_0198), polA (DR_1707), pprA (DR_A0346), pprM (DR_0907) or irrE (DR_0167). The DNA repair proteins RecA, RecF, RecO, RecR and PolA are involved in extended synthesis-dependent strand annealing and recombinational repair (Zahradka et al.2006; Slade et al.2009; Bentchikou et al.2010). PprA is a Deinococcus-specific protein required for accurate chromosome segregation and cell division after exposure of the cells to radiation (Devigne et al.2013). IrrE, also called PprI, is a metalloprotease required for induced expression of recA, pprA and other genes after irradiation (Earl et al.2002a; Hua et al.2003; Ludanyi et al.2014). PprM corresponds to the single cold shock protein homologue in D. radiodurans (Ohba et al.2009). Many other mutants showed more than 10-fold reduced survival compared to the wild type only at higher irradiation doses (> 5 kGy), or were found only slightly IR sensitive with less than 10-fold decreased survival compared to the wild type at the highest dose tested.
Several gene mutant strains have been characterised by two or more research teams, and, remarkably, the reported results are sometimes rather different, with mutants found sensitive to IR or other agents in one study but resistant in another study (for details, see legend of Table S1, Supporting Information). Such contrasting results may be due to differences in the bacterial strains used and/or in the experimental procedures.
Different results have also been reported with respect to obtaining mutant strains: it appeared to be impossible to obtain a recJ (DR_1126, single-stranded-DNA-specific exonuclease) or gyrA (DR_1913, DNA gyrase subunit A) null mutant in one or two studies (Nguyen et al.2009; Bentchikou et al.2010; Cao, Mueller and Julin 2010), suggesting that these genes are essential for viability, whereas others successfully obtained null mutants for these genes (Jiao et al.2012; Kota, Charaka and Misra 2014).
Besides for the naturally transformable D. radiodurans, genetic tools allowing construction of mutant strains have also been developed for D. deserti and D. geothermalis. Like in D. radiodurans, a D. deserti irrE mutant is highly sensitive to gamma and UV radiation (Vujicic-Zagar et al.2009). Deletion of the chromosomal recA gene in D. deserti, the third and last gene of an operon equivalent to that in D. radiodurans, did not lead to radiation sensitivity due to the presence of two additional recA genes located on large plasmids (Dulermo et al.2009). A D. geothermalis cystine ABC transporter mutant showed increased sensitivity to H2O2 (Kim et al.2017).
One might expect that genes important for radiation resistance in D. radiodurans be conserved in other radiation-resistant species within the genus Deinococcus. To investigate this, not only homologues of the gene products listed in Table S1 (Supporting Information) but also other proteins involved in radiation resistance-associated processes such as DNA repair and oxidative stress defence (Tables S2–S6, Supporting Information) were searched in 10 other complete and assembled Deinococcus genome sequences (Table 1). Besides showing presence or absence of protein homologues, we included a comparative analysis of domain composition in multidomain proteins and of functionally important residues in proteins. The results are described in the following sections.
DNA REPAIR IN DEINOCOCCUS
Deinococcus radiodurans DNA repair proteins and comparison with E. coli
The genome sequence of D. radiodurans revealed the presence of homologues of most well-known prokaryotic DNA repair proteins involved in base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR) and recombinational repair, suggesting that the DNA repair machinery of D. radiodurans is globally similar to that of other bacteria (Makarova et al.2001), but that it functions more efficiently than in radiation-sensitive species because of better protection of the (DNA repair) proteins against oxidative damage (Daly 2012). Indeed, at least some DNA repair proteins of E. coli, namely PolA (Gutman, Fuchs and Minton 1994), RadA (Zhou et al.2006) and UvrA (Agostini, Carroll and Minton 1996), can functionally substitute for their counterparts in D. radiodurans.
However, genetic, biochemical and structural studies have shown that several other ‘classical’ DNA repair proteins from D. radiodurans have characteristics different from their E. coli counterparts. Concerning recombinational repair, E. coli recA (Schlesinger 2007) and recO (Xu et al.2008) only partially complement the corresponding gene deletion in D. radiodurans. In contrast to E. coli RecA, purified D. radiodurans RecA preferentially binds to double-stranded DNA when also single-stranded DNA is present in the solution, and initiates DNA-strand exchange primarily from the double-stranded DNA (Kim and Cox 2002; Kim et al.2002). Such inverse DNA-strand exchange pathway has also been observed for D. geothermalis RecA in one biochemical study (Sghaier et al.2010) but not in another (Wanarska et al.2011), possibly because of different experimental conditions. More recent studies have indicated that D. radiodurans RecA forms more frequent but shorter filaments compared to E. coli RecA, and that the specific properties of D. radiodurans RecA contribute to efficient repair of hundreds of double-stranded DNA breaks (Hsu et al.2011; Ngo et al.2013; Pobegalov et al.2015; Warfel and LiCata 2015). Processing of double-stranded DNA ends by the RecFOR pathway requires RecQ helicase in E. coli, but characterisation of mutant strains suggests that D. radiodurans might use UvrD helicase rather than its RecQ protein for this process (Bentchikou et al.2010). In addition, unlike its E. coli counterpart, UvrD of D. radiodurans is a bipolar DNA helicase that can unwind both 3΄- and 5΄-tailed double-stranded DNA in vitro (Stelter et al.2013). Deinococcus radiodurans RecF also interacts with DR_1088, a DNA-binding protein that is encoded by the recF-DR_1088 operon but which is absent in E. coli (Cheng et al.2017). Levels of single-stranded DNA-binding protein (SSB) are higher in D. radiodurans than in E. coli (Bernstein et al.2004). Concerning BER, mismatch-specific uracil DNA glycosylase DR_0715 (MUG) has a modified and broadened substrate specificity compared with MUG from E. coli (Moe et al.2006), DR_0689 uracil DNA glycosylase (Ung, COG0692) of D. radiodurans has high catalytic activity attributed to high substrate affinity (Timmins and Moe 2016), and DNA-3-methyladenine glycosylase 2 family protein DR_2584 (AlkA, COG0122) has altered substrate specificity and a wider DNA-binding cleft compared with E. coli AlkA (Moe et al.2012). Furthermore, D. radiodurans MutS has higher affinity for mismatched DNA than E. coli MutS (Banasik et al.2017), and DnaE polymerase of D. radiodurans, but not that of E. coli, features RecA-dependent DNA polymerase activity (Randi et al.2016). Thus, besides increased protein protection, DNA repair systems may also have evolved to perform better under stress conditions that generate massive DNA damage. This is supported by experiments with E. coli, for which radiation-resistant strains surviving 3 kGy were obtained after repeated exposure to IR (Byrne et al.2014). In these strains, mutations in recA are prominent and contribute to the acquired radiation resistance (Piechura et al.2015).
Deinococcus radiodurans also encodes more than one variant of several DNA repair proteins (e.g. multiple uracil DNA glycosylases and endonuclease III proteins), and these variants may have specialised roles that improve the DNA repair repertoire (Sandigursky et al.2004; Timmins and Moe 2016). Moreover, for various novel proteins more specific to Deinococcus it has been demonstrated or proposed that they contribute to DNA repair or genome preservation (e.g. DdrA to DdrD, PprA, DR_A0282) (Selvam et al.2013; Agapov and Kulbachinskiy 2015; Bouthier de la Tour et al.2017).
Analysis of the genome sequence also revealed that D. radiodurans lacks homologues of several well-known DNA repair proteins, indicating that it does not use some repair mechanisms or that it uses alternative mechanisms. Initiation of homologous recombination in E. coli involves either the RecBCD complex, its major pathway for double-strand break repair, or the RecFOR pathway (Rocha, Cornet and Michel 2005). However, recB and recC are absent in D. radiodurans and it uses the RecFOR pathway for processing of double-stranded DNA ends (Bentchikou et al.2010). In E. coli, the RecFOR pathway is inhibited by SbcB (Exodeoxyribonuclease I) (Kowalczykowski et al.1994), and D. radiodurans lacks SbcB. Overexpression of E. coli RecBC (Khairnar, Kamble and Misra 2008) or SbcB (Misra et al.2006) in D. radiodurans leads to reduced resistance to IR and interferes with DNA double-strand break repair. In addition to homologous recombination, several bacteria use non-homologous end joining (NHEJ) to repair DNA double-strand breaks, but there is no evidence that this generally error-prone repair system exists in D. radiodurans (Slade and Radman 2011). Deinococcus radiodurans also misses specialised translesion synthesis (TLS) DNA polymerases such as UmuCD that, in E. coli, are involved in mutagenic lesion bypass. Thus, the absence of certain DNA repair proteins may be important for efficient and error-free repair of massive DNA damage in D. radiodurans.
DNA repair proteins in 11 Deinococcus species: overview
To get more insight in the DNA repair repertoire in the genus Deinococcus, the DNA repair genes in 10 other radiation-resistant Deinococcus species were searched and compared with that of the well-studied D. radiodurans. The conservation of novel proteins possibly involved in DNA repair is described in the sections ‘The Ddr and Ppr proteins’ and ‘Miscellaneous proteins involved in resistance to radiation and other DNA-damaging agents in Deinococcus’. Most genes for important DNA repair mechanisms are highly conserved, whereas homologues of several DNA repair genes, such as recC and sbcB and genes for the NHEJ proteins LigD and Ku, are absent in each analysed Deinococcus species. Interestingly, we also observed many differences regarding protein presence/absence, domain composition or numbers of protein variants (see Table 2 for the main differences among the 11 Deinococcus species, and Table S2 (Supporting Information) for accession numbers of all DNA repair proteins). Homologues of several D. radiodurans DNA repair proteins are absent in some of the other species, whereas some other proteins lacking in D. radiodurans are present in others. Intriguingly, the latter include three proteins that, compared to D. radiodurans and E. coli, contain novel combinations of two domains within a single protein: AdaA-AlkA, PhrB-Ung and Nth-Dcm (Fig. 3). The conservation or diversity across the Deinococcus species of DNA repair proteins for different DNA repair pathways is described in detail in the following sections. Here, as an overview, the presence or absence of the DNA repair proteins in the Deinococcus species compared with D. radiodurans is as follows:
Present at least once in each species are AlkA, Mpg, MutY, Mug, Ung (fused or not to PhrB), Fpg, Nth, XthA, Mfd, UvrA1, UvrB, UvrC, UvrD, UvsE, MutL, MutS1, MutS2, XseA, XseB, Atl1 (YbaZ), RdgB (YggV), RecA, RecD, RecF, RecF-interacting DR_1088 homologue, RecG, RecJ, RecN, RecO, RecQ (or absent in D. geothermalis), HRDC domain protein, RecX, RadA, RuvA, RuvB, RuvC, SbcC, SbcD, SSB, LigA, GyrA, GyrB, TopA (topoisomerase 1), Top1 (topoisomerase IB), PolA, PolX, RarA.
Absent in each are Tag, Nfo, Cho, MutH, AlkB, RecC, RecE, RecT, SbcB, RadC, LigD, Ku, TopB, UmuCD.
Present in D. radiodurans but not in each of the other species are Udg4, putative Udg DR_0022, Nfi, UvrA2, SSL2 DNA or RNA helicase, HelD (DNA helicase IV), HepA (SNF2 family helicase), DJ-1 family deglycase, nuclease-related domain (NERD) protein.
Absent in D. radiodurans but present in one or more of the other Deinococcus species are AlkD, family 5 Udg, Dam, Dcm, Vsr, Ada, PhrB, SplB, Dut, Dcd, RecB/AddA, RusA, Exo (Xni), NucS, PolB, DnaE2, ImuY, DinP, and the two-domain proteins AdaA-AlkA, PhrB-Ung and Nth-Dcm.
Table 2.
Main differences regarding DNA repair-related proteins in Deinococcus species.
| Protein | Drad | Dgeo | Ddes | Dmar | Dgob | Dpro | Dper | Dswu | Dsol | Dact | Dpun |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BER, MMR, direct reversal and novel two-domain proteins | |||||||||||
| Mpg | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 |
| AlkD | 1 | ||||||||||
| Ung | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| Udg4 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | |||
| Udg DR_0022 | 1 | ||||||||||
| Nfi | 1 | 1 | 1 | 1 | 1 | ||||||
| XthA | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| Dam | 1 | 1 | |||||||||
| Dcm | 1 | 1 | 4 | 2 | 2 | ||||||
| DR_C0020 | 1 | ||||||||||
| Vsr | 1 | 1 | 1 | 1 | |||||||
| Ada | 1 | ||||||||||
| PhrB | 1 | 2 | 2 | 1 | |||||||
| SplB | 1 | 1 | 1 | ||||||||
| AdaA-AlkA | 1 | 1 | 1 | ||||||||
| PhrB-Ung | 1 | 1 | 1 | 1 | 1 | ||||||
| Nth-Dcm | 1 | ||||||||||
| Dut | 1 | 1 | |||||||||
| Dcd | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| Deglycase | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | |
| Nucleotide excision repair | |||||||||||
| UvrA2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | |
| SSL2 helicase | 1 | 1 | 1 | 1 | 2 | 4 | 1 | ||||
| Recombinational repair | |||||||||||
| RecA | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| RecB/AddA | 1 | ||||||||||
| RecQ | 1 | fr | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| RusA (YbcP) | 1 | 2 | |||||||||
| SSB | 1 | 4 | 1 | 1 | 3 | 4 | 1 | 1 | 1 | 1 | 1 |
| Ligases and adjacent genes | |||||||||||
| LigA | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DR_B0100 | 1 | 1 | 1 | 1 | 1 | ||||||
| DR_B0099 | 1 | 1 | |||||||||
| DR_B0098 | 1 | 1 | 1 | 1 | 1 | ||||||
| DR_B0094 | 1 | 1 | 1 | ||||||||
| DR_B0095 | fr | 1 | |||||||||
| Other DNA repair proteins | |||||||||||
| TopA | 1 | 1 | 1 | 2 | 3 | 2 | 2 | 1 | 1 | 1 | 1 |
| Exo (Xni) | 1 | 1 | |||||||||
| NERD domain | 1 | 1 | 1 | 1 | |||||||
| NucS (EndoMS) | 1 | 2 | 1 | fr | 1 | 1 | 1 | ||||
| PolB | 1 | 1 | 1 | 1 | |||||||
| DnaE2 | 1 | 1 | |||||||||
| ImuY | 1 | 1 | |||||||||
| DinP | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| HelD | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |
| HepA | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | ||
fr, frameshift. Names in bold indicate gene inactivation leading to increased sensitivity of D. radiodurans to radiation or oxidative stress in at least one study.
Figure 3.
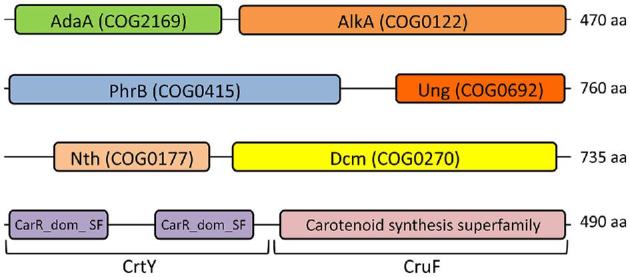
Novel two-domain proteins. The canonical DNA repair proteins AlkA, PhrB, Ung, Nth and Dcm and carotenoid biosynthesis proteins CrtY and CruF are standalone proteins. Genes encoding fusions of two of these proteins were identified in several Deinococcus species. The total number of amino acid residues (aa) of the novel two-domain proteins is indicated at the right.
BER, MMR, direct reversal and novel two-domain proteins
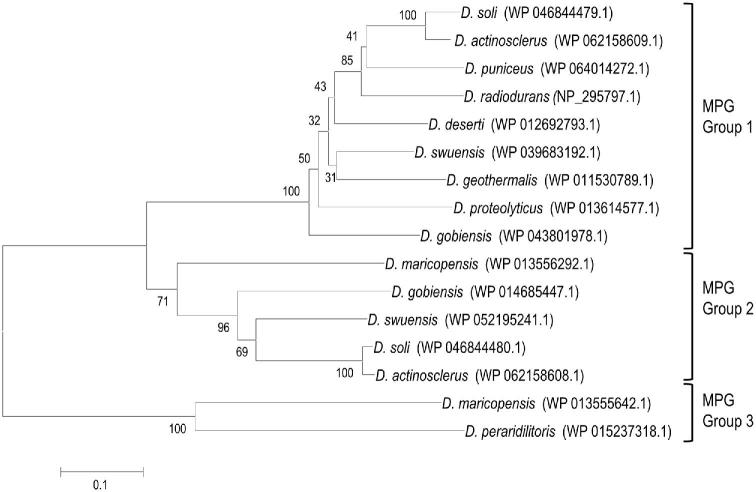
Deinococcus radiodurans and the other Deinococcus species encode multiple DNA glycosylases. Each of these species contains one or two genes for 3-methyladenine DNA glycosylase Mpg (COG2094), one gene (two in D. peraridilitoris) encoding DNA-3-methyladenine glycosylase 2 (AlkA, COG0122) and one (two in D. geothermalis) encoding mismatch-specific uracil DNA-glycosylase (Mug, COG3663) (Table 2 and Table S2, Supporting Information). Interestingly, D. puniceus additionally encodes a 3-methyladenine DNA glycosylase AlkD (COG4912), and three other species have a second AlkA in which the AlkA domain is fused to the AdaA domain (see also below). The single Mpg of D. radiodurans is very similar (more than 70% identity) to an Mpg in eight of the other species, but less similar to others (Fig. 4) (e.g. the single Mpg of D. peraridilitoris shares only 30% identity with D. radiodurans Mpg). The novel catalytic residue (Asp93 in DR_0715) identified in D. radiodurans Mug (Moe et al.2006) is also present in the other Mug proteins except for the second, less-conserved homologue in D. geothermalis. Besides Mug, D. radiodurans possesses three other predicted uracil DNA glycosylases: DR_0689 (Ung, COG0692), DR_1751 (Udg4, COG1573) and DR_0022 (no COG). In vitro uracil DNA glycosylase activity was demonstrated for DR_0689 and DR_1751, but not detected for DR_0022, and the majority of the in vivo uracil DNA glycosylase activity seemed to result from DR_0689 expression (Sandigursky et al.2004). Remarkably, a DR_0689 homologue of similar size was not found in D. proteolyticus, D. actinosclerus, D. soli and D. swuensis. However, these four species, as well as D. gobiensis, do contain a protein in which the Ung domain is fused to a photolyase domain (PhrB, COG0415) (Fig. 3). BLASTP analysis revealed that the PhrB-Ung fusion is unique to Deinococcus species. Besides missing a standalone Ung, D. actinosclerus and D. soli also lack Udg4 uracil DNA glycosylase, indicating that uracil repair in these two organisms depends on Mug and the PhrB-Ung fusion protein. DR_1751 (Udg4) homologues are present in seven other species, and D. peraridilitoris in addition encodes a family 5 Udg. No homologue of the putative uracil DNA glycosylase DR_0022 was found in the other Deinococcus species.
Figure 4.
Three groups of 3-methyladenine DNA glycosylase (MPG) proteins identified in 11 Deinococcus species. The phylogenetic analysis was carried out based on protein sequence alignment of 16 deinococcal MPG proteins (Table S2, Supporting Information) made with Clustal omega. GenBank accession numbers in parentheses follow the species name. The phylogenetic tree was developed using the neighbour-joining algorithm in MEGA 6.0. The scale indicates the number of amino acid substitutions per site, and the node numbers are bootstrap values based on 1000 replications.
Deinococcus radiodurans possesses three endonuclease III proteins (Nth; DR_0289, DR_2438, DR_0928). The nth single, double and triple mutants are as resistant to IR and H2O2 as the wild type, but each single mutant shows slightly elevated levels of spontaneous mutation (Hua et al.2012). In vitro, enzymatic activity has been detected for DR_0289 and DR_2438, but so far not for DR_0928 (Sarre et al.2015). Homologues of these three Nth proteins are present in the other analysed Deinococcus species, except for D. peraridilitoris that lacks a DR_0928 homologue. Deinococcus swuensis has in addition a protein in which the Nth domain is combined with a DNA-cytosine methylase domain (Dcm, COG0270) (Fig. 3). BLASTP analysis revealed only a few Nth-Dcm fusion proteins in other genera (e.g. protein AYO40_02595 of Planctomycetaceae bacterium).
Compared to D. radiodurans, the presence of additional Mpg, AlkA, AlkD, Mug, Udg and two-domain proteins AdaA-AlkA, PhrB-Ung and Nth-Dcm in several species further increases the diversity of DNA glycosylases in Deinococcus. It will be of particular interest to elucidate the precise function(s) of the three novel two-domain proteins.
Besides the Nth-Dcm fusion in D. swuensis, and unlike D. radiodurans, some Deinococcus species possess genes encoding homologues of Dcm and/or DNA-adenine methylase (Dam, COG0338). However, a homologue of DR_C0020 from D. radiodurans, encoding another DNA methylase (COG0863) and whose inactivation results in reduced IR resistance (Table S1, Supporting Information), is not present in the other Deinococcus species.
The bifunctional transcriptional activator/DNA repair enzyme Ada of E. coli is composed of an N-terminal domain AdaA (COG2169, Methylphosphotriester-DNA–protein-cysteine methyltransferase) and a C-terminal domain AdaB (COG0350, O6-methylguanine-DNA–protein-cysteine methyltransferase). D. maricopensis encodes a similar Ada protein. D. gobiensis, D. actinosclerus and D. soli possess the aforementioned novel two-domain protein in which the AdaA domain is not fused with AdaB but with AlkA (Fig. 3). Such AdaA-AlkA fusion is also found in species from various other genera (e.g. protein Rv1317c of Mycobacterium tuberculosis). Other proteins for direct reversal of damage, and which are absent in D. radiodurans but found in others, include homologues of deoxyribodipyrimidine photolyase PhrB (not fused to Ung), photoproduct lyase family protein (SplB, COG1533) and deoxycytidine triphosphate deaminase (Dcd) (Table 2).
Recently, a novel important nucleotide repair system, named guanine glycation repair, has been described (Richarme et al.2017). For this system, it has been shown that the parkinsonism-associated protein DJ-1 and its E. coli homologues Hsp31 (HchA), YhbO and YajL can repair methylglyoxal- and glyoxal-glycated nucleotides, RNA and DNA. These DJ-1/PfpI family proteins, containing domain COG0693 (ThiJ, putative intracellular protease/amidase), are also protein deglycases that can repair methylglyoxal- and glyoxal-glycated proteins. Homologues are present in most Deinococcus species, but remarkably not in D. puniceus. The YhbO homologue DR_1199 of D. radiodurans has been studied previously. Although initially annotated as protease I, no proteolytic or chaperone activity was detected for DR_1199 (Fioravanti et al.2008), in line with the more recently identified deglycase activity of such proteins.
Nucleotide excision repair
Deinococcus radiodurans possesses two NER pathways for repair of UV-induced DNA damage, the UvrA1- and UvsE-dependent pathway, and both are conserved in the other Deinococcus species. A D. radiodurans uvrA1 uvsE mutant is very sensitive to UV (Earl et al.2002b; Tanaka et al.2005). Except for D. geothermalis, the other Deinococcus species have one or two additional UvrA-related proteins, UvrA2. Deinococcus radiodurans UvrA2 has structural similarity with UvrA1 (Timmins et al.2009), but UvrA2 does not contribute to UV resistance in D. radiodurans (Tanaka et al.2005).
Recombinational DNA repair
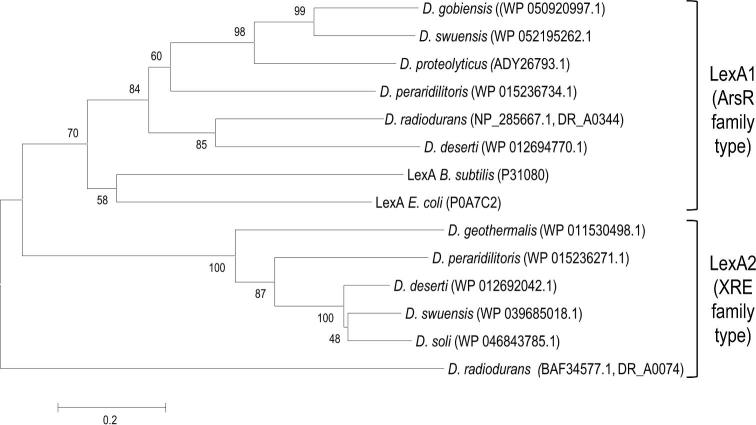
The proteins involved in recombinational DNA repair in D. radiodurans, such as Rec, Ruv and SSB proteins, are highly conserved in the other Deinococcus species. Nevertheless, there is some interesting diversity among the bacteria. Concerning the genetic organisation of the recA gene, each Deinococcus species contains a cinA-ligT-recA gene cluster (probably operon in each), except D. proteolyticus that misses cinA and has a ligT-recA operon (cinA codes for competence/damage-inducible protein A; ligT encodes LigT-like RNA 2΄,3΄-cyclic phosphodiesterase, originally identified as 2΄-5΄ RNA ligase). Whereas the majority of the bacteria possess only one RecA, D. deserti and D. peraridilitoris have two different RecA proteins, encoded by three and two different genes, respectively (Table S2, Supporting Information) (see also the section ‘DNA repair proteins lacking in D. radiodurans but present in other deinococci’). The extra recA genes are not within an operon. Like the cinA-ligT-recA operon, the additional recA genes in D. deserti (de Groot et al.2014), and probably also in D. peraridilitoris (Blanchard et al.2017), are radiation-induced. BLASTP analysis revealed that the extra RecA (RecA2) from both D. deserti and D. peraridilitoris are most similar to RecA proteins from Deinococcus species (e.g. D. radiodurans), suggesting that these RecA2 are of deinococcal origin. However, the two RecA2 proteins do not form a subgroup in a phylogenetic tree (Fig. S1, Supporting Information).
Unlike the other Deinococcus species, D. swuensis codes for a helicase and exonuclease domain-containing protein that is similar to E. coli RecB and Bacillus subtilis AddA, albeit with about 29% identity only. The heterodimer AddAB of B. subtilis, encoded by the addBA operon, is a functional homologue of the E. coli RecBCD enzyme (Kooistra, Haijema and Venema 1993). The RecB/AddA-like protein of D. swuensis shares more than 60% identity with protein fragments from a D. deserti pseudogene that contains two internal stop codons (de Groot et al.2009). Interestingly, both the D. deserti pseudogene and the recB/addA-like gene of D. swuensis are preceded by a gene coding for a protein that is weakly similar to AddB and that includes a nuclease domain. It will be interesting to investigate if this gene pair from D. swuensis encodes a helicase–nuclease complex with a function similar to AddAB in processing of double-stranded DNA ends.
DNA helicase RecQ is important for genome maintenance and DNA repair in a variety of organisms, including E. coli and humans. RecQ contains a catalytic core for ATP-dependent helicase activity and an HRDC (Helicase-and-RNase-D C-terminal) domain involved in DNA binding. Whereas most RecQ proteins have only one HRDC domain, D. radiodurans RecQ has three HRDC domains at its C-terminal region, and in vitro and in vivo studies have shown that all three are involved in RecQ function (Killoran and Keck 2006; Huang et al.2007). The in vivo studies have also shown that a D. radiodurans recQ mutant is slightly sensitive to IR and very sensitive to UV, mitomycin C (MMC) and H2O2 (Huang et al.2007). However, another study has demonstrated that RecQ is not required for IR resistance and for repair of double-strand DNA breaks in D. radiodurans (Bentchikou et al.2010). Therefore, the exact role(s) of RecQ in D. radiodurans is unclear. RecQ-encoding sequences are present in all other Deinococcus species. However, the recQ sequence of D. geothermalis contains a frameshift at one position and an internal stop codon at another position. If these are not DNA sequencing errors, D. geothermalis may not produce an intact RecQ protein. Furthermore, only D. radiodurans RecQ contains three HRDC domains, while one or two HRDC domains are present in the RecQ homologues from the other species (Fig. S2, Supporting Information). Deinococcus peraridilitoris RecQ has in addition a C-terminal helix-turn-helix domain. Another HRDC-domain containing protein (DR_2444 in D. radiodurans), in which the HRDC domain is not associated with a helicase domain, is conserved in Deinococcus, but its function is unknown. RecQ-like proteins containing helicase but not HRDC domains are also present in several Deinococcus species (Fig. S2, Supporting Information).
Deinococcus radiodurans and several bacteria from other genera possess recD although recB and recC are absent. The RecD helicases in these species have an N-terminal extension of about 200 residues compared to E. coli RecD, and have been called RecD2. As for RecQ, conflicting results have been published regarding radiation resistance of a D. radiodurans recD mutant (Table S1, Supporting Information). Although the requirement of RecD for radiation resistance is not clear, it probably has an important in vivo role in Deinococcus species because the protein, including the N-terminal extension, is highly conserved. Other DNA helicases such as UvrD and RecG are also highly conserved. Several Deinococcus species encode additional but less conserved variants of some helicases, including UvrD/REP- and RecD-like proteins (Table S2, Supporting Information).
Ligases
Like in other bacteria, a gene encoding NAD-dependent DNA ligase (LigA) is present in each Deinococcus. Deinococcus deserti expresses two different LigA proteins that share 57% identity (de Groot et al.2009). Deinococcus radiodurans also contains a gene (DR_B0100, also known as ligB or ddrP) predicted to encode an ATP-dependent DNA ligase (Liu et al.2003). DR_B0100 is the first gene of a radiation-induced operon also encoding DR_B0099 (poly ADP-ribose glycohydrolase) and DR_B0098 (polynucleotide kinase) (Blasius et al.2007; Slade et al.2011). A DR_B0100 mutant is IR resistant as the wild type according to one study (Makarova et al.2007), but sensitive according to another (Kota et al.2010). The latter study has also reported that functional complementation of the DR_B0100 deletion requires expression in trans of the entire operon, and that in vitro DNA ligase activity by DR_B0100 requires the presence of DR_B0098 as well as another radiation-induced protein, PprA (pleiotropic protein promoting DNA repair; see also section ‘The Ddr and Ppr proteins’). Recently, it has also been described that DR_B0098 is required for IR resistance, and that the DR_B0098 mutant is equally IR sensitive as the mutant lacking the entire operon (Schmier and Shuman 2018). Homologues of DR_B0100, DR_B0099 and/or DR_B0098 are present in only a few other Deinococcus (Table 2), with D. gobiensis, D. actinosclerus and D. soli possessing a putative operon composed of DR_B0100 and DR_B0098 homologues (Fig. S3, Supporting Informaton). In D. maricopensis, homologues of all three genes are present, but at different locations on its chromosome. Not far downstream of the ligB operon in D. radiodurans is another gene with a reported role in radiation resistance. This gene, DR_B0094 (rnl), encodes a nick-sealing RNA ligase. Recent results indicate that Rnl is involved in DNA repair. Inactivation of rnl sensitises D. radiodurans to radiation and also results in a delay of genome reconstitution following exposure to IR (Schmier et al.2017). However, a DR_B0094 homologue is present in only two of the other Deinococcus species, D. puniceus and D. gobiensis. In the latter, rnl is located adjacent to ligB. D. radiodurans rnl is the first gene of a probable operon also containing DR_B0095. Inactivation of DR_B0095 also results in increased sensitivity to IR (Schmier et al.2017). Sequence analysis suggests that DR_B0095 contains a frameshift, and that the entire gene is predicted to encode an exonuclease (homologue in D. maricopensis).
Multiple variants of a DNA repair protein
For a dozen DNA repair proteins with only one variant in D. radiodurans, more than one variant exists in several other Deinococcus species. Besides some of multiple variants mentioned above (e.g. for RecA, RecD, Mpg), another example is DNA topoisomerase I. Escherichia coli DNA topoisomerase I (TopA, 865 amino acids) contains the conserved domains COG0550 (TopA, DNA topoisomerase IA) and COG0551 (YrdD, ssDNA-binding Zn-finger and Zn-ribbon domains of topoisomerase 1) at the N- and C-terminal region, respectively. Deinococcus radiodurans DNA topoisomerase I (DR_1374) is conserved in the other Deinococcus species, but in these deinococcal proteins (of about 1000 residues) the COG0550 domain is not followed by COG0551 but by COG1754 (uncharacterised C-terminal domain of topoisomerase IA). Four Deinococcus species encode one or two additional topoisomerase I proteins (of about 670 amino acids) that contain only the COG0550 domain (Table S2, Supporting Information). Two of these additional topoisomerase I genes, DGo_PC0276 in D. gobiensis and Deipr_2353 in D. proteolyticus, are directly followed by a gene encoding a UvrD/REP-like helicase, indicating a possible functional link.
DNA repair proteins lacking in D. radiodurans but present in other deinococci
About 20 DNA repair-related genes are present in one or several Deinococcus species but absent in D. radiodurans (Table 2). These include genes for error-prone DNA polymerases PolB, DinP, ImuY and DnaE2. For D. deserti it has been shown that an operon containing lexA-imuY-dnaE2 is involved in UV-induced mutagenesis (Dulermo et al.2009). This operon, which is similar to a RecA/LexA-controlled mutagenesis cassette identified in various bacteria (Erill et al.2006), is also present in desert isolate D. peraridilitoris. If induced mutations are advantageous, for example by changing characteristics of a protein or by generating trancripts encoding small peptides (see section ‘Oxidative stress defence in Deinococcus’), they may contribute to adaptation to harsh environments such as deserts. Therefore, unlike believed earlier (Sale 2007), absence of error-prone TLS DNA polymerases is not crucial for extreme radiation resistance.
Endonuclease NucS (COG1637) is another protein absent in D. radiodurans but found in seven of the other analysed Deinococcus species (although nucS of D. peraridilitoris has a frameshift) (Table 2). NucS has initially been described in the archaeon Pyrococcus abyssi and identified as a novel, DNA structure-dependent endonuclease for single-stranded DNA (Ren et al.2009). However, more recently it has been demonstrated that NucS is a mismatch-specific endonuclease acting on double-stranded DNA containing mismatched bases, and that it is required for a non-canonical MMR pathway in prokaryotes as an alternative to the canonical MutSL-based MMR (Ishino et al.2016; Castaneda-Garcia et al.2017). Therefore, NucS has also been named EndoMS (mismatch-specific Endonuclease) (Ishino et al.2016). Scanning of 3942 reference proteomes has revealed the presence of NucS in 60 archaeal and 310 bacterial species, with the majority of NucS-containing bacterial species (303) belonging to the phylum Actinobacteria and the remaining to the phylum Deinococcus-Thermus (Castaneda-Garcia et al.2017). Most NucS-encoding species lack MutS and MutL. The presence of both NucS and MutS-MutL has been found in only 28 species (Castaneda-Garcia et al.2017), and these include the seven nucS-containing Deinococcus species in Table 2. In the archaeon Halobacterium salinarum, which encodes both NucS and MutS-MutL, inactivation of mutS or mutL produced no hypermutability (Busch and DiRuggiero 2010), suggesting redundancy of the two MMR pathways, which may also be the case in the Deinococcus bacteria possessing both systems. Interestingly, the Deinococcus species with nucS are the same as those possessing a dinP gene encoding error-prone DNA polymerase IV (Table 2), suggesting the possibility that NucS might counteract potential replication errors introduced by PolIV.
In summary, the comparison of the 11 genomes shows a remarkable diversity regarding DNA repair genes among Deinococcus species. As has been proposed for the unusually high number of DNA glycosylases in D. radiodurans, additional DNA repair proteins or variants in other Deinococcus species may contribute to efficient error-free DNA repair and stress survival in these organisms. Interestingly, the presence of different error-prone DNA polymerases indicates that there is also diversity in mechanisms generating genetic variability in Deinococcus species.
OXIDATIVE STRESS DEFENCE IN DEINOCOCCUS
IR induces DNA damage in cells either via direct (energy deposition in the deoxyribose moiety) or indirect (water radiolysis generating ROS) action. Since ROS including superoxide (O2·−), hydrogen peroxide (H2O2) and hydroxyl radicals (OH·) can damage not only DNA but also other macromolecules such as proteins, radiation-resistant organisms should develop efficient anti-oxidative system to cope with oxidative stress. The model organism D. radiodurans has some metabolic configurations that suppress endogenous ROS production, such as the relatively low number of respiratory chain enzymes, the import of peptides and amino acids, and the induction of the glyoxylate bypass of the tricarboxylic acid cycle following IR (Ghosal et al.2005; Slade and Radman 2011). In addition, D. radiodurans is well equipped with enzymatic and non-enzymatic systems to curb ROS levels (Slade and Radman 2011). Here, we compared proteins involved in these direct anti-oxidative systems across 11 Deinococcus species. The differences regarding oxidative stress defence-related proteins between these species are shown in Table 3. Besides these differences, each of the analysed Deinococcus genomes encodes one homologue of SodA, MsrA, MsrB, HslO (Hsp33), DR_B0067 extracellular nuclease, FrnE (truncated in D. maricopensis), Dps1, MsrP, MsrQ, Ppk1 (frameshift in D. maricopensis), Ppk2, Ppx, CrtE, -B, -I, -D, -O, BshA, -B, -C, YpdA, YtxJ (Table S3, Supporting Information). Genes that are absent in each of the 11 genomes include sodB, katG, tpx, grxB, grxD, ahpC and ahpF.
Table 3.
Differences regarding oxidative stress defence-related proteins in Deinococcus species.
| Protein | Drad | Dgeo | Ddes | Dmar | Dgob | Dpro | Dper | Dswu | Dsol | Dact | Dpun |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Superoxide dismutases and catalases | |||||||||||
| SodC (DR_1546) | 1 | 1 | 1 | 1 | |||||||
| SodC (DR_A0202) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| KatE (clade 1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| KatE (clade 2) | 1 | 1 | |||||||||
| KatE (clade 3) | 1 | 1 | 1 | ||||||||
| MnCat | 1 | 1 | 1 | 1 | |||||||
| DR_A0146 (kat-like) | 1 | 2 | 1 | ||||||||
| Peroxiredoxins, Prx-related proteins, and other peroxidases | |||||||||||
| BCP | 3 | 3 | 2 | 3 | 3 | 2 | 3 | 3 | 4 | 3 | 4 |
| AhpE | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| AhpD | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 1 |
| DR_A0145 (EfeB) | 1 | ||||||||||
| CCP | 1 | 1 | |||||||||
| OsmC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Ohr | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | |||
| YhfA | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| Thioredoxins, Trx reductase and glutaredoxin-like proteins | |||||||||||
| TrxA | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 |
| TrxC | 1 | 1 | 1 | ||||||||
| TrxR | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| Grx/NrdH-like | 4 | 4 | 2 | 2 | 5 | 4 | 2 | 4 | 3 | 3 | 2 |
| Carotenoid | |||||||||||
| CrtLm | 1 | 1 | 1 | 1 | 1 | 1 | fr | 1 | |||
| CrtY-CruF | 1 | 1 | 1 | ||||||||
| CruF | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| CYP287A1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Manganese transport | |||||||||||
| MntH | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | ||
| MntA | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 |
| MntB | 1 | 1 | 5 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 |
| MntC | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 |
| MntE | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Others | |||||||||||
| Dps2 | 1 | 1 | |||||||||
| FrnE | 1 | 1 | 1 | 1* | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DsbA-like (DR_2335) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| PqqE | 1 | ||||||||||
FrnE from D. maricopensis lacks an extended C-terminal tail.
See further the legend to Table 2.
Enzymatic systems for oxidative stress defence
Superoxide dismutases
Superoxide dismutases (SODs) are metalloenzymes that catalyse the disproportionation of O2·− to give H2O2 and O2 using a redox-active metal. SOD is classified according to metal cofactor into the manganese-containing SOD (MnSOD), the iron-containing SOD (FeSOD) and the copper/zinc-cofactored type (CuZnSOD) (Imlay 2013). The accepted nomenclature for bacterial SODs is SodA, SodB and SodC for the Mn-, Fe- and Cu/Zn-SODs, respectively (Broxton and Culotta 2016). Escherichia coli contains three SODs: two cytoplasmic SODs, SodA and SodB, and the periplasmic SodC (Imlay 2013). Deinococcus radiodurans has one cytoplasmic SodA (DR_1279) and two periplasmic SodCs (DR_1546 and DR_A0202). FeSOD (SodB) is not present in D. radiodurans or any of the other Deinococcus species analysed. SodA is present constitutively at high levels in D. radiodurans (Basu and Apte 2012) and is well conserved across all Deinococcus species analysed, suggesting that SodA plays an important role in superoxide detoxification in Deinococcus. However, a sodA mutant strain of D. radiodurans is only slightly sensitive to very high doses of IR (Table S1, Supporting Information). Escherichia coli SodA and D. radiodurans SodA share a positively charged region located at the dimer interface providing DNA-anchoring loops (Dennis et al.2006; Smolik et al.2014). It has been proposed that interaction of SodA with DNA would be helpful in protecting a genome against radiation or oxidative attack in bacterial cells (Smolik et al.2014). Because O2·− cannot cross membranes (Imlay 2013), the extracytoplasmic location of the two D. radiodurans SodCs (Farci et al.2014) implies that these enzymes may play a role in defending bacteria against oxidative stress from the surrounding environment. However, D. soli, D. peraridilitoris and D. geothermalis do not contain SodC. Besides its N-terminal SodC domain, the amino acid sequence of the DR_A0202-type SodC predicts a beta-propeller fold in the C-terminal domain (Fig. S4, Supporting Information). Moreover, DR_A0202 and the homologue in each of the other Deinococcus species are directly preceded, and probably forming an operon, by a gene encoding a predicted exported glucose/arabinose dehydrogenase, which also contains a beta-propeller fold. This conserved gene organisation suggests that their gene products may functionally interact.
Catalases
Catalase is a metalloenzyme that converts H2O2 to water and O2. Catalases are divided into three families, namely typical (monofunctional) heme catalases (KatEs), (bifunctional) heme catalase-peroxidases (KatGs) and (non-heme) manganese catalases (MnCats) (Zamocky et al.2012). Although D. radiodurans encodes two KatE-type catalases, DR_1998 (KatE1) and DR_A0259 (KatE2), and a eukaryotic-type catalase, DR_A0146, it was recently reported that DR_A0146 does not have catalase activity and that the highly expressed DR_1998 has a more critical role in detoxifying H2O2 than DR_A0259 (Jeong et al.2016a). Most, but not all, of the Deinococcus species encode catalase, but there is remarkable diversity in the number and type of catalases across these species (Table 3). KatEs are subdivided into clades 1, 2 and 3 (Zamocky et al.2012). As reported previously (Jeong et al.2016a), clade 1, which includes DR_1998, is the most common catalase in Deinococcus, but KatE catalases of clade 2 (e.g. DR_A0259) and clade 3 are also present in some Deinococcus species. The KatG-type catalase was not found in Deinococcus, but MnCat was found in D. proteolyticus, D. peraridilitoris, D. maricopensis and D. gobiensis.
Interestingly, neither heme catalase (KatE and KatG) nor non-heme catalase (MnCat) was found in D. puniceus even though the radiation resistance of this species was comparable to that of D. radiodurans (Lee et al.2015), indicating that catalase is not crucial for IR resistance in Deinococcus. Indeed, another study showed that there is no significant correlation between levels of catalase activity and IR resistance within seven Deinococcus species (Shashidhar et al.2010). Nevertheless, moderately increased sensitivity, albeit at high doses only, has been reported for D. radiodurans mutants in which either DR_1998 or DR_A0259 was disrupted (Table S1, Supporting Information). To our knowledge, a strain lacking both DR_1998 and DR_A0259 has not been characterised. In contrast to the moderate effect at most on resistance to acute high IR dose, disruption of DR_1998 strongly sensitises D. radiodurans to high H2O2 concentrations (> 20 mM) (Jeong et al.2016a). Interestingly, very recent data have indicated that DR_1998 is required for resistance of D. radiodurans to chronic IR (36 Gy/h) (Shuryak et al.2017).
Peroxiredoxins
Peroxiredoxins (Prxs) represent a family of thiol-dependent peroxidases catalysing the reduction of H2O2 or organic hydroperoxides (Meyer et al.2009), and include bacterioferritin comigratory protein (BCP), thiol peroxidase (Tpx) and alkyl hydroperoxide reductase (AhpC) (Mishra and Imlay 2012). RPS-BLAST was used to search the Deinococcus genomes for members of the PRX family (cd02971). Compared to E. coli encoding BCP, Tpx and AhpC (Meyer et al.2009), Tpx and AhpC are absent in the 11 Deinococcus species. Besides BCP, however, the Deinococcus species analysed encode AhpE (DR_2242 in D. radiodurans), which is an atypical type of AhpC (Fig. S5, Supporting Information). AhpC is classified as 2-Cys Prx with conserved N-terminal peroxidatic CysP and C-terminal resolving CysR, but AhpE, which has been characterised in M. tuberculosis, contains no CysR (Perkins et al.2015). Ahp is a two-component (AhpC/AhpF) peroxidase, and AhpF restores the disulphide in AhpC to its reduced form (Mishra and Imlay 2012). However, there is no AhpF homologue in Deinococcus species. In M. tuberculosis encoding AhpE, AhpD is known to substitute for AhpF (Lu and Holmgren 2014). Deinococcus radiodurans possesses the AhpD homologue (DR_1765), which is alkyl hydroperoxidase D-like protein (COG2128, YciW), and this protein is observed in all other Deinococcus species. Surprisingly, D. peraridilitoris encodes five AhpD homologues. A comparison of AhpDs from M. tuberculosis and Pseudomonas aeruginosa, whose crystal structures have been determined (Koshkin et al.2003; Clarke et al.2011), shows that the important residues involved in the proton relay system, Glu118, Cys130, Cys133 and His137 (numbering based on AhpD from M. tuberculosis), are invariant for deinococcal AhpDs (except for the less conserved second AhpD-like protein of D. deserti that lacks the equivalent Glu and His) (Fig. S6, Supporting Information). These data suggest that AhpD may play a role as a reducing partner of AhpE in Deinococcus as shown in M. tuberculosis (Lu and Holmgren 2014).
BCP is a 1-Cys Prx, and its peroxidatic CysP is contained within a universally conserved PxxxT(S)xxC motif (Lu and Holmgren 2014). Deinococcal BCPs can be divided into two groups (Fig. S5, Supporting Information), with the proteins in group 2 showing more sequence variation of the PxxxT(S)xxC motif including a less conserved T(S). Group 1 including D. radiodurans DR_0846 is closer to the typical BCP from E. coli or B. subtilis than group 2, which includes DR_1208 and DR_1209. All Deinococcus species analysed encode one or two BCPs from each group (Table 3 and Fig. S5, Supporting Information).
Other peroxidases
In terms of antioxidant defences, a few peroxidases, such as catalase and Prx, play important roles because they have a primary purpose of reducing peroxides. In contrast, for a second group of peroxidases the primary purpose is to use the peroxide as an oxidising agent to oxidise a second molecule (Karplus 2015). Cytochrome c peroxidases (CCP) are heme enzymes that catalyse the two-electron reduction of H2O2 to water by accepting electrons from a soluble cytochrome c. CCP displays peroxidase activity in vitro but its physiological function seems to enable H2O2 to serve as an alternative respiratory electron acceptor in the absence of oxygen (Mishra and Imlay 2012). Only two Deinococcus species, D. radiodurans and D. proteolyticus, possess CCP (DR_A0301 in D. radiodurans). The D. radiodurans DR_A0145 protein, which was predicted to be an iron-dependent peroxidase (Slade and Radman 2011) and which is encoded by a gene located next to the catalase-like gene DR_A0146, is assigned to the COG2837 category for periplasmic deferrochelatase/peroxidase EfeB. In E. coli, EfeB showed a deferrochelation activity, releasing iron from heme leaving the tetrapyrrol intact (Letoffe et al.2009). Homologues of DR_A0145 were not found in other Deinococcus species.
The OsmC (osmotically inducible protein C) family is divided into three subgroups: Ohr (organic hydroperoxide resistance protein), OsmC and YhfA (Shin et al.2004). Among them, Ohr and OsmC have been identified as a new family of 2-Cys peroxidases (Zhang and Baseman 2014) because two additional residues (an Arg and a Glu) required for the peroxidatic activity are absent in YhfA (Shin et al.2004). Deinococcus radiodurans encodes OsmC family proteins: DR_1857, DR_1538 and DR_1177 are homologues of Ohr, OsmC and YhfA, respectively. A sequence alignment of the deinococcal proteins belonging to the OsmC family revealed that the catalytic Arg and Glu residues are conserved in Ohr and OsmC but not in YhfA proteins (Fig. S7, Supporting Information). Although the conserved Arg residue is present at different positions in the sequences of Ohr and OsmC proteins, in the tertiary structures they occupy a similar orientation between the conserved Glu and CysP (Meireles et al.2017). Ohr protein exhibits peroxidatic activity that is much more substantial against organic peroxides than against H2O2 itself (Mishra and Imlay 2012). In constrast, H2O2 appears to be a good substrate for OsmC (Zhang and Baseman 2014). The OsmC protein is conserved in Deinococcus species except for D. proteolyticus, while the Ohr homologue is absent in D. soli, D. deserti and D. actinosclerus. Two members of the Ohr subfamily are present in D. proteolyticus, D. maricopensis and D. gobiensis. The conservation of OsmC and the activity of OsmC towards H2O2 suggest that it plays a more important role in the resistance of Deinococcus to oxidative stress compared to Ohr as well as to CCP and DR_A0145.
Thioredoxins
The thioredoxin system, comprising NADPH, thioredoxin reductase (TrxR) and thioredoxin (Trx), is a major disulphide reductase system, which can provide electrons to a large range of enzymes, and is found to be critical for DNA synthesis and defence against oxidative stress in diverse organisms (Lu and Holmgren 2014). CDD search with RPS-BLAST showed that D. radiodurans contains two Trxs (cd02947) with classic active site motif CGPC, DR_0944 and DR_A0164, which are similar to thioredoxin 1 (trxA) and 2 (trxC) of E. coli, respectively. TrxC contains two additional CXXC motifs that are involved in zinc ion binding (Collet et al.2003), and these motifs are conserved in DR_A0164. The DR_0944-type Trx is present in each of the other Deinococcus species (three homologues in D. peraridilitoris), but the DR_A0164-type in only two other species (Table 3). The antioxidant activity of the Trx system is manifested when electrons are transferred to antioxidative proteins such as BCP. In addition, AhpE in M. tuberculosis can be reduced by the Trx system (Lu and Holmgren 2014). This suggests that deinococcal AhpE can obtain electrons from AhpD and/or Trx. The methionine sulfoxide reductase enzymes MsrA and MsrB catalyse the reduction of protein-bound methionine sulfoxide to methionine in the presence of Trx and have been shown to have an important role in protecting organisms against oxidative damage (Cabreiro et al.2006). Hsp33 (heat shock protein 33) is a chaperone specialised for oxidative stress protection (Dahl, Gray and Jakob 2015). Hsp33 is rapidly activated through formation of a homodimer in response to severe oxidative stress to prevent aggregation of unfolding proteins, and oxidised Hsp33 dimers are converted into reduced Hsp33 dimers by the Trx system (Meyer et al.2009; Mayer 2012). MsrA, MsrB and Hsp33 are well conserved across all Deinococcus species analysed. Thioredoxin reductase TrxR is known to reduce oxidised Trx in the presence of NADPH. DR_1982 of D. radiodurans is a homologue of E. coli TrxR (Fig. S8, Supporting Information), and is able to revert DR_A0164 (TrxC) to its reduced form (Obiero et al.2010). Each Deinococcus species contains one or two DR_1982 homologues (Table 3).
Glutaredoxin-like proteins
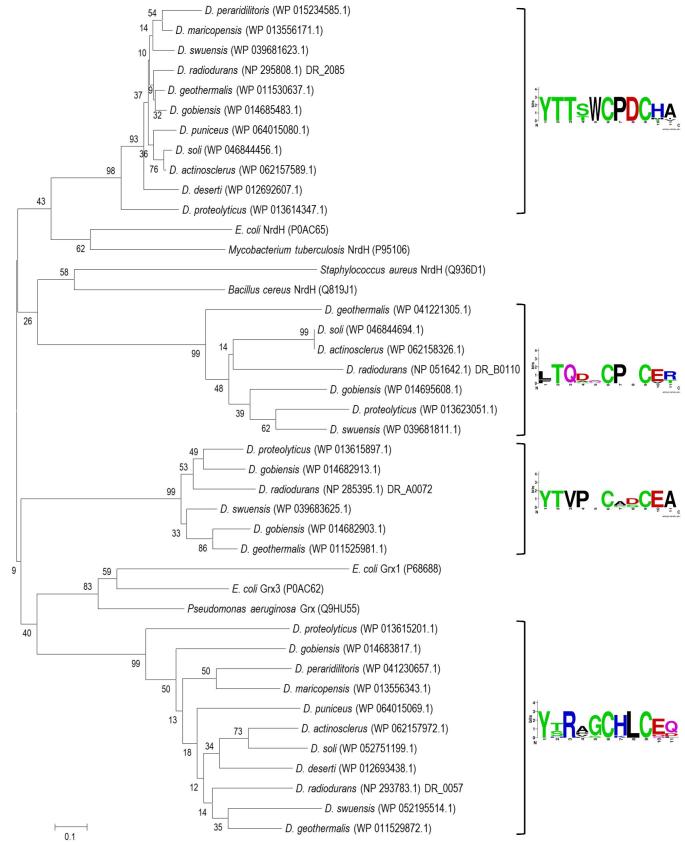
The glutathione (GSH) system, which, besides GSH, comprises GSH peroxidase, GSH reductase and glutaredoxin (Grx), is one of the major antioxidant systems, but is lacking in some bacteria such as Helicobacter pylori, M. tuberculosis and B. subtilis (Lu and Holmgren 2014). It has been previously reported that D. radiodurans has three small (80 to 100 residues) CXXC motif-containing Grx-like proteins (i.e. DR_2085, DR_A0072 and DR_0057) (de Groot et al.2009; Yuan et al.2012), while GSH, GSH reductase and GSH peroxidase are absent (Slade and Radman 2011). Of these three Grx-like proteins, the DR_2085- and DR_0057-type proteins are present in all other Deinococcus species, but the DR_A0072-type in only 5 of the 11 species (Fig. 5 and Table S3, Supporting Information). A multiple-sequence alignment of the deinococcal Grx-like proteins shows that ‘CPDC’ and ‘CHLC’ motifs are well conserved in the DR_2085-type and DR_0057-type proteins, respectively (Fig. 5), whereas canonical Grx contains a CPYC redox-active motif. Interestingly, the C-terminal regions of the DR_2085-type proteins contain the SGFRP structural motif that is present in canonical NrdH proteins (Rabinovitch et al.2010) (Fig. S9, Supporting Information). NrdH is a Grx-like protein disulphide oxidoreductase of the Trx superfamily, typically containing the redox-active C(M/V)QC motif and functioning primarily as the electron donor for the NrdEF ribonucleotide reductase, and is reduced by the Trx system but not by GSH (Rabinovitch et al.2010).
Figure 5.
Phylogenetic relationship of glutaredoxin-like proteins identified in 11 Deinococcus species. The phylogenetic analysis was carried out based on protein sequence alignment of 35 deinococcal Grx/NrdH-like proteins (Table S3, Supporting Information) with some representative proteins taken from Uniprot: NrdH from E. coli (Uniprot Number P0AC65), M. tuberculosis (P95106), S. aureus (Q936D1) and B. cereus (Q819J1), and Grx from E. coli (P68688 and P0AC262) and P. aeruginosa (Q9HU55). Weblogo plots show sequences around the CxxC motif extracted from deinococcal proteins of each group. See further the legend to Figure 4.
Ribonucleotide reductases (RNRs) are essential enzymes catalysing conversion of the four ribonucleotide triphosphates (NTPs) into their corresponding dNTPs necessary for DNA replication and repair (Torrents 2014). Until now, three different RNR classes have been described (I, II and III), and class I is further subdivided into Ia (NrdAB) and Ib (NrdEF) (Torrents 2014). Class Ib genes are organised as an nrdHIEF operon in E. coli and Mycobacterium, but in Bacillus and Staphylococcus the class Ib-specific nrdH gene is located elsewhere on the chromosome (Rabinovitch et al.2010). Deinococcus radiodurans (genes DR_B0107-DR_B0109) and six other Deinococcus species also have an nrdIEF gene cluster. In each of these Deinococcus species, nrdIEF is followed, probably in the same operon, by a gene encoding a small protein (86 to 100 residues) containing a characteristic CPXC redox motif (Fig. 5). These small proteins (DR_B0110 in D. radiodurans), which have been annotated as Trx, Trx-related or hypothetical proteins, presumably function as electron donor for the NrdEF RNR encoded by the same gene cluster. Class II RNRs encoded by a single nrdJ gene are represented not only in the four species lacking NrdIEF (i.e. D. deserti, D. maricopensis, D. peraridilitoris and D. puniceus), but also in five Deinococcus species (D. actinosclerus, D. geothermalis, D. radiodurans, D. soli and D. swuensis) having NrdIEF (Table S7, Supporting Information). Class II RNRs are also reduced by Trx (Jordan et al.1997; Torrents 2014). Although NrdH can act as a hydrogen donor for the class Ib NrdEF RNR (Jordan et al.1997), it has been supposed to be one of the candidates in the antioxidant system of bacteria lacking GSH. In Corynebacterium glutamicum, overexpression of NrdH increased the resistance to oxidative stress by reducing ROS accumulation (Si et al.2014). Together, Deinococcus species encode two to five Grx/NrdH-like proteins (Table 3). Further research is needed to investigate the physiological function of these proteins in Deinococcus.
Other proteins involved in ROS protection
In Gram-negative bacteria, disulphide bond formation occurs in the periplasm and is catalysed by Dsb proteins. In Gram-positive bacteria, however, disulphide bond formation is not fully understood due to lack of periplasmic space (Reardon-Robinson and Ton-That 2015). Deinococcus radiodurans DR_0659 encodes a DsbA-like protein (COG2761) designated FrnE, which belongs to the Trx superfamily of proteins, and the frnE mutant strain shows reduced tolerance to IR and H2O2 (Table S1, Supporting Information). It has been recently reported that DR_0659 represents a novel cytoplasmic thiol-disulphide oxidoreductase system that could be functional in eubacteria under conditions where Trx/Grx systems are inhibited or absent (Bihani et al.2018). This protein contains a canonical 22-CPWC-25 active site motif embedded in the Trx fold and an additional, functionally important 239-CxxxxC-244 motif in a unique extended C-terminal tail. A protein homologous to DR_0659 is observed in all of the Deinococcus species analysed. However, the FrnE proteins from D. proteolyticus and D. maricopensis have a CPFC motif instead of CPWC and, moreover, the CxxxxC motif is absent in FrnE from D. maricopensis, which is shorter (about 20 residues) than the other FrnE proteins (Fig. S10, Supporting Information). Some Deinococcus species encode an additional, predicted cytoplasmic DsbA-like protein (DR_2335 in D. radiodurans) (Table 3), but the characteristic motifs (both CPWC/CPFC and CxxxxC) are not present in DR_2335 and its homologues. Instead, they contain a single conserved Cys residue.
In D. radiodurans, deletion of DR_B0067, which encodes an extracellular nuclease (COG2374), might slightly decrease the survival ability after H2O2 or IR treatment (Li et al.2013). The degradation of extracellular DNA into dNMPs (especially, dGMP) by extracellular nuclease DR_B0067 might enhance D. radiodurans tolerance to oxidative stress (Li et al.2013). Homologues of DR_B0067 are found in all other Deinococcus species. However, their C-terminal regions have different combinations and arrangements of a few domains (Fig. S11, Supporting Information).
Iron is essential for the life processes of all living organisms, but the element is toxic when in excess of that needed for cellular homeostasis. The primary role of ferritin family proteins is to sequester iron to protect cells from the damage caused by Fenton reaction, where free ferrous ions react with H2O2 to produce ·OH (Smith 2004). Three subfamilies of proteins representing the ferritin fold are observed in bacteria: ferritin, bacterioferritin (Bfr) and the ferritin-like Dps (DNA-binding proteins during stationary phase). Among them, Dps plays an important role in the detoxification of ROS, in iron scavenging and in the mechanical protection of DNA (Zeth 2012). Whereas E. coli produces the three ferritin family proteins (Smith 2004), the Deinococcus species analysed contain only Dps. Deinococcus radiodurans encodes two Dps proteins, Dps1 (DR_2263) and Dps2 (DR_B0092). Dps1 is well conserved across all of the Deinococcus species, but Dps2 is present only in D. radiodurans and D. gobiensis. Dps1 has a longer N-terminal extension (54 amino acids) before the ferritin fold compared to other Dps, which is essential for stabilising the protein–DNA complex (Santos et al.2017). The N-terminal extension region is observed in all of the Dps1 homologues (Fig. S12, Supporting Information). Although there is some variation of sequences and amino acid composition, the N-terminal extension is rich in positively charged residues, in particular lysine and has been proposed to be involved in the association with DNA (Santos et al.2017). Similar N-terminal lysine-rich extensions are present in nucleoid-associated HU proteins of Deinococcus (Bouthier de la Tour et al.2015). Remarkably, the Dps2 proteins of D. radiodurans and D. gobiensis possess a predicted N-terminal signal peptide, indicating translocation of Dps2 across the cytoplasmic membrane. Experimental evidence for the extracytoplasmic localisation of D. radiodurans Dps2 has indeed been obtained, suggesting that Dps2 may have an iron-sequestering role outside the cytoplasm and, like SodC, may protect against exogenously derived ROS (Reon et al.2012). Reduced resistance to H2O2 has been reported for a D. radiodurans dps2 mutant, whereas resistance to IR appeared unaffected for dps1 and dps2 single and double mutants (Table S1, Supporting Information). Interestingly, both in D. radiodurans and D. gobiensis the dps2 gene is directly adjacent to genes encoding the two-component signal transduction system (TCS) RadS/RadR (Fig. S13, Supporting Information), which, like Dps2, is only found in these two species (see section ‘Radiation and oxidative stress resistance-associated regulatory proteins’).
While non-cytoplasmic SodC and, in particular, Dps2 are not strictly conserved across the Deinococcus species, MsrP and MsrQ homologues of the periplasmic MsrPQ system for repair of methionine sulfoxide damage in bacterial cell envelope proteins (Ezraty et al.2017) are present in each Deinococcus (Table S3, Supporting Information).
Non-enzymatic systems for oxidative stress defence
Carotenoid
Carotenoids are widespread natural pigments and act as ROS scavengers in non-phototrophic bacteria for cellular protection. One important group of non-phototrophic bacteria that produce carotenoids is the phylum Deinococcus–Thermus (Tian and Hua 2010). The major carotenoid in D. radiodurans is deinoxanthin, a unique ketocarotenoid, which gives the bacterium its characteristic red color. Deinoxanthin shows higher scavenging activity on H2O2 than carotenes (lycopene and β-carotene) and xanthophylls (zeaxanthin and lutein) and has a protective effect in vitro on DNA and protein (Tian et al.2007, 2009). In vivo, decreased resistance to radiation or H2O2 has been observed for D. radiodurans crt gene mutants lacking deinoxanthin (Table S1, Supporting Information). The biosynthetic pathway for deinoxanthin includes the reactions catalysed by geranylgeranyl diphosphate synthase (CrtE, DR_1395), phytoene synthase (CrtB, DR_0862), phytoene desaturase (CrtI, DR_0861), lycopene cyclase (CrtLm, DR_0801), carotenoid 3΄,4΄-desaturase (CrtD, DR_2250), carotenoid 1,2-hydratase (CruF, DR_0091), carotenoid ketolase (CrtO, DR_0093) and carotenoid 2-β-hydroxylase (cytochrome P450 CYP287A1, DR_2473) (Zhou et al.2015). Most of these proteins are well conserved in the Deinococcus species analysed except for CrtLm and CYP287A1 (Table 3). Following synthesis of lycopene, carotenoid biosynthesis is diversified into acyclic or cyclic carotenoids. The cyclisation of lycopene on one or both Ψ-ends of lycopene is usually catalysed by CrtL- or CrtY-type lycopene β-cyclase (Tian and Hua 2010). An asymmetrically acting lycopene β-cyclase (CrtLm), which catalyses the production of monocyclic carotenoids, is encoded by eight of the analysed Deinococcus species, although the gene in D. actinosclerus contains a frameshift. For the three species that lack CrtLm (i.e. D. proteolyticus, D. peraridilitoris and D. deserti), genes encoding CrtY-CruF fusion proteins were detected (Fig. 3). A single gene encoding a bifunctional enzyme with lycopene cyclase (CrtY) and phytoene synthase (CrtB) activities in fungi has been reported previously (Velayos, Eslava and Iturriaga 2000; Guo, Tang and Zhang 2014). BLASTP analysis revealed that the CrtY-CruF fusion protein is unique to the class Deinococci. Deinoxanthin is a unique C2-hydroxylated monocyclic ketocarotenoid, and the 2-β-hydroxylase CYP287A1 catalyses β-ring hydroxylation at the C2 position of 2-deoxydeinoxanthin in D. radiodurans (Zhou et al.2015). The 2-β-hydroxylase, which completes the biosynthetic pathway of deinoxanthin, was not found in D. deserti and D. geothermalis. Taken together, these results suggest that D. proteolyticus, D. peraridilitoris, D. deserti and D. geothermalis are likely to produce different kinds of carotenoid distinct from deinoxanthin. This may explain the observed colony colours of these four Deinococcus species, which are orange-red, light pink, whitish/light pink and orange, respectively (Brooks and Murray 1981; Ferreira et al.1997; de Groot et al.2005; Rainey et al.2007).
Bacillithiol
Bacillithiol (BSH), the α-anomeric glycoside of l-cysteinyl-d-glucosamine with l-malic acid, is a low-molecular-weight thiol analogous to GSH and is found in several Firmicutes (e.g. Bacillus, Staphylococcus) and in D. radiodurans, which lack GSH (Newton et al.2009; Perera, Newton and Pogliano 2015). Some other bacteria produce the low-molecular-weight thiol mycothiol (MSH) instead of GSH or BSH (Rosario-Cruz and Boyd 2016). The biosynthesis and roles of BSH have been studied in Bacillus and Staphylococcus species. Similar to GSH, BSH protects against H2O2, hypochlorite and thiol/disulphide stress. BSH also plays a role in metal ion buffering and thereby protects cells from metal ion intoxication (Rosario-Cruz and Boyd 2016; Chandrangsu et al.2018). BSH synthesis initiates with a glycosyltransferase (BshA) that couples N-acetylglucosamine (GlcNAc) and l-malate. The BshB deacetylase hydrolyses the acetyl group from GlcNAc-Mal to generate GlcN-Mal. Subsequent addition of cysteine, catalysed by BshC, generates the final product, BSH (Gaballa et al.2010). Homologues of these three enzymes are encoded within the genomes of each of the analysed Deinococcus species (Table S3, Supporting Information). In D. radiodurans, the levels of bsh gene expression and BSH were slightly reduced during irradiation and increased again in the recovery period (Luan et al.2014).
In analogous systems, in which enzymes function with GSH as cofactor, Grx is reduced by the oxidation of GSH, and the oxidised GSH is then regenerated by GSH reductase. Phylogenomic profiling identified a putative BSH reductase, YpdA, which is related to Trx reductase (24% identity with B. subtilis TrxB), and three putative bacilliredoxin (Brx) proteins (i.e. YqiW, YphP and YtxJ) in B. subtilis (Gaballa et al.2010). Bacilliredoxin activity has been demonstrated for YphP (renamed BrxA) and YqiW (BrxB) (Gaballa et al.2014). Homologues of YpdA and YtxJ (but not BrxA and BrxB) were found in the analysed Deinococcus species. However, a putative monothiol active site (TCPIS) observed in Bacillus YtxJ is replaced with TCHKT in all Deinococcus YtxJ homologues, and the deinococcal YtxJs (more than 200 residues) are longer than B. subtilis YtxJ (108 residues). Under oxidising conditions, BSH has been shown to form mixed disulphides with the Cys residues of several proteins, termed S-bacillithiolation, which is a widespread thiol protection and redox-regulatory mechanism in Firmicutes (Loi, Rossius and Antelmann 2015). The organic hydroperoxide regulator OhrR is a redox-sensing transcriptional repressor that is bacillithiolated by BSH in Bacillus, but the 11 Deinococcus species lack this protein. BSH transferases (Bst) are enzymes that catalyse the transfer of BSH to target substrates. In B. subtilis, the YfiT (BstA) protein, which is a member of the DinB superfamily, functions as Bst to conjugate reactive electrophiles with BSH, such as monobromobimane (Newton et al.2011). One of the largest protein expansions observed in D. radiodurans is the DinB/YfiT protein family (Makarova et al.2001). Among them, DR_0841 and DR_1024 were found to be close sequence homologues (>40% identity) of B. subtilis BstA and belong to the DinB_2 family (PF12867) (Newton et al.2011). One or two homologues of B. subtilis BstA are detected in each of the other Deinococcus species except for D. proteolyticus.
BSH is proposed to have a role in Fe–S cluster biogenesis (Rosario-Cruz and Boyd 2016). Three Fe-S biogenesis systems have been identified and characterised in bacteria, namely Isc (iron-sulphur cluster), Suf (sulphur formation) and Nif (nitrogen fixation) systems (Roche et al.2013). The role of BSH in Fe–S biogenesis has functional overlap with Fe–S cluster carriers in Gram-positive bacteria, such as Staphylococcus aureus and B. subtilis, in which Suf is the sole Fe–S cluster biosynthetic machinery system (Rosario-Cruz and Boyd 2016). Deinococcus radiodurans carries the Suf system constituted by the SufSE complex, which serves as the sulphur donor, the Fe-S scaffold SufBCD and the SufA Fe-S carrier protein. All of the Deinococcus species have these Suf proteins, and five of these species contain additionally the alternate Fe-S scaffold SufU, which is an IscU-like protein (Table S3, Supporting Information).
Manganese and peptides
In biology, Fe and Mn play important roles in cellular adaptation to oxidative stress. Whereas Fe2+ is known as a pro-oxidant because of its reactivity with peroxide, generating the highly reactive OH· through Fenton reaction, Mn2+ is considered an antioxidant (Aguirre and Culotta 2012). In E. coli, oxidative stress induces import of Mn2+ that can replace Fe2+ from sensitive targets in metalloenzymes (Faulkner and Helmann 2011). Manganese ions can also form antioxidant complexes with small metabolites such as orthophosphate, carboxylates, amino acids and small peptides. These manganese antioxidants can scavenge or catalytically remove ROS (Culotta and Daly 2013). Deinococcus radiodurans and other radiation-resistant bacteria have a very high intracellular Mn content leading to a high Mn/Fe ratio (0.24 in D. radiodurans) compared to that of radiation-sensitive bacteria (<0.01 in E. coli) (Daly et al.2004). It was established that protein-free D. radiodurans cell extracts prevent protein oxidation at high doses of radiation, and that these extracts are enriched in Mn2+, inorganic phosphate, peptides and nucleosides (Daly et al.2010). When reconstituted in vitro in phosphate buffer, peptides or Mn-peptide complexes appeared to be the most protective (Daly et al.2010; Berlett and Levine 2014). The accumulated small peptides in cells may be derived from proteolysis, peptide import and/or peptide-encoding transcripts. Indeed, many novel leaderless transcripts predicted to code for small peptides were discovered in D. deserti, a species in which 60% of the mRNAs were found to be leaderless (de Groot et al.2014). Regarding manganese, D. radiodurans has three types of predicted Mn transport systems involved in maintenance of the intracellular Mn concentration homeostasis: Mn2+ efflux protein MntE (DR_1236), Nramp H-dependent Mn2+ transporter MntH (DR_1709) and the ATP-binding cassette (ABC)-type Mn2+ transporter (DR_2283, DR2284 and DR_2523 encoding transmembrane subunit/permease component MntB, ATPase component MntC and solute-binding component MntA, respectively) (Sun et al.2010; Ul Hussain Shah et al.2014,). The D. radiodurans mntE mutant exhibited higher intracellular Mn concentrations and lower protein oxidation level than wild type under oxidative stress. Consistent with these characteristics, the mntE mutant was reported to be more resistant to H2O2, UV and IR, supporting the involvement of Mn in the radiation resistance of D. radiodurans (Sun et al.2010). All the other Deinococcus species encode one MntE homologue except for D. maricopensis encoding an additional homologue. MntH is essential in D. radiodurans (Makarova et al.2007), and the heterozygous mntH mutant shows increased sensitivity to IR (Dulermo et al.2015). However, D. radiodurans MntH homologues sharing 42–77% identity are found in only seven of the other Deinococcus species analysed. Deinococcus peraridilitoris and D. puniceus lack the MntH homologue. The MntH homologue from D. deserti and two of the three MntH homologues from D. proteolyticus share only 26% identity with D. radiodurans MntH. ABC transporters act in conjunction with extracytoplasmic solute-binding proteins that feed the solute into the transmembrane channel, which is energised by the cytoplasmic ATP-binding cassette protein. Both the transmembrane protein and the ATPase are usually present as dimers, each consisting either of a single gene product, yielding a homodimer, or two gene products, yielding a heterodimer (Lorca et al.2007). Each Deinococcus species has a predicted ABC-type Mn2+ transporter encoded by an mntACB gene cluster or by separate mntA and mntCB genes. Deinococcus peraridilitoris and D. deserti additionally possess respectively one and two operons containing four ABC-type Mn2+ transporter genes encoding MntA, MntC and two different MntB homologues (Fig. S14, Supporting Information), which may improve their Mn2+ import capacity or compensate for the absence of MntH.
Bacteria often replace some proteins that have Fe-dependent functions with alternative proteins that do not require iron. This adaptive response to oxidative stress and Fe limitation is well understood in E. coli. In this organism, FeSod (SodB) is replaced by MnSod (SodA) during periods of Fe starvation. At the same time, E. coli also replaces the Fe-dependent RNR NrdAB with its Mn-dependent isozyme, NrdEF, which is crucial for survival under conditions of H2O2 stress (Chandrangsu, Rensing and Helmann 2017). Neither SodB nor NrdAB is found in the Deinococcus species analysed here. The constitutively high intracellular Mn/Fe ratio as an adaptation to harsh environmental conditions might drive the loss of such iron-dependent proteins during evolution. Notably, the radiation-sensitive bacterium Shewanella oneidensis (Mn/Fe ratio < 0.001) encodes 65% more proteins containing Fe-S clusters than D. radiodurans (Ghosal et al.2005).
Other antioxidants: pyrroloquinoline quinone and polyphosphate
Pyrroloquinoline quinone (PQQ) was initially characterised as a redox cofactor for membrane-bound dehydrogenases in bacteria. Subsequently, PQQ was shown to be an antioxidant protecting cells from oxidative damage (Misra, Rajpurohit and Khairnar 2012). PQQ neutralises ROS including O2·− and OH· in vitro without producing reactive molecular products and protects plasmid DNA and proteins in vitro from oxidative damage caused by IR (Misra et al.2004). Several prokaryotes are able to produce PQQ, and its biosynthesis involves the gene products of a specific pqq operon consisting of five or six genes. For example, expression of the pqqA-F operon from Klebsiella pneumoniae in the non-PQQ producer E. coli led to PQQ synthesis, and genetic studies revealed that four of the six pqq gene products (PqqA, PqqC, PqqD and PqqE) are absolutely required for this PQQ production in E. coli (Klinman and Bonnot 2014). Deinococcus radiodurans has a gene (DR_C0034) encoding a homologue of PqqE. It has been reported that DR_C0034 (pqqE) mediates PQQ production in D. radiodurans and also in transgenic E. coli, despite absence of other pqq gene homologues (Khairnar, Misra and Apte 2003; Rajpurohit, Gopalakrishnan and Misra 2008). Moreover, E. coli expressing DR_C0034 showed an increased tolerance to oxidative stress, possibly by scavenging of ROS by PQQ and/or by stimulation, through an unknown mechanism, of catalase and SOD activities (Khairnar, Misra and Apte 2003). Disruption of pqqE in D. radiodurans decreases the resistance not only to oxidative stress (H2O2) but also to the DNA-damaging agents MMC and IR (Rajpurohit, Gopalakrishnan and Misra 2008). Recently, the involvement of PQQ in signal transduction mechanisms involved in radiation resistance and DNA double-strand break repair has also been reported (see the section ‘Radiation and oxidative stress resistance-associated regulatory proteins’). However, a homologue of PqqE is not found in the other Deinococcus species analysed.
Inorganic polyphosphate (Poly P) found as unbranched chains up to 1000 residues long in cells is a universally conserved biopolymer. Poly P functions as a protein-protective chaperone that is able to protect a broad spectrum of proteins from aggregation, so its accumulation significantly increases bacterial oxidative stress resistance (Dahl, Gray and Jakob 2015; Gray and Jakob 2015). Deinococcus radiodurans exponential-phase cells contain electron-dense granules for the likely storage of Poly Ps (Slade and Radman 2011). Bacterial polyphosphate kinases (PPK) reversibly catalyse the generation of Poly P directly from ATP, whereas exopolyphosphatases (PPX) can degrade Poly P into Pi molecules (Dahl, Gray and Jakob 2015). PPKs are subdivided into two families: PPK1 is responsible for Poly P synthesis and PPK2 preferentially catalyses the reverse reaction (utilisation of Poly P) (Rao, Gomez-Garcia and Kornberg 2009). Deinococcus radiodurans has PPK1 and 2 homologues (Zhang, Ishige and Kornberg 2002), and its PPK2 belongs to the class III subfamily that is characterised by being able to catalyse both ADP and AMP phosphorylation, enabling synthesis of ATP from AMP and Poly P by a single enzyme (Motomura et al.2014). PPK1, PPK2 and PPX are well conserved across the 11 Deinococcus species (Table S3, Supporting Information), although the PPK1 gene of D. maricopensis, downstream of gene Deima_3207, has a frameshift. Hydrolysis of Poly P by PPX could generate orthophosphate that may be used for the generation of Mn2+-phosphate complexes (Slade and Radman 2011).
THE DDR AND PPR PROTEINS
Transcriptomics experiments with D. radiodurans revealed the induced expression of various novel genes of unknown function in cells recovering from IR and desiccation (Tanaka et al.2004). These DNA damage response genes have been named ddrA to ddrP. An additional radiation- and desiccation-induced gene, pprA (pleiotropic protein promoting DNA repair), has also been identified in another study (Narumi et al.2004). The name ppr has been given to two other D. radiodurans genes that are required for radiation resistance, but which are not radiation induced. One has been designated pprI (inducer of pleiotropic proteins promoting DNA repair) (Hua et al.2003), and this gene is identical to irrE, identified independently in another study (Earl et al.2002a). The other has been named pprM (a modulator of the PprI-dependent DNA damage response), although it encodes a homologue of cold shock protein usually designated Csp in bacteria (Ohba et al.2009). The five genes most highly induced in response to each stress were ddrA, ddrB, ddrC, ddrD and pprA (Tanaka et al.2004). More recently, additional radiation-induced genes of unknown function were identified in D. deserti and designated ddrQ to ddrX, with ddrT to ddrX organised in an operon (Blanchard et al.2017). It has also been reported that the genuine DdrC proteins of D. radiodurans and D. geothermalis, and the real DdrH protein of D. radiodurans, are encoded by the DNA strand opposite to the initially annotated gene, which is of course crucial for characterisation of these proteins (de Groot et al.2009). Together, 23 ddr and ppr loci have been described (Table 4).
Table 4.
The ddr and ppr genes in Deinococcus species.
| Gene | Drad | Dgeo | Ddes | Dmar | Dgob | Dpro | Dper | Dswu | Dsol | Dact | Dpun |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ddrA | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | fr | 1 | |
| ddrB | 1 | 1 | 1 | 1 | 3 | 2 | 2 | 1 | 1 | 1 | 1 |
| ddrC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ddrD | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| ddrE | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| ddrF | 1 | ||||||||||
| ddrG | 1 | ||||||||||
| ddrH | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ddrI | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ddrJ | 1 | 1 | 1 | 1 | |||||||
| ddrK | 1 | ||||||||||
| ddrL | 1 | 1 | 1 | ||||||||
| ddrM | 1 | 1 | 1 | 1 | 1 | ||||||
| ddrN | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ddrO | 1 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 1 |
| ddrP (ligB) | 1 | 1 | 1 | 1 | 1 | ||||||
| ddrQ | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| ddrR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | |
| ddrS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| ddrTUVWX | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| pprA | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| pprI (irrE) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| pprM (csp) | 1 | 2 | 3 | 2 | 2 | 1 | 1 | 1 | 3 | 3 | 2 |
See the legend to Table 2.
The (highly) radiation-induced expression of pprA and ddr genes in D. radiodurans (Tanaka et al.2004), and also in D. deserti (de Groot et al.2014), suggests a role of these genes in radiation resistance. This has been confirmed for the genes that have been studied in more detail (i.e. ddrA to ddrD, ddrI, ddrO, ddrP, ddrR, pprA). DdrI, DdrO and PprI (IrrE) are involved in transcriptional regulation and are described in the section ‘Radiation and oxidative stress resistance-associated regulatory proteins’. DdrP (DR_B0100) corresponds to the putative DNA ligase LigB and is described in section ‘DNA repair in Deinococcus’. Strongly or moderately decreased resistance of D. radiodurans to radiation, MMC and/or oxidative stress has been observed in one or more studies after deletion of ddrA (DR_0423), ddrB (DR_0070), ddrR (DR_0053), pprA (DR_A0346) or pprM (DR_0907) (Table S1, Supporting Information). Mutant strains lacking either ddrC (reversed DR_0003) or ddrD (DR_0326) were found as resistant as the wild type to IR, UV and MMC in earlier studies (Tanaka et al.2004; Selvam et al.2013), but more recent work reported slight sensitivity of a ddrC mutant to high doses of UV (Bouthier de la Tour et al.2017). Larger effects on resistance have been observed when the ddrC or ddrD deletion is combined with another gene deletion (Tanaka et al.2004; Selvam et al.2013; Bouthier de la Tour et al.2017). However, the results are rather complex and depend on the genetic background and on the applied stress. For example, whereas ΔddrC ΔddrB and ΔddrD ΔddrB double mutant strains are more sensitive to MMC than the ΔddrB strain, the ΔddrC ΔpprA and ΔddrD ΔpprA strains appear less sensitive to gamma rays than the ΔpprA strain (Tanaka et al.2004; Selvam et al.2013). Compared to ΔddrC and ΔddrD single deletion mutants, the ΔddrC ΔddrD double mutant is slightly more sensitive to IR and MMC, but not to UV (Tanaka et al.2004; Selvam et al.2013). Remarkably, unlike the ΔddrC and ΔddrD single deletion mutants, the ΔddrC ΔddrD double mutant is very slow growing, indicating that the presence of either ddrC or ddrD is required for normal growth. However, the functions or DdrC and DdrD do not seem redundant because deletion of ddrD, but not of ddrC, dramatically increases UV sensitivity of a ΔpprA strain (Selvam et al.2013).
Massive DNA damage is generated after exposure to high doses of radiation, and extensive DNA degradation and formation of potentially lethal DNA repair intermediates must be avoided. DdrA, DdrB, DdrC and PprA proteins are able to bind DNA in vitro, and may contribute to repair of heavily damaged DNA and/or to preserving genome integrity by preventing DNA degradation. DdrA is a Rad52 family protein, but unlike eukaryotic Rad52, DdrA does not display DNA-strand annealing activity (Harris et al.2004). DdrA forms ringlike oligomers (Gutsche et al.2008), and it binds to 3΄ single-stranded DNA ends and protects those ends from nuclease degradation (Harris et al.2004). Deletion of ddrA in a D. radiodurans strain expressing a low constitutive level of RecA dramatically decreases resistance to IR, supporting the proposed role of DdrA in preserving DNA ends for recombinational repair (Jolivet et al.2006). DdrB forms pentameric rings that bind single-stranded DNA but not double-stranded DNA (Norais et al.2009; Sugiman-Marangos and Junop 2010). DdrB promotes high-fidelity DNA annealing and is involved in an early step of DNA double-strand break repair (Xu et al.2010; Bouthier de la Tour et al.2011; Sugiman-Marangos, Weiss and Junop 2016). DdrC binds to DNA with a preference for single-stranded DNA, protects DNA from nucleases and exhibits single-strand annealing activity (Bouthier de la Tour et al.2017). It also promotes circularisation but not ligation of linear plasmid DNA. The DdrD protein has not been characterised in vitro yet, but its recruitment to the nucleoid and dynamics of localisation comparable to that of RecA after irradiation have been observed, suggesting a possible functional interaction between DdrD and RecA (Bouthier de la Tour et al.2013). Regulation of ddrR (DR_0053) has been investigated and the DR_0053 protein has been purified, but its specific function is still unknown (Appukuttan et al.2015). DdrR belongs to the DinB superfamily of predicted metalloenzymes, of which the protein encoded by the DNA damage-inducible dinB gene of B. subtilis is the founding member (the DinB superfamily proteins are not to be confused with DNA polymerase IV encoded by E. coli gene dinB, also called dinP). The DinB superfamily includes bacillithiol transferases that can detoxify toxic molecules such as reactive electrophiles (Chandrangsu et al.2018) (see also the section ‘Bacillithiol’). Besides homologues of DdrR, Deinococcus species contain several additional genes encoding DinB superfamily proteins. Further research is needed to determine if DdrR and other deinococcal DinB superfamily proteins are bacillithiol transferases, and if so, to identify their target compounds in vivo.
According to the literature, PprA is a protein with various functions and able to stimulate activity of several, diverse enzymes. In vitro, PprA preferentially binds to double-stranded DNA with strand breaks, inhibits exonuclease activity and stimulates DNA end-joining by T4 DNA ligase and E. coli DNA ligase, which has led to the suggestion that PprA plays a role in a possible NHEJ-like DNA repair mechanism in vivo (Narumi et al.2004). Moreover, PprA is required for the in vitro activity of the putative ATP-dependent DNA ligase LigB (DdrP) from D. radiodurans (Kota et al.2010) (see also section ‘DNA repair in Deinococcus’). However, some Deinococcus species possess PprA but not LigB, or possess LigB but not PprA (Table 4), strongly suggesting that stimulating LigB activity cannot be the major function of PprA. Moreover, there is no evidence that NHEJ occurs in Deinococcus. It has also been reported that PprA is able to stimulate activity of E. coli catalase in vivo and in vitro (Kota and Misra 2006). More recent results indicate that PprA plays a major role in chromosome segregation via its physical and functional interaction with DNA gyrase in D. radiodurans after IR exposure and DNA repair (Devigne et al.2013, 2016). In vitro, PprA stimulates the decatenation activity of DNA gyrase (Devigne et al.2016). PprA also interacts with D. radiodurans topoisomerase IB in a bacterial two-hybrid system, and enhances the topoisomerase IB-mediated relaxation activity of supercoiled DNA in vitro (Kota et al.2014).
Cold shock proteins (Csps) are small (less than 100 amino acid residues), single-strand nucleic acid-binding proteins widely conserved in bacteria. Csps are known for being induced during cold shock, and they are thought to prevent the formation of secondary structures in mRNA at low temperature. However, Csps are not only produced during cold stress. Experimental data have indicated that Csps are also required under standard growth conditions and for adaptation to stresses other than cold, during which Csps may influence transcription or translation of many genes (Keto-Timonen et al.2016). Many bacteria encode several Csps (e.g. nine Csps in E. coli) with at least some of these having overlapping functions. Deinococcus radiodurans has only one csp gene (pprM), whereas two or three csp genes are present in some other Deinococcus species (Table 4 and Fig. 6). Compared to Csp from other bacteria, Csps from Deinococcus contain a C-terminal extension of 10–20 residues of unknown function (Fig. S15, Supporting Information), which appears to be unique to the class Deinococci (LaGier 2017). Each deinococcal Csp ends with the residues RW, and a subgroup (type A) has a highly conserved C-terminal sequence RD(N)DRW. Under normal growth conditions, increased amount of PprA protein (Ohba et al.2009), decreased levels of catalase KatE1 mRNA and protein, and increased transcription of three proteins (i.e. PerR-like, DtxR-like and RecG) that negatively affect katE1 expression have been observed in a D. radiodurans pprM mutant (Jeong et al.2016b). PprM also confers higher oxidative stress tolerance to E. coli (Park et al.2017). Further studies are needed to elucidate how PprM and other deinococcocal Csps influence expression of various genes and proteins.
Figure 6.
Phylogenetic relationship between two types of cold-shock proteins (Csp) identified in 11 Deinococcus species. The phylogenetic analysis was carried out based on protein sequence alignment of 19 deinococcal Csp homologues (Table S4, Supporting Information) made with Clustal omega. Two Csps of D. soli are identical to Csps of D. actinosclerus. See further the legend to Figure 4.
Of the 23 ddr and ppr loci (Table 4 and Table S4, Supporting Information), only 8 are conserved in each of the 11 analysed Deinococcus genomes: ddrB, ddrC, ddrH, ddrI, ddrN, ddrO, pprI (irrE) and pprM (csp). The other 15 loci are absent in one, several or even most of the Deinococcus species. Remarkably, these non-conserved genes include ddrA, ddrD, ddrP (ligB), ddrR and pprA, for which an important or moderate contribution to radiation and oxidative stress resistance in D. radiodurans has been reported.
MISCELLANEOUS PROTEINS INVOLVED IN RESISTANCE TO RADIATION AND OTHER DNA-DAMAGING AGENTS
Widely conserved proteins
In addition to the genes described in the preceding sections, many other genes appear to be required for full resistance of D. radiodurans to radiation and other DNA-damaging agents (Table S1, Supporting Information). These include genes widespread in other bacterial genera and that are also present in each of the analysed Deinococcus species (Table S5, Supporting Information), for example homologues of DR_0199 (encoding nucleoid-associated protein, YbaB/EbfC family), DR_0382 (YgjD/TsaD), DR_0756 (YeaZ/TsaB), DR_1321 (signal peptidase I), DR_1525 (fructokinase RbsK), DR_1972 (ClpP, ATP-dependent Clp protease proteolytic subunit), DR_1973 (ClpX, ATP-dependent Clp protease ATP-binding subunit), DR_2417 (DncA/RNase J), DR_2462 (RNase Y). Homologues of DR_0342 (Rieske-like Fe-S protein) and DR_1471 (chromosome partition protein Smc) are also present in each Deinococcus, although these have a frameshift in D. gobiensis and D. actinosclerus, respectively (Table 5). The smc mutant of D. radiodurans is not affected in growth and IR resistance, but has an increased sensitivity to gyrase inhibitors (Bouthier de la Tour et al.2009).
Table 5.
Differences regarding miscellaneous proteins involved in resistance to radiation and other DNA-damaging agents in Deinococcus species.
| Protein | Drad | Dgeo | Ddes | Dmar | Dgob | Dpro | Dper | Dswu | Dsol | Dact | Dpun |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DR_0342 | 1 | 1 | 1 | 1 | fr | 1 | 1 | 1 | 1 | 1 | 1 |
| DR_1471 (fr) Smc | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | fr | 1 |
| DR_0679 | 1 | ||||||||||
| DR_0392 | 1 | 1 | 1 | 2 | 1 | 1 | |||||
| DR_1262 Rsr | 1 | 1 | |||||||||
| DR_1631 RelQ | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| DR_1790 | 1 | 1 | |||||||||
| DR_2310 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| DR_A0018 (fr) | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| DR_A0281-DR_A0282 (fr) | 2 | 1 | 1 | 1 | 1 | 1 | 2 | ||||
| DR_A0283 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | ||||
| DR_B0118 | 1 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | |
| DR_1172 / DR_0105 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| Deide_15148 | 1 | 1 | 1 | 1 | 1 | 1 |
Bold, gene inactivation leading to increased sensitivity of D. radiodurans to radiation, desiccation or oxidative stress in at least one study. fr, frameshift.
The yeaZ and ygjD mutants of D. radiodurans are very sensitive to MMC, but not or only marginally sensitive to radiation and H2O2, and therefore a role of YeaZ and YgjD in repair of DNA cross-links has been proposed (Onodera et al.2013). However, other studies have reported that YeaZ and YgjD are required for an essential and universal modification of ANN-decoding tRNAs (Deutsch et al. 2012), and therefore these proteins have been renamed TsaB (tRNA threonylcarbamoyladenosine biosynthesis protein TsaB) and TsaD (tRNA N6-adenosine threonylcarbamoyltransferase), respectively. Remarkably, yeaZ and ygjD are not essential for viability in D. radiodurans, unlike in other species (e.g. E. coli) (Onodera et al.2013). It remains to be established how YeaZ and YgjD are specifically required for MMC resistance in D. radiodurans.
DR_2417 corresponds to RNase J that belongs to the β-CASP family of nucleases. A DR_2417 (rnj) null mutant could not be obtained, and cells having reduced copy number of DR_2417 are affected in growth and radiation resistance (Das and Misra 2012). Remarkably, one study has reported that DR_2417 has strong DNase but poor RNase activity, and therefore the protein has also been called DncA (Das and Misra 2012), whereas in another study a clear preference for RNA has been observed (Zhao et al.2015).
Several other miscellaneous proteins required for radiation or oxidative stress resistance in D. radiodurans are not conserved in other Deinococcus species (Table 5). DR_0679 encodes a small putative nucleotidyltransferase (COG1669). DR_0392 contains a BON (bacterial OsmY and nodulation) domain that is found in a family of osmotic shock protection proteins. DR_1262 (rsr) codes for a 60 kDa SS-A/Ro ribonucleoprotein homologue. DR_1838 (conserved in the other Deinococcus species) and DR_1631 are homologues of RelA and RelQ, respectively, and are predicted to be involved in the synthesis and hydrolysis of the stringent response signal molecule (p)ppGpp in D. radiodurans (Wang et al.2016a).
Signal-peptide containing proteins
Interestingly, some proteins with a reported role in radiation and/or oxidative stress resistance contain a predicted N-terminal signal peptide, although the presence of this signal peptide and its role (translocation of the protein across the cytoplasmic membrane) has not always been realised or considered. These proteins include Dps2 (as described above), DR_1790, DR_2310, DR_A0018, DR_A0282 and DR_A0283 (Fig. S16, Supporting Information), which have homologues in some but not all of the other Deinococcus species (Table 5). Except for the hydrophobic region in the signal peptide, which is cleaved off by signal peptidase I, or by signal peptidase II in case of a lipoprotein, none of these proteins possess a predicted transmembrane helix in their mature region. These proteins are thus expected to have their function in the cell wall/periplasm or extracellularly. Some of these proteins have indeed been identified in the cell wall, for example DR_A0282 and DR_A0283, and also (see below) DR_0505 (Farci et al.2014).
DR_1790 is known as yellow-related protein and belongs to the major royal jelly protein family. In addition to increased sensitivity to H2O2 and IR, decreased growth rate of the DR_1790 mutant compared to the wild type has been reported (Cheng et al.2015). The molecular mechanism by which DR_1790 contributes to growth and resistance to radiation and oxidative stress in D. radiodurans has not been elucidated.
DR_2310 is a reported serralysin metalloprotease that is secreted (Basu and Apte 2008). Our sequence and comparative analysis strongly suggest a re-annotation of the start codon position of DR_2310, resulting in a protein that, like its homologues, has an N-terminal lipoprotein signal peptide. Together with other extracellular proteases, DR_2310 probably has a role in amino acid nutrition. The DR_2310 mutant and wild-type strains grow equally well in rich medium; nevertheless, the mutant shows a marginal sensitivity to radiation when irradiated in rich medium (Basu and Apte 2008).
The protein encoded by the corrected DR_A0018 gene, which showed a frameshift in the first genome sequence but not after resequencing (Hua and Hua 2016), is a 5΄-nucleotidase family protein (COG0737). DR_0505, which is conserved in the other Deinococcus species, is another 5΄-nucleotidase family protein. No radiation sensitivity was observed for the DR_0505 mutant (Kota, Kumar and Misra 2010) unlike for the DR_A0018 mutant (Lu et al.2009). A role for DR_0505 in DNA double-strand break processing and repair has been suggested (Kota, Kumar and Misra 2010). However, DR_A0018 and DR_0505, as well as their homologues in the other Deinococcus species, possess a predicted N-terminal signal peptide or lipoprotein signal peptide, respectively.
DR_A0282 is another protein with a suggested role in DNA repair; recombinant DR_A0282 was found to bind DNA and to protect it from exonuclease digestion (Das and Misra 2011). Homologues of DR_A0282 are present in some other Deinococcus species and also in D. radiodurans itself (DR_B0068). However, resequencing of the D. radiodurans genome and our protein comparisons and sequence analyses show that the original DR_A0282 gene contains a frameshift and that the N-terminal region of the actual gene product corresponds to the protein that was annotated as DR_A0281 in the first genome sequence. This corrected DR_A0281/0282 protein (Fig. S16, Supporting Information), as well as each homologue, contains an N-terminal lipoprotein signal peptide. Conserved domain analysis indicates a carboxypeptidase regulatory-like domain in these proteins. The DR_A0281/0282 gene is directly followed by DR_A0283 encoding a probable subtilase-type serine protease. Interestingly, each DR_A0281/0282 homologue is followed by a DR_A0283 homologue, suggesting a functional link of this conserved gene pair. Also DR_A0283 and homologues contain a lipoprotein signal peptide.
Although these various signal-peptide containing proteins may have a role in radiation or oxidative stress resistance, a direct role in DNA repair, as suggested for DR_0505 and DR_A0282, seems unlikely because of their predicted extracytoplasmic location.
Desiccation tolerance-associated proteins
Radiation resistance is probably a consequence of adaptation to natural stress conditions such as desiccation. Various D. radiodurans mutants that are sensitive to IR are also sensitive to desiccation (Mattimore and Battista 1996). A common pool of genes is induced after exposure of D. radiodurans to either IR or desiccation (Tanaka et al.2004). Both radiation and desiccation generate oxidative stress, DNA damage and, especially in radiation/desiccation-sensitive species, protein damage (Fredrickson et al.2008).
Genes encoding proteins that may specifically contribute to desiccation tolerance have been identified in D. radiodurans, and include homologues of plant desiccation resistance-associated proteins (Makarova et al.2001) and proteins containing large unstructured low-complexity regions (Krisko et al.2010). Four of these genes have been studied for their role in desiccation tolerance: DR_1172, DR_1372, DR_1769, DR_B0118 (Table S1, Supporting Information). Inactivation of DR_1172 and DR_B0118 sensitises D. radiodurans to desiccation, but not to IR. The DR_1769 mutant is sensitive to desiccation and also slightly sensitive to IR. The DR_1372 (drwH) mutant, however, has increased sensitivity to oxidative (H2O2) stress, but not to desiccation. In D. deserti, Deide_15148 encodes a protein almost entirely consisting of a low complexity region, and it may contribute to protein and membrane protection and prevention of protein aggregation during desiccation (de Groot et al.2014). Homologues of these various proteins are present in all (DR_1372, DR_1769) or most other Deinococcus species (Table 5). Some of these proteins (e.g. DR_1372, DR_B0118) possess an N-terminal signal peptide, indicating an extracytoplasmic location.
RADIATION AND OXIDATIVE STRESS RESISTANCE-ASSOCIATED REGULATORY PROTEINS
Like other bacteria, Deinococcus species encode many proteins with a predicted role in signal transduction and gene regulation, and a requirement for radiation and oxidative stress resistance in D. radiodurans has been reported for several of these proteins (Table 6 and Table S6, Supporting Information). Expression or activity of DNA repair and other stress response proteins may be controlled by more than one regulatory protein.
Table 6.
Radiation and oxidative stress resistance-associated regulator proteins in Deinococcus species.
| Protein | Drad | Dgeo | Ddes | Dmar | Dgob | Dpro | Dper | Dswu | Dsol | Dact | Dpun |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA repair | |||||||||||
| DdrO | 1 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 1 |
| IrrE (PprI) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| LexA-ArsR | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| LexA-XRE | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| RqkA | 1 | 1 | 1 | 1 | fr | 1 | 1 | 1 | 1 | 1 | |
| Oxidative stress reponse and Mn/Fe homeostatis | |||||||||||
| OxyR1-like (LysR) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OxyR2-like (LysR) | 1 | 1 | 1 | ||||||||
| Mur-like (FUR 1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| PerR-like (FUR 2) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Irr-like (FUR 3) | 1 | 1 | 1 | 1 | |||||||
| DtxR | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| MntR | 1 | 1 | |||||||||
| SoxR | 1 | 1 | 1 | fr | |||||||
| Two-component systems | |||||||||||
| DrRRA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| DR_2419 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| RadS | 1 | 1 | |||||||||
| RadR | 1 | 1 | |||||||||
| DrtS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DrtR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DR_1556 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| DR_A0205 | 1 | 1 | 1 | 1 | 1 | ||||||
| Quorum sensing | |||||||||||
| DqsI-1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| DqsI-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DqsR | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | fr | 1 |
| LuxS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| LsrB-like | 1 | 1 | |||||||||
| Others | |||||||||||
| DdrI | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DR_0171 | 1 | ||||||||||
| DR_0265 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | |
See the legend to Table 2.
Regulators of DNA repair system
The metallopeptidase/repressor pair IrrE/DdrO
Based on the currently available data, the IrrE/DdrO pair appears to be the most important regarding gene regulation in response to radiation and DNA-damaging agents. Indeed, D. radiodurans and D. deserti irrE deletion mutants are very sensitive to radiation (Earl et al.2002a; Hua et al.2003; Vujicic-Zagar et al.2009), and the most highly induced genes in D. radiodurans and D. deserti following exposure to IR are regulated by IrrE/DdrO (Tanaka et al.2004; Makarova et al.2007; Blanchard et al.2017). The crystal structure of D. deserti IrrE showed structural similarity with zinc metallopeptidases (Vujicic-Zagar et al.2009). IrrE (PprI) is indeed a metalloprotease (COG2856) that, after exposure of the cells to radiation, cleaves and inactivates DdrO, the XRE family transcriptional repressor of pprA and several ddr and DNA repair genes (e.g. recA) (Fig. 7) (Ludanyi et al.2014; Devigne et al.2015; Wang et al.2015; Blanchard et al.2017). This proteolytic activity of IrrE is essential for radiation resistance because strains expressing IrrE with point mutations in the active site that abolish its protease activity, or a strain expressing uncleavable DdrO, are as sensitive to radiation as irrE deletion mutants (Vujicic-Zagar et al.2009; Ludanyi et al.2014; Wang et al.2015). The molecular mechanism by which radiation triggers DdrO cleavage has not been elucidated, but may be related to oxidative stress and metal ion availability for IrrE (Blanchard et al.2017). The genes regulated by IrrE/DdrO are preceded by a 17-bp palindromic motif named RDRM (radiation/desiccation-response motif) (Makarova et al.2007), which overlaps with or is located very close to the promoter of these genes (de Groot et al.2014). Binding of repressor protein DdrO to RDRM-containing fragments has been demonstrated in vitro (Wang et al.2015; Blanchard et al.2017), and constitutive, derepressed expression has been observed in vivo for genes carrying point mutations in their preceding RDRM (Devigne et al.2015; Anaganti et al.2017). Cleaved DdrO does not bind the RDRM (Blanchard et al.2017). IrrE, DdrO and the RDRM are highly conserved in Deinococcus species, but there is diversity in the RDR regulon composition (Blanchard et al.2017). Demonstrated or predicted RDR regulon members in Deinococcus bacteria include ddrA, ddrB, ddrC, ddrD, ddrF, ddrO, ddrQ, ddrR, ddrS, ddrTUVWX, pprA, recA operon, extra recA, ssb, gyrA, gyrB, recD, recQ, ruvB, uvrA, uvrB and uvrD, but some of these genes are not present in each Deinococcus species (Table 4). The RDR regulon includes ddrO itself, allowing DdrO to re-accumulate and re-repress the RDR regulon when the stress is alleviated. Interestingly, DdrO is essential for viability in at least D. deserti (Ludanyi et al.2014) and D. radiodurans (Devigne et al.2015), and its prolonged depletion in D. radiodurans results in apoptotic-like cell death (Fig. 7) (Devigne et al.2015). The genes/proteins that provoke this apoptotic-like death are currently unknown. It is also noteworthy that three Deinococcus species encode one or two additional DdrO homologues (Table 6). Protein sequence comparisons suggest that these extra DdrO proteins can be cleaved and inactivated by IrrE (Fig. S17, Supporting Information), and in vivo evidence for this has indeed been obtained for the second DdrO in D. deserti (Ludanyi et al.2014). Either one or the other ddrO gene is required for viability in D. deserti, suggesting that its two DdrO proteins have at least partially overlapping function regarding the genes they regulate. The different DdrO proteins in D. gobiensis and D. peraridilitoris may also have overlapping functions or, alternatively, regulate different genes.
Figure 7.
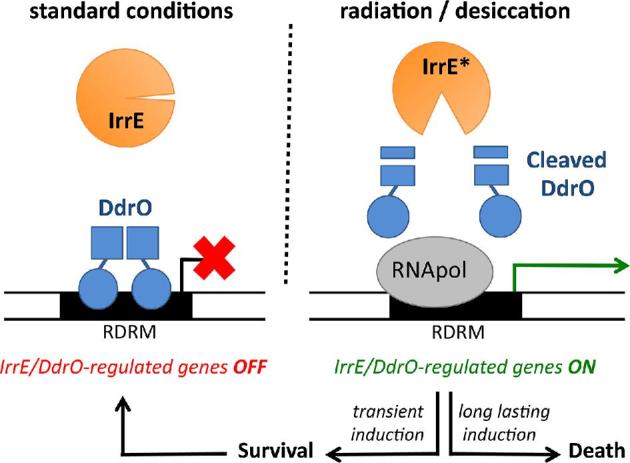
Model for the radiation/desiccation response (RDR) mechanism in Deinococcus: induction of the RDR regulon after cleavage of a repressor by a separate and specific metalloprotease. Under standard conditions, DdrO probably binds as a dimer to the two half-sites of the palindromic motif RDRM, thereby inhibiting transcription of IrrE/DdrO-regulated genes. Exposure of Deinococcus to radiation or desiccation stimulates cleavage of DdrO by IrrE. Cleaved DdrO may not form dimers and no longer binds the RDRM, allowing rapid induction of RDR regulon genes (e.g. DNA repair genes and ddrO itself). Persistence of the signal leading to DdrO cleavage may induce, directly or indirectly, one or more pro-apoptotic genes.
Potential metalloprotease/repressor pairs related to IrrE/DdrO have been identified in other bacterial genera, for example in Meiothermus spp. and some other species belonging to the phylum Deinococcus-Thermus and also in bacteria more distant to Deinococcus, indicating that a similar regulatory mechanism involving cleavage of an XRE family repressor by a specific COG2856 protease may also occur in these species (Ludanyi et al.2014).
RecA/LexA
Induction of DNA repair genes in Deinococcus after cleavage of repressor DdrO by the separate protease IrrE is different from the well-known SOS response in E. coli and many other bacteria, in which recA and other DNA repair genes are induced after RecA-stimulated autocleavage of repressor LexA (Butala et al.2011). Nevertheless, D. radiodurans encodes two LexA-related proteins, LexA1 (DR_A0344) and LexA2 (DR_A0074). Moreover, RecA-dependent cleavage of both these proteins has been demonstrated in vitro and in vivo (Narumi et al.2001; Sheng et al.2004; Satoh et al.2006), strongly suggesting that expression of some currently unknown genes is under direct control of these LexA proteins, and induced following DNA damage. Indeed, the lexA1 mutant forms cell aggregates (Bonacossa de Almeida et al.2002). However, LexA1 and LexA2 are not involved in recA induction and at least LexA1 does not play a major role in radiation resistance in D. radiodurans (Jolivet et al.2006). A lexA2 mutant has increased IR resistance according to one study (Satoh et al.2006), but not according to another (Sheng et al.2004). One or two LexA-related proteins are encoded by most, but not all, Deinococcus bacteria (Table 6 and Fig. 8). However, the similarities between these LexA proteins are rather low, and therefore the LexA-regulated genes might be different in these species. For example, the best hit in BLASTP analysis of D. radiodurans LexA1 against the other 10 Deinococcus species is Deide_1p01870 from D. deserti with only 50% identity. BLASTP of D. radiodurans LexA2 against these 10 other species does not give any hit at all. For comparison, D. radiodurans DdrO has 86 to 97% identity with a DdrO from each of these other species. The deinococcal LexA-related proteins contain either an ArsR-type (e.g. DR_A0344 and Deide_1p01870) or an XRE-type (e.g. DR_A0074) DNA-binding domain (Fig. S18, Supporting Information).
Figure 8.
Phylogenetic relationship of LexA homologues identified in Deinococcus species. The phylogenetic analysis was carried out based on protein sequence alignment of 12 deinococcal LexA homologues (Table S6, Supporting Information) with some representative proteins taken from Uniprot: LexA from B. subtilis (Uniprot Number P31080) and E. coli (P0A7C2). See further the legend to Figure 4.
RecA-mediated cleavage has also been demonstrated for Deide_1p01870, which controls expression of an operon that is radiation-induced in a RecA-dependent but IrrE-independent manner and that encodes Deide_1p01870 itself and the TLS DNA polymerases ImuY and DnaE2 (Dulermo et al.2009). This operon is also found in D. peraridilitoris (see the section ‘DNA repair in Deinococcus’), strongly suggesting that both D. deserti and D. peraridilitoris possess RecA/LexA-dependent regulation of TLS DNA polymerases besides the IrrE/DdrO-dependent regulation of other DNA repair genes (Fig. 9). Interestingly, both D. deserti and D. peraridilitoris encode two different RecA, and for D. deserti it has been shown that only one of its two RecA proteins mediates induction of the lexA-imuY-dnaE2 operon.
Figure 9.
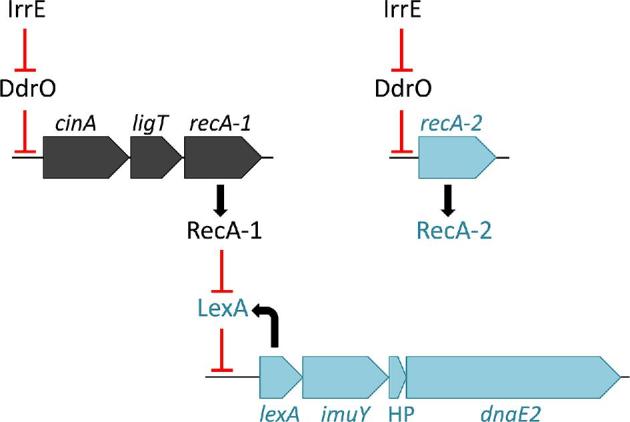
IrrE/DdrO- and RecA/LexA-regulated expression of DNA repair genes within the same bacterium. The cinA-ligT-recA operon (ligT-cinA in D. proteolyticus) and genes for IrrE and DdrO are present in each Deinococcus species. Additional RecA (RecA-2) and the lexA-imuY-dnaE2 operon are found in D. deserti and D. peraridilitoris. Experimental data, obtained for D. deserti only, have shown that both recA genes are radiation-induced in an IrrE-dependent way, and that the presence of either recA-1 or recA-2 is sufficient for radiation resistance. Radiation exposure also induces expression of the lexA-imuY-dnaE2 operon leading to induced mutagenesis mediated by the translesion polymerases ImuY and DnaE2, and this induction requires recA-1 but not recA-2. The recA-2 product is thus functional for recombinational repair but not for induction of mutagenic lesion bypass. The lexA-imuY-dnaE2 operon is also radiation induced in the irrE mutant, indicating that basal level of RecA-1 is sufficient for this induction. The red symbols indicate transcriptional repression by DdrO or LexA, or repressor inactivation by IrrE- or RecA-mediated cleavage. HP, hypothetical protein.
Serine/threonine-protein kinase RqkA
It has been reported that transgenic E. coli cells expressing D. radiodurans pqqE (DR_C0034), encoding coenzyme PQQ synthesis protein E, showed an improved tolerance to DNA-damaging agents (UV, IR and MMC) (Khairnar et al.2007), and that the D. radiodurans pqqE mutant lacking PQQ was sensitive to IR and MMC (but not to UV) (Rajpurohit, Gopalakrishnan and Misra 2008). The higher radiation tolerance possibly mediated by PQQ in E. coli was suggested to require YfgL, which contains PQQ-binding motifs and a Ser/Thr kinase domain (Khairnar et al.2007). To identify one or more proteins through which PQQ could contribute to IR resistance in D. radiodurans, five genes predicted to encode PQQ-binding proteins were inactivated. Among the five mutants, the strain lacking DR_2518 showed a similar phenotype as the pqqE mutant, that is, increased radiation sensitivity and impaired DNA double-strand break repair (Rajpurohit and Misra 2010). Increased sensitivity to DNA-damaging agents upon DR_2518 inactivation was also found independently in another study (Dulermo et al.2015). DR_2518 encodes a protein containing a eukaryotic-type Ser/Thr protein kinase domain in addition to the multiple PQQ-binding motifs, suggesting that, like in E. coli, the PQQ contribution to radiation tolerance is functionally linked to a protein kinase. Moreover, PQQ stimulated kinase activity of DR_2518 in vitro, and IR exposure induced autophosphorylation of DR_2518 in vivo (Rajpurohit and Misra 2010), and therefore DR_2518 was designated RqkA, radiation and PQQ-inducible protein kinase (Rajpurohit and Misra 2013). Remarkably, only D. radiodurans possesses both pqqE and rqkA, whereas most other IR-resistant Deinococcus species have only rqkA (Tables 3 and 6), suggesting that RqkA may play a role in IR resistance without PQQ. However, D. proteolyticus lacks rqkA and the homologous sequence in D. gobiensis contains a frameshift. To find potential target proteins for RqkA, conserved phosphorylation motifs have been identified using bioinformatics in many D. radiodurans proteins, including PprA and RecA (Rajpurohit and Misra 2013). Subsequently, in vitro phosphorylation of D. radiodurans PprA (the majority at residue T72, and also at S112 and T144) and D. radiodurans RecA (at Y77 and T318) by RqkA has been reported, although at other residues than predicted (Rajpurohit and Misra 2013; Rajpurohit et al.2016). However, T72 of PprA and T318 of RecA are not conserved among deinococcal homologues, and understanding the role of phosphorylation of these proteins by RqkA requires further investigation.
Regulators of oxidative stress response and Mn/Fe homeostasis
Several studies have revealed that when bacteria are exposed to H2O2 and other ROS, they not only induce ROS detoxification enzymes such as catalases and SOD, but also adapt to the oxidative stress by modifications of metal ion homeostasis with the net effect of reducing the damage caused by reactive ferrous iron (Faulkner and Helmann 2011). Generally, bacteria reduce levels of free Fe2+ in the cell, for example by sequestration in storage proteins (e.g. Dps) and repression of Fe2+ uptake, and elevate cytosolic levels of Mn2+, which can replace Fe2+ from sensitive sites in enzymes, by inducing Mn2+ import. For D. radiodurans, which has a high Mn/Fe ratio under standard laboratory conditions, five regulators with a proposed role in oxidative stress response and Mn/Fe homeostatis have been described. These are two LysR family regulators (proposed to be 1-Cys-OxyR proteins), two FUR family regulators (possible PerR and Mur proteins) and a DtxR-like regulator (see below for more details). Of these, the possible Mur seems the most important under standard conditions, because its inactivation results in a growth defect (Ul Hussain Shah et al.2014), while such defect was not observed or reported for mutant strains lacking one of the other four regulator proteins. Except for the mutant strain lacking the PerR-like protein, transcriptomics experiments have been performed to analyse global gene expression in these D. radiodurans regulator mutants compared to the wild-type strain, which revealed differential expression (more than 2-fold up- or downregulation) of dozens to hundreds of genes. However, more work is needed to determine which genes are regulated directly by these regulators and to decipher if and how these regulators respond to oxidative stress or levels of specific metals.
Hydrogen peroxide sensor OxyR
Escherichia coli encodes dozens of LysR-type transcriptional regulators. One of these has been characterised as the H2O2-sensing regulator OxyR. Escherichia coli OxyR contains two Cys residues that are essential for H2O2 sensing: upon oxidation by H2O2 of the sensing cysteine C199 followed by intramolecular disulphide bond formation with the resolving cysteine C208, OxyR is locked in a conformation that activates the protein as a transcription factor (Dubbs and Mongkolsuk 2012; Imlay 2013, 2015). Only two LysR-type regulators, DR_0615 and DR_A0336, are found in D. radiodurans. The E. coli LysR-type proteins most similar to both DR_0615 and DR_A0336 are YnfL and HcaR (34–38% identity), while E. coli OxyR is about 30% identical to DR_0615 and DR_A0336. Nevertheless, DR_0615 and DR_A0336, which share 38% identity, have been described as OxyR1 and OxyR2, respectively, each containing only one cysteine residue that was reported to be essential for protein function in vivo (Chen et al.2008; Yin et al.2010). The single Cys residue (C210) of DR_0615 could be oxidised to sulfenic acid in vitro (Chen et al.2008).
The OxyR regulon of E. coli is comprised of more than 20 genes, including genes involved in H2O2 scavenging (katG, ahpCF), Fe-S cluster assembly (sufABCDE), iron scavenging (dps), Mn import (mntH) and disulphide reduction (trxC, grxA, etc.) (Imlay 2015). The genes directly regulated by DR_A0336 (OxyR2) are unknown, and DR_A0336 has clear homologues (60% identity) only in two of the analysed Deinococcus species (Table 6). Under standard conditions, hundreds of genes showed either increased or decreased expression in the DR_0615 (OxyR1) deletion mutant compared to the wild-type strain (Chen et al.2008). DR_0615 may directly regulate the genes DR_B0125 (iron transporter), katE1, mntH and dps1, because binding of DR_0615 to DNA regions upstream of these genes has been observed in vitro. The same DNA-binding pattern was observed with oxidised DR_0615, reduced DR_0615 or DR_0615 with the C210A mutation. However, the precise mechanism by which DR_0615 may regulate genes either positively or negatively upon H2O2 exposure has not been elucidated. The katE1 expression was induced 4-fold by H2O2 in wild type, but an approximately 3-fold induction was still observed in DR_0615 mutants. In addition, although DR_0615 is proposed to be a negative regulator of mntH and dps1, mntH expression is reduced, whereas dps1 is highly induced by H2O2 stress in DR_0615 mutants (Chen et al.2008). DR_0615 is highly similar (74–80%) to one protein in each of the other 10 Deinococcus species (Fig. S19, Supporting Information), indicating a crucial conserved role. Importantly, however, the single and proposed sensing Cys of DR_0615 is not strictly conserved. The homologues from D. peraridilitoris and D. deserti lack Cys, and therefore at least these Cys-less DR_0615 homologues cannot function as canonical or 1-Cys-OxyR. Clearly, further research is necessary to elucidate the precise role and mode of function of DR_0615 and homologues.
FUR family regulators
In most bacteria, proteins of the FUR (ferric uptake regulator) family act as metal sensors that regulate genes connected to metal homeostasis and reponse to oxidative stress. Although the iron-responsive regulator Fur is a well-known member of the FUR family, there is diversity in metal selectivity and biological function within the FUR family which includes manganese uptake regulator Mur (responding to Mn2+), Zur (responding to Zn2+), Nur (responding to Ni2+), PerR (responding to peroxide stress) and Irr (heme-dependent iron responsive) (Lee and Helmann 2007; Fillat 2014). Initially, only one gene encoding a FUR family protein was predicted in D. radiodurans (DR_0865, possible mur). Later, a second gene was identified (de Groot et al.2009) and proposed to encode PerR (Liu et al.2014).
The FUR family protein DR_0865 of D. radiodurans was proposed to correspond to Mur (Ul Hussain Shah et al.2014). To date, Mur proteins have been found in some α-proteobacteria, such as Rhizobium and Sinorhizobium, and the only known target for Mur is the ABC-type Mn2+ transporter (Johnston et al.2007; Fillat 2014). Of the five Mn2+-transport-related genes of D. radiodurans, the expression of mntA, mntB and mntH increased, while mntE was repressed in the DR_0865 mutant strain compared to the wild-type strain under standard laboratory conditions. Moreover, expression of mntA and mntE in the DR_0865 mutant under Mn2+ stress was significantly higher and lower, respectively, compared to the Mn2+-stressed wild type (Ul Hussain Shah et al.2014). Differential gene expression in Mn2+-stressed cells versus non-stressed cells was not reported. DR_0865 was found to bind to DNA fragments containing the promoters of MntABC transporter genes in vitro but not to the mntH promoter (Sun et al.2012). However, there is a controversy about mutant phenotypes: the DR_0865 mutant strain was reported to be sensitive to Mn2+ and H2O2 (Ul Hussain Shah et al.2014), but other studies mentioned that the mutant strain exhibited greater resistance to H2O2 than wild type (Chen et al.2008) and showed that its Mn2+ resistance was comparable that of wild type (Sun et al.2012). Because the Mur and Fur proteins have similar sequences, and Murs from Rhizobiales can complement E. coli fur mutants (Johnston et al.2007; Hohle and O’Brian 2016), further research is needed to define the exact role of DR_0865, especially in terms of metal homeostasis. Moreover, phylogenetic analysis indicates that DR_0865 and homologues (FUR group 1), present in each Deinococcus, are more related to some of the Zur proteins than to the Mur of Rhizobium (Fig. 10).
Figure 10.
Three groups of FUR family proteins identified in 11 Deinococcus species. The phylogenetic analysis was carried out based on protein sequence alignment of 26 deinococcal FUR family proteins (Table S6, Supporting Information) with some representative proteins taken from Uniprot: Fur proteins from Vibrio cholerae (Uniprot Number P0C6C8), E. coli (P0A9A9), P. aeruginosa (Q03456), Campylobacter jejuni (P0C631) and H. pylori (O25671); Mur from Rhizobium leguminosarum (Q1MMB4); Zur from Streptomyces coelicolor (Q9L2H5), M. tuberculosis (P9WN85), B. subtilis (P54479), and E. coli (P0AC51); Nur from S. coelicolor (Q9K4F8); PerR from B. subtilis (P71986) and Streptococcus pyogenes (Q1JIU5); Irr from R. leguminosarum (I9X7E3) and Bradyrhizobium japonicum (O85719). GenBank accession numbers in parentheses follow the species name. The phylogenetic consensus tree was developed using the neighbour-joining algorithm in MEGA 6.0. The node numbers are bootstrap values based on 1000 replications.
PerR is a global regulator that responds primarily to H2O2, and substitutes for OxyR in many Gram-positive bacteria, although it may also coexist with OxyR (Dubbs and Mongkolsuk 2012). In, for example, B. subtilis, PerR is inactivated by H2O2 stress, leading to derepression of the PerR regulon, including the genes for Prx, catalase and the ferritin-like proteins (Hillion and Antelmann 2015). In D. radiodurans, disruption of the putative perR gene increased katE1 and dps1 expression and resistance to H2O2 stress (Liu et al.2014). However, it is necessary to investigate if the derepression of these genes, especially dps1, occurs via inactivation of the PerR-like protein under oxidative (H2O2) stress condition, because the dps1 gene expression is not induced by H2O2 (Liu et al.2014). Homologues of the putative PerR of D. radiodurans are present in all other Deinococcus species (FUR group 2) (Fig. 10). PerR is a metal-dependent transcriptional regulator, and the Fe2+-containing form is thought to be responsible for H2O2 sensing, in which Fe2+ bound at the regulatory site is coordinated by three His (H37, H91, H93) and two Asp (D104, D85) residues in B. subtilis (Dubbs and Mongkolsuk 2012). Exposure to H2O2 leads to oxidation of Fe2+ in the regulatory site by a Fenton reaction, which causes oxidation of H37 and H91 to 2-oxo-histidine and inactivation of PerR (Hillion and Antelmann 2015). The five amino acid residues (H23, H79, H81, D71 and D92 in the PerR-like protein from D. radiodurans) are strictly conserved in all FUR group 2 proteins (Fig. S20, Supporting Information). Phylogenetic analysis also indicates that these deinococcal proteins are related to PerR (Fig. 10).
An additional, third FUR family protein (FUR group 3) is found in four of the analysed Deinococcus species (Table 6), increasing the diversity of FUR metalloregulators compared to D. radiodurans. Like the deinococcal PerR-like proteins, these proteins contain the three His and two Asp residues that are involved in metal ion binding at the regulatory site of B. subtilis PerR (Fig. S20, Supporting Information). However, phylogenetic analysis shows that these proteins of FUR group 3 are more closely related to the heme-dependent iron responsive regulator Irr (Fig. 10). Irr binds heme at a HXH motif, conserved in most FUR family proteins, and acts as both positive and negative regulator of gene expression modulating a number of genes related to iron metabolism in Rhizobiales (Fillat 2014). However, unlike Irr proteins but similar to many other FUR family proteins (including the deinococcal FUR group 1 and 2, PerR, Zur and some Fur proteins), this third group of deinococcal FUR proteins contain two CXXC motifs. The Cys residues in these motifs may be required for activity and protein stability, and probably coordinate a structural zinc ion (Lee and Helmann 2007; Fillat 2014).
DtxR and MntR regulators
The control of iron metabolism and its coupling with regulation of defences against oxidative stress is carried out by Fur in most prokaryotes, but high-GC Gram positive bacteria, such as Mycobacterium, tend to use the DtxR (diphtheria toxin repressor) family for iron homeostasis (Fillat 2014). In D. radiodurans, DR_2539 was proposed to be a novel DtxR-like regulator (Chen et al.2010). Compared to the wild-type strain, the iron transporter genes DR_1219 and DR_B0125 were downregulated in the DR_2539 mutant under standard growth conditions. Under these conditions, the manganese transporter genes mntBC were found (slightly) upregulated in the DR_2539 mutant, while expression of the manganese transporter gene mntH and of the two dps genes was not affected by the DR_2539 disruption (Chen et al.2010). In another study, mntH expression was found to be reduced in wild-type strain, but not in the DR_2539 mutant, by the addition of Mn2+ or Fe2+ to the growth medium. In vitro DNA-binding experiments indicated that this Mn2+/Fe2+-dependent mntH repression occurs through the direct binding of DR_2539 to the mntH promoter, while binding of DR_2539 to the promoters of MntABC transporter genes was not observed (Sun et al.2012). Although DR_2539 seems to function as a repressor of mntH only upon addition of Mn2+/Fe2+, the data indicate that DR_2539 is also functional under standard conditions because expression of dozens of genes is affected in the DR_2539 mutant in standard growth medium (Chen et al.2010). The other Deinococcus species produce one DR_2539 homologue except D. deserti encoding two homologues. Metal-binding site 1 (MBS1) of DR_2539 from D. radiodurans is composed of His79, Glu83, His98, Arg176 and Pro179, and MBS2 is composed of Asp11, Glu102, Glu105 and His106 (Chen et al.2010). Sequence alignment analysis shows that most of the residues located in MBS are conserved in the deinococcal DR_2539 homologues except Arg176 in MBS1 and Glu102 in MBS2 (Fig. S21, Supporting Information). DtxR is composed of two domains: the N-terminal domain is involved in metal binding, dimerisation and DNA recognition, and the C-terminal Src homology 3 (SH3)-like domain is involved in metal ion binding at MBS1, thereby affecting repressor activity (Love, VanderSpek and Murphy 2003). MntR is a DtxR homologue regulated by Mn2+. DtxR and MntR share similar structure and metal-binding residues, but MntR lacks the C-terminal SH3-like domain (Stoll et al.2009). Interestingly, an MntR-like regulator is found in D. deserti and D. peraridilitoris (Fig. S21, Supporting Information), and in both species this regulator is encoded by a gene located directly after and likely in operon with four ABC-type Mn2+ transporter genes encoding MntA, MntC and two different MntB homologues.
SoxR: redox-sensitive transcriptional activator
Escherichia coli and several other bacterial species encode the MerR family regulator protein SoxR, involved in induction of sodA and other defensive genes upon exposure to superoxide-generating redox-cycling compounds such as phenazines and quinones. SoxR contains a [2Fe-2S] cluster, which involves cysteine residues that are present in the conserved motif CIGCGCxxxxxC located in the C-terminal region of the protein. Oxidation of the iron–sulphur cluster activates SoxR and transcription of target genes. Initially, it was expected that superoxide directly oxidises the [2Fe-2S] clusters, but experiments have indicated that the redox-cycling compounds themselves activate SoxR. In E. coli, SoxR induces a second transcription factor, SoxS, which then induces expression of sodA and other target genes. In non-enterics, however, SoxR directly controls expression of SoxR regulon genes, which are different from those of E. coli and may encode pumps to excrete redox-cycling compounds but generally do not include sodA (Imlay 2013, 2015).
Deinococcus radiodurans does not possess a soxR gene. However, homologues of E. coli SoxR (54–60% identity), including the CIGCGCxxxxxC motif, are found in D. deserti, D. proteolyticus, D. soli and D. actinosclerus (although the gene in D. actinosclerus has a frameshift). In these Deinococcus species, SoxR may regulate currently unknown target genes involved in defence against redox-active compounds.
Other radiation and oxidative stress resistance-associated regulators
Two-component signal transduction systems
TCSs, composed of a histidine kinase (HK) and a response regulator (RR), are major means by which bacteria adapt to changing environments. Typically, the environmental signal triggers HK autophosphorylation at one His residue, followed by phosphoryl transfer from the phospho-His to an Asp residue in the RR, thereby regulating expression of genes and/or modulating activity of proteins (Casino, Rubio and Marina 2010; Agrawal, Sahoo and Saini 2016). DrRRA (DR_2418) was the first RR identified as contributing to the resistance of D. radiodurans not only to IR and H2O2 but also to desiccation (Wang et al.2008). Compared to the wild type, the expression of numerous genes, including stress response and DNA repair genes as well as many uncharacterised genes (e.g. katE1, katE2, sodA, sodC, dps1, recA, uvrA, gyrB, ddrC, pprA, ddrI, ddrP), is lower in the drRRA mutant, both under standard growth conditions and after irradiation (Wang et al.2008). However, it appears that at least several genes that are IR induced in the wild type are still IR induced in the drRRA mutant (e.g. recA, pprA, ddrC). Binding of DrRRA protein to a ddrI promoter-containing DNA fragment has been observed in vitro (Wang et al.2008), but the DrRRA–DNA interaction has not been studied in more detail. At the protein level, one study suggested reduction of RecA and PprA levels in the drRRA mutant compared to the wild type (Wang et al.2008), but this was not observed in another study (Wang et al.2016b). DrRRA is conserved in the other Deinococcus species except for D. peraridilitoris. Concerning the genetic organisation, drRRA in D. radiodurans is adjacent to the HK gene DR_2419, suggesting that DrRRA might be the cognate RR for DR_2419, but the DR_2419 disruption has a less strong effect on IR resistance than the drRRA disruption (Wang et al.2008; Im et al.2013). Moreover, several DrRRA-containing Deinococcus species do not encode the homologue of HK DR_2419 (Table 6). In addition, DR_2420 encoding another RR is adjacent to DR_2419, and the ‘RR-HK-RR’ gene cluster is also found in some other Deinococcus species (Fig. S22, Supporting Information). Further research is needed to identify HKs that can phosphorylate DrRRA.
The RadS/RadR (DR_B0090/DR_B0091) TCS contributes to radiation resistance in D. radiodurans (Desai et al.2011; Im et al.2013). However, homologues are only present in D. gobiensis. Both in D. radiodurans and D. gobiensis this radSR gene pair is directly adjacent to the divergently oriented gene encoding extracytoplasmic Dps2, indicating a possible functional link (see also the section ‘Other proteins involved in ROS protection’).
In D. radiodurans, inactivation of either DR_2416 encoding a HK or DR_2415 encoding the probable cognate RR of HK DR_2416 resulted in slightly reduced resistance to radiation, MMC and/or oxidative stress; hence, DR_2415 and DR_2416 were designated as DrtR and DrtS (DNA damage response TCS regulator and sensor), respectively (Im et al.2013). Contrary to DrRRA and RadR/RadS, both DrtR and DrtS are conserved in the other analysed Deinococcus species.
In addition to radS and drtS, 10 other HK genes have been inactivated separately in D. radiodurans, resulting in slightly reduced resistance to radiation and/or oxidative stress for each mutant (Im et al.2013). Except for DR_1556 and DR_A0205 (Table 6), these HKs are conserved in the other analysed Deinococcus species.
Quorum-sensing systems
Quorum sensing (QS) is a cell-to-cell communication process that enables bacteria to behave coordinately and to regulate gene expression in response to changes in the cell density. QS involves the production, release and detection of extracellular signalling molecules called autoinducers (AIs) (Papenfort and Bassler 2016). There are several QS systems used by bacteria: the LuxR/I-type systems, primarily used by Gram-negative bacteria, in which the signaling molecule is an acyl-homoserine lactone (AHL or AI-1); the peptide signaling systems used primarily by Gram-positive bacteria; and the LuxS/furanone metabolites (collectively called AI-2) signaling used for interspecies communication (Reading and Sperandio 2006). A few studies have indicated that QS may also contribute to the resistance phenotype of D. radiodurans (Lin et al.2016a,b). Slightly reduced resistance to radiation and/or oxidative stress has been reported for strains carrying disruptions of genes involved in AHL- and AI-2-mediated QS systems: DR_2587 and DR_0090 encoding homologues of AHL synthase, designated dqsI-1 and dqsI-2 for Deinococcusquorum sensing autoinducer-1 and -2, respectively, DR_0987 encoding the AHL-responsive regulator DqsR, and DR_2387 encoding the LuxS enzyme responsible for the synthesis of AI-2. H2O2 treatment was found to induce AHL accumulation in D. radiodurans. Deinococcus radiodurans also possesses the quorum quenching enzymes AHL-acylase (QqaR, DR_A0255) and AHL-lactonase (QqlR, DR_0172), which are able to inactivate foreign AHLs (Koch et al.2014), and AHL levels are higher in the qqaR and qqlR mutants (Lin et al.2016a). The expression of many genes was affected in luxS and dqsR mutants compared to the wild-type strain, including stress response-related genes (Lin et al.2016a,b). DqsR-binding sites have been predicted in the upstream regions of various genes that are downregulated in the dqsR mutant, and in vitro binding of DqsR to three selected regions has been observed: upstream of DR_1436 (ABC transporter), DR_A0158 (phosphate ABC transporter) and DR_B0067 (extracellular nuclease) (Lin et al.2016a).
The DqsI and DqsR proteins involved in the AHL-mediated QS system are conserved in the other analysed Deinococcus species, but D. proteolyticus lacks the AI-2 synthesis protein LuxS. The AI-2 signal molecule is detected by LuxP that functions in conjunction with the two-component sensor kinase LuxQ in Vibrionaceae, or is imported by the LsrABC transporter, in which LsrB acts as an AI-2 receptor, in Enterobacteriaceae (Rezzonico, Smits and Duffy 2012). LuxP homologues were not found in the Deinococcus species. An LsrB-like protein is detected only in D. deserti and D. geothermalis. Given that the supernatant of the D. radiodurans wild-type strain restores the radioresistance phenotype of the luxS mutant strain (Lin et al.2016b), however, it is possible that additional, yet undiscovered, AI-2 receptors exist in D. radiodurans and other deinococci (Rezzonico, Smits and Duffy 2012).
Other DNA-binding transcriptional regulators
The cyclic AMP receptor protein (CRP) is a global regulator that regulates over 490 genes in E. coli, especially in relation to carbon metabolism, and can indirectly mediate the expression of a large number of stress response proteins (Geng and Jiang 2015). Of the four genes encoding putative CRP family proteins (DR_0997, DR_1646, DR_2362 and DR_0834) in D. radiodurans, DR_0997 (also referred to as ddrI) is highly induced by IR (Tanaka et al.2004), and its disruption, but not that of any of the other predicted CRP genes, results in increased sensitivity to H2O2, MMC, UV and IR (Yang et al.2016). The transcriptional levels of a series of genes involved in DNA repair, oxidative resistance and other cellular pathways were measured, and expression of several of these genes (e.g. katE1, DR_A0202 sodC, pprA, uvsE, uvrC, ruvC, recA, recF, recN, ddrB, ddrC, ddrD, Lon protease genes, glycometabolism gene glgC) was found to be lower in the ddrI mutant than in the wild-type strain under both normal and stress (IR or H2O2) conditions (Yang et al.2016). Moreover, the ddrI deletion mutant grows slower than the wild-type under standard conditions. The upstream region of at least 18 genes in D. radiodurans contains sequences similar to the E. coli CRP-binding site, and binding of the CRP family protein DdrI to these regions has been demonstrated in vitro (Yang et al.2016). These 18 genes, including pprA, uvsE and recN, may be regulated directly by DdrI, whereas the reduced expression of many other genes in the ddrI mutant may be caused indirectly (e.g. katE1, recA, ddrC). A recent study indicated that DdrI expression is DrRRA dependent in D. radiodurans, and that DdrI may regulate hundreds of genes involved in various cellular processes, underlining its important role in cell physiology under normal and stress conditions (Meyer et al.2018). The DdrI homologue, but not DrRRA, is present in each Deinococcus species (Table 6). The studies on DdrI and other data (Kamble et al.2010) suggest that cyclic AMP signalling contributes to expression of stress response and DNA repair genes. Also cyclic di-AMP signalling may contribute in the recovery of D. radiodurans cells from genotoxic stresses, because inactivation of DR_0007, encoding a homologue of B. subtilis CdaA that catalyses cyclic di-AMP synthesis, sensitises D. radiodurans to radiation (Table S1, Supporting Information). Homologues of DR_0007, as well as of the adjacent DR_0008 gene encoding a homologue of CdaR that stimulates CdaA activity in B. subtilis, are present in each of the analysed Deinococcus species (Table S5, Supporting Information).
DR_0171 is a predicted DNA-binding protein that is induced after exposure to high doses of IR (Liu et al.2003; Lu et al.2011). Compared to the wild type, a lower expression of various genes, encoding proteins belonging to different functional categories as well as proteins of unknown function, has been reported in the DR_0171 mutant after exposure to IR (Lu et al.2011). DR_0171 is not conserved in Deinococcus (Table 6), and understanding how it directly or indirectly regulates gene expression in D. radiodurans requires further work.
DR_0265 is another predicted DNA-binding transcription factor (GntR family). Its contribution to radiation resistance has been found after screening transposon mutants (Dulermo et al.2015). The target genes for DR_0265 are currently unknown. Deinococcus proteolyticus lacks a DR_0265 homologue, while three other Deinococcus species possess two DR_0265 homologues (Table 6).
CONCLUDING REMARKS
Repair of massive DNA damage is not given to everyone. Deinococcus bacteria have this astonishing skill when facing high doses of radiation, desiccation and oxidative stress-generating conditions. To decipher the underlying mechanisms, D. radiodurans appeared to be an excellent model organism. Its thorough characterisation over the last decades has led to many important discoveries, and indicated that its extreme resistance results from a combination of multiple factors and well-regulated mechanisms that limit oxidative protein damage and enable repair of massive DNA damage. At a first glance, the DNA repair machinery of D. radiodurans seems globally similar to the one of other bacteria like E. coli, but detailed studies revealed several specificities that may contribute to radiation resistance (proteins like RecA may have evolved to perform better under stress conditions, multiple variants of some DNA repair proteins such as DNA glycosylases, novel Deinococcus-specific proteins such as DdrB and PprA). Although the various resistance and repair mechanisms are not fully understood, these pioneering results obtained with D. radiodurans have greatly advanced our understanding of radiation resistance in general, with some common aspects (but also differences) observed in other radiation-resistant organisms that have evolved either naturally (i.e. archaea, small invertebrates) or after repeated irradiation in the laboratory (i.e. E. coli mutants), and may pave the way to better understand radiation resistance in cancer cells emerging after radiotherapy.
Since the description of D. radiodurans, many other radiation-resistant Deinococcus species have been isolated offering potential sources of new discoveries. In this paper, we explored this large biodiversity by analysing and comparing the genomes of 11 Deinococcus species. For this, we have investigated the conservation of more than 250 genes, including genes with a reported contribution to radiation or oxidative stress resistance in D. radiodurans and other genes with an expected role in radiation resistance-associated mechanisms such as DNA repair, oxidative stress defence and their regulation. Conservation was indeed observed for many genes encoding proteins that are also important or even essential in non-Deinococcus bacteria (RecA, SSB, GyrAB, SodA, etc.). Several proteins with currently unknown precise role are also present in each Deinococcus (e.g. RecD, glutaredoxin-like proteins, DdrC). Striking specificities also emerged, with a huge diversity with respect to the presence/absence or number of variants of radiation resistance-associated proteins. The radiation/desiccation response regulon is partly constituted of Deinococcus-specific proteins, of which only a few are conserved in each Deinococcus, including its regulator pair IrrE/DdrO (essential for radiation resistance) and DdrB (single-stranded DNA-binding protein). Several genes encoding DNA repair or oxidative stress response proteins are present in one or several of the more recently sequenced Deinococcus species but not in D. radiodurans (e.g. DNA repair and carotenoid biosynthesis proteins composed of novel two-domain combinations, endonuclease NucS, photolyase, TLS polymerases, SoxR, manganese-containing catalase). Conversely, and remarkably, dozens of genes encoding proteins with a reported contribution to radiation and oxidative stress resistance in D. radiodurans are absent in one, several or even most other Deinococcus species (e.g. the RNA or DNA ligases Rnl and LigB, PprA, PqqE, the RadS/RadR two-component system, one or both heme-containing catalases). The high sensitivity to IR of the pprA mutant in particular has been demonstrated in several independent studies. Why are these genes absent in other Deinococcus species? One could argue that the presence of these genes in D. radiodurans makes this species more radiation resistant than all the other Deinococcus species, but this seems unlikely. Indeed, D. radiodurans, D. geothermalis and D. actinosclerus are equally resistant to IR under identical experimental conditions (Makarova et al. 2007; Joo et al. 2016). The absence of a gene homologue in another Deinococcus species may be compensated by another gene that has little sequence similarity but may encode a protein with similar function, or the gene is not required because the species employs other molecular or regulatory mechanism(s). As some non-conserved genes contribute to radiation resistance in D. radiodurans, it is reasonable to propose that the other Deinococcus species also contain genes that have an important role in radiation resistance but which are absent in D. radiodurans or others. These may be genes of currently unknown function, for example genes under control of the important regulator pair IrrE/DdrO, or some of the additional DNA repair or oxidative stress defence genes identified in several of the Deinococcus species.
It is clear from this comprehensive analysis that there is not only one winning combination that leads to radiation resistance even in the Deinococcus genus. These bacteria not only possess common protection and repair systems but also molecular mechanisms that are different between species, including diversity in DNA repair mechanisms and oxidative stress response. These results open the way to deciphering new protein functions and new mechanisms.
Supplementary Material
Acknowledgements
We are grateful to David Pignol and Christophe Carles for constant support and interest.
FUNDING
This work was supported by the Nuclear Research & Development program of Ministry of Science and Information and Communications Technologies (Republic of Korea) to SL and JHJ, and by the transverse division number 4 of the French Alternative Energies and Atomic Energy Commission (Segment number 4 Radiobiology) to LB and AdG.
Conflict of interest. None declared.
REFERENCES
- Agapov AA, Kulbachinskiy AV. Mechanisms of stress resistance and gene regulation in the radioresistant bacterium Deinococcus radiodurans. Biochemistry 2015;80:1201–16. [DOI] [PubMed] [Google Scholar]
- Agostini HJ, Carroll JD, Minton KW. Identification and characterization of uvrA, a DNA repair gene of Deinococcus radiodurans. J Bacteriol 1996;178:6759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R, Sahoo BK, Saini DK. Cross-talk and specificity in two-component signal transduction pathways. Future Microbiol 2016;11:685–97. [DOI] [PubMed] [Google Scholar]
- Aguirre JD, Culotta VC. Battles with iron: manganese in oxidative stress protection. J Biol Chem 2012;287:13541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaganti N, Basu B, Mukhopadhyaya R et al. Proximity of radiation desiccation response motif to the core promoter is essential for basal repression as well as gamma radiation-induced gyrB gene expression in Deinococcus radiodurans. Gene 2017;615:8–17. [DOI] [PubMed] [Google Scholar]
- Anderson AW, Nordan HC, Cain RF et al. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol 1956;10:575–8. [Google Scholar]
- Appukuttan D, Seo HS, Jeong S et al. Expression and mutational analysis of DinB-like protein DR0053 in Deinococcus radiodurans. PLoS One 2015;10:e0118275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasik M, Stanislawska-Sachadyn A, Hildebrandt E. et al. In vitro affinity of Deinococcus radiodurans MutS towards mismatched DNA exceeds that of its orthologues from Escherichia coli and Thermus thermophilus. J Biotechnol 2017;252:55–64. [DOI] [PubMed] [Google Scholar]
- Basu B, Apte SK. A novel serralysin metalloprotease from Deinococcus radiodurans. Biochim Biophys Acta 2008;1784:1256–64. [DOI] [PubMed] [Google Scholar]
- Basu B, Apte SK. Gamma radiation-induced proteome of Deinococcus radiodurans primarily targets DNA repair and oxidative stress alleviation. Mol Cell Proteomics 2012;11:M111 011734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista JR. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol 1997;51:203–24. [DOI] [PubMed] [Google Scholar]
- Bentchikou E, Servant P, Coste G et al. A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet 2010;6:e1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett BS, Levine RL. Designing antioxidant peptides. Redox Report 2014;19:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Eggington JM, Killoran MP et al. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc Natl Acad Sci USA 2004;101:8575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihani SC, Panicker L, Rajpurohit YS et al. drFrnE represents a hitherto unknown class of eubacterial cytoplasmic disulfide oxido-reductases. Antioxid Redox Signal 2018;28:296–310. [DOI] [PubMed] [Google Scholar]
- Blanchard L, Guerin P, Roche D et al. Conservation and diversity of the IrrE/DdrO-controlled radiation response in radiation-resistant Deinococcus bacteria. MicrobiologyOpen 2017;6:e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius M, Buob R, Shevelev IV et al. Enzymes involved in DNA ligation and end-healing in the radioresistant bacterium Deinococcus radiodurans. BMC Mol Biol 2007;8:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacossa de Almeida C, Coste G, Sommer S et al. Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol Genet Genomics 2002;268:28–41. [DOI] [PubMed] [Google Scholar]
- Bouthier de la Tour C, Blanchard L, Dulermo R et al. The abundant and essential HU proteins in Deinococcus deserti and Deinococcus radiodurans are translated from leaderless mRNA. Microbiology 2015;161:2410–22. [DOI] [PubMed] [Google Scholar]
- Bouthier de la Tour C, Boisnard S, Norais C et al. The deinococcal DdrB protein is involved in an early step of DNA double strand break repair and in plasmid transformation through its single-strand annealing activity. DNA Repair (Amst) 2011;10:1223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C, Mathieu M, Meyer L. et al. In vivo and in vitro characterization of DdrC, a DNA damage response protein in Deinococcus radiodurans bacterium. PLoS One 2017;12:e0177751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C, Passot FM, Toueille M et al. Comparative proteomics reveals key proteins recruited at the nucleoid of Deinococcus after irradiation-induced DNA damage. Proteomics 2013;13:3457–69. [DOI] [PubMed] [Google Scholar]
- Bouthier de la Tour C, Toueille M, Jolivet E et al. The Deinococcus radiodurans SMC protein is dispensable for cell viability yet plays a role in DNA folding. Extremophiles 2009;13:827–37. [DOI] [PubMed] [Google Scholar]
- Brooks BW, Murray RGE. Nomenclature for "Micrococcus radiodurans" and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int J Syst Bacteriol 1981;31:353–60. [Google Scholar]
- Broxton CN, Culotta VC. SOD enzymes and microbial pathogens: surviving the oxidative storm of infection. PLoS Pathog 2016;12:e1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch CR, DiRuggiero J. MutS and MutL are dispensable for maintenance of the genomic mutation rate in the halophilic archaeon Halobacterium salinarum NRC-1. PLoS One 2010;5:e9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butala M, Klose D, Hodnik V et al. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res 2011;39:6546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Klingele AJ, Cabot EL et al. Evolution of extreme resistance to ionizing radiation via genetic adaptation of DNA repair. Elife 2014;3:e01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Picot CR, Friguet B et al. Methionine sulfoxide reductases: relevance to aging and protection against oxidative stress. Ann N Y Acad Sci 2006;1067:37–44. [DOI] [PubMed] [Google Scholar]
- Cao Z, Mueller CW, Julin DA. Analysis of the recJ gene and protein from Deinococcus radiodurans. DNA Repair (Amst) 2010;9:66–75. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol 2010;20:763–71. [DOI] [PubMed] [Google Scholar]
- Castaneda-Garcia A, Prieto AI, Rodriguez-Beltran J et al. A non-canonical mismatch repair pathway in prokaryotes. Nat Commun 2017;8:14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S, Srinivasan S, Seo T. et al. Deinococcus soli sp. nov., a gamma-radiation-resistant bacterium isolated from rice field soil. Curr Microbiol 2014;68:777–83. [DOI] [PubMed] [Google Scholar]
- Chandrangsu P, Loi VV, Antelmann H et al. The role of bacillithiol in Gram-positive Firmicutes. Antioxid Redox Signal 2018;28:445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 2017;15:338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wu R, Xu G et al. DR2539 is a novel DtxR-like regulator of Mn/Fe ion homeostasis and antioxidant enzyme in Deinococcus radiodurans. Biochem Bioph Res Co 2010;396:413–8. [DOI] [PubMed] [Google Scholar]
- Chen H, Xu G, Zhao Y et al. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One 2008;3:e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Wang H, Xu X et al. Characteristics of dr1790 disruptant and its functional analysis in Deinococcus radiodurans. Braz J Microbiol 2015;46:601–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Xu G, Xu H. et al. Deinococcus radiodurans DR1088 is a novel RecF-interacting protein that stimulates single-stranded DNA annealing. Mol Microbiol 2017;106:518–29. [DOI] [PubMed] [Google Scholar]
- Clarke TE, Romanov V, Chirgadze YN et al. Crystal structure of alkyl hydroperoxidase D like protein PA0269 from Pseudomonas aeruginosa: homology of the AhpD-like structural family. BMC Struct Biol 2011;11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet JF, D'Souza JC, Jakob U et al. Thioredoxin 2, an oxidative stress-induced protein, contains a high affinity zinc binding site. J Biol Chem 2003;278:45325–32. [DOI] [PubMed] [Google Scholar]
- Confalonieri F, Sommer S. Bacterial and archaeal resistance to ionizing radiation. J Phys: Conf Ser 2011;261:012005. [Google Scholar]
- Copeland A, Zeytun A, Yassawong M et al. Complete genome sequence of the orange-red pigmented, radioresistant Deinococcus proteolyticus type strain (MRPT). Stand Genomic Sci 2012;6:240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Battista JR. Deinococcus radiodurans - the consummate survivor. Nat Rev Microbiol 2005;3:882–92. [DOI] [PubMed] [Google Scholar]
- Culotta VC, Daly MJ. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid Redox Signal 2013;19:933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JU, Gray MJ, Jakob U. Protein quality control under oxidative stress conditions. J Mol Biol 2015;427:1549–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ. Death by protein damage in irradiated cells. DNA Repair (Amst) 2012;11:12–21. [DOI] [PubMed] [Google Scholar]
- Daly MJ, Gaidamakova EK, Matrosova VY et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 2004;306:1025–8. [DOI] [PubMed] [Google Scholar]
- Daly MJ, Gaidamakova EK, Matrosova VY et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol 2007;5:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Gaidamakova EK, Matrosova VY et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 2010;5:e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AD, Misra HS. Characterization of DRA0282 from Deinococcus radiodurans for its role in bacterial resistance to DNA damage. Microbiology 2011;157:2196–205. [DOI] [PubMed] [Google Scholar]
- Das AD, Misra HS. DR2417, a hypothetical protein characterized as a novel beta-CASP family nuclease in radiation resistant bacterium, Deinococcus radiodurans. Biochim Biophys Acta 2012;1820:1052–61. [DOI] [PubMed] [Google Scholar]
- de Groot A, Chapon V, Servant P. et al. Deinococcus deserti sp. nov., a gamma-radiation-tolerant bacterium isolated from the Sahara Desert. Int J Syst Evol Microbiol 2005;55:2441–6. [DOI] [PubMed] [Google Scholar]
- de Groot A, Dulermo R, Ortet P et al. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet 2009;5:e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot A, Roche D, Fernandez B et al. RNA sequencing and proteogenomics reveal the importance of leaderless mRNAs in the radiation-tolerant bacterium Deinococcus deserti. Genome Biol Evol 2014;6:932–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis RJ, Micossi E, McCarthy J et al. Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms. Acta Crystallogr F 2006;62:325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SS, Rajpurohit YS, Misra HS et al. Characterization of the role of the RadS/RadR two-component system in the radiation resistance of Deinococcus radiodurans. Microbiology 2011;157:2974–82. [DOI] [PubMed] [Google Scholar]
- Deutsch C, El Yacoubi B, de Crecy-Lagard V et al. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 2012;287:13666–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devigne A, Guerin P, Lisboa J et al. PprA protein is involved in chromosome segregation via its physical and functional interaction with dna gyrase in irradiated Deinococcus radiodurans bacteria. mSphere 2016;1:e00036–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devigne A, Ithurbide S, Bouthier de la Tour C et al. DdrO is an essential protein that regulates the radiation desiccation response and the apoptotic-like cell death in the radioresistant Deinococcus radiodurans bacterium. Mol Microbiol 2015;96:1069–84. [DOI] [PubMed] [Google Scholar]
- Devigne A, Mersaoui S, Bouthier-de-la-Tour C et al. The PprA protein is required for accurate cell division of gamma-irradiated Deinococcus radiodurans bacteria. DNA Repair (Amst) 2013;12:265–72. [DOI] [PubMed] [Google Scholar]
- Dubbs JM, Mongkolsuk S. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol 2012;194:5495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulermo R, Fochesato S, Blanchard L et al. Mutagenic lesion bypass and two functionally different RecA proteins in Deinococcus deserti. Mol Microbiol 2009;74:194–208. [DOI] [PubMed] [Google Scholar]
- Dulermo R, Onodera T, Coste G et al. Identification of new genes contributing to the extreme radioresistance of Deinococcus radiodurans using a Tn5-based transposon mutant library. PLoS One 2015;10:e0124358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Mohundro MM, Mian IS et al. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J Bacteriol 2002;184:6216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Rankin SK, Kim KP et al. Genetic evidence that the uvsE gene product of Deinococcus radiodurans R1 is a UV damage endonuclease. J Bacteriol 2002;184:1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erill I, Campoy S, Mazon G et al. Dispersal and regulation of an adaptive mutagenesis cassette in the bacteria domain. Nucleic Acids Res 2006;34:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezraty B, Gennaris A, Barras F et al. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 2017;15:385–96. [DOI] [PubMed] [Google Scholar]
- Farci D, Bowler MW, Kirkpatrick J et al. New features of the cell wall of the radio-resistant bacterium Deinococcus radiodurans. Biochim Biophys Acta 2014;1838:1978–84. [DOI] [PubMed] [Google Scholar]
- Faulkner MJ, Helmann JD. Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid Redox Signal 2011;15:175–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AC, Nobre MF, Rainey FA. et al. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int J Syst Bacteriol 1997;47:939–47. [DOI] [PubMed] [Google Scholar]
- Fillat MF. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 2014;546:41–52. [DOI] [PubMed] [Google Scholar]
- Fioravanti E, Dura MA, Lascoux D et al. Structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily. Biochemistry 2008;47:11581–9. [DOI] [PubMed] [Google Scholar]
- Fredrickson JK, Li SM, Gaidamakova EK et al. Protein oxidation: key to bacterial desiccation resistance? ISME J 2008;2:393–403. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Chi BK, Roberts AA et al. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 2014;21:357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Newton GL, Antelmann H et al. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci USA 2010;107:6482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Jiang R. cAMP receptor protein (CRP)-mediated resistance/tolerance in bacteria: mechanism and utilization in biotechnology. Appl Microbiol Biot 2015;99:4533–43. [DOI] [PubMed] [Google Scholar]
- Ghosal D, Omelchenko MV, Gaidamakova EK et al. How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev 2005;29:361–75. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Jakob U. Oxidative stress protection by polyphosphate—new roles for an old player. Curr Opin Microbiol 2015;24:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Tang H, Zhang L. Lycopene cyclase and phytoene synthase activities in the marine yeast Rhodosporidium diobovatum are encoded by a single gene crtYB. J Basic Microbiol 2014;54:1053–61. [DOI] [PubMed] [Google Scholar]
- Gutman PD, Fuchs P, Minton KW. Restoration of the DNA damage resistance of Deinococcus radiodurans DNA polymerase mutants by Escherichia coli DNA polymerase I and Klenow fragment. Mutat Res 1994;314:87–97. [DOI] [PubMed] [Google Scholar]
- Gutsche I, Vujicic-Zagar A, Siebert X et al. Complex oligomeric structure of a truncated form of DdrA: a protein required for the extreme radiotolerance of Deinococcus. Biochim Biophys Acta 2008;1784:1050–8. [DOI] [PubMed] [Google Scholar]
- Harris DR, Tanaka M, Saveliev SV et al. Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLoS Biol 2004;2:e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillion M, Antelmann H. Thiol-based redox switches in prokaryotes. Biol Chem 2015;396:415–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. Metal-specific control of gene expression mediated by Bradyrhizobium japonicum Mur and Escherichia coli Fur is determined by the cellular context. Mol Microbiol 2016;101:152–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HF, Ngo KV, Chitteni-Pattu S et al. Investigating Deinococcus radiodurans RecA protein filament formation on double-stranded DNA by a real-time single-molecule approach. Biochemistry 2011;50:8270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Hua Y. Improved complete genome sequence of the extremely radioresistant bacterium Deinococcus radiodurans R1 obtained using pacbio single-molecule sequencing. Genome Announc 2016;4:e00886–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Xu X, Li M et al. Three nth homologs are all required for efficient repair of spontaneous DNA damage in Deinococcus radiodurans. Extremophiles 2012;16:477–84. [DOI] [PubMed] [Google Scholar]
- Hua Y, Narumi I, Gao G et al. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem Bioph Res Co 2003;306:354–60. [DOI] [PubMed] [Google Scholar]
- Huang L, Hua X, Lu H et al. Three tandem HRDC domains have synergistic effect on the RecQ functions in Deinococcus radiodurans. DNA Repair (Amst) 2007;6:167–76. [DOI] [PubMed] [Google Scholar]
- Im S, Song D, Joe M et al. Comparative survival analysis of 12 histidine kinase mutants of Deinococcus radiodurans after exposure to DNA-damaging agents. Bioprocess Biosyst Eng 2013;36:781–9. [DOI] [PubMed] [Google Scholar]
- Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 2013;11:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Transcription factors that defend bacteria against reactive oxygen species. Annu Rev Microbiol 2015;69:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino S, Nishi Y, Oda S et al. Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea. Nucleic Acids Res 2016;44:2977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SW, Jung JH, Kim MK et al. The three catalases in Deinococcus radiodurans: only two show catalase activity. Biochem Bioph Res Co 2016a;469:443–8. [DOI] [PubMed] [Google Scholar]
- Jeong SW, Seo HS, Kim MK et al. PprM is necessary for up-regulation of katE1, encoding the major catalase of Deinococcus radiodurans, under unstressed culture conditions. J Microbiol 2016b;54:426–31. [DOI] [PubMed] [Google Scholar]
- Jiao J, Wang L, Xia W et al. Function and biochemical characterization of RecJ in Deinococcus radiodurans. DNA Repair (Amst) 2012;11:349–56. [DOI] [PubMed] [Google Scholar]
- Johnston AW, Todd JD, Curson AR et al. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals 2007;20:501–11. [DOI] [PubMed] [Google Scholar]
- Jolivet E, Lecointe F, Coste G et al. Limited concentration of RecA delays DNA double-strand break repair in Deinococcus radiodurans R1. Mol Microbiol 2006;59:338–49. [DOI] [PubMed] [Google Scholar]
- Joo ES, Kim EB, Jeon SH et al. Complete genome sequence of Deinococcus soli N5T, a gamma-radiation- resistant bacterium isolated from rice field in South Korea. J Biotechnol 2015;211:115–6. [DOI] [PubMed] [Google Scholar]
- Joo ES, Lee JJ, Kang MS. et al. Deinococcus actinosclerus sp. nov., a novel bacterium isolated from soil of a rocky hillside. Int J Syst Evol Microbiol 2016;66:1003–8. [DOI] [PubMed] [Google Scholar]
- Jordan A, Aslund F, Pontis E et al. Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J Biol Chem 1997;272:18044–50. [DOI] [PubMed] [Google Scholar]
- Kamble VA, Rajpurohit YS, Srivastava AK et al. Increased synthesis of signaling molecules coincides with reversible inhibition of nucleolytic activity during postirradiation recovery of Deinococcus radiodurans. FEMS Microbiol Lett 2010;303:18–25. [DOI] [PubMed] [Google Scholar]
- Karplus PA. A primer on peroxiredoxin biochemistry. Free Radic Biol Med 2015;80:183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keto-Timonen R, Hietala N, Palonen E et al. Cold shock proteins: a minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front Microbiol 2016;7:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairnar NP, Kamble VA, Mangoli SH et al. Involvement of a periplasmic protein kinase in DNA strand break repair and homologous recombination in Escherichia coli. Mol Microbiol 2007;65:294–304. [DOI] [PubMed] [Google Scholar]
- Khairnar NP, Kamble VA, Misra HS. RecBC enzyme overproduction affects UV and gamma radiation survival of Deinococcus radiodurans. DNA Repair (Amst) 2008;7:40–7. [DOI] [PubMed] [Google Scholar]
- Khairnar NP, Misra HS, Apte SK. Pyrroloquinoline–quinone synthesized in Escherichia coli by pyrroloquinoline–quinone synthase of Deinococcus radiodurans plays a role beyond mineral phosphate solubilization. Biochem Bioph Res Co 2003;312:303–8. [DOI] [PubMed] [Google Scholar]
- Killoran MP, Keck JL. Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J Biol Chem 2006;281:12849–57. [DOI] [PubMed] [Google Scholar]
- Kim JI, Cox MM. The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proc Natl Acad Sci USA 2002;99:7917–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Sharma AK, Abbott SN et al. RecA Protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. J Bacteriol 2002;184:1649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Jeong S, Lim S et al. Oxidative stress response of Deinococcus geothermalis via a cystine importer. J Microbiol 2017;55:137–46. [DOI] [PubMed] [Google Scholar]
- Kim MK, Kang MS, Lee DH et al. Complete genome sequence of Deinococcus actinosclerus BM2T, a bacterium with Gamma-radiation resistance isolated from soil in South Korea. J Biotechnol 2016;224:53–54. [DOI] [PubMed] [Google Scholar]
- Klinman JP, Bonnot F. Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem Rev 2014;114:4343–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobatake M, Tanabe S, Hasegawa S. New Micrococcus radioresistant red pigment, isolated from Lama glama feces, and its use as microbiological indicator of radiosterilization. C R Seances Soc Biol Fil 1973;167:1506–10. [PubMed] [Google Scholar]
- Koch G, Nadal-Jimenez P, Cool RH. et al. Deinococcus radiodurans can interfere with quorum sensing by producing an AHL-acylase and an AHL-lactonase. FEMS Microbiol Lett 2014;356:62–70. [DOI] [PubMed] [Google Scholar]
- Kooistra J, Haijema BJ, Venema G. The Bacillus subtilis addAB genes are fully functional in Escherichia coli. Mol Microbiol 1993;7:915–23. [DOI] [PubMed] [Google Scholar]
- Koshkin A, Nunn CM, Djordjevic S et al. The mechanism of Mycobacterium tuberculosis alkylhydroperoxidase AhpD as defined by mutagenesis, crystallography, and kinetics. J Biol Chem 2003;278:29502–8. [DOI] [PubMed] [Google Scholar]
- Kota S, Charaka VK, Misra HS. PprA, a pleiotropic protein for radioresistance, works through DNA gyrase and shows cellular dynamics during postirradiation recovery in Deinococcus radiodurans. J Genet 2014;93:349–54. [DOI] [PubMed] [Google Scholar]
- Kota S, Charaka VK, Ringgaard S et al. PprA contributes to Deinococcus radiodurans resistance to nalidixic acid, genome maintenance after DNA damage and interacts with deinococcal topoisomerases. PLoS One 2014;9:e85288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota S, Kamble VA, Rajpurohit YS et al. ATP-type DNA ligase requires other proteins for its activity in vitro and its operon components for radiation resistance in Deinococcus radiodurans in vivo. Biochem Cell Biol 2010;88:783–90. [DOI] [PubMed] [Google Scholar]
- Kota S, Kumar CV, Misra HS. Characterization of an ATP-regulated DNA-processing enzyme and thermotolerant phosphoesterase in the radioresistant bacterium Deinococcus radiodurans. Biochem J 2010;431:149–57. [DOI] [PubMed] [Google Scholar]
- Kota S, Misra HS. PprA: A protein implicated in radioresistance of Deinococcus radiodurans stimulates catalase activity in Escherichia coli. Appl Microbiol Biot 2006;72:790–6. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK et al. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev 1994;58:401–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci USA 2010;107:14373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Smole Z, Debret G et al. Unstructured hydrophilic sequences in prokaryotic proteomes correlate with dehydration tolerance and host association. J Mol Biol 2010;402:775–82. [DOI] [PubMed] [Google Scholar]
- LaGier MJ. Predicted cold shock proteins from the extremophilic bacterium Deinococcus maricopensis and related Deinococcus species. Int J Microbiol 2017;2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Lee HJ, Jang GS. et al. Deinococcus swuensis sp. nov., a gamma-radiation-resistant bacterium isolated from soil. J Microbiol 2013;51:305–11. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Srinivasan S, Lim S. et al. Deinococcus puniceus sp. nov., a bacterium isolated from soil-irradiated gamma radiation. Curr Microbiol 2015;70:464–9. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals 2007;20:485–99. [DOI] [PubMed] [Google Scholar]
- Letoffe S, Heuck G, Delepelaire P et al. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc Natl Acad Sci USA 2009;106:11719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Zaidman S, Englander J, Shimoni E et al. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 2003;299:254–6. [DOI] [PubMed] [Google Scholar]
- Li M, Sun H, Feng Q et al. Extracellular dGMP enhances Deinococcus radiodurans tolerance to oxidative stress. PLoS One 2013;8:e54420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Dai S, Tian B et al. DqsIR quorum sensing-mediated gene regulation of the extremophilic bacterium Deinococcus radiodurans in response to oxidative stress. Mol Microbiol 2016a;100:527–41. [DOI] [PubMed] [Google Scholar]
- Lin L, Li T, Dai S et al. Autoinducer-2 signaling is involved in regulation of stress-related genes of Deinococcus radiodurans. Arch Microbiol 2016b;198:43–51. [DOI] [PubMed] [Google Scholar]
- Liu C, Wang L, Li T et al. A PerR-like protein involved in response to oxidative stress in the extreme bacterium Deinococcus radiodurans. Biochem Bioph Res Co 2014;450:575–80. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhou J, Omelchenko MV et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci USA 2003;100:4191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi VV, Rossius M, Antelmann H. Redox regulation by reversible protein S-thiolation in bacteria. Front Microbiol 2015;6:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca GL, Barabote RD, Zlotopolski V et al. Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim Biophys Acta 2007;1768:1342–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JF, VanderSpek JC, Murphy JR. The src homology 3-like domain of the diphtheria toxin repressor (DtxR) modulates repressor activation through interaction with the ancillary metal ion-binding site. J Bacteriol 2003;185:2251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gao G, Xu G. et al. Deinococcus radiodurans PprI switches on DNA damage response and cellular survival networks after radiation damage. Mol Cell Proteomics 2009;8:481–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xia W, Chen H et al. Characterization of the role of DR0171 in transcriptional response to radiation in the extremely radioresistant bacterium Deinococcus radiodurans. Arch Microbiol 2011;193:741–50. [DOI] [PubMed] [Google Scholar]
- Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med 2014;66:75–87. [DOI] [PubMed] [Google Scholar]
- Luan H, Meng N, Fu J et al. Genome-wide transcriptome and antioxidant analyses on gamma-irradiated phases of Deinococcus radiodurans R1. PLoS One 2014;9:e85649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludanyi M, Blanchard L, Dulermo R et al. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol Microbiol 2014;94:434–49. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol R 2001;65:44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Omelchenko MV, Gaidamakova EK. et al. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS One 2007;2:e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattimore V, Battista JR. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol 1996;178:633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. The unfolding story of a redox chaperone. Cell 2012;148:843–4. [DOI] [PubMed] [Google Scholar]
- Meireles DA, Domingos RM, Gaiarsa JW et al. Functional and evolutionary characterization of Ohr proteins in eukaryotes reveals many active homologs among pathogenic fungi. Redox Biol 2017;12:600–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer L, Coste G, Sommer S et al. DdrI, a cAMP receptor protein family member, acts as a major regulator for adaptation of Deinococcus radiodurans to various stresses. J Bacteriol 2018;200:e00129–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F et al. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 2009;43:335–67. [DOI] [PubMed] [Google Scholar]
- Mishra S, Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys 2012;525:145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra HS, Khairnar NP, Barik A et al. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett 2004;578:26–30. [DOI] [PubMed] [Google Scholar]
- Misra HS, Khairnar NP, Kota S et al. An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol Microbiol 2006;59:1308–16. [DOI] [PubMed] [Google Scholar]
- Misra HS, Rajpurohit YS, Khairnar NP. Pyrroloquinoline-quinone and its versatile roles in biological processes. J Biosci 2012;37:313–25. [DOI] [PubMed] [Google Scholar]
- Moe E, Hall DR, Leiros I et al. Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans. Acta Crystallogr D 2012;68:703–12. [DOI] [PubMed] [Google Scholar]
- Moe E, Leiros I, Smalas AO et al. The crystal structure of mismatch-specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans reveals a novel catalytic residue and broad substrate specificity. J Biol Chem 2006;281:569–77. [DOI] [PubMed] [Google Scholar]
- Motomura K, Hirota R, Okada M et al. A new subfamily of polyphosphate kinase 2 (class III PPK2) catalyzes both nucleoside monophosphate phosphorylation and nucleoside diphosphate phosphorylation. Appl Environ Microb 2014;80:2602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi I, Satoh K, Cui S et al. PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol Microbiol 2004;54:278–85. [DOI] [PubMed] [Google Scholar]
- Narumi I, Satoh K, Kikuchi M et al. The LexA protein from Deinococcus radiodurans is not involved in RecA induction following gamma irradiation. J Bacteriol 2001;183:6951–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Leung SS, Wakabayashi JI et al. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry 2011;50:10751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Rawat M, La Clair JJ et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 2009;5:625–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KV, Molzberger ET, Chitteni-Pattu S et al. Regulation of Deinococcus radiodurans RecA protein function via modulation of active and inactive nucleoprotein filament states. J Biol Chem 2013;288:21351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HH, de la Tour CB, Toueille M et al. The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol Microbiol 2009;73:240–52. [DOI] [PubMed] [Google Scholar]
- Norais CA, Chitteni-Pattu S, Wood EA et al. DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J Biol Chem 2009;284:21402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiero J, Pittet V, Bonderoff SA et al. Thioredoxin system from Deinococcus radiodurans. J Bacteriol 2010;192:494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba H, Satoh K, Sghaier H et al. Identification of PprM: a modulator of the PprI-dependent DNA damage response in Deinococcus radiodurans. Extremophiles 2009;13:471–9. [DOI] [PubMed] [Google Scholar]
- Onodera T, Satoh K, Ohta T. et al. Deinococcus radiodurans YgjD and YeaZ are involved in the repair of DNA cross-links. Extremophiles 2013;17:171–9. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bassler BL. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol 2016;14:576–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Singh H, Appukuttan D et al. PprM, a cold shock domain-containing protein from Deinococcus radiodurans, confers oxidative stress tolerance to Escherichia coli. Front Microbiol 2017;7:2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera VR, Newton GL, Pogliano K. Bacillithiol: a key protective thiol in Staphylococcus aureus. Expert Rev Anti Infe 2015;13:1089–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Nelson KJ, Parsonage D et al. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci 2015;40:435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechura JR, Tseng TL, Hsu HF et al. Biochemical characterization of RecA variants that contribute to extreme resistance to ionizing radiation. DNA Repair (Amst) 2015;26:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobegalov G, Cherevatenko G, Alekseev A. et al. Deinococcus radiodurans RecA nucleoprotein filaments characterized at the single-molecule level with optical tweezers. Biochem Bioph Res Co 2015;466:426–30. [DOI] [PubMed] [Google Scholar]
- Pukall R, Zeytun A, Lucas S et al. Complete genome sequence of Deinococcus maricopensis type strain (LB-34T). Stand Genomic Sci 2011;4:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch I, Yanku M, Yeheskel A. et al. Staphylococcus aureus NrdH redoxin is a reductant of the class Ib ribonucleotide reductase. J Bacteriol 2010;192:4963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey FA, Ferreira M, Nobre MF. et al. Deinococcus peraridilitoris sp. nov., isolated from a coastal desert. Int J Syst Evol Microbiol 2007;57:1408–12. [DOI] [PubMed] [Google Scholar]
- Rainey FA, Ray K, Ferreira M et al. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl Environ Microb 2005;71:5225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit YS, Bihani SC, Waldor MK et al. Phosphorylation of Deinococcus radiodurans RecA regulates its activity and may contribute to radioresistance. J Biol Chem 2016;291:16672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit YS, Gopalakrishnan R, Misra HS. Involvement of a protein kinase activity inducer in DNA double strand break repair and radioresistance of Deinococcus radiodurans. J Bacteriol 2008;190:3948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit YS, Misra HS. Characterization of a DNA damage-inducible membrane protein kinase from Deinococcus radiodurans and its role in bacterial radioresistance and DNA strand break repair. Mol Microbiol 2010;77:1470–82. [DOI] [PubMed] [Google Scholar]
- Rajpurohit YS, Misra HS. Structure-function study of deinococcal serine/threonine protein kinase implicates its kinase activity and DNA repair protein phosphorylation roles in radioresistance of Deinococcus radiodurans. Int J Biochem Cell Biol 2013;45:2541–52. [DOI] [PubMed] [Google Scholar]
- Randi L, Perrone A, Maturi M et al. The DnaE polymerase from Deinococcus radiodurans features RecA-dependent DNA polymerase activity. Biosci Rep 2016;36:e00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 2009;78:605–47. [DOI] [PubMed] [Google Scholar]
- Reading NC, Sperandio V. Quorum sensing: the many languages of bacteria. FEMS Microbiol Lett 2006;254:1–11. [DOI] [PubMed] [Google Scholar]
- Reardon-Robinson ME, Ton-That H. Disulfide-bond-forming pathways in Gram-positive bacteria. J Bacteriol 2016;198:746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Kuhn J, Meslet-Cladiere L et al. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J 2009;28:2479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reon BJ, Nguyen KH, Bhattacharyya G et al. Functional comparison of Deinococcus radiodurans Dps proteins suggests distinct in vivo roles. Biochem J 2012;447:381–91. [DOI] [PubMed] [Google Scholar]
- Rezzonico F, Smits TH, Duffy B. Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors 2012;12:6645–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richarme G, Liu C, Mihoub M et al. Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science 2017;357:208–11. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet 2005;1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Aussel L, Ezraty B et al. Reprint of: Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 2013;1827:923–37. [DOI] [PubMed] [Google Scholar]
- Rosario-Cruz Z, Boyd JM. Physiological roles of bacillithiol in intracellular metal processing. Curr Genet 2016;62:59–65. [DOI] [PubMed] [Google Scholar]
- Sale JE. Radiation resistance: resurrection by recombination. Curr Biol 2007;17:R12–4. [DOI] [PubMed] [Google Scholar]
- Sandigursky M, Sandigursky S, Sonati P et al. Multiple uracil-DNA glycosylase activities in Deinococcus radiodurans. DNA Repair (Amst) 2004;3:163–9. [DOI] [PubMed] [Google Scholar]
- Santos SP, Cuypers MG, Round A et al. SAXS structural studies of Dps from Deinococcus radiodurans highlights the conformation of the mobile N-terminal extensions. J Mol Biol 2017;429:667–87. [DOI] [PubMed] [Google Scholar]
- Sarre A, Okvist M, Klar T et al. Structural and functional characterization of two unusual endonuclease III enzymes from Deinococcus radiodurans. J Struct Biol 2015;191:87–99. [DOI] [PubMed] [Google Scholar]
- Satoh K, Ohba H, Sghaier H et al. Down-regulation of radioresistance by LexA2 in Deinococcus radiodurans. Microbiology 2006;152:3217–26. [DOI] [PubMed] [Google Scholar]
- Schlesinger DJ. Role of RecA in DNA damage repair in Deinococcus radiodurans. FEMS Microbiol Lett 2007;274:342–7. [DOI] [PubMed] [Google Scholar]
- Schmier BJ, Chen X, Wolin S et al. Deletion of the rnl gene encoding a nick-sealing RNA ligase sensitizes Deinococcus radiodurans to ionizing radiation. Nucleic Acids Res 2017;45:3812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmier BJ, Shuman S. Deinococcus radiodurans HD-Pnk, a nucleic acid end-healing enzyme, abets resistance to killing by ionizing radiation and mitomycin C. J Bacteriol 2018;200:e00151–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvam K, Duncan JR, Tanaka M et al. DdrA, DdrD, and PprA: components of UV and mitomycin C resistance in Deinococcus radiodurans R1. PLoS One 2013;8:e69007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sghaier H, Satoh K, Ohba H et al. Assessing the role of RecA protein in the radioresistant bacterium Deinococcus geothermalis. Afr J Biochem Res 2010;4:111–8. [Google Scholar]
- Shashidhar R, Kumar SA, Misra HS et al. Evaluation of the role of enzymatic and nonenzymatic antioxidant systems in the radiation resistance of Deinococcus. Can J Microbiol 2010;56:195–201. [DOI] [PubMed] [Google Scholar]
- Sheng D, Zheng Z, Tian B et al. LexA analog (dra0074) is a regulatory protein that is irrelevant to recA induction. J Biochem (Tokyo) 2004;136:787–93. [DOI] [PubMed] [Google Scholar]
- Shin DH, Choi IG, Busso D et al. Structure of OsmC from Escherichia coli: a salt-shock-induced protein. Acta Crystallogr D 2004;60:903–11. [DOI] [PubMed] [Google Scholar]
- Shuryak I, Matrosova VY, Gaidamakova EK et al. Microbial cells can cooperate to resist high-level chronic ionizing radiation. PLoS One 2017;12:e0189261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si MR, Zhang L, Yang ZF et al. NrdH Redoxin enhances resistance to multiple oxidative stresses by acting as a peroxidase cofactor in Corynebacterium glutamicum. Appl Environ Microb 2014;80:1750–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D, Dunstan MS, Barkauskaite E et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 2011;477:616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D, Lindner AB, Paul G et al. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 2009;136:1044–55. [DOI] [PubMed] [Google Scholar]
- Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol R 2011;75:133–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL. The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol 2004;30:173–85. [DOI] [PubMed] [Google Scholar]
- Smolik AC, Bengez-Pudja L, Cheng I et al. Characterization of E. coli manganese superoxide dismutase binding to RNA and DNA. Biochim Biophys Acta 2014;1844:2251–6. [DOI] [PubMed] [Google Scholar]
- Stelter M, Acajjaoui S, McSweeney S et al. Structural and mechanistic insight into DNA unwinding by Deinococcus radiodurans UvrD. PLoS One 2013;8:e77364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll KE, Draper WE, Kliegman JI et al. Characterization and structure of the manganese-responsive transcriptional regulator ScaR. Biochemistry 2009;48:10308–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiman-Marangos S, Junop MS. The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins. Nucleic Acids Res 2010;38:3432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiman-Marangos SN, Weiss YM, Junop MS. Mechanism for accurate, protein-assisted DNA annealing by Deinococcus radiodurans DdrB. Proc Natl Acad Sci USA 2016;113:4308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li M, Xu G et al. Regulation of MntH by a dual Mn(II)- and Fe(II)-dependent transcriptional repressor (DR2539) in Deinococcus radiodurans. PLoS One 2012;7:e35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Xu G, Zhan H et al. Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC Microbiol 2010;10:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Earl AM, Howell HA et al. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 2004;168:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Narumi I, Funayama T et al. Characterization of pathways dependent on the uvsE, uvrA1, or uvrA2 gene product for UV resistance in Deinococcus radiodurans. J Bacteriol 2005;187:3693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hua Y. Carotenoid biosynthesis in extremophilic Deinococcus–Thermus bacteria. Trends Microbiol 2010;18:512–20. [DOI] [PubMed] [Google Scholar]
- Tian B, Sun Z, Shen S et al. Effects of carotenoids from Deinococcus radiodurans on protein oxidation. Lett Appl Microbiol 2009;49:689–94. [DOI] [PubMed] [Google Scholar]
- Tian B, Xu Z, Sun Z et al. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim Biophys Acta 2007;1770:902–11. [DOI] [PubMed] [Google Scholar]
- Timmins J, Gordon E, Caria S et al. Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs. Structure 2009;17:547–58. [DOI] [PubMed] [Google Scholar]
- Timmins J, Moe E. A Decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: Major findings, functional and mechanistic insight and challenges. Comput Struct Biotechnol J 2016;14:168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents E. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol 2014;4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul Hussain Shah AM, Zhao Y, Wang Y et al. A Mur regulator protein in the extremophilic bacterium Deinococcus radiodurans. PLoS One 2014;9:e106341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos A, Eslava AP, Iturriaga EA. A bifunctional enzyme with lycopene cyclase and phytoene synthase activities is encoded by the carRP gene of Mucor circinelloides. Eur J Biochem 2000;267:5509–19. [DOI] [PubMed] [Google Scholar]
- Vujicic-Zagar A, Dulermo R, Le Gorrec M et al. Crystal structure of the IrrE protein, a central regulator of DNA damage repair in Deinococcaceae. J Mol Biol 2009;386:704–16. [DOI] [PubMed] [Google Scholar]
- Wanarska M, Krawczyk B, Hildebrandt P et al. RecA proteins from Deinococcus geothermalis and Deinococcus murrayi - cloning, purification and biochemical characterisation. BMC Mol Biol 2011;12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian Y, Zhou Z et al. Identification and functional analysis of RelA/SpoT homolog (RSH) genes in Deinococcus radiodurans. J Microbiol Biotechn 2016a;26:2106–15. [DOI] [PubMed] [Google Scholar]
- Wang L, Hu J, Liu M et al. Proteomic insights into the functional basis for the response regulator DrRRA of Deinococcus radiodurans. Int J Radiat Biol 2016b;92:273–80. [DOI] [PubMed] [Google Scholar]
- Wang L, Xu G, Chen H et al. DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol Microbiol 2008;67:1211–22. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu Q, Lu H et al. Protease activity of PprI facilitates DNA damage response: Mn(2+)-dependence and substrate sequence-specificity of the proteolytic reaction. PLoS One 2015;10:e0122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfel JD, LiCata VJ. Enhanced DNA binding affinity of RecA protein from Deinococcus radiodurans. DNA Repair (Amst) 2015;31:91–96. [DOI] [PubMed] [Google Scholar]
- White O, Eisen JA, Heidelberg JF et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 1999;286:1571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Lu H, Wang L et al. DdrB stimulates single-stranded DNA annealing and facilitates RecA-independent DNA repair in Deinococcus radiodurans. DNA Repair (Amst) 2010;9:805–12. [DOI] [PubMed] [Google Scholar]
- Xu G, Wang L, Chen H et al. RecO is essential for DNA damage repair in Deinococcus radiodurans. J Bacteriol 2008;190:2624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Xu H, Wang J et al. Cyclic AMP receptor protein acts as a transcription regulator in response to stresses in Deinococcus radiodurans. PLoS One 2016;11:e0155010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang L, Lu H et al. DRA0336, another OxyR homolog, involved in the antioxidation mechanisms in Deinococcus radiodurans. J Microbiol 2010;48:473–9. [DOI] [PubMed] [Google Scholar]
- Yuan M, Chen M, Zhang W et al. Genome sequence and transcriptome analysis of the radioresistant bacterium Deinococcus gobiensis: insights into the extreme environmental adaptations. PLoS One 2012;7:e34458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Zhang W, Dai S. et al. Deinococcus gobiensis sp. nov., an extremely radiation-resistant bacterium. Int J Syst Evol Microbiol 2009;59:1513–7. [DOI] [PubMed] [Google Scholar]
- Zahradka K, Slade D, Bailone A et al. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 2006;443:569–73. [DOI] [PubMed] [Google Scholar]
- Zamocky M, Gasselhuber B, Furtmuller PG et al. Molecular evolution of hydrogen peroxide degrading enzymes. Arch Biochem Biophys 2012;525:131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K. Dps biomineralizing proteins: multifunctional architects of nature. Biochem J 2012;445:297–311. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ishige K, Kornberg A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc Natl Acad Sci USA 2002;99:16678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Baseman JB. Functional characterization of osmotically inducible protein C (MG_427) from Mycoplasma genitalium. J Bacteriol 2014;196:1012–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lu M, Zhang H et al. Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J. Nucleic Acids Res 2015;43:5550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang X, Xu H et al. RadA: A protein involved in DNA damage repair processes of Deinococcus radiodurans R1. Chin Sci Bull 2006;51:2993–9. [Google Scholar]
- Zhou Z, Zhang W, Su S et al. CYP287A1 is a carotenoid 2-beta-hydroxylase required for deinoxanthin biosynthesis in Deinococcus radiodurans R1. Appl Microbiol Biot 2015;99:10539–46. [DOI] [PubMed] [Google Scholar]
- Zimmerman JM, Battista JR. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol 2005;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.