The gammaproteobacterium Legionella pneumophila is the causative agent of Legionnaires’ disease, an atypical pneumonia that manifests itself with severe lung damage. L. pneumophila, a common inhabitant of freshwater environments, replicates in free-living amoebae and persists in biofilms in natural and man-made water systems.
KEYWORDS: Legionella pneumophila, PpiB, human lung tissue explants, intracellular infection, macrophage infectivity potentiator, motility, peptidyl-prolyl cis/trans isomerase, sliding, temperature adaptation
ABSTRACT
The gammaproteobacterium Legionella pneumophila is the causative agent of Legionnaires’ disease, an atypical pneumonia that manifests itself with severe lung damage. L. pneumophila, a common inhabitant of freshwater environments, replicates in free-living amoebae and persists in biofilms in natural and man-made water systems. Its environmental versatility is reflected in its ability to survive and grow within a broad temperature range as well as its capability to colonize and infect a wide range of hosts, including protozoa and humans. Peptidyl-prolyl-cis/trans-isomerases (PPIases) are multifunctional proteins that are mainly involved in protein folding and secretion in bacteria. In L. pneumophila the surface-associated PPIase Mip was shown to facilitate the establishment of the intracellular infection cycle in its early stages. The cytoplasmic PpiB was shown to promote cold tolerance. Here, we set out to analyze the interrelationship of these two relevant PPIases in the context of environmental fitness and infection. We demonstrate that the PPIases Mip and PpiB are important for surfactant-dependent sliding motility and adaptation to suboptimal temperatures, features that contribute to the environmental fitness of L. pneumophila. Furthermore, they contribute to infection of the natural host Acanthamoeba castellanii as well as human macrophages and human explanted lung tissue. These effects were additive in the case of sliding motility or synergistic in the case of temperature tolerance and infection, as assessed by the behavior of the double mutant. Accordingly, we propose that Mip and PpiB are virulence modulators of L. pneumophila with compensatory action and pleiotropic effects.
INTRODUCTION
The human pathogen Legionella pneumophila ubiquitously occurs in freshwater habitats, where it persists as a free-living bacterium and replicates intracellularly in amoebae residing mainly in biofilms (1–4). The transmission to humans occurs via man-made water systems that are colonized by L. pneumophila and its natural hosts. Legionella-containing aerosols that are generated by showers, cooling towers, and air-conditioning systems enable the entry of the bacteria into the human lung. Here, they replicate within human alveolar macrophages and destroy the lung tissue, causing Legionnaires’ disease, a severe atypical pneumonia with mortality rates of up to 15% (5, 6).
Being considered an accidental human pathogen, L. pneumophila stands out with its environmental versatility and remarkably broad protozoan host spectrum that extends to simple metazoans like nematodes as well as human alveolar macrophages (7–9). The host spectrum can mainly be attributed to a type IVB secretion system (TIVBSS) with its more than 300 described and putative effectors (10–13). However, the contribution of factors other than the TIVBSS and its associated effectors on environmental fitness and infection processes has been only rudimentarily evaluated. Among those factors are the flagellum; pili; a type II secretion system (TIISS); several hydrolytic enzymes, including a chitinase; outer membrane vesicles; and peptidyl-proly-cis/trans-isomerases (PPIases) (14–20).
PPIases constitute a superfamily of proteins that is divided into three structural and inhibitor-based classes, (i) cyclophilins, (ii) parvulins, and (iii) FK506-binding proteins (FKBPs), which are inhibited by cyclosporine (CsA), juglone, and FK506 or rapamycin, respectively (21, 22). Nevertheless, all members are characterized by their ability to convert peptidyl-prolyl bonds from cis to trans or vice versa, as in these bonds no intrinsic preference for either of the isomers exists due to the unique side chain of proline residues (23, 24). By this, they primarily contribute to protein folding, stability, and activity. Furthermore, many PPIases have been shown to be associated with virulence in pathogenic microorganisms (25).
L. pneumophila possesses six PPIases, two cyclophilins (PpiA and PpiB), two parvulins (PpiD and SurA), and two FKBPs (trigger factor and Mip). Among these, the macrophage infectivity potentiator (Mip) is the best-characterized member (21). This outer membrane-associated protein, with a size of 25 kDa and a homodimeric structure, is important during the early stages of intracellular replication in U937 cells, explanted human alveolar macrophages, and environmental amoebae like Acanthamoeba castellanii (26–29). Mip-deficient strains are also attenuated in the guinea pig infection model, failing to spread to organs like the spleen, where a relationship between the PPIase activity and the outcome of the infection was demonstrated (30). In accordance with this, Mip binds via its PPIase domain to collagen IV in the extracellular matrix (ECM) and thereby contributes to transmigration of the bacteria through a barrier consisting of NCI-H292 epithelial cells and ECM (31, 32). Moreover, a secreted phospholipase C activity was shown to be dependent on Mip (33).
PpiB of L. pneumophila was shown to accumulate in culture supernatants and to contribute to growth at 17°C (34). Infection studies in A. castellanii showed that PpiB, previously also known as Lcy, has a weak effect on intracellular replication (35). Considering the impact of Mip on Legionella replication and the preliminary work on PpiB regarding the stress response as well as a possible implication in infection, we attempted to extend our knowledge relating to the involvement of the PPIases Mip and PpiB of L. pneumophila in extracellular fitness and virulence by using single- and double-knockout mutants.
RESULTS
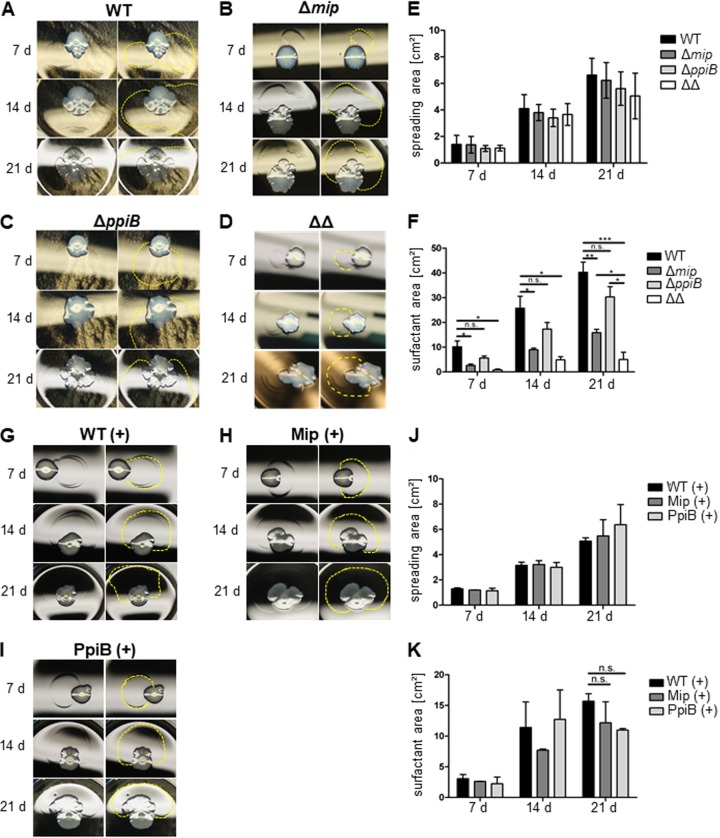
The PPIases Mip and PpiB together facilitate surfactant formation and surface translocation of L. pneumophila.
L. pneumophila displays sliding motility through a flagellum- and pilus-independent surface translocation mechanism. It was shown that the type I secretion system (TISS) and the TIISS contribute to the secretion of surfactant, which promotes surface translocation (36). Since PPIases can also be involved in secretion processes in bacteria, we aimed at clarifying whether the PPIases of L. pneumophila also play a role in surfactant-mediated surface translocation (33, 37, 38). For this, an mip-negative strain (the Δmip strain), a ppiB-negative strain (the ΔppiB strain), and an mip- and ppiB-negative double mutant strain (the Δmip ΔppiB strain) were tested for their spreading phenotype on 0.5% (wt/vol) agar containing buffered charcoal yeast extract (BCYE) plates over 21 days at 30°C (Fig. 1A to E). Over the course of the experiment, all strains displayed comparable surface spreading with no significant differences. However, deletion of mip resulted in a reduction of the surfactant area by 60% (Fig. 1B and F). In the absence of ppiB, this area was reduced by 25% compared to that for the wild type; however, this difference was not statistically significant (Fig. 1C and F). Interestingly, deleting both PPIases resulted in a surfactant film 90% smaller than that of the wild type after 21 days (Fig. 1D and F). This phenotype was not dependent on different growth rates, as the surface spreading of all mutants was comparable over the course of the experiment (Fig. 1E and F). Introduction of the respective genes into the mip or ppiB deletion mutants reverted the phenotype of surfactant film formation (Fig. 1G to I) and resulted in comparable spreading and surfactant film areas in all strains (Fig. 1J and K). In order to exclude the possibility of the involvement of the second annotated cyclophilin of L. pneumophila, PpiA, in sliding motility, we also created a ppiA single deletion mutant and a ppiA ppiB double deletion mutant. The ΔppiA mutant was as effective as the wild type in forming a surfactant film, and the ΔppiA ΔppiB double mutant showed no significant difference from the ΔppiB single mutant (see Fig. S1 in the supplemental material).
FIG 1.
The surface translocation of L. pneumophila depends on Mip and PpiB. (A to D) L. pneumophila wild-type (WT) (A), Δmip (B), ΔppiB (C), and Δmip ΔppiB (ΔΔ) (D) strains were grown on BCYE plates containing 0.5% agar at 30°C for 7, 14, and 21 days (d). For each time point and strain, a representative picture from three independent experiments performed in triplicate is presented on the left, while the boundaries of the surfactant film are highlighted on the right. (E) Quantification of the spreading area of the strains revealed no significant differences over 21 days. (F) In the case of the surfactant area, PPIase-negative Δmip and Δmip ΔppiB strains showed a significant decrease in the secretion of surfactants. (G to I) The defect in the formation of a surfactant film was complemented by introducing the respective gene in the single-gene-knockout mutants. (J and K) This was confirmed by quantitative measurement of the spreading and surfactant film areas. Shown are the means and standard deviations from three experiments performed in duplicate. Statistical significance was calculated using an unpaired Student's t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., not significant).
In L. pneumophila, the biosynthesis of surfactant depends on two separate operons, bbcABCDEF and bbcGHIJK (36). In order to evaluate whether the deletion of mip and/or ppiB influences the expression of these gene clusters, we performed quantitative reverse transcription-PCR (qRT-PCR), using primers targeting the overlapping regions of the first two genes in the respective operons, on RNA isolated from bacteria that were grown on 0.5% (wt/vol) agar containing BCYE medium. The comparison of the target mRNA concentration by the threshold cycle (CT) method showed that in all PPIase mutants the transcript level for both operons was negatively affected. However, the levels of the bbcABCDEF and bbcGHIJK transcripts were significantly reduced by only approximately half in the Δmip ΔppiB mutant (Fig. 2).
FIG 2.
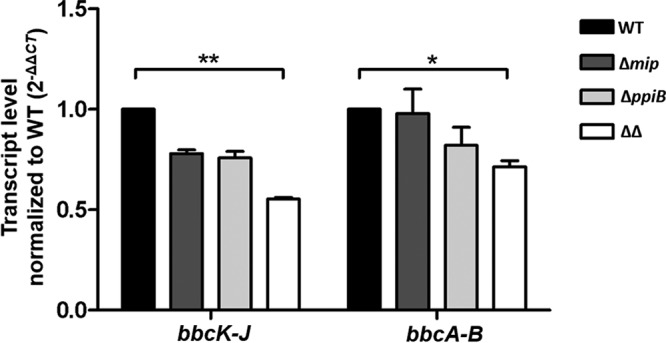
Transcription of the bbc loci is affected by Mip and PpiB. Twenty nanograms of total RNA isolated from each strain grown on low-agar BCYE plates was subjected to qRT-PCR. As assessed by the transcript level normalized to that for the wild type, a significant decrease in the transcript level for both surfactant biosynthesis operons was measured in the Δmip ΔppiB mutant. Shown are the mean and standard deviation of three independent measurements from at least two separate RNA isolations. Statistical significance was calculated using an unpaired Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01).
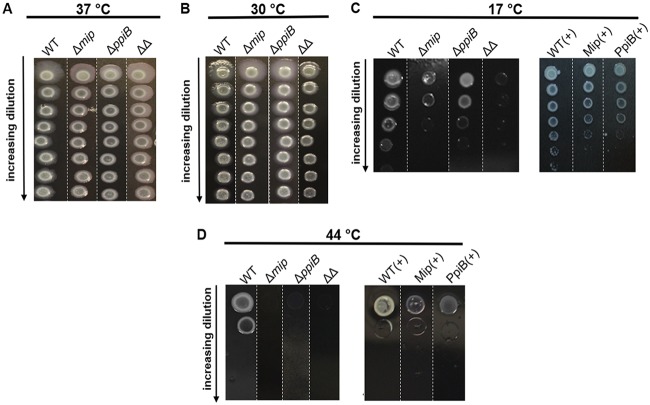
Mip and PpiB confer tolerance to suboptimal temperatures in L. pneumophila.
The cyclophilin PpiB of L. pneumophila was found to promote growth at low temperatures (34). In this work, we analyzed if the PPIase Mip also plays a role in growth at low temperatures. Moreover, we tested whether PpiB and Mip influence growth at high temperatures. For this, serial dilutions of each strain were applied onto BCYE agar plates and incubated at various temperatures. No growth difference was observed between the knockout strains and their isogenic wild type at 37°C or 30°C (Fig. 3A and B). However, at 17°C, both the Δmip and ΔppiB strains displayed a reduced growth capability compared to that of the wild type. While the single deletion mutants were affected in their growth to the same degree, the Δmip ΔppiB double mutant showed almost no growth at the highest bacterial density (Fig. 3C). Complementing the single deletion mutants with the respective gene reverted the phenotype to a level comparable to that for the wild type (Fig. 3C). At 44°C, deleting either of the PPIases caused a severe growth deficiency, which could again be complemented by the reintroduction of mip or ppiB (Fig. 3D). Similar to the findings for sliding motility, no substantial differences in growth at different temperatures, with the exception of that at 44°C, were observed between the ΔppiA and the ΔppiA ΔppiB mutants. The mutant lacking both cyclophilins was also impaired in its growth at high temperatures (Fig. S2).
FIG 3.
Growth of different L. pneumophila PPIase-negative mutants at suboptimal temperatures. Bacteria grown in liquid culture were spotted in increasing 10-fold dilutions onto BCYE agar starting at 109 bacteria/ml and grown at 37°C (A), 30°C (B), 17°C (C), or 44°C (D). Shown are representative pictures from three separate experiments performed in triplicate. Representative sections of bacterial plates from three separate experiments are shown and aligned for a better comparison of the strains. Single sections originating from different plates are separated by white dashed lines.
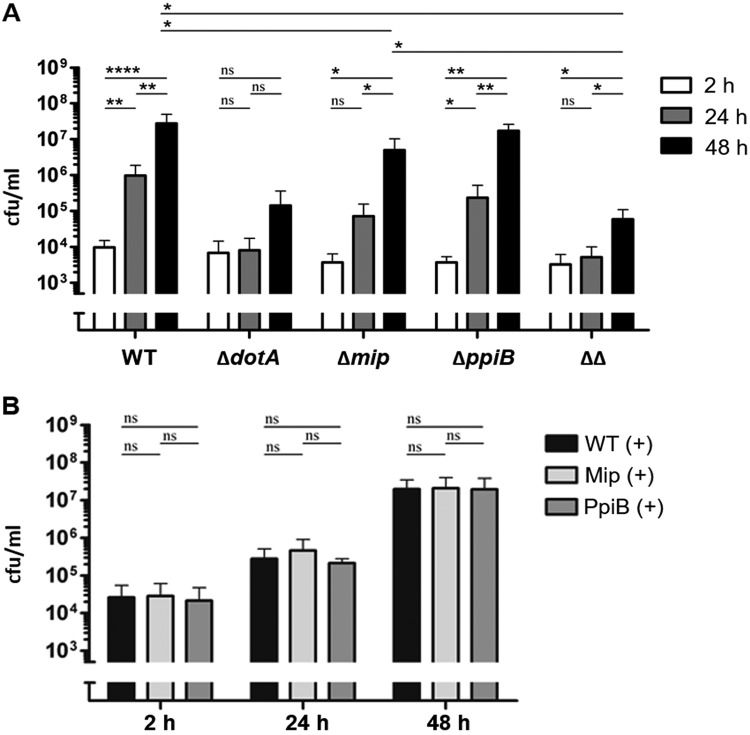
Intracellular replication in the environmental host A. castellanii depends on the concerted action of PPIases Mip and PpiB.
In previous studies, decreased intracellular growth in A. castellanii was shown for the Mip-negative mutant strain (39, 40), while a ppiB-negative strain had a reduced rate of intracellular replication due to a decrease in invasion (35). In this study, we analyzed if the Δmip ΔppiB mutant strain shows an enhanced effect in intracellular replication compared to the single mutant strains. As a control, the ΔdotA type IV secretion system-deficient mutant strain was used (Fig. 4). We could confirm the reduced intracellular replication for the Δmip strain (Fig. 4A). After 24 h of replication, the number of CFU of the ΔppiB strain was reduced 5 times compared to that of the wild type and was 2.9 times higher than the number of CFU of the Δmip strain. After 24 h the double-knockout strain showed a 14 times reduced number of CFU compared to that of the mip deletion strain, and after 48 h it showed a 200 times reduced number of CFU compared to that of the mip deletion strain (Fig. 4A). In its final outcome, the replication of the double mutant strain was comparable to that of the avirulent ΔdotA-negative mutant strain. Complementation of the strains in trans restored the intracellular replication ability (Fig. 4B). Deleting ppiA had no detrimental effect on the intracellular replication of L. pneumophila in A. castellanii (Fig. S3A).
FIG 4.
Replication of PPIase mutant strains in A. castellanii. A. castellanii was infected with different L. pneumophila mutant strains. After 2 h, 24 h, and 48 h, the amoebae were lysed and dilutions were plated out on BCYE agar plates. (A) Infection with the wild-type strain (WT), the ΔdotA mutant (ΔdotA), the mip-negative strain (Δmip), the ppiB-negative strain (ΔppiB), and the mip- and ppiB-negative double mutant strain (ΔΔ). The PPIase mutants showed decreased intracellular growth. (B) Complementing the single-knockout mutants with the respective gene restored the wild-type phenotype. Shown are the means and standard deviations from three experiments performed in triplicate. Statistical significance was calculated by an unpaired Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001; n.s., not significant).
Mip and PpiB contribute to bacterial replication in HLTEs and THP-1 macrophages.
In order to evaluate the impact of Mip and PpiB in human lung tissue, we infected tumor-free human lung tissue explants (HLTEs) from five different donors with the wild-type, ΔdotA, Δmip, ΔppiB, or Δmip ΔppiB strain. The infected tissue was weighed, homogenized, and plated on BCYE medium at different time points. After 24 h, all mutant strains grew more slowly than the wild-type strain. This trend continued over the following 24 h, whereby the ΔppiB mutant reached a higher number of CFU per gram of tissue than the Δmip mutant and the Δmip ΔppiB mutant. The wild-type strain replicated at 48 h postinfection (p.i.), on average, 17.5-fold, whereas the Δmip mutant replicated only 4-fold, the ΔppiB mutant 5.7-fold, and the Δmip ΔppiB mutant 6.6-fold (Fig. 5A).
FIG 5.
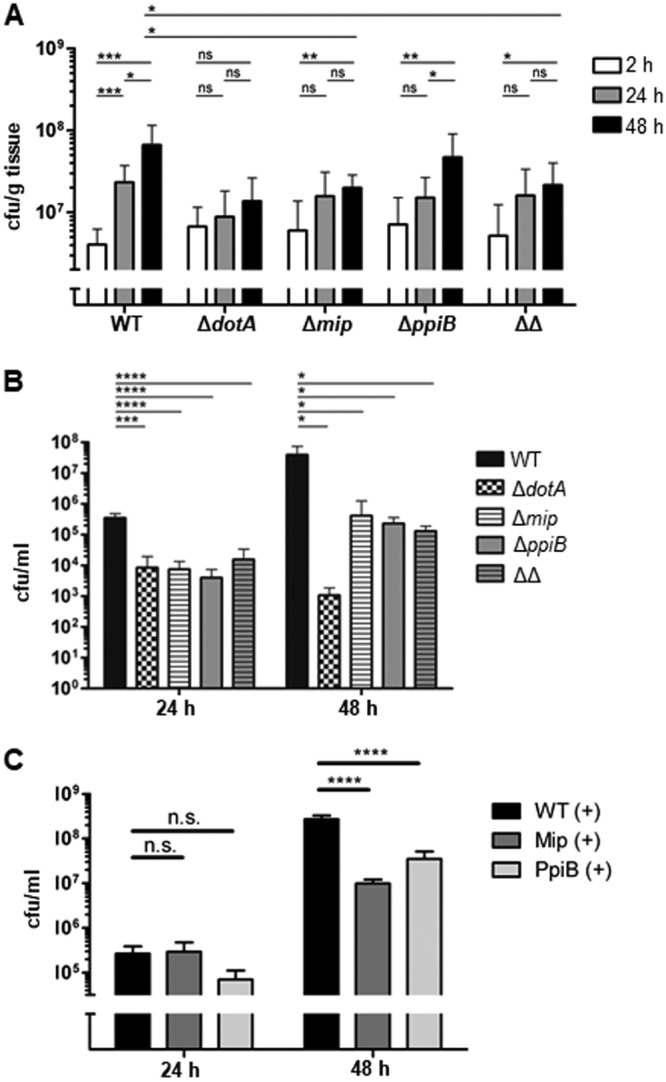
Replication of PPIase mutant strains in human lung tissue explants (HLTEs) and THP-1 macrophages. (A) HLTEs were infected with the wild-type strain (WT) and the ΔdotA, Δmip, ΔppiB, and Δmip ΔppiB (ΔΔ) L. pneumophila mutants. Shown are the means and standard deviations from experiments performed in duplicate with tissue from five different donors. (B) Infection of THP-1 macrophages with wild-type, ΔdotA, Δmip, ΔppiB, and Δmip ΔppiB (ΔΔ) L. pneumophila strains. PPIase mutants showed decreased intracellular growth. Shown are the means and standard deviations from three experiments performed in triplicate. Statistical significance was calculated by an unpaired Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., not significant).
Alveolar macrophages constitute the main host cell type for L. pneumophila in human lungs (34). Accordingly, in order to verify whether the observed replication defect in HLTEs is due to the impairment in the infection of macrophages, we tested the PPIase mutants in the human macrophage-like cell line THP-1. The comparison of the total replication at 48 h p.i. showed that, although they were not as attenuated as the ΔdotA mutant, all PPIase mutants were impaired in their intracellular replication. The Δmip mutant replicated about 100 times less than the wild type, while the ΔppiB mutant was about 200 times less replicative in the macrophages than the wild type. Interestingly, the strongest replication defect was observed in the Δmip ΔppiB mutant with, on average, a 400 times reduced final replication rate (Fig. 5B). The complementation of the mutant strains with the respective gene restored the growth in the initial 24 h (Fig. 5C). Also, in contrast to the deletion of ppiB, the deletion of ppiA had no effect on the intracellular replication of L. pneumophila in THP-1 cells (Fig. S3B).
DISCUSSION
Initially, PPIases were postulated and subsequently identified to be enzymes that catalyze the cis/trans isomerization of peptidyl-prolyl bonds (23, 24, 41). Nowadays, it is known that the functions of PPIases are not restricted to this isomerization alone but also cover folding, transport, and the activity of proteins by means of chaperoning and protein-protein interactions (25). By this, they are connected to many physiological processes and diseases (42). Nevertheless, the decryption of the physiological role of PPIases is difficult due to their pleiotropic phenotypes and the absence of strong phenotypes in single deletion mutants of the respective PPIases (21, 43–45). The latter suggests that their activity is redundant with that of other PPIases and, hence, may be compensated for fully or partially by the activity of the others PPIases (42).
The Mip protein of L. pneumophila was the first microbial PPIase shown to promote virulence and as such is a prototypic PPIase (14, 29, 32). Studies about the contribution of the other five PPIases to the virulence and fitness of L. pneumophila have remained preliminary. Only the cyclophilin PpiB and, to a lesser extent, its homolog, PpiA, have been analyzed in the context of cold adaptation, as the former accumulates in culture supernatants of L. pneumophila isolates that are grown at 17°C (34). Accordingly, we aimed in our current study at expanding our understanding of the influence of PpiB on extra- and intracellular virulence-related attributes, including temperature tolerance, sliding motility, and infection of protozoan and human hosts. Also, we evaluated to what extent PpiB and Mip are redundant in their action and can compensate for the absence of the other.
We first focused on the extracellular virulence properties sliding motility and temperature sensitivity. Motility is an important feature that contributes to the colonization and dissemination of bacteria in different habitats, and several PPIases were shown to be involved in bacterial motility. The parvulin PrsA2 contributes to the swimming motility of Listeria monocytogenes, as a PrsA2-deficient mutant displays reduced motility (46). A similar influence was observed for the Mip-like protein in Burkholderia pseudomallei during swarming motility (47). Recently, PpiB of Escherichiacoli was shown to be a negative regulator of swarming and swimming motility as well as biofilm formation, and several cytosolic interaction partners that reverted the observed phenotypes to varying degrees were identified (48). However, for none of these bacteria are the molecular mechanisms by which the corresponding PPIase affects the degree of motility understood. In this study, we could show that the sliding motility of L. pneumophila is promoted by the FKBP Mip and the cyclophilin PpiB. Sliding motility is a special kind of bacterial movement that occurs on a secreted surfactant film where a reduced surface tension is utilized in order to facilitate passive spreading (49, 50). Hence, our studies show for the first time that PPIases influence flagellum-independent motility in L. pneumophila.
Deleting Mip or PpiB individually resulted in a significant decrease of the surfactant film size which was stronger than the reduction in colony size. Among the single mutants, the Δmip strain produced a smaller surfactant film than the ΔppiB strain. Interestingly, deletion of both PPIases resulted in the smallest surfactant film, which suggests that both PPIases contribute in an additive manner to the formation of the surfactant film. In L. pneumophila, TolC-dependent secretion and the TIISS were shown to act jointly, where the lipids forming the surfactant were secreted in a TolC-dependent manner, while the TIISS delivered the signal for secretion (36). Accordingly, it would be interesting for future studies to determine whether Mip or PpiB influences the formation of the surfactant film by affecting secretion systems or translocated factors. Interestingly, for the first time we were able to show that deleting mip and ppiB also has transcriptional consequences, as the transcript levels of the two main biosynthetic gene clusters were significantly reduced in the mutant lacking both genes. Until now, no link between bacterial PPIases and gene regulation has been reported, to the best of our knowledge. Here is the question: is the downregulation directly dependent on one or both of the PPIases, or is it a result of secondary effects due to other, PPIase-dependent but not yet identified regulators?
Next, we monitored the influence of Mip and PpiB on growth at suboptimal temperatures. The temperature optimum for L. pneumophila growth is 37°C, but the bacteria are versatile in this respect, since they can colonize freshwater habitats as well as technical water systems covering a broad temperature range (9, 51). At temperatures above 44°C, L. pneumophila forfeits its proliferative and respiratory capacity. At between 48.7 and 50°C, proliferation stops, but the metabolic activity still remains (52). In this study, we showed that both Mip and PpiB are important for the growth of L. pneumophila at 17 and 44°C. In the case of growth at 17°C, Mip and PpiB most likely compensate for the deficiency of each other, as single deletion mutants were affected to the same degree in their ability to grow. Only the deletion of both PPIases resulted in complete growth failure. This finding is supported by the findings of previous studies, where an accumulation of PpiB in the culture supernatants of L. pneumophila at 17°C was observed (34). At elevated temperatures, on the other hand, both PPIases seem to be equally important, as deleting either one of them diminished bacterial growth completely. The involvement of PPIases in growth at suboptimal temperatures is also known from other species. The PPIase PpiB from Bacillus subtilis works as a cold shock protein (53). On the contrary, PpiD of Escherichia coli plays a role under heat shock conditions. Additionally, the deletion of PpiD leads to an upregulation of the σE stress response due to an accumulation of misfolded proteins in the periplasm (54). Furthermore, in previous studies with E. coli, a functional redundancy of the FKBP FkpA and the parvulin SurA was shown. These PPIases with chaperone activity play an important role in outer membrane protein biogenesis and compensate for each other in order to circumvent unfolded protein stress under heat shock (55).
Previous studies already established the importance of Mip in the early phases of the intracellular infection cycle in amoebae as well as human macrophages and in a guinea pig infection model (29, 30, 39). Here, we show that out of the two PPIases, Mip seems to be more relevant for the infection of a wide range of hosts, including the complex human lung tissue, which is demonstrated here for the first time. PpiB on its own seems to have a minor role or might be fully compensated for by Mip in the context of the intracellular infection of amoebae. However, especially in the case of the natural host A. castellanii, a clear additive effect of both PPIases on infection efficiency could be observed, as the double-knockout mutant was severely impaired in its replication. Surfactant production is dispensable for infecting amoebae, human macrophages, or epithelial cells as well as A/J mice (36). Hence, attenuation of the PPIase mutants in infection models must be due to additional defects in protein folding or secretion. Thus, the fact that the replication levels of the Δmip ΔppiB mutant remained comparable to those of the ΔdotA TIVSS-deficient mutant raises the question of whether there might be an interconnection between these PPIases and the type IV secretion apparatus or the effectors that are secreted by it.
Taken together, we show for the first time that the PPIases Mip and PpiB together are important for extra- and intracellular virulence properties. They do so either in a synergistic manner, as in the case of sliding motility, or in a compensatory manner, as in the case of cold adaptation. This, furthermore, impacts the ability of L. pneumophila to infect its host cells and has implications for the course and outcome of the human lung infection.
MATERIALS AND METHODS
Bacterial strains and culture.
L. pneumophila Corby and all mutant strains (listed in Table 1) were cultured in buffered yeast extract broth (YEB) to the early stationary phase at 37°C and 200 rpm or on buffered charcoal yeast extract (BCYE) agar at 37°C for 3 days (30). When needed, 12.5 µg/ml chloramphenicol or 20 µg/ml kanamycin was used for selection.
TABLE 1.
Wild-type strains and mutant L. pneumophila strains used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| L. pneumophila Corby | Wild-type strain | 61 |
| L. pneumophila Corby ΔdotA | dotA::Tn5 mutant of L. pneumophila Corby | Kindly provided by Antje Flieger, Robert-Koch-Institut, Wernigerode, Germany |
| L. pneumophila Corby Δmip | mip::nptI mutant of L. pneumophila Corby | 40 |
| L. pneumophila Corby ΔppiB | ppiB::nptI mutant of L. pneumophila Corby | This study |
| L. pneumophila Corby Δmip ΔppiB (ΔΔ) | mip::nptI and ppiB::cat mutant of L. pneumophila Corby | This study |
| L. pneumophila Corby ΔppiA | ppiA::aac1 mutant of L. pneumophila Corby | This study |
| L. pneumophila Corby ΔppiA ΔppiB | ppiA::aac1 and ppiB::nptI mutant of L. pneumophila Corby | This study |
| Complemented strains | ||
| WT(+) | Wild-type strain carrying empty pBCKS plasmid | This study |
| Mip(+) | mip::nptI mutant of L. pneumophila Corby carrying pEWM103 | 40 |
| PpiB(+) |
ppiB::nptI mutant of L. pneumophila Corby carrying pJRA1 ppiB
cloned in pBCKS with SacI and KpnI |
This study |
| pBCKS complementation vector | pUC ori Cmr (cat) | Stratagene (62) |
Construction of isogenic L. pneumophila knockout mutants.
For analysis of L. pneumophila PpiB, isogenic knockout mutants of the wild type and the Mip mutant strain were generated via homologous recombination as previously described (56). Up- and downstream flanking regions of ppiB with overhangs of 1 to 1.5 kb complementary to the 5′ and 3′ regions of the open reading frame of the respective resistance gene were amplified and merged together in a joining PCR. Naturally competent bacteria were transformed by adding 2 µg of the purified linear knockout construct to 1 ml of a bacterial suspension with an optical density at 600 nm (OD600) of 0.8 to 1.0 and incubating at 30°C for 3 days. Depending on the selection marker, transformants were selected on BCYE agar containing 20 µg/ml kanamycin and 12.5 µg/ml chloramphenicol or 4 µg/ml gentamicin. The resulting clones were checked via PCR and sequencing.
RNA isolation and qRT-PCR.
For comparing the expression levels of genes that are involved in surfactant production, the L. pneumophila Corby wild type and its isogenic mutants were grown on BCYE plates with 0.5% (wt/vol) agar for 3 weeks at 30°C. The RNA of the bacteria was isolated, using an RNeasy kit from Qiagen, by resuspending a loopful of bacteria in 350 µl RLT+ buffer (Qiagen) and proceeding by following the instructions in the manufacturer’s manual. The expression levels of the bbc operons, which are responsible for the production of Legionella surfactant, were compared between the strains by quantitative reverse transcription-PCR (qRT-PCR). For this, 20 ng total RNA of each strain was reverse transcribed and amplified in 40 cycles using a Luna Universal one-step reverse transcription-quantitative PCR kit (NEB) following the manufacturer’s instructions. The primer sequences for the reverse transcription of parts of the bbc operons were published previously (36) (Table 2). Primers targeting gapA (the gene for glyceraldehyde 3-phosphate dehydrogenase) were used in order to confirm template uniformity between samples and in order to calculate the relative change in gene expression as the ratio of normalized target concentrations by the threshold cycle (ΔΔCT) method (57).
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence | Reference or source |
|---|---|---|
| PpiB_F1 | CATGAGCTCCTGATGATTTGGCACCTGTTATG | This study |
| PpiB_R1 | AAGGTACCAGGAAGATCCCTCTTCCATACT | This study |
| PpiB_Cm_F | ATGGAGAAAAAAATCACTGGA | This study |
| PpiB_Cm_R | TTACGCCCCGCCCTG | This study |
| PpiB_UFF | ATTCCTGGCATTAAACGCGTG | This study |
| PpiB_DFR | CCTGCGACAAATAGTATTGCAG | This study |
| PpiB_UFR_Kan | GTTTCCCGTTGAATATGGCTCATCATGGATGTAGAAATTAAAACCATTATT | This study |
| PpiB_DFF_Kan | ATTTGATGCTCGATGAGTTTTTCTAAGAGGTTAGAGAACTGGCTTAG | This study |
| PpiB_UFR_Cm | TCCAGTGATTTTTTTCTCCATCATGGATGTAGAAATTAAAACCATTATT | This study |
| PpiB_DFF_Cm | CAGGGCGGGGCGTAAGAGGTTAGAGAACTGGCTTAG | This study |
| PpiA_UFF | GGCAAAGGCGATAAGTCATACTC | This study |
| PpiA_UFR_Gm | CATCGTTGCTGCTGCGTAACATAATAAGCTCTCCATAGAATGGCTAAG | This study |
| PpiA_DFF_Gm | GACCCAAGTACCGCCACCTAAAAGATCAGTGCCTGTGTCAGATTAG | This study |
| PpiA_DFR | CGAAAAAGAGTTGGTTAATGAGCC | This study |
| PpiA_Gm_F | ATGTTACGCAGCAGCAACGATG | This study |
| PpiA_Gm_R | TTAGGTGGCGGTACTTGGGTC | This study |
| Lpc1693_rtF | CTGGCGAAGGCTCTCACTCA | 36 |
| Lpc1694_rtR | TCACGCAGCACAAATTCGCA | 36 |
| Lpc1699_rtF | CAGTCAGTGGTGGTCTGCCT | 36 |
| Lpc1700_rtR | TGTACGAGAGGGCTGCTTGG | 36 |
| gapA_rtF | CGCATCGCAAATCCGTCCAA | This study |
| gapA_rtR | GCGTGATCCAGCCAAACTGC | This study |
Sliding motility.
Sliding motility assays with the L. pneumophila wild type and its isogenic mutant strains were performed as described previously (58). All strains were cultured to the early stationary phase (OD600, 2.8 to 3), and 10 µl of the cultures was spotted on BCYE agar plates containing 0.5% agar and incubated at 30°C for 7, 14, and 21 days. To assess the impact of PPIases on the surface translocation of L. pneumophila, images were taken with a Canon 450D camera and an EFS 18- to 55-mm macro lens. The sizes of the surfactant film area and colony area in relation to the area of the petri dish were calculated using Adobe Photoshop CS5 extended (version 12.0.4).
Growth temperature assay.
The growth of the L. pneumophila wild type and its isogenic PPIase mutants at different temperatures was monitored as previously described (34). Briefly, stationary-phase cultures were adjusted to 1 × 109 bacteria/ml and diluted to 108, 107, 106, 105 104, 103, and 102 bacteria/ml. An amount of 10 µl of all dilutions was dropped on a BCYE agar plate and incubated for 3 to 5 days at 17°C, 37°C, and 44°C. Growth on the agar plates was documented by taking pictures with a Canon 450D camera and an EFS 18- to 55-mm macro lens.
Infection of Acanthamoeba castellanii with L. pneumophila.
A. castellanii (ATCC 30234) was grown and passaged every 3 to 4 days in peptone-yeast extract-glucose medium at 22°C and was used for infection as previously described, with slight modifications (40). To analyze the intracellular replication of the L. pneumophila mutant strains in A. castellanii, 5 ml of the amoeba was seeded into 25-cm2 cell culture flasks (TPP) at a concentration of 5 × 105 cells/ml in amoeba buffer and infected at a multiplicity of infection (MOI) of 0.5. After 2, 24, and 48 h, the cells were detached by knocking the flask. A 200-µl aliquot was transferred from each flask into 1.5-ml tubes, and the cells were lysed mechanically by centrifuging at 20,000 × g for 5 min, followed by vigorous vortexing for 15 s. Serial dilutions were prepared with water and plated on BCYE agar plates. Bacterial colonies were counted after cultivation for 4 days at 37°C, and the number of CFU per milliliter was plotted in relation to time.
Infection of a THP-1 macrophage-like cell line with L. pneumophila.
To analyze the intracellular replication of L. pneumophila mutant strains in human macrophages, the acute monocytic leukemia cell line THP-1 (DSMZ ACC16) was used (59). The cells were adjusted to 5 × 105 cells/ml in cell culture medium (RPMI 1640, 2 mM l-glutamine, 10% fetal calf serum [FCS]) supplemented with 100 nM phorbol-12-myristate-13-acetate (PMA) for differentiation into macrophage-like cells. Two hundred microliters of this suspension was seeded into each well of a 96-well plate (TPP) and incubated at 37°C in 5% CO2 for 48 h. Early-stationary-phase L. pneumophila wild-type and mutant strains were adjusted to 1 × 106 bacteria/ml in cell culture medium. Differentiated THP-1 cells were washed one time with prewarmed cell culture medium and infected at an MOI of 1. After 2, 24, and 48 h, the cells were lysed by adding Triton X-100 at a final concentration of 0.1% (vol/vol), and several dilutions were plated on BCYE agar plates and incubated at 37°C for 4 days. Bacterial colonies were counted, and the numbers of CFU per milliliter were plotted in relation to time.
Infection of human tissue lung explants (HLTEs) with L. pneumophila.
Lung infections were performed as described previously (60). Briefly, tumor-free pulmonary tissue samples were obtained from surgery patients and infected with the L. pneumophila strains. For infection, the bacteria were adjusted to 107 bacteria/ml in RPMI 1640 (Gibco, Darmstadt, Germany) with 10% fetal calf serum (FCS), 20 mM HEPES, and 1 mM sodium pyruvate. The samples were incubated at 37°C in 5% CO2 for 2, 24, and 48 h. For determination of the number of CFU, duplicate samples from five donors were infected. At the indicated time points, samples were weighed and homogenized in deionized water. Dilutions were plated on BCYE agar and incubated at 37°C with 5% CO2 for 4 days. The number of CFU per gram of tissue was calculated; means and standard deviations of the results for samples were compared by using an unpaired Student’s t test.
Ethics statement.
This study was conducted in accordance with the Helsinki Declaration and was approved by the Ethics Committee of the Hannover Medical School (no. 2235-2014).
Supplementary Material
ACKNOWLEDGMENT
This work received financial support from the Deutsche Forschungsgemeinschaft (DFG; STE 838/8-1).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00939-17.
REFERENCES
- 1.Alleron L, Merlet N, Lacombe C, Frère J. 2008. Long-term survival of Legionella pneumophila in the viable but nonculturable state after monochloramine treatment. Curr Microbiol 57:497–502. doi: 10.1007/s00284-008-9275-9. [DOI] [PubMed] [Google Scholar]
- 2.Atlas RM. 1999. Legionella: from environmental habitats to disease pathology, detection and control. Environ Microbiol 1:283–293. doi: 10.1046/j.1462-2920.1999.00046.x. [DOI] [PubMed] [Google Scholar]
- 3.Declerck P. 2010. Biofilms: the environmental playground of Legionella pneumophila. Environ Microbiol 12:557–566. doi: 10.1111/j.1462-2920.2009.02025.x. [DOI] [PubMed] [Google Scholar]
- 4.Taylor M, Ross K, Bentham R. 2009. Legionella, protozoa, and biofilms: interactions within complex microbial systems. Microb Ecol 58:538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- 5.Muder RR, Yu VL. 2002. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis 35:990–998. doi: 10.1086/342884. [DOI] [PubMed] [Google Scholar]
- 6.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, Chereshsky A. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis 186:127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 7.Brassinga AKC, Kinchen JM, Cupp ME, Day SR, Hoffman PS, Sifri CD. 2010. Caenorhabditis is a metazoan host for Legionella. Cell Microbiol 12:343–361. doi: 10.1111/j.1462-5822.2009.01398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilbi H, Weber S, Finsel I. 2011. Anchors for effectors: subversion of phosphoinositide lipids by Legionella. Front Microbiol 2:91. doi: 10.3389/fmicb.2011.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasch J, Krüger S, Fontvieille D, Ünal CM, Michel R, Labrosse A, Steinert M. 2016. Legionella-protozoa-nematode interactions in aquatic biofilms and influence of Mip on Caenorhabditis elegans colonization. Int J Med Microbiol 306:443–451. doi: 10.1016/j.ijmm.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 12.Schroeder GN. 2017. The toolbox for uncovering the functions of Legionella Dot/Icm type IVb secretion system effectors: current state and future directions. Front Cell Infect Microbiol 7:528. doi: 10.3389/fcimb.2017.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Valero L, Rusniok C, Rolando M, Neou M, Dervins-Ravault D, Demirtas J, Rouy Z, Moore RJ, Chen H, Petty NK, Jarraud S, Etienne J, Steinert M, Heuner K, Gribaldo S, Médigue C, Glöckner G, Hartland EL, Buchrieser C. 2014. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome Biol 15:505. doi: 10.1186/PREACCEPT-1086350395137407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer G, Bang H, Ludwig B, Mann K, Hacker J. 1992. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPIase) activity. Mol Microbiol 6:1375–1383. doi: 10.1111/j.1365-2958.1992.tb00858.x. [DOI] [PubMed] [Google Scholar]
- 15.Pruckler JM, Benson RF, Moyenuddin M, Martin WT, Fields BS. 1995. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect Immun 63:4928–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone BJ, Abu Kwaik Y. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun 66:1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossier O, Starkenburg SR, Cianciotto NP. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infect Immun 72:310–321. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galka F, Wai SN, Kusch H, Engelmann S, Hecker M, Schmeck B, Hippenstiel S, Uhlin BE, Steinert M. 2008. Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect Immun 76:1825–1836. doi: 10.1128/IAI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhle K, Flieger A. 2013. Legionella phospholipases implicated in virulence. Curr Top Microbiol Immunol 376:175–209. doi: 10.1007/82_2013_348. [DOI] [PubMed] [Google Scholar]
- 20.DebRoy S, Dao J, Söderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103:19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasch J, Ünal CM, Steinert M. 2014. Peptidylprolyl cis-trans isomerases of Legionella pneumophila: virulence, moonlighting and novel therapeutic targets. Biochem Soc Trans 42:1728–1733. doi: 10.1042/BST20140202. [DOI] [PubMed] [Google Scholar]
- 22.Schiene-Fischer C, Aumüller T, Fischer G. 2013. Peptide bond cis/trans isomerases: a biocatalysis perspective of conformational dynamics in proteins. Top Curr Chem 328:35–67. doi: 10.1007/128_2011_151. [DOI] [PubMed] [Google Scholar]
- 23.Fischer G, Bang H. 1985. The refolding of urea-denatured ribonuclease A is catalyzed by peptidyl-prolyl cis-trans isomerase. Biochim Biophys Acta 828:39–42. doi: 10.1016/0167-4838(85)90006-8. [DOI] [PubMed] [Google Scholar]
- 24.Lang K, Schmid FX, Fischer G. 1987. Catalysis of protein folding by prolyl isomerase. Nature 329:268–270. doi: 10.1038/329268a0. [DOI] [PubMed] [Google Scholar]
- 25.Ünal CM, Steinert M. 2014. Microbial peptidyl-prolyl cis/trans isomerases (PPIases): virulence factors and potential alternative drug targets. Microbiol Mol Biol Rev 78:544–571. doi: 10.1128/MMBR.00015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riboldi-Tunnicliffe A, König B, Jessen S, Weiss MS, Rahfeld J, Hacker J, Fischer G, Hilgenfeld R. 2001. Crystal structure of Mip, a prolylisomerase from Legionella pneumophila. Nat Struct Biol 8:779–783. doi: 10.1038/nsb0901-779. [DOI] [PubMed] [Google Scholar]
- 27.Engleberg NC, Carter C, Weber DR, Cianciotto NP, Eisenstein BI. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect Immun 57:1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helbig JH, König B, Knospe H, Bubert B, Yu C, Lück CP, Riboldi-Tunnicliffe A, Hilgenfeld R, Jacobs E, Hacker J, Fischer G. 2003. The PPIase active site of Legionella pneumophila Mip protein is involved in the infection of eukaryotic host cells. Biol Chem 384:125–137. doi: 10.1515/BC.2003.013. [DOI] [PubMed] [Google Scholar]
- 29.Cianciotto NP, Eisenstein BI, Mody CH, Engleberg NC. 1990. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis 162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 30.Köhler R, Fanghänel J, König B, Lüneberg E, Frosch M, Rahfeld J-U, Hilgenfeld R, Fischer G, Hacker J, Steinert M. 2003. Biochemical and functional analyses of the Mip protein: influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila. Infect Immun 71:4389–4397. doi: 10.1128/IAI.71.8.4389-4397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner C, Khan AS, Kamphausen T, Schmausser B, Unal C, Lorenz U, Fischer G, Hacker J, Steinert M. 2007. Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell Microbiol 9:450–462. doi: 10.1111/j.1462-5822.2006.00802.x. [DOI] [PubMed] [Google Scholar]
- 32.Ünal C, Schwedhelm KF, Thiele A, Weiwad M, Schweimer K, Frese F, Fischer G, Hacker J, Faber C, Steinert M. 2011. Collagen IV-derived peptide binds hydrophobic cavity of Legionella pneumophila Mip and interferes with bacterial epithelial transmigration. Cell Microbiol 13:1558–1572. doi: 10.1111/j.1462-5822.2011.01641.x. [DOI] [PubMed] [Google Scholar]
- 33.Debroy S, Aragon V, Kurtz S, Cianciotto NP. 2006. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect Immun 74:5152–5160. doi: 10.1128/IAI.00484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Söderberg MA, Cianciotto NP. 2008. A Legionella pneumophila peptidyl-prolyl cis-trans isomerase present in culture supernatants is necessary for optimal growth at low temperatures. Appl Environ Microbiol 74:1634–1638. doi: 10.1128/AEM.02512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt B, Tradler T, Rahfeld JU, Ludwig B, Jain B, Mann K, Rücknagel KP, Janowski B, Schierhorn A, Küllertz G, Hacker J, Fischer G. 1996. A cyclophilin-like peptidyl-prolyl cis/trans isomerase from Legionella pneumophila—characterization, molecular cloning and overexpression. Mol Microbiol 21:1147–1160. doi: 10.1046/j.1365-2958.1996.00061.x. [DOI] [PubMed] [Google Scholar]
- 36.Stewart CR, Burnside DM, Cianciotto NP. 2011. The surfactant of Legionella pneumophila is secreted in a TolC-dependent manner and is antagonistic toward other Legionella species. J Bacteriol 193:5971–5984. doi: 10.1128/JB.05405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Perez F, Henderson IR, Nataro JP. 2010. Interaction of FkpA, a peptidyl-prolyl cis/trans isomerase with EspP autotransporter protein. Gut Microbes 1:339–344. doi: 10.4161/gmic.1.5.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlapschy M, Skerra A. 2011. Periplasmic chaperones used to enhance functional secretion of proteins in E. coli. Methods Mol Biol 705:211–224. doi: 10.1007/978-1-61737-967-3_12. [DOI] [PubMed] [Google Scholar]
- 39.Cianciotto NP, Fields BS. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci U S A 89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, Hacker J. 1995. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect Immun 63:4576–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer G, Bang H, Mech C. 1984. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed Biochim Acta 43:1101–1111. [PubMed] [Google Scholar]
- 42.Dunyak BM, Gestwicki JE. 2016. Peptidyl-proline isomerases (PPIases): targets for natural products and natural product-inspired compounds. J Med Chem 59:9622–9644. doi: 10.1021/acs.jmedchem.6b00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolinski K, Muir S, Cardenas M, Heitman J. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obi IR, Nordfelth R, Francis MS. 2011. Varying dependency of periplasmic peptidylprolyl cis-trans isomerases in promoting Yersinia pseudotuberculosis stress tolerance and pathogenicity. Biochem J 439:321–332. doi: 10.1042/BJ20110767. [DOI] [PubMed] [Google Scholar]
- 45.Shou W, Aghdasi B, Armstrong DL, Guo Q, Bao S, Charng MJ, Mathews LM, Schneider MD, Hamilton SL, Matzuk MM. 1998. Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature 391:489–492. doi: 10.1038/35146. [DOI] [PubMed] [Google Scholar]
- 46.Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol 191:3950–3964. doi: 10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norville IH, Harmer NJ, Harding SV, Fischer G, Keith KE, Brown KA, Sarkar-Tyson M, Titball RW. 2011. A Burkholderia pseudomallei macrophage infectivity potentiator-like protein has rapamycin-inhibitable peptidylprolyl isomerase activity and pleiotropic effects on virulence. Infect Immun 79:4299–4307. doi: 10.1128/IAI.00134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skagia A, Zografou C, Vezyri E, Venieraki A, Katinakis P, Dimou M. 2016. Cyclophilin PpiB is involved in motility and biofilm formation via its functional association with certain proteins. Genes Cells 21:833–851. doi: 10.1111/gtc.12383. [DOI] [PubMed] [Google Scholar]
- 49.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 50.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fields BS, Benson RF, Besser RE. 2002. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kusnetsov JM, Ottoila E, Martikainen PJ. 1996. Growth, respiration and survival of Legionella pneumophila at high temperatures. J Appl Bacteriol 81:341–347. [DOI] [PubMed] [Google Scholar]
- 53.Graumann P, Schröder K, Schmid R, Marahiel MA. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol 178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dartigalongue C, Raina S. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J 17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge X, Lyu Z-X, Liu Y, Wang R, Zhao XS, Fu X, Chang Z. 2014. Identification of FkpA as a key quality control factor for the biogenesis of outer membrane proteins under heat shock conditions. J Bacteriol 196:672–680. doi: 10.1128/JB.01069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoppe J, Ünal CM, Thiem S, Grimpe L, Goldmann T, Gaßler N, Richter M, Shevchuk O, Steinert M. 2017. PilY1 promotes Legionella pneumophila infection of human lung tissue explants and contributes to bacterial adhesion, host cell invasion, and twitching motility. Front Cell Infect Microbiol 7:63. doi: 10.3389/fcimb.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Stewart CR, Rossier O, Cianciotto NP. 2009. Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J Bacteriol 191:1537–1546. doi: 10.1128/JB.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasch J, Theuerkorn M, Ünal C, Heinsohn N, Tran S, Fischer G, Weiwad M, Steinert M. 2015. Novel cycloheximide derivatives targeting the moonlighting protein Mip exhibit specific antimicrobial activity against Legionella pneumophila. Front Bioeng Biotechnol 3:41. doi: 10.3389/fbioe.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jäger J, Marwitz S, Tiefenau J, Rasch J, Shevchuk O, Kugler C, Goldmann T, Steinert M. 2014. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect Immun 82:275–285. doi: 10.1128/IAI.00703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jepras RI, Fitzgeorge RB, Baskerville A. 1985. A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea-pigs. J Hyg (Lond) 95:29–38. doi: 10.1017/S0022172400062252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobi S, Schade R, Heuner K. 2004. Characterization of the alternative sigma factor sigma54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J Bacteriol 186:2540–2547. doi: 10.1128/JB.186.9.2540-2547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.