Schistosome worms infect over 200 million people worldwide. They live in the host’s bloodstream and alter host immunity.
KEYWORDS: schistosomiasis, transcriptomics
ABSTRACT
Schistosome worms infect over 200 million people worldwide. They live in the host’s bloodstream and alter host immunity. Epidemiological data suggest that males and females have different responses to schistosome infection, but the effect of sex on systemic response is undetermined. Our objective was to characterize differences in peripheral blood transcriptional profiles in people with or without active Schistosoma haematobium infection and to determine whether this signature differs between males and females. mRNA was isolated using poly(A) selection and sequenced on an Illumina Hi-Seq4000 platform. Transcripts were aligned to the human hg19 reference genome and counted with the HTSeq package. Genes were compared for differential expression using DESeq2. Ingenuity Pathway Analysis (IPA) was used to identify gene networks altered in the presence of S. haematobium. We enrolled 33 participants from villages in rural Tanzania where S. haematobium is endemic. After correction for multiple comparisons, we observed 383 differentially expressed genes between those with or without S. haematobium infection when sex was included as a covariate. Heat-mapping of the genes with >1.5-fold differences in gene expression revealed clustering by S. haematobium infection status. The top networks included development, cell death and survival, cell signaling, and immunologic disease pathways. We observed a distinct whole blood transcriptional profile, as well as differences in men and women, with S. haematobium infection. Additional studies are needed to determine the clinical effects of these divergent responses. Attention to sex-based differences should be included in studies of human schistosome infection.
INTRODUCTION
Schistosome worms infect over 200 million people worldwide. The highest burden of disease is in Africa, where 90% of infections occur (1). Schistosomiasis is a chronic infection. The parasitic worms live in host blood vessels and induce upregulation of Th 17 cells, T regulatory cells, and monocytes systemically (2–5). Schistosome eggs laid by adult worms migrate into host organs and mucosal tissues, where they provoke a chronic inflammatory granulomatous response locally, with subsequent systemic responses (6). Chronic schistosomiasis leads to long-term scarring and fibrosis in the organs most affected by egg migration, typically the bladder, kidneys, and genital organs (Schistosoma haematobium) and the intestine and liver (S. mansoni and S. japonicum).
In our recent work in Tanzania, we reported that when men and women had the same schistosome worm burden as quantified by serum schistosome antigen testing, women excreted fewer parasite eggs than did men (7). Whether differential responses to schistosome infection by sex are mediated by local or systemic changes, or a combination of both, is not clear. Significantly, our group showed that schistosome infection increased rates of HIV acquisition in Tanzanian women, but not men (8). Few studies have investigated whether the sex of the infected host affects the host response to schistosome infection.
We hypothesized that gene expression in whole blood would be impacted by schistosome infection and that these effects would differ with the sex of the host. Specifically, we hypothesized that women would exhibit an increased immune response to schistosome infection compared to men and that the genes that would be differentially expressed in those with versus without S. haematobium infection would be related to inflammation and host immunity.
RESULTS
Between April and July 2015, we provided screening for schistosome and HIV infections to 322 people and invited 48 men and women of reproductive age (18 to 45 years) living in four villages in rural northwest Tanzania to participate in this study. Four of the invited people were excluded in the initial screening. One person had S. mansoni eggs in the stool, two did not consent to all study procedures, and one was tested and found to be pregnant after reporting a delayed last menstrual period. Among the remaining 44 individuals, we excluded 3 people who had discordant circulating anodic antigen (CAA) and microscopy results and 5 who had inconclusive CAA values. Among the remaining 36 individuals, all met the threshold for RNA quality to continue to transcriptome sequencing (RNA-Seq). In total, 36 people had RNA-Seq completed. Gene expression in three of these samples, as indicated by the library size, was significantly lower than the other 33 samples, and these were removed from further analysis (Fig. 1).
FIG 1.
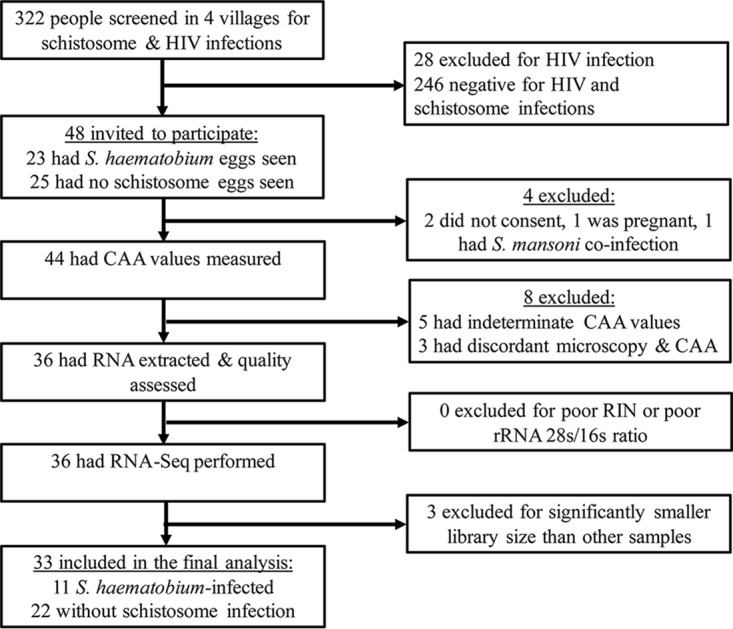
Flow chart of study screening and enrollment.
Therefore, in total, we analyzed peripheral blood gene expression in 33 individuals: 6 women and 5 men with S. haematobium infection and 14 women and 8 men without schistosome infection. All women were premenopausal. There were no significant demographic or clinical differences between those with and those without S. haematobium infection (Table 1). Men more frequently reported ever receiving prior antischistosome treatment (54% versus 15%, P = 0.03). Only one man and one woman had received treatment for schistosome infection in the past 5 years. After log transformation [log10(CAA+1)], the median CAA values were slightly higher in males than in females (1.03 versus 0.84, rank sum P = 0.027). Within the 11 schistosome-infected people, the log10 CAA levels were also higher in males than females (4.04 versus 3.72, rank sum P = 0.001).
TABLE 1.
Demographic and clinical characteristics of study participants
| Characteristic | No. (%) or median [IQR] |
P (for difference) | |
|---|---|---|---|
| S. haematobium infected (n = 11) | S. haematobium uninfected (n = 22) | ||
| Mean age in yrs | 25 [21–38] | 29 [25–33] | 0.42 |
| Female gender | 6 (54.6) | 14 (63.6) | 0.61 |
| Currently breastfeeding | 3/6 (50.0) | 5/14 (38.5) | 0.64 |
| Marital status | |||
| Married | 9 (81.8) | 18 (81.8) | |
| Single/divorced/widowed | 2 (18.2) | 4 (18.2) | 1.0 |
| Yrs of school completed | 7 [2–7] | 7 [0–7] | 0.96 |
| History of receiving treatment for schistosome infection | 4 (36.4) | 6 (27.3) | 0.70 |
| C. trachomatis PCR positive (genital tract/urine) | 1 (9.1) | 2 (9.1) | 1.0 |
| N. gonorrhoeae PCR positive (genital tract/urine) | 0 | 1 (4.5) | 1.0 |
| S. haematobium ova/10 ml of urine | 4 [2–13] | 0 | <0.001 |
| Serum schistosome circulating anodic antigen (pg/ml)a | 919 [327–50,000] | 2 [0–6] | <0.001 |
The schistosome-infected group included individuals who had serum antigen levels of ≥40 pg/ml, and the uninfected group included those with a CAA of <25 pg/ml.
Principal component analyses of the overall gene expression suggested separation between men and women (see Fig. S1 in the supplemental material). There were 29 differentially expressed genes with adjusted P values of <0.05 in males and 2,142 genes with adjusted P values of <0.05 in females. Because of these findings, subsequent analysis included sex as a covariate.
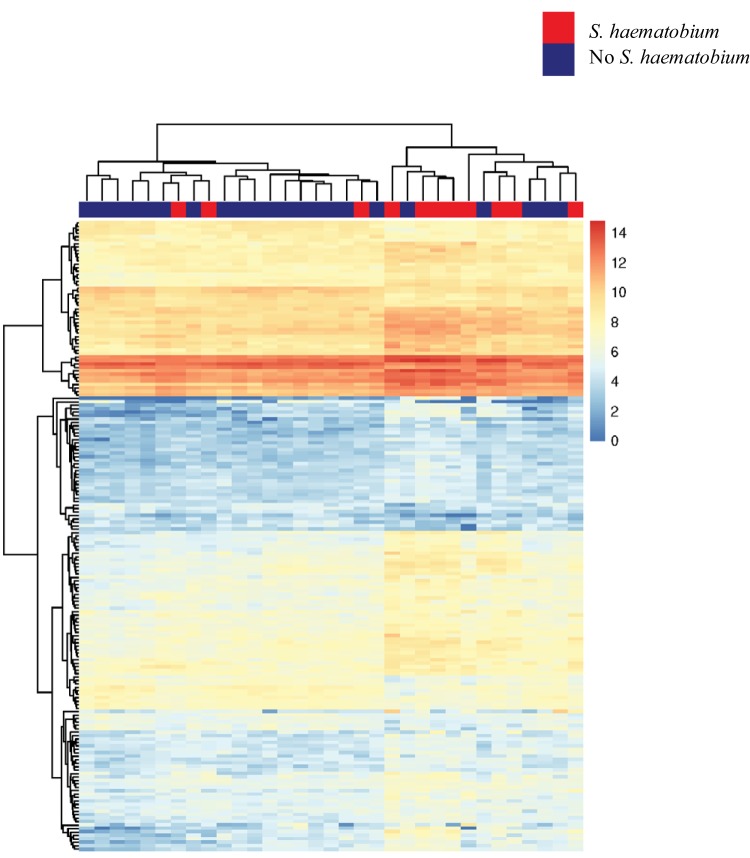
After correction for multiple comparisons, we observed 383 differentially expressed genes between those with and those without S. haematobium infection when sex was included as a covariate. A heat map of the genes that were significantly differentially expressed with at least ≥1.5-fold changes in expression between those with and those without S. haematobium is shown in Fig. 2. Comparison of the magnitude of log2-fold changes in the 383 genes that were differentially expressed in S. haematobium infection revealed significant differences between men and women (Fig. 3). The median absolute value of the fold change in gene expression in men was 0.39, while that in women was 0.69 (P < 0.001 as determined by a Wilcoxon rank sum test).
FIG 2.
Genes with at least 1.5-fold changes in expression between individuals with and without S. haematobium infection. In this heat map generated with unsupervised clustering, data for individuals with S. haematobium infection are indicated in red along the top horizontal bar, and data for individuals without S. haematobium infection are indicated in blue.
FIG 3.
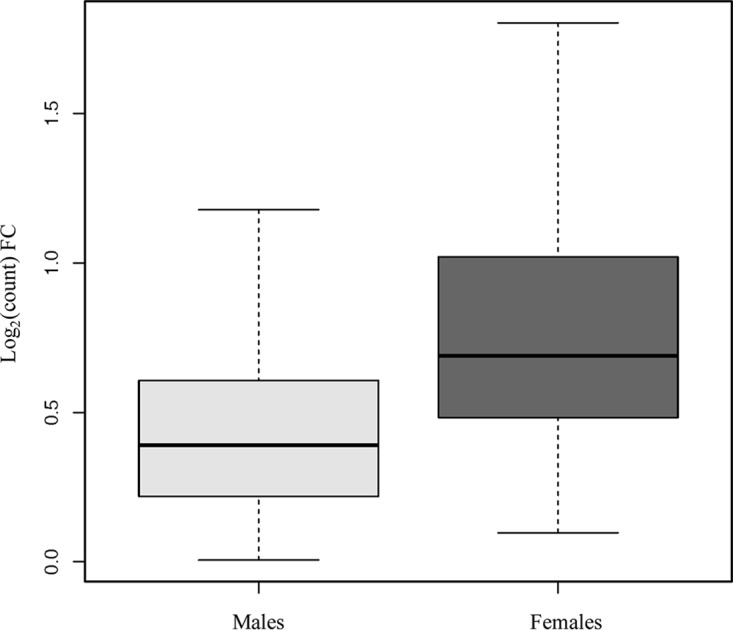
Fold changes of the 383 genes that were differentially expressed in S. haematobium infection, by host sex. Among the 383 genes that were differentially expressed between those with and without S. haematobium infection with a Padj of <0.05, the median absolute fold change in gene expression was significantly higher in females than in males (0.39 versus 0.69, P < 0.0001, as determined by Wilcoxon rank sum test). The boxplot includes the median (dark horizontal line) and interquartile range (box), with error bars representing 1.5 times the interquartile range or the minimum/maximum value.
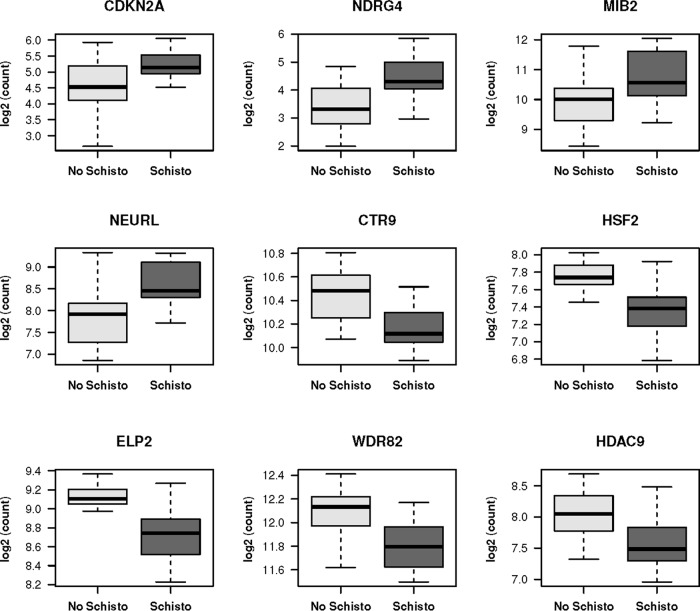
The 20 significantly differentially expressed genes between those with and without S. haematobium infection with the greatest changes (both positive and negative) are listed in Table S1, and representative plots showing the differences in gene expression by S. haematobium infection status are shown in Fig. 4.
FIG 4.
Differences in transcript counts in peripheral blood for nine representative genes found to be significantly differentially expressed in S. haematobium infection. The plots include median (dark horizontal line) and interquartile range (box), with error bars representing 1.5 times the interquartile range or the minimum/maximum value. The P value for differences was <0.05 after adjustment for multiple comparisons for all genes shown.
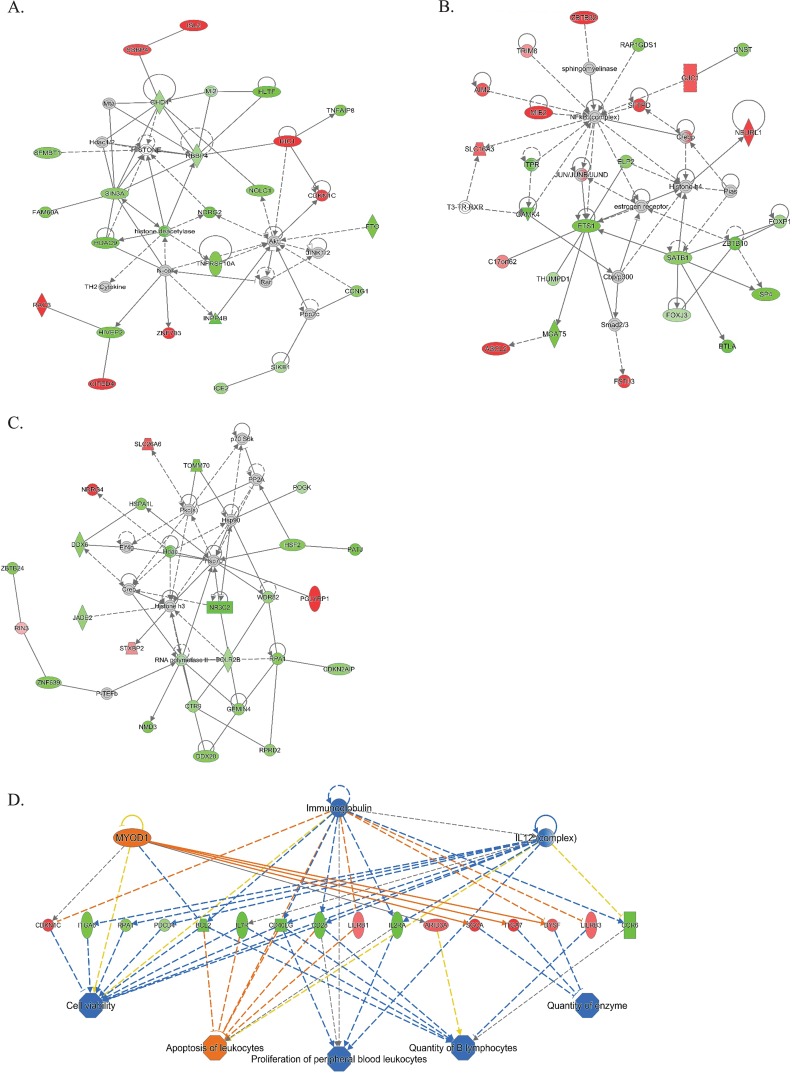
Of the 383 differentially expressed genes, 379 mapped to gene records in Ingenuity. Pathways analysis identified networks that reflect the differential gene expression in peripheral blood of people with or without S. haematobium. The networks were associated with development, cell death and survival, cell signaling, and immunologic disease pathways (Fig. 5). In addition, seven of the differentially expressed genes were associated with p53 signaling, including BCL2 (bcl2, 0.57-fold decrease, Padj = 0.024) caspase 6 (casp6, 0.34-fold decrease, Padj = 0.042), and histone deacetylase 9 (hdac9, 0.43-fold decrease, Padj = 0.05). In addition to hdac9, other molecules related to histone transcription that had significantly decreased expression included ctr9 (0.28-fold decrease, Padj = 0.04), hsf2 (0.34-fold decrease, Padj = 0.03), elp2 (0.32-fold decrease, Padj = 0.04), and wdr82 (0.27-fold decrease, Padj = 0.04). We also found increased expression of genes from the Notch signaling pathway: mindbomb ubiquitin ligase (mib2, 0.65-fold increase, Padj = 0.04) and neuralized E3 ubiquitin protein ligase 1 (neurl1, 0.61-fold increase, Padj = 0.03).
FIG 5.
(A to C) Top networks identified by IPA as differentially regulated in individuals with versus without schistosome infection, adjusting for sex as a covariate. The top networks were identified by IPA as being differentially affected in the setting of S. haematobium infection. Green indicates genes that have decreased expression in people with S. haematobium, and red indicates increased expression. Higher-intensity red or green indicates a greater magnitude fold change in expression. Solid lines represent direct interactions, and dotted lines represent indirect interactions. Gray indicates that a relationship is not predicted. (D) Top regulator effect network for the gene expression changes. Yellow indicates that findings are inconsistent. Orange indicates the predicted activation, while blue indicates the predicted inhibition, with intensity of color reflecting increased or decreased confidence in the prediction.
There was sufficient RNA remaining after RNA-Seq to complete quantitative PCR of 8 transcripts comparing 5 schistosoma-infected and 11 schistosoma-uninfected people from the original cohort. In this small sample, comparison of the ΔCT values for 4 of the 8 genes yielded P values of <0.30 and showed fold changes (FC) similar to those obtained by RNA-Seq. These were neurl1 (FC = 2.05, P = 0.03), mib2 (FC = 1.54, P = 0.11), bcl2 (FC = 0.83, P = 0.28), and casp6 (FC = 0.84, P = 0.23). The four genes that were tested but not confirmed were elongator acetyltransferase complex subunit 2 (elp2), WD repeat domain 82 (wdr82), histone deacetylase 9 (hdac9), and heat shock transcription factor 2 (hsf2).
Of the 383 differentially expressed genes identified in the initial analysis, 270 genes correlated with CAA values (Spearman correlation, P < 0.05). To determine whether gene expression correlated with the burden of infection, we then limited the correlation analysis to schistosome-positive samples only. Five genes correlated with CAA level in schistosome-infected people: leucine-rich repeats and calponin homology domain containing 1 (lrch1), Rho guanine nucleotide exchange factor 25 (arhgef25), zinc finger and BTB domain containing 47 (zbtb47), yippee-like 4 (ypel4), and ABL proto-oncogene 2 nonreceptor tyrosine kinase (abl2).
The top regulator effect network identified by Ingenuity Pathway Analysis (IPA) was related to interleukin-12 (IL-12), immunoglobulin, and myogenic differentiation 1 (myod1). IPA identified T-cell receptor (P = 1.48 × 10−5) and zinc finger and BTB domain containing 16 (ZBTB16, P = 8.2 × 10−5) as the most significant upstream regulators of differential gene expression between S. haematobium-infected and uninfected people.
DISCUSSION
We identified 383 genes that were differentially expressed in the peripheral blood of people with versus without S. haematobium infection, while controlling for sex as a covariate. The fold changes were of higher magnitude in women. Our work draws attention to two important aspects of S. haematobium infection that are not widely studied: (i) S. haematobium is associated with important systemic shifts in gene expression that are detectable in the peripheral blood, and (ii) the host response to S. haematobium infection differs by sex and should be incorporated into the study design and analysis of future research on schistosomiasis. Of note, our study was not powered to test the statistical significance of the sex-schistosoma status interaction term in the DESeq2 model of RNA-Seq analysis.
Our findings extend prior findings from human and mouse studies that have demonstrated the importance of host sex in determining the response to schistosome infection. In a study that investigated peripheral blood mononuclear cell (PBMC) response to schistosome antigens in men and women in Senegal who had low-level S. haematobium infection, PBMCs from women generated higher levels of transforming growth factor β (TGF-β) and IL-10 after schistosome antigen stimulation than PBMCs from men, which had higher levels of the inflammatory cytokines tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ) (9). Women have also been shown to have higher serum IgA antischistosome antibody levels in both S. haematobium (9) and S. mansoni (10) infections. In mice with chronic S. mansoni infection that had schistosome antigens injected into their ear pinnae and then underwent pinna measurements after 20 and 44 h, females exhibited significantly more pinna swelling than males (11). Our findings suggest that sex differences in response to S. haematobium infection exist not only in immune response but also in other processes, including cell-cell interactions and regulation of the cell cycle. These are intriguing findings, especially given the association of S. haematobium and urologic malignancies.
Schistosome infection is known to augment the Th2-predominant immune response typical of parasites. The adult worms reside in the vasculature, and worm-derived antigens circulate in the blood, including the antigens secreted from the worm digestive tract that are detected by CAA measurement (12). Eggs in tissue of the urogenital tract induce a local immune response that can evolve into metaplasia and malignant transformation, particularly in the bladder wall. Ray et al. studied the transcriptomes of female mice with urogenital schistosomiasis induced by egg injection into the bladder wall and found that in addition to increased expression of genes related to the Th2 immune response, there was differential expression of genes in carcinogenesis pathways and oncogenes (13). The group then histologically examined bladder walls of male and female mice following S. haematobium egg injection and found that female mice more frequently developed urothelial ulceration and metaplasia (14). Our top networks included multiple regulators of transcription and translation, which may relate to the pathways by which schistosome eggs could modify cellular proliferation and morphology in tissues. One candidate is myogenic differentiation 1 (MYOD1), which is involved in cell differentiation and was a component of the top regulator effect network.
Pathways analysis also showed the centrality of genes related to the cell cycle, including differential expression of ndrg4, an N-myc alpha/beta hydrolase required for cell cycle progression. Interestingly, there was increased expression of mindbomb ubiquitin ligase and neuralized E3 ubiquitin protein ligase 1, which are genes from the Notch signaling pathway, in both RNA-Seq and quantitative reverse transcription-PCR (qRT-PCR) methods. In a mouse model of S. japonicum-induced hepatic fibrosis, blockade of Notch signaling reversed macrophage M2 polarization and resolved the hepatic granulomas (15). The increased expression of Notch pathway-associated genes in the peripheral blood of people with S. haematobium may be a marker of the systemic inflammation associated with chronic schistosomiasis (16).
Networks 2 and 3 include molecules related to heat shock protein regulation and histones. Multiple molecules related to histone transcription had decreased expression in RNA-Seq analysis. These include CTR9, which associates with RNA polymerase II; HSF2, which activates transcription of heat shock proteins; a histone acetylator, ELP2, that remodels chromatin; the histone methyltransferase WDR82; and histone deacetylase 9. Histone deacetylases have been investigated as targets for chemotherapy adjuvants. Intriguingly, blockade of schistosomal histone deacetylase 8 reduced worm and egg burden in S. mansoni-infected mice (17). Future studies could investigate the role of human and schistosomal histone deacetylases in the pathogenesis of schistosomiasis.
The limitations of the study include a relatively small sample size, making it difficult to control for all potential confounders that could contribute to the observed differences between men and women. These could include differences in prior praziquantel treatment, in levels of exposure to schistosomes throughout the life span, or in concomitant unmeasured infections. Since this study did not include preinfection longitudinal specimens, we were not able to assess whether the observed differences in gene expression are caused by S. haematobium infection or are merely associated with it. Longitudinal studies that characterize whole blood transcriptomes prior to, during, and after treatment of schistosome infection would be highly informative. Such studies would be possible in cohorts that have archived whole blood or dried blood spots prior to, during, and after S. haematobium infection and praziquantel treatment (18).
In conclusion, our work highlights the necessity of accounting for host sex as a biological variable in future studies of schistosomiasis. Host sex had a major impact on gene expression and should be considered in studies of schistosomiasis treatment and prevention, particularly given the known differential effects of vaccines in men and women (19). In addition, as has been documented in the literature, the pathogenesis of schistosome-related malignancy differs by sex in mouse models (14). We must ensure that in studies of this neglected tropical disease, sex as a biological variable is not neglected as well.
MATERIALS AND METHODS
Study design.
We identified HIV-uninfected people who were confirmed to be positive for S. haematobium infection by both egg visualization on urine microscopy and elevated Schistosoma circulating anodic antigen (CAA) in the blood and compared them to HIV-uninfected individuals with confirmed negative urine and serum Schistosoma studies. All participants had stool screened and were confirmed to be negative for S. mansoni ova.
Study sites and population.
We invited a community-based sample of adults of reproductive age living in rural villages in northwest Tanzania, in which we have previously documented a high prevalence of S. haematobium, to receive free screening for schistosomiasis and HIV as part of a community outreach project. Individuals provided urine samples that were filtered and examined microscopically in the field and received same-day results. In the villages where we worked, the prevalence of S. haematobium among adults is approximately 5% by urine microscopy and 30% by serum CAA measurement (8, 20). Due to the known poor sensitivity of urine screening for S. haematobium infection in adults, all individuals screened received free praziquantel treatment in accordance with World Health Organization (WHO) recommendations (21).
While their urine samples were being examined, men and women received free individual voluntary HIV counseling and testing provided by a trained nurse. HIV testing was conducted on whole blood using two separate point-of-care tests in accordance with the Tanzanian national guidelines, with those testing positive for HIV via a screening Determine HIV-1/2 test (Alere, Waltham, MA) undergoing confirmatory testing using a Uni-Gold HIV 1/2 test (Trinity Biotech, Wicklow, Ireland). Participants received their HIV test results immediately and those who were given a first-time diagnosis of HIV were given a referral letter to obtain free HIV care at the nearest HIV care and treatment center.
We invited HIV-uninfected individuals who were found to have S. haematobium ova on microscopic screening to provide written informed consent for participation in the current research study. We also invited a random sample of HIV-uninfected individuals who were negative for S. haematobium infection to participate. Women were asked about their last menstrual periods to confirm that they were not pregnant due to the need to sample the cervix. Participants provided written informed consent and underwent a structured interview in a private setting with a nurse fluent in the local language.
Sample collection.
Following the interview, study participants provided additional peripheral blood for transcriptional analysis and schistosome circulating anodic antigen quantification, as well as stool to rule out S. mansoni infection using five Kato Katz slides. Gonorrhea and chlamydia were tested in urine from men. Women underwent a gynecologic examination by the study physician (J.A.D.), which included endocervical swab sampling for gonorrhea/chlamydia testing, collection of cervical lavage and cervical cytobrush samples, and screening for cervical cancer using acetic acid according to the Tanzanian national guidelines.
Peripheral blood was collected from the antecubital fossa into Tempus RNA isolation tubes (Applied Biosystems), shaken vigorously according to the manufacturer’s instructions, and transported from the field site to the central laboratory in Mwanza at 4°C. Upon arrival in the laboratory, tubes were stored at −20°C until transport on dry ice to the Weill Cornell Global Health laboratory in New York.
Serum CAA testing was performed in the reference laboratory in Mwanza as previously described (8, 22). People with CAA results greater than 40 pg/ml were considered to be definitive positives. People with CAA values less than 25 pg/ml were negative. People with CAA values between 25 and 40 pg/ml or with discrepant CAA and egg microscopy results were excluded.
RNA extraction and purification.
RNA was extracted from Tempus tubes with the Tempus Spin RNA isolation kit (Invitrogen, Carlsbad, CA) with on-column DNase digestion according to the manufacturer’s instructions. RNA integrity was assessed with a Bioanalyzer 2100 (Agilent Technologies, Carpinteria, CA) and concentration was measured with the NanoDrop 8000 system (Thermo Fisher Scientific, Waltham, MA). Samples with RNA integrity numbers of >6.4 and 28S/18S rRNA ratios of >1.2 were submitted for RNA sequencing.
RNA library preparation and RNA-Seq.
RNA sample library preparation and next-generation sequencing were performed by the Weill Cornell Genomics Core laboratory. mRNA was prepared using TruSeq stranded mRNA sample preparation kit (Illumina, San Diego, CA) in accordance with the manufacturer’s instructions. Prior to the sequencing run on the HiSeq 4000 (Illumina), samples were hybridized onto a patterned flow cell and amplified using a cBot fluidics device (Illumina). Patterned flow cells were sequenced on a HiSeq 4000 sequencer (Illumina) with single-end 50 bp. Illumina bcl2fastq2 Conversion Software was used to demultiplex samples into individual samples and to convert per-cycle BCL base call files into FASTQ files for downstream data analysis. FastQC (Babraham Bioinformatics, Babraham, UK) was used to determine sequencing quality. Transcripts were aligned to the human hg19 reference genome using Tophat2 (23) and counted with the HTSeq package (24).
Quantitative RT-PCR.
We completed qRT-PCR for a subset of statistically significantly differentially expressed genes identified by RNA-Seq analysis on RNA prepared for RNA-Seq. A total of 20 ng of RNA was reverse transcribed to cDNA using Superscript IV VILO (Applied Biosystems/Thermo Fisher, Waltham, MA) reverse transcriptase according to the manufacturer’s instructions. Quantitative PCRs were prepared using TaqMan Fast Advanced Master Mix (Applied Biosystems), 1 ng of cDNA, and FAM-MGB TaqMan (Applied Biosystems) gene expression assay for the particular gene (Table S2). All assays were performed in triplicate and run on the QuantStudio 6 qPCR instrument (Applied Biosystems). Samples with a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) CT standard deviation of >0.5 were excluded from analysis. ΔΔCT values were calculated with GAPDH as the endogenous control CT.
Statistical methods.
For demographic and clinical characteristics, variables were expressed as the number (percentage) or median (interquartile range) and compared using a Fisher exact test or with a Wilcoxon rank sum test, as appropriate. To compare CAA values, the absolute values were log transformed and compared between men and women using a rank sum test. Correlation of CAA values with gene expression was calculated with the Spearman correlations. ΔCT values from qRT-PCR were compared by using a Wilcoxon rank sum test as previously described (25).
In order to assess sample variability in transcript data, count data were normalized by library size using DESeq2 (v3.4.1) (27), and multidimensional-scaling plots were created to summarize between-sample distances of log2 count data in two dimensions. Genes were compared for differential expression in blood from people with or without S. haematobium using DESeq2 while including sex as a covariate. The resulting P values were adjusted for multiple comparisons using the procedure of Benjamini and Hochberg (26). Differentially expressed genes between those with and without S. haematobium with a false detection rate of ≤0.05 were considered for subsequent analysis.
Heat maps were generated with unsupervised clustering using all genes with at least a 1.5-fold difference in gene expression between those with and without schistosome infection. Gene functions were determined by a search on the National Center for Biotechnology Information’s gene database.
All genes identified as being differentially expressed with a Padj of ≤0.05 in those with versus without S. haematobium were submitted for Ingenuity Pathway Analysis (IPA) to generate representative pathways (Qiagen).
Ethical considerations.
This study was approved by Bugando Medical Centre and the National Institute for Medical Research (both in Tanzania) and by Weill Cornell Medical College. All individuals screened for schistosome infection were provided with praziquantel treatment free of charge on the day of screening according to WHO guidelines. Study participants provided written informed consent. Those found to have sexually transmitted infections received free treatment for themselves and their sexual partners in accordance with Tanzanian national guidelines. Those with HIV infection were referred for ongoing free care at the nearest HIV care and treatment center. Women with abnormalities during the acetic acid-based cervical cancer screening examination were given a referral appointment at the Bugando Medical Centre and money for transport to the appointment.
Supplementary Material
ACKNOWLEDGMENTS
We thank the study participants for their enthusiastic participation in this study. We thank Laura Kirkman and Xu Zhang for assistance with the QuantStudio 6 qPCR instrument.
This study was supported by National Institutes of Health grant K23 AI 110238 (to J.A.D.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We have no conflicts of interest related to this research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00291-18.
REFERENCES
- 1.World Health Organization. 2017. Schistosomiasis fact sheet no. 115. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Watanabe K, Mwinzi P, Black C, Muok E, Karanja D, Secor W, Colley D. 2007. T regulatory cell levels decrease in people infected with Schistosoma mansoni on effective treatment. Am J Trop Med Hyg 77:676–682. doi: 10.4269/ajtmh.2007.77.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mbow M, Larkin BM, Meurs L, Wammes LJ, de Jong SE, Labuda LA, Camara M, Smits HH, Polman K, Dieye TN, Mboup S, Stadecker MJ, Yazdanbakhsh M. 2013. T-helper 17 cells are associated with pathology in human schistosomiasis. J Infect Dis 207:186–195. doi: 10.1093/infdis/jis654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prodger JL, Ssemaganda A, Ssetaala A, Kitandwe PK, Muyanja E, Mpendo J, Nanvubya A, Wambuzi M, Nielsen L, Kiwanuka N, Kaul R. 2015. Schistosoma mansoni infection in Ugandan men is associated with increased abundance and function of HIV target cells in blood, but not the foreskin: a cross-sectional study. PLoS Negl Trop Dis 9:e0004067. doi: 10.1371/journal.pntd.0004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleppa E, Ramsuran V, Zulu S, Karlsen GH, Bere A, Passmore J-AS, Ndhlovu P, Lillebø K, Holmen SD, Onsrud M, Gundersen SG, Taylor M, Kjetland EF, Ndung’u T. 2014. Effect of female genital schistosomiasis and anti-schistosomal treatment on monocytes, CD4+ T-cells and CCR5 expression in the female genital tract. PLoS One 9:e98593. doi: 10.1371/journal.pone.0098593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colley DG, Bustinduy AL, Secor WE, King CH. 2014. Human schistosomiasis. Lancet 383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombe S, Lee MH, Masikini PJ, van Lieshout L, de Dood CJ, Hoekstra PT, Corstjens PLAM, Mngara J, van Dam GJ, Downs JA. 2018. Decreased sensitivity of Schistosoma sp. egg microscopy in women and HIV-infected individuals. Am J Trop Med Hyg 98:1159–1164. doi: 10.4269/ajtmh.17-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downs JA, de Dood CJ, Dee HE, McGeehan M, Khan H, Marenga A, Adel PE, Faustine E, Issarow B, Kisanga EF, Kisigo GA, Ngahyolerwa S, Zahoro F, Miyaye D, Magawa RG, Mngara J, Lee MH, Corstjens PLAM, van Dam GJ, Fitzgerald DW. 2017. Schistosomiasis and human immunodeficiency virus in men in Tanzania. Am J Trop Med Hyg 96:856–862. doi: 10.4269/ajtmh.16-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remoué F, To Van D, Schacht AM, Picquet M, Garraud O, Vercruysse J, Ly A, Capron A, Riveau G. 2001. Gender-dependent specific immune response during chronic human Schistosomiasis haematobia. Clin Exp Immunol 124:62–68. doi: 10.1046/j.1365-2249.2001.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remoué F, Rogerie F, Gallissot M, Guyatt HL, Neyrinck J, Diakkhate M, Niang M, Butterworth AE, Auriault C, Capron A, Riveau G. 2000. Sex-dependent neutralizing humoral response to Schistosoma mansoni 28GST antigen in infected human populations. J Infect Dis 181:1855–1859. doi: 10.1086/315454. [DOI] [PubMed] [Google Scholar]
- 11.Boissier J, Chlichlia K, Digon Y, Ruppel A, Moné H. 2003. Preliminary study on sex-related inflammatory reactions in mice infected with Schistosoma mansoni. Parasitol Res 91:144–150. doi: 10.1007/s00436-003-0943-1. [DOI] [PubMed] [Google Scholar]
- 12.de Water R, Fransen JA, Deelder AM. 1986. Ultrastructural localization of the circulating anodic antigen in the digestive tract of Schistosoma mansoni using monoclonal antibodies in an immunogold labeling procedure. Am J Trop Med Hyg 35:549–558. doi: 10.4269/ajtmh.1986.35.549. [DOI] [PubMed] [Google Scholar]
- 13.Ray D, Nelson T, Fu C, Patel S, Gong D, Odegaard J, Hsieh M. 2012. Transcriptional profiling of the bladder in urogenital schistosomiasis reveals pathways of inflammatory fibrosis and urothelial compromise. PLoS Negl Trop Dis 6:e1912. doi: 10.1371/journal.pntd.0001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honeycutt J, Hammam O, Hsieh M. 2015. Schistosoma haematobium egg-induced bladder urothelial abnormalities dependent on p53 are modulated by host sex. Exp Parasitol 158:55–60. doi: 10.1016/j.exppara.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng S, Zhang P, Chen Y, Zheng S, Zheng L, Weng Z. 2016. Inhibition of Notch signaling attenuates schistosomiasis hepatic fibrosis via blocking macrophage M2 polarization. PLoS One 11:e0166808. doi: 10.1371/journal.pone.0166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert DR, Hölscher C, Mohrs M, Arendse B, Schwegmann A, Radwanska M, Leeto M, Kirsch R, Hall P, Mossmann H, Claussen B, Förster I, Brombacher F. 2004. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 20:623–635. doi: 10.1016/S1074-7613(04)00107-4. [DOI] [PubMed] [Google Scholar]
- 17.Marek M, Kannan S, Hauser A-T, Moraes Mourão M, Caby S, Cura V, Stolfa DA, Schmidtkunz K, Lancelot J, Andrade L, Renaud J-P, Oliveira G, Sippl W, Jung M, Cavarelli J, Pierce RJ, Romier C. 2013. Structural basis for the inhibition of histone deacetylase 8 (HDAC8), a key epigenetic player in the blood fluke Schistosoma mansoni. PLoS Pathog 9:e1003645. doi: 10.1371/journal.ppat.1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reust MJ, Lee MH, Xiang J, Zhang W, Xu D, Batson T, Zhang T, Downs JA, Dupnik KM. 2018. Dried blood spot RNA transcriptomes correlate with transcriptomes derived from whole blood RNA. Am J Trop Med Hyg 98:1541–1546. doi: 10.4269/ajtmh.17-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein SL, Flanagan KL. 2016. Sex differences in immune responses. Nat Rev Immunol 16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 20.Downs JA, Mguta C, Kaatano GM, Mitchell KB, Bang H, Simplice H, Kalluvya SE, Changalucha JM, Johnson WD, Fitzgerald DW. 2011. Urogenital schistosomiasis in women of reproductive age in Tanzania’s Lake Victoria region. Am J Trop Med Hyg 84:364–369. doi: 10.4269/ajtmh.2011.10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. 2006. Preventive chemotherapy in human helminthiasis. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 22.Corstjens P, De Dood CJ, Kornelis D, Tjon Kon Fat E, Wilson R, Kariuki TM, Nyakundi RK, Loverde PT, Abrams WR, Tanke HJ, Van Lieshout L, Deelder AM, Van Dam GJ. 2014. Tools for diagnosis, monitoring and screening of Schistosoma infections utilizing lateral-flow based assays and upconverting phosphor labels. Parasitology 141:1841–1855. doi: 10.1017/S0031182014000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan JS, Reed A, Chen F, Stewart CN Jr. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. [Google Scholar]
- 27.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.