Members of the genus Brucella are Gram-negative pathogens that cause chronic systemic infection in farm animals and zoonotic infection in humans. Study of the genus Brucella has been hindered by the need for biosafety level 3 select agent containment.
KEYWORDS: Brucella, Brucella neotomae, brucellosis, bioluminescent imaging, murine model, pathogenesis, pathology, type IV secretion system, undulant fever
ABSTRACT
Members of the genus Brucella are Gram-negative pathogens that cause chronic systemic infection in farm animals and zoonotic infection in humans. Study of the genus Brucella has been hindered by the need for biosafety level 3 select agent containment. Brucella neotomae, originally isolated from the desert pack rat, presented an opportunity to develop an alternative, non-select agent experimental model. Our prior in vitro work indicated that the cell biology and type IV secretion system (T4SS) dependence of B. neotomae intracellular replication were similar to observations for human-pathogenic select agent Brucella species. Therefore, here, we investigated the pathobiology of B. neotomae infection in the BALB/c mouse. During a sustained infectious course, B. neotomae replicated and persisted in reticuloendothelial organs. Bioluminescent imaging and histopathological and PCR-based analysis demonstrated that the T4SS contributed to efficient early infection of the liver, spleen, and lymph nodes; granuloma formation and hepatosplenomegaly; and early induction of Th1-associated cytokine gene expression. The infectious course and pathologies in the murine model showed similarity to prior observations of primate and native host infection with zoonotic Brucella species. Therefore, the B. neotomae BALB/c infection model offers a promising system to accelerate and complement experimental work in the genus Brucella.
INTRODUCTION
Members of the genus Brucella, Gram-negative coccobacilli, cause systemic bacterial infections in domesticated farm animals and other vertebrates (1). Humans may be infected through ingestion of unpasteurized dairy products, inhalation of organisms from birthing farm animals, and during occupational exposure in abattoir and meat packing industries. Disease in humans is characterized by chronicity and a waxing and waning fever (so-called undulant fever). Lymph node infection and enlargement are common (2); endovascular and osteoarticular infection are unwelcome and not infrequent complications (3, 4).
Based on the low aerosol infectious dose and debilitating disease, the major zoonotic species, Brucella melitensis, Brucella abortus, and Brucella suis, are classified as biosafety level 3, category B overlap select agents (5). Biosafety containment and select agent regulations represent significant impediments to experimental investigation.
Brucella organisms have been studied in several in vitro and in vivo model systems (6–9). In vitro assays have demonstrated the capability of Brucella species to grow within specialized vacuoles inside of macrophages, endothelial cells, and other cell types. In particular, Brucella species alter the normal process of phagosomal maturation. Remodeling of the phagosome occurs in a temporal fashion in which organisms are first found within endocytic vacuoles that subsequently take on properties of the endoplasmic reticulum and autophagosomes and permit intracellular replication (10–14). These events have been found to depend on a bacterial VirB-based type IV secretion system (T4SS) (10–14). Presumably, effectors translocated into the eukaryotic cytoplasm by this secretion apparatus alter phagosome biogenesis to establish an intracellular replication niche. Murine infection by human-pathogenic zoonotic Brucella species has also been explored (10, 15–18). In particular, select agents B. abortus, B. melitensis, and B. suis cause sustained T4SS-dependent, high-burden infection in BALB/c mice.
Our specific goal was to establish a non-select agent Brucella infection model that would accelerate and complement experimental work in the genus. Brucella neotomae was originally isolated from the desert wood rat (19), a pack rat species in the genus Neotoma, and has been previously classified as a biosafety level 2 pathogen (13). Importantly, our prior in vitro work indicated that the organism replicated efficiently in macrophages in a T4SS-dependent manner and showed temporal phagosome maturation dynamics similar to those of human-pathogenic species (13).
However, the infectious course of B. neotomae in laboratory mice remains insufficiently characterized. One prior study found that B. neotomae caused sustained infection in BALB/c mice but not in C57BL/6 mice (20), a murine host previously noted to be less hospitable to infection by Brucella species (7); however, infectious pathogenesis was not further investigated. Therefore, here, we characterized the microbiology, pathology, and T4SS dependence of B. neotomae infection in the BALB/c murine host.
RESULTS
Assessment of virulence of B. neotomae in BALB/c mice.
The pathogenicity of B. neotomae in the BALB/c mouse was initially evaluated during intraperitoneal (i.p.) infection with either 1.0 × 106 or 1.0 × 108 CFU (Fig. 1A). Mice infected with the higher dose showed 75% mortality within 8 days postinfection. In contrast, mice infected at the lower dose all survived (P = 0.002 for difference in survival; log-rank Mantel-Cox test). In all the mice surviving high-dose infections, punctate white lesions were observed on the liver surface, consistent with granulomas or microabscesses (Fig. 1B).
FIG 1.
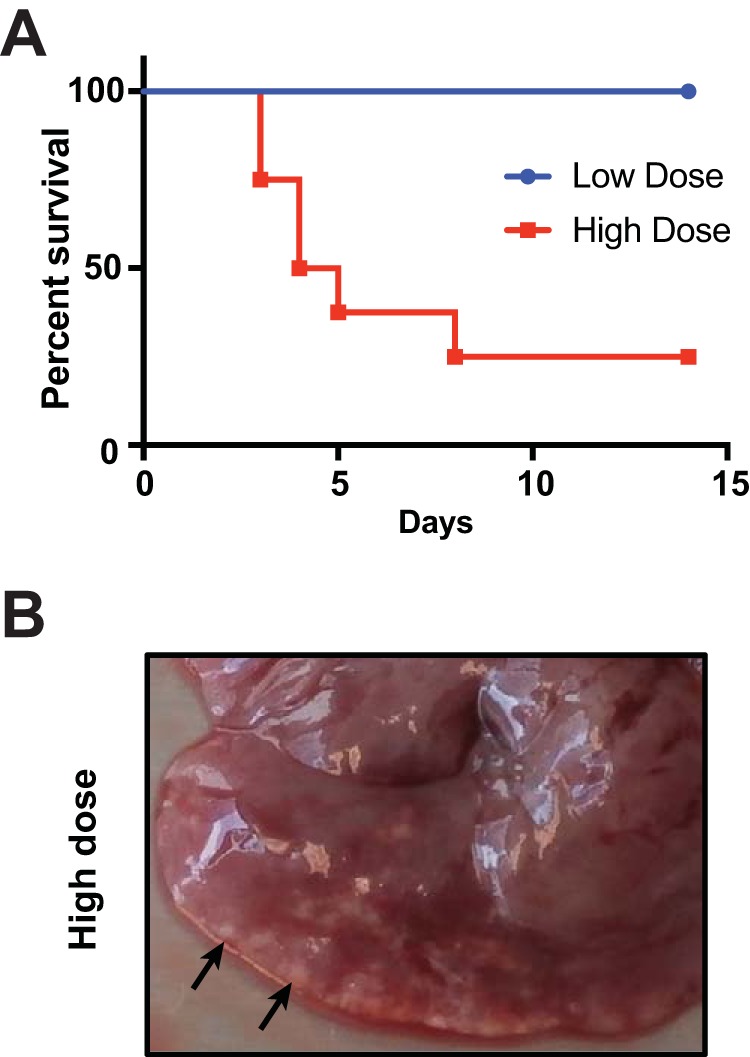
Virulence of B. neotomae in BALB/c mice. (A) Two groups of eight mice each were infected i.p. with a low dose (106 CFU) or a high dose (108 CFU) of B. neotomae. Survival was monitored for 2 weeks. (B) Representative image of liver 14 days after high-dose infection in a surviving mouse. The arrows indicate white-tinged granulomas or microabscesses on the liver surface.
Type IV secretion system-dependent Brucella virulence.
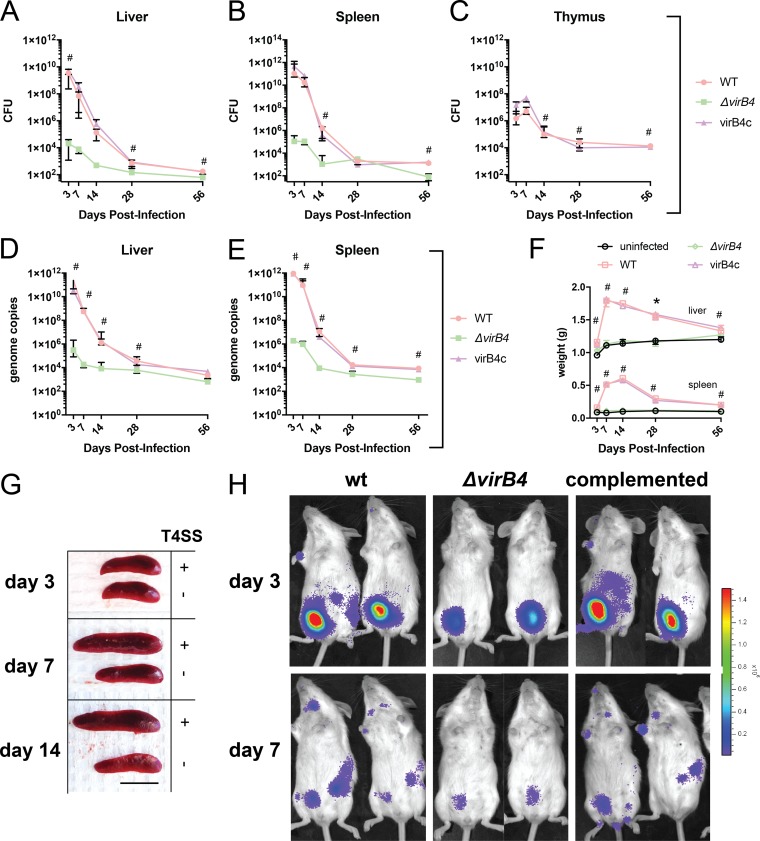
To investigate the role of the T4SS in B. neotomae pathogenesis, we infected mice via intraperitoneal injection with either 107 CFU of B. neotomae wild type (wt) or ΔvirB4, a previously described T4SS in-frame deletion mutant that is unable to replicate in cultured J744A.1 macrophages (13). At time intervals spanning 2 months, we monitored CFU in liver, spleen, and thymus. In wt bacterial infection, peak CFU counts were observed at 3 days postinfection in spleen and liver, at levels considerably above the initial inoculum, and 7 days postinfection in thymus, suggesting significant in vivo replication during the early infectious period (Fig. 2A to C). CFU decreased thereafter. However, infection was sustained for at least 2 months, with levels of approximately 103 CFU at 8 weeks postinfection. During ΔvirB4 bacterial infection, CFU counts at early time points were lower than for wild-type bacterial infection during early infection and were sustained at lower levels at later time points. Notably, ΔvirB4 bacteria were not isolated from the thymus at any time point. These differences reached statistical significance at several time points in liver, spleen, and thymus (as shown in Fig. 2A to C). In contrast, results for the complemented ΔvirB4 strain (virB4c) were not significantly different from wild-type bacterial infection for all comparisons. Taken together, these results suggested that an intact T4SS provided a significant growth advantage during all stages of infection and at all the sites examined. The organism burden was separately quantified by quantitative PCR (qPCR) of the multicopy B. neotomae IS711 insertion sequence for liver and spleen with similar results (Fig. 2D and E). Calculated genome copy numbers were roughly 10-fold higher than CFU counts, suggesting a substantial fraction of unculturable or nonviable organisms.
FIG 2.
VirB T4SS contributes to pathogenesis. (A to C) Bacterial CFU in liver, spleen, and thymus. (D and E) B. neotomae genome copies in liver and spleen. The data shown are the medians and interquartile ranges for five mice per group. #, significant difference between wild-type and ΔvirB4 bacterial infections using Dunn’s posttest. There were no statistically significant differences for either CFU or genome copies between wild-type and complemented virB4 (virB4c) infections at any time point. (F) Liver and spleen weights during infection. The data shown are medians and interquartile ranges for five mice per group. #, significant difference between both wild-type and virB4c infections and uninfected controls. Liver and spleen weights were not statistically different between ΔvirB4 strain-infected mice and uninfected control mice at any time point. (G) Representative spleens from infections with wild-type (T4SS+) or ΔvirB4 (T4SS−) strains. Significant splenic enlargement was noted 7 and 14 days following wild-type bacterial infection. Scale bar = 1 cm. (H) Small-animal imaging following infection with bacterial strains with a luciferase operon reporter. The color bar is scaled to a minimum and maximum of 2.0 × 104 and 1.5 × 106 p/s/cm2/sr, respectively. Two of three mice per infecting strain with maximal signal are shown. Bacterial inocula in all experiments were 107 per mouse by intraperitoneal injection.
During infection with the wild-type strain, hepatosplenomegaly was most pronounced at 1 to 2 weeks postinfection (Fig. 2F and G). Liver and splenic organ weights during both wild-type and virB4c infection were statistically greater than uninfected control organ weights at all time points. In contrast, there was no significant difference in liver and spleen weights during infection with the ΔvirB4 mutant in comparison to mock-infected controls of the same age.
To further assess infection dynamics, isogenic wild-type, ΔvirB4, and complemented ΔvirB4 strains expressing a bacterial luciferase operon (13) were used to infect mice, followed by whole-body bioluminescence imaging (BLI) (Fig. 2H). Wild-type and complemented ΔvirB4 strain signals were maximal at 3 days postinfection and were concentrated in the peritoneal cavity (the inoculation site). On days 3 and 7, there was partial redistribution of bacterial signal to liver, spleen, and lymph nodes. Scattered lymph node positivity was observed at day 14, and no signal was detected at day 28 (data not shown). In contrast, the signal from ΔvirB4 bacterial infection was notably lower than that from infection by the wild type or complemented mutants at the inoculation site from day 1 postinfection onward (Fig. 2E and data not shown). Furthermore, there was no detectable signal in liver, spleen, or lymph nodes at all time points.
Pathologies.
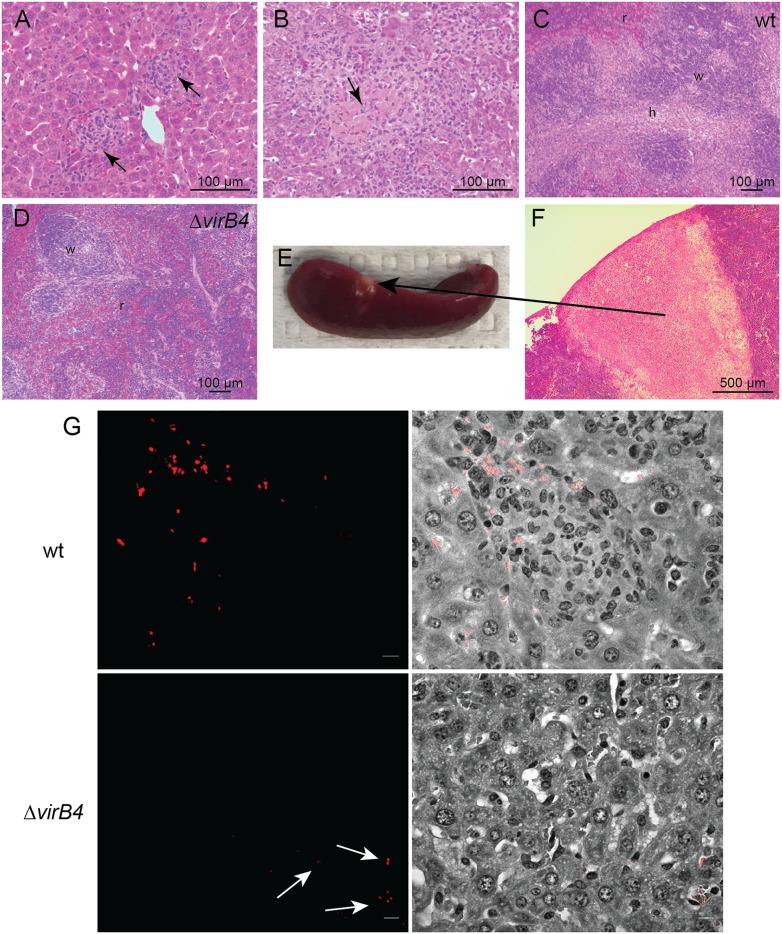
Liver and spleen showed significant pathology. Following infection with wild-type B. neotomae, there was a prominent granulomatous response in the liver consisting of nodular clusters of histiocytes with occasional lymphocytes and granulocytes (Fig. 3A), with central necrosis observed in granulomas in several mice (Fig. 3B). The spleens demonstrated significant white-pulp expansion (Fig. 3C), most prominent 7 to 14 days postinfection, with one mouse at 14 days postinfection demonstrating a wedge-shaped area of necrosis under the capsule (Fig. 3E), suggestive of an infarct (Fig. 3F). In contrast, in ΔvirB4 bacterial infections, granulomas and splenic white-pulp expansion were not observed (Fig. 3D). As the strains used in the experiments constitutively expressed a tdTomato fluorescent-protein reporter, association of organisms with pathologies could also be assessed by fluorescence microscopy. Specifically, fluorescent organisms were detected within granulomas during wild-type bacterial infection, often in large intracellular clusters suggestive of robust intracellular replication (Fig. 3G). However, during ΔvirB4 bacterial infection, fluorescent organisms were infrequently observed and, when present, were found only individually or in small clusters.
FIG 3.
Histopathology of B. neotomae infection. (A and B) Granulomatous inflammation (arrows) 3 days (A) and necrotizing granuloma (arrow) 7 days (B) postinfection with wild-type B. neotomae. (C) Splenic white-pulp expansion 7 days postinfection with wild-type organisms (w, white pulp; r, red pulp; h, histiocytes). (D) In contrast, white-pulp expansion was not evident following ΔvirB4 bacterial infection. (E and F) Splenic infarct gross pathology (E) and associated microscopic pathology with evidence of tissue necrosis (F) 14 days postinfection with wild-type organisms. (G) (Top) Immunofluorescence microscopy demonstrating intracellular clusters of B. neotomae organisms within hepatic granulomas 3 days postinfection with the wild-type strain. (Left) Fluorescent image of bacteria expressing tdTomato fluorescent reporter. (Right) Merged bright-field and gray-scale images. (Bottom) In contrast, only very rare organisms were observed following infection with the ΔvirB4 strain, generally as single organisms or very small clusters (arrows), likely within Kupffer cells lining sinusoidal spaces. Bars = 5 µm.
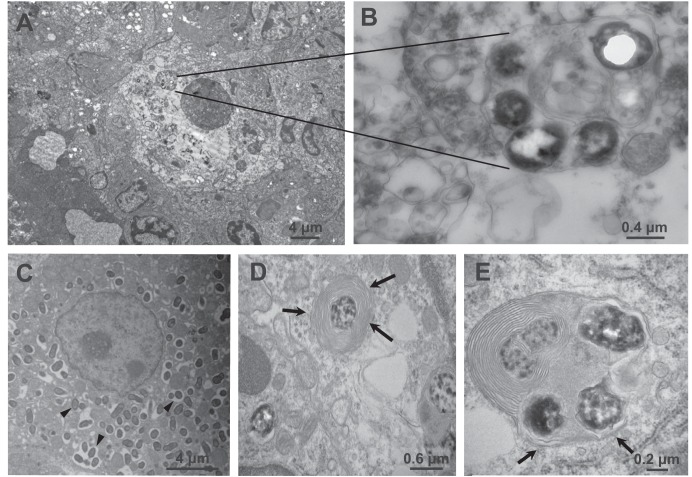
Transmission electron microscopy (TEM) was also used to visualize subcellular localization of bacteria in tissues. Bacteria were detected only during infections with the wild-type strain, presumably based on the difficulty in finding a smaller number of ΔvirB4 organisms in ultrathin sections. Figure 4A shows a hepatic granuloma 3 days postinfection consisting of concentric macrophages with a single central necrotic macrophage containing numerous intracellular bacteria in a multilaminar phagosome (Fig. 4B). More generally, phagosomes containing bacteria surrounded by single (Fig. 4C) or multiple (Fig. 4D and E) smooth, concentric layers of membrane were observed, suggestive of autophagosomes (21–23). Rough endoplasmic reticulum was observed in proximity to, but not as a constituent of, phagosomal membranes.
FIG 4.
Transmission electron microscopy images of hepatic infection. (A) Hepatic granuloma 3 days postinfection, composed of concentric histiocytes. (B) A central histiocyte appears necrotic and contains a replicative phagosome, inside of which are multiple bacteria. (C to E) B. neotomae bacteria replicating inside membrane-bound phagosomes (arrowheads) 7 days postinfection (C) and also found in multilaminar phagosomes (D, arrows) suggestive of autophagosomes or autophagolysosomes in close association with rough endoplasmic reticulum (E, arrows).
Induction of cytokine gene expression during infection.
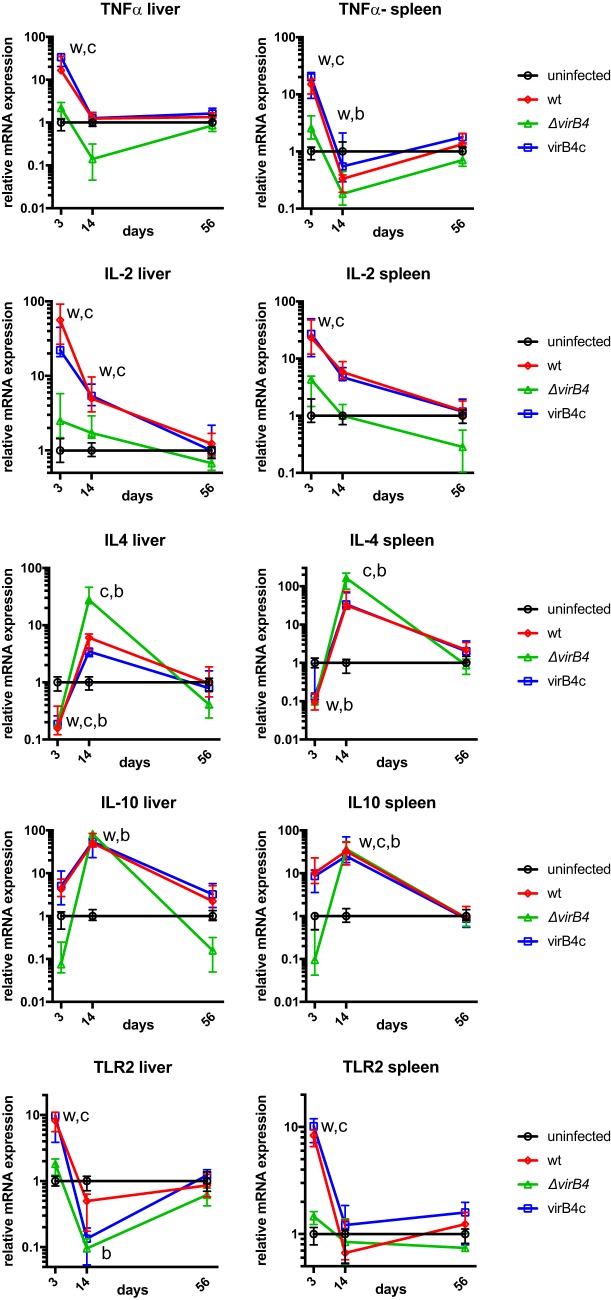
To determine whether B. neotomae infection was associated with a predominant Th1- or Th2-mediated immune response, cytokine gene expression levels in liver and spleen were measured during infection (Fig. 5). Notably, at 3 days postinfection, tumor necrosis factor alpha (TNF-α) and interleukin 2 (IL-2) expression, typically associated with a Th1 response, were significantly upregulated over 10-fold during wt and virB4c infection compared to uninfected controls. Similarly, Toll-like receptor 2 (TLR2), but not TLR4, expression (data not shown) was significantly induced by wt and virB4c at 3 days postinfection but not at later time points. In contrast, ΔvirB4 bacterial infection did not induce significant upregulation of TNF-α, IL-2, or TLR2. At 3 days postinfection, IL-4 expression levels were significantly lower than those of uninfected controls during both wt and ΔvirB4 bacterial infections. However, at day 14 postinfection, expression levels induced by wt, virB4c, and ΔvirB4 strains exceeded those of uninfected controls, reaching statistical significance for ΔvirB4 and virB4c infections. A similar statistically significant increase of IL-10 expression was present at day 14 postinfection. Taken together, TNF-α, IL-2, and TLR2 data were consistent with an early T4SS-dependent Th1 response; IL-4 and IL-10 data were consistent with a delayed, T4SS-independent Th2 response.
FIG 5.
Cytokine gene expression. Cytokine and Toll-like receptor mRNA gene expression was determined in liver and spleen at the indicated times postinfection by reverse transcriptase qPCR. Expression was normalized to actin gene expression within each sample to control for efficiency of mRNA extraction. The data points are plotted relative to the median gene expression from uninfected mice and represent the medians and interquartile ranges of values from five mice per group. w, significant difference between wild-type bacterial infection and a mock-infected control; c, significant difference between virB4c infection and an uninfected control; b, significant difference between ΔvirB4 bacterial infection and an uninfected control.
DISCUSSION
Here, we describe microbiological and pathological features of murine B. neotomae infection. Several aspects of T4SS-driven pathogenesis were evident by complementary analyses, including persistent infection of the reticuloendothelial organs, intracellular replication within granulomas, and early induction of a Th1 cytokine transcriptional response.
It is worthwhile discussing results in the context of prior observations for the species and genus. Five independent strains of B. neotomae were originally isolated from tissues of 108 desert wood rats from Utah (19), including the type strain, 5K33, used in our studies. Five additional isolates from desert wood rats and a sixth from a desert wood rat flea were also described separately by Thorpe et al. (24). These observations indicate a substantial prevalence (∼5%) of B. neotomae in U.S. desert pack rats (genus Neotomae). Transmission among animals was not observed under laboratory conditions, and therefore, the natural modes of transmission are far from clear (24).
In prior experimental work, B. neotomae was noted to cause sustained infection in Mus musculus, Neotoma lepida (desert wood rat), and guinea pigs for periods as long as 18 to 24 months (24). The minimal infectious dose during intraperitoneal inoculation of mice was noted to be only several organisms (19). In contrast, in limited experiments, rabbits, cows, sheep, swine, and mule deer were noted to be relatively resistant to infection (24, 25). Low-inoculum infectivity and sustained infection by B. neotomae, therefore, occurred most readily in rodents.
Although not previously thought capable of infecting humans, B. neotomae was recently isolated from the cerebrospinal fluid of two patients from Costa Rica (26). Potential epidemiological associations and patient comorbidities were not investigated. Therefore, the mechanism of infection and risk factors are uncertain. It is notable that human contact with pack rat middens (nests) is presumably not insignificant, yet there have been only two documented infections, and none in the area of known endemicity, potentially consistent with lower pathogenic potential in humans. Nevertheless, B. neotomae now assumes importance as a rare, and likely underrecognized, zoonotic pathogen capable of causing serious disease in humans.
In our experiments, the B. neotomae infectious course in BALB/c mice showed similarity to B. abortus, B. melitensis, and B. suis infection of domesticated farm animals and primates. For example, during aerosol infection of rhesus monkeys with B. melitensis with inocula ranging generally from 105 to 6 × 106 organisms, both liver and spleen CFU were prominent early in infection (27–29); liver counts dropped from 106 to 104 CFU from days 15 to 45 (29); and organisms were isolated at low levels from liver and spleen 8 to 9 weeks after infection (27, 28), similar to observations in the B. neotomae murine model. Additionally, significant quantities of organisms were isolated from lymph nodes during B. melitensis infection of rhesus monkeys; during B. abortus strain 1308 and/or 544 McEwen infection of cows and bison (30–32); and during human infection, especially in children (33, 34). Therefore, the infectious course of B. neotomae in BALB/c mice and, in particular, the sustained low organism burden and lymph node colonization observed by bioluminescent imaging, appear similar to other examples of native host infection. These findings may reflect the relative relatedness of M. musculus to the native desert pack rat host.
It has been observed previously that Brucella species highly pathogenic for humans display a distinct infectious course in BALB/c mice. Specifically, high-level splenic infection of at least 106 CFU per organ peaks at 2 to 3 weeks postinfection and is sustained for months (7, 35). In contrast, organisms are cleared from the liver within 1 month (7). As mentioned previously, these findings differ from the chronic lower burden of infection observed in native hosts (and humans infected with B. abortus, B. melitensis, and B. suis [1]) and from B. neotomae in the BALB/c model. Findings for B. neotomae, however, were similar to observations of BALB/c infection with the attenuated B. abortus S19 vaccine strain and Brucella ceti (7, 36, 37), a cetacean pathogen. S19 (7) and B. neotomae murine infection (this study) also are both associated with maximum splenic enlargement at 2 weeks. This contrasted with virulent B. abortus 2308 murine infection, in which spleen weight gradually increases during the course of prolonged infection (7). Therefore, the absence of sustained splenic infection and splenomegaly during murine B. neotomae infection was consistent with a pattern associated with Brucella strains and species with lower infectivity and pathogenic potential for humans (7).
The T4SS plays an important role during murine infection with Brucella species (10–14, 38–40). Interestingly, B. neotomae ΔvirB4 infection showed both quantitative and qualitative attenuation, the latter, as observed during BLI studies, suggesting that T4SS contributes to colonization of the reticuloendothelial system (lymph nodes, spleen, and liver). We did not sample lymph nodes for CFU analysis; however, the absence of ΔvirB4 CFU isolated from the thymus, a site of long-term Brucella colonization in animals where examined (34, 41, 42), at any time point also supports a dissemination defect. This qualitative phenotype is reminiscent of the previously observed VirB-dependent colonization of mesenteric lymph nodes, spleen, and liver after gastrointestinal infection of BALB/c mice with B. melitensis 16 M (39) and the VirB-dependent reticuloendothelial system colonization of C57BL/6 mice by B. melitensis after intraperitoneal infection (40).
B. neotomae infection was associated with a granulomatous immune response most evident in the liver, also observed during human infection (43, 44) and infection of mice with zoonotic and attenuated Brucella species and strains (7). Related to these observations, the pattern of inflammatory cytokine transcriptional induction by B. neotomae during early infection was consistent with a virB4-dependent Th1 response previously observed during murine infection (45–47), a Th1 response observed during human infection (48), and the known association of Th1 cytokine induction with a granulomatous response to chronic bacterial infections (49). Expression of TLR2, but not TLR4, was also markedly induced during early infection with T4SS+ organisms. This finding was notable, given the previously described TLR2 dependence of the Th1 cytokine response to B. abortus in mice (50). The selective induction of TLR2 may potentially be explained by the previously described induction of TLR2, but not TLR4, gene expression in murine macrophages by inflammatory cytokines and lipopolysaccharide (51).
There were several limitations to our studies. For BLI, bacteria were labeled with a constitutively expressed bacterial luciferase operon. Luminescence depends on substrate production and the cellular ATP level. Therefore, discrepancies between BLI and CFU counts may be partially accounted for by the culturability and metabolism of organisms, which may differ by body site. Furthermore, BLI intrinsically has much lower sensitivity than both CFU counts and qPCR and therefore cannot provide information on low-burden chronic infection. In addition, the peritoneal cavity and lymph nodes, areas of BLI intensity, were not systematically sampled for CFU counts and histopathological analysis, a goal for future work with this model. Furthermore, bacteria were observed only rarely by TEM, and therefore, ultrastructural observations of phagosomal properties may have been subject to sampling bias. Relatively high inocula were used for infections, and it is possible that the disease course and pathologies would have been different with lower inocula. Cytokine gene expression was measured in whole organs. Systemic cytokine loads and expression in individual cell types were not measured and might have provided a different and more nuanced readout. Nevertheless, our results are consistent with the Th1-type predominant early cytokine response observed in other models. Finally, only one of the original B. neotomae strains isolated by Stoenner and Lackman (19) and Thorpe et al. (24) remains available: strain 5K33, used in our studies. Therefore, extension of our findings to other B. neotomae isolates would be of interest as future strains become available.
In summary, we found that the B. neotomae murine model recapitulates attributes of infection caused by Brucella species that are highly pathogenic for humans and native hosts. Importantly, B. neotomae is neither a biosafety level 3 pathogen nor a select agent. Therefore, we believe it can be safely handled with enhanced biosafety level 2 practices (13), thereby greatly simplifying logistics and accelerating scientific investigation. As such, the B. neotomae murine infection model should provide an informative and tractable in vivo model system for investigation of Brucella pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. neotomae 5K33 (BEI Resources, Manassas, VA) and isogenic ΔvirB4 mutant strains constitutively expressing a chromosomally integrated tdTomato fluorescent protein reporter or a lux operon reporter, both linked to a nourseothricin resistance cassette, were described previously (13). A complemented ΔvirB4 mutant (virB4c) with the virB4 gene expressed constitutively from the broad-host-range, stable pBMTL2 plasmid, was also previously described and characterized in vitro (13). Prior to the experiments, Brucella strains, stored at −80°C, were passaged on sheep blood agar and grown overnight in tryptic soy broth (BD Biosciences, Franklin Lakes, NJ) at 37°C with 5% CO2 in a shaking incubator.
Murine infection.
Mouse experiments were performed under an institutional animal care and use committee-approved protocol. BALB/c mice (Charles River Laboratories, Wilmington, MA), 4 to 6 weeks of age, were infected through i.p. inoculation. To prepare infectious inocula, 1 ml of overnight Brucella culture was pelleted, washed twice with phosphate-buffered saline (PBS), and adjusted to the desired inoculum in 100 µl PBS based on an empirically determined correspondence of an optical density at 600 nm (OD600) of 1.0 to approximately 3 × 109 CFU/ml. At the indicated time points, mouse organs were harvested and homogenized in 1 ml of PBS (pH 7.2) using a sterile 15-ml conical tissue grinder (Biomedical Polymers, Gardner, MA). Serial dilutions of the homogenates were plated on tryptic soy agar (TSA) containing 50 µg/ml nourseothricin for CFU determination.
Whole-mouse imaging.
Whole-mouse imaging was performed on a Xenogen IVIS-50TM imaging system with Living Image software using a 3-min exposure and a field of view of 12 cm. The system software quantifies the total luminescence flux signal (photons [p] per second) and displays average radiance intensity (photons per second per square centimeter per steradian [sr]) using a color-coded histogram with user-defined minimum and maximum signal thresholds.
qPCR analysis.
For qPCR experiments to quantify B. neotomae in tissues, 50 mg of each tissue sample was extracted using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Primers GCCTCCAGTCGATTGTTGGGGCAC and GTGACTGTCCGCAAGCTTCAAGCC amplify a 177-bp fragment within IS711 and were identified based on multiple-sequence alignment of IS711 elements from Brucella sp. strain B14/94, B. ceti B1/94, B. pinnipedialis B2/94, B. abortus B12, and B. abortus B16 (GenBank accession numbers AF242534, FJ376557, FJ376556, JF345125, and JF345126, respectively) using the Lasergene software package (DNAStar, Madison, WI). The reagents in the DyNAmo HS SYBR Green qPCR kit (Thermo Scientific) were used to set up qPCRs. Cycling and detection were performed on a CFX 384 real-time PCR detection system (Bio-Rad, Hercules, CA) with an initial 95°C denaturation for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 40 s.
To convert quantification cycle (Cq) values obtained during real-time PCR detection to B. neotomae genome equivalents, a 354-bp fragment of IS711 from B. neotomae was amplified using primers CGCGAATTCGAGACCCATGAATGCGGTCAA and CGCGTCGACACGACCAAGCTGCATGCTGTT and cloned into the EcoRI/SalI restriction sites of the pMOD3 vector (Epicentre, Madison, WI). Plasmid DNA was extracted using a QIAprep miniprep kit (Qiagen, Valencia, CA), quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA), serially diluted, and analyzed by qPCR to establish a correspondence between Cq values and IS711 quantity. Following qPCR of tissue samples, the IS711 quantity was then determined based on the calibration curve and converted to B. neotomae genome equivalents by dividing by the IS711 copy number in B. neotomae 5K33 (n = 8) inferred from previously published Southern blots (52) and the genome sequence (accession number KN046827.1).
Frozen-section fluorescence microscopy.
Tissue immersed in OCT compound (Sakura Finetek USA, Inc., Torrance, CA) was frozen in liquid nitrogen on a Tissue-Tek cryomold (Sakura Finetek). Frozen sections (10 μm thick) were then cut using a Leica CM3050s cryostat (Leica Biosystems, Buffalo Grove, IL), applied to Superfrost Plus glass slides (Fisher Scientific, Hampton, NH), and fixed with 4% formaldehyde at 4°C for 12 h. The fixed sections were washed twice with PBS, stained in Gill’s hematoxylin solution no. 3 (Fisher Scientific) for 3 min, washed in tap water for 5 min, immersed in Scott’s bluing solution for 3 min (53, 54), and counterstained with alcoholic Eosin Y (Fisher Scientific) for 3 min. Tissue sections were subsequently dehydrated with graded series of ethanol, cleared with xylene, and coverslipped.
Visualization of tdTomato-expressing bacteria was performed using a Zeiss LSM 880 confocal microscope (Carl Zeiss Microscopy, Thornwood, NY) with a 562.5- to 587.5-nm bandpass filter as previously described (13). The microscope PMT (photomultiplier tube) setting was used to detect corresponding bright-field images.
Electron microscopy.
Organ specimens were fixed in Trump's fixative containing 4% formaldehyde and 1% glutaraldehyde, washed in 0.1 M cacodylate buffer, and postfixed in 1% OsO4 as described previously (55). Samples were then dehydrated in ascending alcohol washes and embedded in Eponate 12 resin (Ted Pella, Inc., Redding, CA). Digital images of ultrathin sections were acquired with a Jeol (Tokyo, Japan) MEM-1011 transmission electron microscope.
Cytokine mRNA expression.
Total RNA was extracted from liver and spleen tissue using an RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. After RNA elution, reverse transcription-quantitative PCR (RT-qPCR) was performed to determine relative transcription of cytokine-related murine genes normalized to β-actin expression. The following primer sets designed with the Integrated DNA Technologies (Skokie, IL) PrimerQuest tool were used (forward primer, reverse primer, and murine gene accession number): IL-2 (TGAGCAGGATGGAGAATTACAG, GAGGTCCAAGTTCATCTTCTAGG, NM_008366), IL-4 (TTGAGAGAGATCATCGGCATTT, CTCACTCTCTGTGGTGTTCTTC, NM_021283), IL-10 (AACATACTGCTAACCGACTCC, TCCTTGATTTCTGGGCCATG, NM_010548.2), TNF-α (GTTGTACCTTGTCTACTCCCAG, GGTTGACTTTCTCCTGGTATGAG, NM_013693.3), TLR2 (CACTATCCGGAGGTTGCATATC, GGAAGACCTTGCTGTTCTCTAC, NM_011905), TLR4 (CAGCAGAGGAGAAAGCATCTAT, GTAGTGAAGGCAGAGGTGAAA, NM_021297), and β-actin (GTGGGAATGGGTCAGAAGG, AGCTCATTGTAGAAGGTGTGG, NM_007393.5). Briefly, cDNA synthesis was performed using 2 µg of total RNA, a random primer mixture, and Moloney murine leukemia virus (MMuLV) reverse transcriptase (New England Biolabs). Subsequent qPCR was performed with the DyNAmo HS SYBR Green qPCR kit, 1 µl of cDNA, and 0.2 μM primers. Cycling condition were 3 min at 95°C, followed by 35 cycles of 45 s at 95°C, 45 s at 55°C, and 45 s at 72°C, followed by 10 min at 72°C.
Statistics.
Unless otherwise specified, the nonparametric Kruskal-Wallis analysis of variance (ANOVA) test was performed, followed by Dunn’s test for post hoc comparisons with control groups, using Prism 7 for Mac OSX (GraphPad Software, Inc., La Jolla, CA). A P value of ≤0.050 was considered statistically significant.
ACKNOWLEDGMENTS
We acknowledge Meaghan Fox and John-Mac Mroczka from the Beth Israel Deaconess Medical Small Animal Imaging Facility for their expertise. We also thank Katelyn Zulauf and Kenneth Smith for thoughtful comments on the manuscript.
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01 AI099122 to J.E.K.
The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Dalrymple-Champneys W. 1960. Brucella infection and undulant fever in man. Oxford University Press, London, United Kingdom. [Google Scholar]
- 2.Varona JF, Guerra JM, Guillen V, Guillen S, Menassa A, Palenque E. 2002. Isolated cervical lymphadenopathy as unique manifestation of brucellosis. Scand J Infect Dis 34:538–540. doi: 10.1080/003655402320208802. [DOI] [PubMed] [Google Scholar]
- 3.Jeroudi MO, Halim MA, Harder EJ, Al-Siba’i MB, Ziady G, Mercer EN. 1987. Brucella endocarditis. Br Heart J 58:279–283. doi: 10.1136/hrt.58.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajapakse CN. 1995. Bacterial infections: osteoarticular brucellosis. Baillieres Clin Rheumatol 9:161–177. doi: 10.1016/S0950-3579(05)80153-0. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grillo MJ, Blasco JM, Gorvel JP, Moriyon I, Moreno E. 2012. What have we learned from brucellosis in the mouse model? Vet Res 43:29. doi: 10.1186/1297-9716-43-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myeni S, Child R, Ng TW, Kupko JJ III, Wehrly TD, Porcella SF, Knodler LA, Celli J. 2013. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog 9:e1003556. doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otin C, Virgin HW, Celli J. 2012. Selective subversion of autophagy complexes facilitates completion of the brucella intracellular cycle. Cell Host Microbe 11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna N, Jiménez de Bagüés MP, Ouahrani-Bettache S, El Yakhlifi Z, Köhler S, Occhialini A. 2011. The virB operon is essential for lethality of Brucella microti in the Balb/c murine model of infection. J Infect Dis 203:1129–1135. doi: 10.1093/infdis/jiq163. [DOI] [PubMed] [Google Scholar]
- 11.Sun YH, den Hartigh AB, Santos RL, Adams LG, Tsolis RM. 2002. virB-Mediated survival of Brucella abortus in mice and macrophages is independent of a functional inducible nitric oxide synthase or NADPH oxidase in macrophages. Infect Immun 70:4826–4832. doi: 10.1128/IAI.70.9.4826-4832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolán HG, Tsolis RM. 2007. Mice lacking components of adaptive immunity show increased Brucella abortus virB mutant colonization. Infect Immun 75:2965–2973. doi: 10.1128/IAI.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang YS, Kirby JE. 2017. Promotion and rescue of intracellular Brucella neotomae replication during coinfection with Legionella pneumophila. Infect Immun 85:e00991-16. doi: 10.1128/IAI.00991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palomares-Resendiz E, Arellano-Reynoso B, Hernandez-Castro R, Tenorio-Gutierrez V, Salas-Tellez E, Suarez-Guemes F, Diaz-Aparicio E. 2012. Immunogenic response of Brucella canis virB10 and virB11 mutants in a murine model. Front Cell Infect Microbiol 2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montaraz JA, Winter AJ. 1986. Comparison of living and nonliving vaccines for Brucella abortus in BALB/c mice. Infect Immun 53:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edmonds MD, Cloeckaert A, Elzer PH. 2002. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet Microbiol 88:205–221. doi: 10.1016/S0378-1135(02)00110-4. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez de Bagues MP, Iturralde M, Arias MA, Pardo J, Cloeckaert A, Zygmunt MS. 2014. The new strains Brucella inopinata BO1 and Brucella species 83-210 behave biologically like classic infectious Brucella species and cause death in murine models of infection. J Infect Dis 210:467–472. doi: 10.1093/infdis/jiu102. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez de Bagüés MP, Ouahrani-Bettache S, Quintana JF, Mitjana O, Hanna N, Bessoles S, Sanchez F, Scholz HC, Lafont V, Köhler S, Occhialini A. 2010. The new species Brucella microti replicates in macrophages and causes death in murine models of infection. J Infect Dis 202:3–10. doi: 10.1086/653084. [DOI] [PubMed] [Google Scholar]
- 19.Stoenner HG, Lackman DB. 1957. A new species of brucella isolated from the desert wood rat, Neotoma lepida Thomas. Am J Vet Res 18:947–951. [PubMed] [Google Scholar]
- 20.Moustafa DA, Jain N, Sriranganathan N, Vemulapalli R. 2010. Identification of a single-nucleotide insertion in the promoter region affecting the sodC promoter activity in Brucella neotomae. PLoS One 5:e14112. doi: 10.1371/journal.pone.0014112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn WA. 1994. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol 4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 22.Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, Song CH, Lee SH, Lee ZW, Cho SN, Kim JM, Friedman RL, Jo EK. 2010. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog 6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Detilleux PG, Deyoe BL, Cheville NF. 1990. Entry and intracellular localization of Brucella spp. in Vero cells: fluorescence and electron microscopy. Vet Pathol 27:317–328. doi: 10.1177/030098589002700503. [DOI] [PubMed] [Google Scholar]
- 24.Thorpe BD, Sidwell RW, Lundgren DL. 1967. Experimental studies with four species of Brucella in selected wildlife, laboratory, and domestic animals. Am J Trop Med Hyg 16:665–674. doi: 10.4269/ajtmh.1967.16.665. [DOI] [PubMed] [Google Scholar]
- 25.Beal GA, Lewis RE, McCullough NB, Claflin RM. 1959. Experimental infection of swine with Brucella neotomae. Am J Vet Res 20:872–875. [PubMed] [Google Scholar]
- 26.Suárez-Esquivel M, Ruiz-Villalobos N, Jiménez-Rojas C, Barquero-Calvo E, Chacón-Díaz C, Víquez-Ruiz E, Rojas-Campos N, Baker KS, Oviedo-Sánchez G, Amuy E, Chaves-Olarte E, Thomson NR, Moreno E, Guzmán-Verri C. 2017. Brucella neotomae infection in humans, Costa Rica. Emerg Infect Dis 23:997–1000. doi: 10.3201/eid2306.162018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mense MG, Borschel RH, Wilhelmsen CL, Pitt ML, Hoover DL. 2004. Pathologic changes associated with brucellosis experimentally induced by aerosol exposure in rhesus macaques (Macaca mulatta). Am J Vet Res 65:644–652. doi: 10.2460/ajvr.2004.65.644. [DOI] [PubMed] [Google Scholar]
- 28.Henning LN, Miller SM, Pak DH, Lindsay A, Fisher DA, Barnewall RE, Briscoe CM, Anderson MS, Warren RL. 2012. Pathophysiology of the rhesus macaque model for inhalational brucellosis. Infect Immun 80:298–310. doi: 10.1128/IAI.05878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell-Lodrigue KE, Killeen SZ, Ficht TA, Roy CJ. 2018. Mucosal bacterial dissemination in a rhesus macaque model of experimental brucellosis. J Med Primatol 47:75–77. doi: 10.1111/jmp.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xavier MN, Paixão TA, Poester FP, Lage AP, Santos RL. 2009. Pathological, immunohistochemical and bacteriological study of tissues and milk of cows and fetuses experimentally infected with Brucella abortus. J Comp Pathol 140:149–157. doi: 10.1016/j.jcpa.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Davis DS, Templeton JW, Ficht TA, Williams JD, Kopec JD, Adams LG. 1990. Brucella abortus in captive bison. I. Serology, bacteriology, pathogenesis, and transmission to cattle. J Wildl Dis 26:360–371. doi: 10.7589/0090-3558-26.3.360. [DOI] [PubMed] [Google Scholar]
- 32.Payne JM. 1959. The pathogenesis of experimental brucellosis in the pregnant cow. J Pathol Bacteriol 78:447–463. doi: 10.1002/path.1700780211. [DOI] [PubMed] [Google Scholar]
- 33.Poston MA, Parsons PB. 1940. Isolation of Brucella from lymph nodes. J Infect Dis 66:86–90. doi: 10.1093/infdis/66.1.86. [DOI] [Google Scholar]
- 34.von Bargen K, Gagnaire A, Arce-Gorvel V, de Bovis B, Baudimont F, Chasson L, Bosilkovski M, Papadopoulos A, Martirosyan A, Henri S, Mège JL, Malissen B, Gorvel JP. 2015. Cervical lymph nodes as a selective niche for Brucella during oral infections. PLoS One 10:e0121790. doi: 10.1371/journal.pone.0121790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford RM, Van De Verg L, Yuan L, Hadfield TL, Warren RL, Drazek ES, Houng HH, Hammack C, Sasala K, Polsinelli T, Thompson J, Hoover DL. 1996. Deletion of purE attenuates Brucella melitensis infection in mice. Infect Immun 64:2188–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackaness GB. 1964. The immunological basis of acquired cellular resistance. J Exp Med 120:105–120. doi: 10.1084/jem.120.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nymo IH, Arias MA, Pardo J, Álvarez MP, Alcaraz A, Godfroid J, Jiménez de Bagüés MP. 2016. Marine mammal Brucella reference strains are attenuated in a BALB/c mouse model. PLoS One 11:e0150432. doi: 10.1371/journal.pone.0150432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Hartigh AB, Rolán HG, de Jong MF, Tsolis RM. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J Bacteriol 190:4427–4436. doi: 10.1128/JB.00406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paixão TA, Roux CM, den Hartigh AB, Sankaran-Walters S, Dandekar S, Santos RL, Tsolis RM. 2009. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect Immun 77:4197–4208. doi: 10.1128/IAI.00417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajashekara G, Glover DA, Krepps M, Splitter GA. 2005. Temporal analysis of pathogenic events in virulent and avirulent Brucella melitensis infections. Cell Microbiol 7:1459–1473. doi: 10.1111/j.1462-5822.2005.00570.x. [DOI] [PubMed] [Google Scholar]
- 41.Tessaro SV, Forbes LB. 2004. Experimental Brucella abortus infection in wolves. J Wildl Dis 40:60–65. doi: 10.7589/0090-3558-40.1.60. [DOI] [PubMed] [Google Scholar]
- 42.Ozkaraca M, Ceribasi S, Ceribasi AO, Kilic A, Ongor H. 2016. The role of apoptosis and autophagy in bovine abortions associated with Brucella spp. Acta Veterinaria 66:37–50. doi: 10.1515/acve-2016-0003. [DOI] [Google Scholar]
- 43.Hunt AC, Bothwell PW. 1967. Histological findings in human brucellosis. J Clin Pathol 20:267–272. doi: 10.1136/jcp.20.3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen FB, Robins B, Lipstein W. 1957. Isolation of Brucella abortus by percutaneous liver biopsy. N Engl J Med 257:228–230. doi: 10.1056/NEJM195708012570508. [DOI] [PubMed] [Google Scholar]
- 45.Im YB, Park WB, Jung M, Kim S, Yoo HS. 2016. Evaluation of Th1/Th2-related immune response against recombinant proteins of Brucella abortus infection in mice. J Microbiol Biotechnol 26:1132–1139. doi: 10.4014/jmb.1512.12046. [DOI] [PubMed] [Google Scholar]
- 46.Agranovich I, Scott DE, Terle D, Lee K, Golding B. 1999. Down-regulation of Th2 responses by Brucella abortus, a strong Th1 stimulus, correlates with alterations in the B7.2-CD28 pathway. Infect Immun 67:4418–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rolan HG, Tsolis RM. 2008. Inactivation of the type IV secretion system reduces the Th1 polarization of the immune response to Brucella abortus infection. Infect Immun 76:3207–3213. doi: 10.1128/IAI.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodríguez-Zapata M, Matías MJ, Prieto A, Jonde MA, Monserrat J, Sánchez L, Reyes E, De la Hera A, Alvarez-Mon M. 2010. Human brucellosis is characterized by an intense Th1 profile associated with a defective monocyte function. Infect Immun 78:3272–3279. doi: 10.1128/IAI.01385-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. 2002. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol 168:4620–4627. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 50.Ferrero MC, Hielpos MS, Carvalho NB, Barrionuevo P, Corsetti PP, Giambartolomei GH, Oliveira SC, Baldi PC. 2014. Key role of Toll-like receptor 2 in the inflammatory response and major histocompatibility complex class II downregulation in Brucella abortus-infected alveolar macrophages. Infect Immun 82:626–639. doi: 10.1128/IAI.01237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuguchi T, Musikacharoen T, Ogawa T, Yoshikai Y. 2000. Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J Immunol 165:5767–5772. doi: 10.4049/jimmunol.165.10.5767. [DOI] [PubMed] [Google Scholar]
- 52.Bricker BJ, Ewalt DR, MacMillan AP, Foster G, Brew S. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J Clin Microbiol 38:1258–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheehan DC, Hrapchak BB. 1980. Theory and practice of histotechnology, 2nd ed Battelle Press, Columbus, OH. [Google Scholar]
- 54.Crookham JN. 1991. Hazardous chemicals in the histopathology laboratory: regulations risks handling and disposal, 2nd ed ANATECH, Battle Creek, MI. [Google Scholar]
- 55.Chiaraviglio L, Duong S, Brown DA, Birtles RJ, Kirby JE. 2010. An immunocompromised murine model of chronic Bartonella infection. Am J Pathol 176:2753–2763. doi: 10.2353/ajpath.2010.090862. [DOI] [PMC free article] [PubMed] [Google Scholar]