Summary
The microbiome is represented by microorganisms which live in a symbiotic way with the mammalian. Microorganisms have the ability to influence different physiological aspects such as the immune system, metabolism and behaviour. In recent years, several studies have highlighted the role of the microbiome in the pathogenesis of autoimmune diseases. Notably, in systemic lupus erythematosus an alteration of the intestinal flora (lower Firmicutes/Bacteroidetes ratio) has been described. Conversely, changes to the gut commensal and periodontal disease have been proposed as important factors in the pathogenesis of rheumatoid arthritis. At the same time, other autoimmune diseases (i.e. systemic sclerosis, Sjögren’s syndrome and anti‐phospholipid syndrome) also share modifications of the microbiome in the intestinal tract and oral flora. Herein, we describe the role of the microbiome in the maintenance homeostasis of the immune system and then the alterations of the microorganisms that occur in systemic autoimmune diseases. Finally, we will consider the use of probiotics and faecal transplantation as novel therapeutic targets.
Keywords: anti‐phospholipid, autoimmune diseases, faecal transplantation, microbiome, probiotics, rheumatoid arthritis, syndrome systemic lupus erythematosus, systemic sclerosis, Sjögren’s syndrome
Introduction
The human body is densely populated by commensal and symbiotic microbes, the majority of the constituent microorganisms being bacteria. These microbes occupy different habitats such as gut, skin, vagina and oral. Not only are the types and abundance of microbes different in different organs, but these may also differ in different individuals. The genome of these microorganisms and their ecosystems constitute a microbiome. Factors such as diet, environment, host genetics and mode of delivery may be the reason behind the wide microbial diversity 1. The presence of the microbiome and microbial products regulate the development and function of the immune system in the host. Furthermore, other physiological aspects of the mammalian (metabolism, behaviour) are affected by commensal microorganisms 2. Recently, many scientists have focused on the importance of the commensal bacteria in the pathogenesis of several diseases, including autoimmune diseases. The aim of this review is to describe the main alterations of the microbiome that occur in autoimmune diseases 1.
Alterations of the microbiome in pregnancy and childhood
During pregnancy, the microbiome undergoes profound changes, in particular in the vagina and gut. In a recent study, Koren et al. 2 found that there were differences between the microbiome in women in the first trimester of pregnancy with that of the third trimester: in the last months of pregnancy an abundance of Proteobacteria and Actinobacteria and a depletion of Faecalibacterium (bacterium butyrate‐producer with anti‐inflammatory effects) resulted. These alterations of microbiome, known as dysbiosis, can induce weight gain, insulin‐resistant and metabolic inflammation if the microorganisms contained in the gut of the mice at the third trimester have been transferred into germ‐free mice 2, 3. The fetal gastrointestinal tract is believed to be sterile, and microorganisms colonize the intestine of the fetus during delivery through the birth canal (Fig. 1) 4, 5. During childhood, the gut microbiome can be influenced by several environmental factors, such as geographic area, breast feeding, solid food and ways of delivery. Vaginally delivered infants acquire bacterial communities resembling their own mother’s vaginal microbiota (Lactobacillus, Prevotella, Sneathia spp.). Conversely, Caesarean‐section infants harbour bacterial communities similar to those found on the skin surface (Staphylococcus, Corynebacterium and Propionibacterium) 4, 6. The ability of the gastrointestinal mucosa of the newborn to adapt to the colonization of microorganisms is not entirely understood. Realistically, the colostrum and breast milk are rich in immunoglobulin (Ig)A, which are important to neutralize pathogens and to avoid translocation through the intestinal epithelia, therefore ensuring the homeostasis between symbiotic gut bacteria and the mucosa epithelium. Maternal milk contains several metabolites, such as gangliosides, lactoferrin (Lf) and human milk oligosaccharides (HMOs) that provide protection against anti‐infective agents 1, 7. Furthermore, HMOs exhibit some prebiotic effects contributing to the colonization of specific bacteria, i.e. Bifidobacterium longum, thus preserving the integrity of the intestinal barrier. Moreover, existing dendritic cells in breast milk might contribute to neonatal immunological imprinting by influencing the nature of the immune response to the commensal antigens 8, 9, 10, 11. The immaturity of the immune system of the newborn and tolerogenic environment factors could explain how the microbiome has been accepted by the neonate gut. Therefore, the dialogue between commensals and host plays a crucial role in the development and homeostasis of the immune system 1, 12, 13. This is possible through pattern recognition (PRR) receptors, that include Toll‐like receptor (TLR) families, nucleotide‐binding oligomerization domains (NOD) as receptors (NLR), type C lectin receptors (CLR), cytosolic DNA receptors (CDR) and retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs) 14. In part, TLRs are present mainly on the membrane of immune and epithelial cells and are able to recognize conserved molecular motifs, such as molecular models associated with microbes (MAMPS) expressed by the resident microbiota or molecular models associated with pathogens (PAMPS) produced by microbial invaders 15, 16. The microorganism commensals are also implicated in the development both of the lymphoid structure in the intestinal tract (i.e. Payer’s patch, isolated lymphoid follicles) and for the promotion of epithelial cell maturation and angiogenesis of gastrointestinal mucosa 1, 17, 18. Furthermore, the symbiotic bacteria (Bacteroides fragilis), through the production of sphingolipids, can influence activation of the invariant natural killer T (iNK T) and then conditioning the immune system into adulthood 19, 20. It is now clear that the microbiome induces regulatory responses and consequently the immunological tolerance through manifold pathways. In this regard, the regulatory T cell (Treg) forkhead box protein 3 (FoxP3+), which manifests itself both in the thymus and in the extrathymus through (i) production of regulatory cytokines [transforming growth factor (TGF)‐β, interleukin (IL)‐10 and IL‐35], (ii) modulations of the antigen‐presenting cell (APC) function [lymphocyte‐activation gene 3 (LAG‐3), cytotoxic T lymphocyte antigen (CTLA)‐4, granzyme/perforin] and (iii) alteration of the cellular metabolism [CD25, CD39/CD73, indoleamine 2,3‐dioxygenase (IDO) induction in dendritic cells (DC) ], have all been associated with the function of Treg cells in distinct inflammatory contexts 21, 22. In the intestinal tract, the commensal microorganisms could modulate the Treg function through the production of IL‐10 and TGF‐β. B. fragilis produces polysaccharide A (PSA) capable of inducing IL‐10 production by the intestinal T cells, and limit T helper type 17 (Th17) responses during intestinal inflammation 23. At the same time, other bacterial metabolites, such as short‐chain fatty acids (SCFAs), and in particular butyrate, increase the function of Treg cells and macrophages by inhibiting the expression of the histone deacetylases gene (HDAC) 23, 24. However, alterations in the microbiome (dysbiosis) can result from the exposure to various environmental factors, including diet, toxins, drugs and pathogens. Of these, enteric pathogens have the greatest potential to cause microbial dysbiosis and can trigger both local and systemic inflammation. The alteration of the composition of the microbiome and the barrier function can allow the development of autoimmune and chronic inflammatory diseases, metabolic dysfunction and cancer 1, 5.
Figure 1.
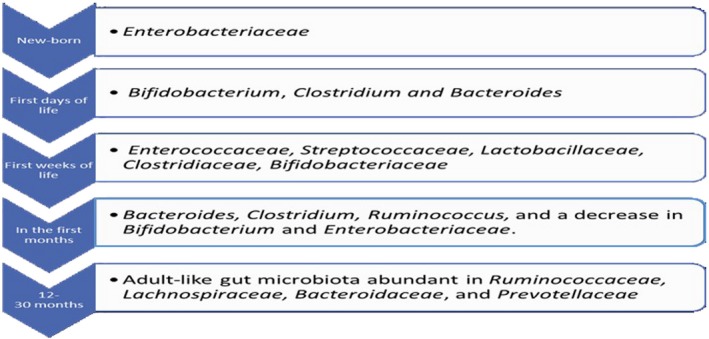
The composition of the gut microbiota throughout life.
Microbiome and autoimmune diseases
Autoimmune diseases (AIDs) result from an individual’s immune system attacking self‐tissues, with an estimated incidence of approximately 3–5% worldwide. The pathogenesis is not understood completely, but environmental factors (life‐style, diet, drugs, infections) and certain genetic backgrounds have been proposed 25, 26. The human microbiome might be a major player in autoimmunity, as the loss of immune tolerance can be caused by microbial composition changes 1, 5. Microorganisms can elicit the immune response against the host if the mechanisms of tolerance fail for several reasons (Fig. 2) 27, 28, 29, 30, 31. Recently, Rinaldi et al. 32 found that autoantibodies directed against the cell wall mannan of the yeast Saccharomyce cerevisiae (phosphopeptidomannan), a ubiquitous commensal microorganism, were detected in several autoimmune diseases with different sensibilities (i.e. rheumatoid arthritis, systemic lupus erythematosus, anti‐phospholipid syndrome). More specifically, anti‐S. cerevisiae antibodies (ASCAs) are a specific serological marker of Crohn’s disease (CD) by appearing before CD onset in 32% of cases. In addition, S. cerevisiae is used as an adjuvant in vaccines, and this has led scientists to think of a hypothetical risk of developing abnormal immune activation that may be associated with an autoimmune/inflammatory syndrome induced by adjuvants (ASIA) 32, 33. Inflammatory bowel diseases (IBD) such as CD and ulcerative colitis (UC) represent an example of how the alteration of gut microbiome could induce disease. Notably, numerous studies have shown that both CD and UC are associated with a reduced complexity of the commensal microbiota and consistent shifts to a dysbiotic state. In a similar manner to that observed during acute mucosal infections, both CD and UC are characterized by the outgrowth of the phyla proteobacteria, in particular the Enterobacteriaceae family and Fusobacteriaceae 34, 35, 36. Moreover, adherent‐invasive E. coli, Yersinia and Clostridium difficile are much more common in patients affected by Crohn’s disease than healthy individuals and, in some mouse models, these bacteria have been shown to be key contributors to IBD 37, 38, 39. Evidence from neuroscience research suggests that the microbiome is essential for development and maturation of the central nervous system, as well for behavioural and cognitive functions. Communication between the central nervous system and gut is bidirectional, and is referred to as the ‘gut microbiota–brain axis’. The gut can interact with the brain through several pathways and through commensal metabolism, such as short‐chain fatty acid (SCAFs), 5‐hydroxytryptamine (5‐HT) and gamma‐aminobutyric acid (GABA) 40, 41. Multiple sclerosis (MS) is an autoimmune disease characterized by the invasion of the central nervous system by immune cells (i.e. CD4 and CD8 T cells, B cells and activated monocytes), resulting in the demyelination of neurones and subsequent pathology 42. Patients affected by MS exhibit a decrease in the percentage of several Bacteroides (i.e. B. stercoris, B. coprocola, B. coprophilus), Faecalibacterium and SCAFs producing bacteria and increase of Methanobrevibacter, Enterobacteriaceae and Akkermansia 43. Conversely, treatment with disease‐modifying drugs induces an increase of Prevotella compared with untreated patients 44. Furthermore, intestinal colonization by the C. perfringens type B is associated with relapse in MS. Toxins produced by C. perfringens can induce microvascular complications leading to neuronal and oligodendrocyte damage 43, 44, 45. According to many researchers, dysbiosis also seems to be involved in the pathogenesis of type 1 diabetes mellitus (T1DM). In an interesting Chinese study, the faecal samples of children with T1DM were lower in abundance of bacteria than healthy controls, in particular Intestinimonas, a newly isolated Gram‐positive and anaerobic bacteria producing butyrate. Comparatively, an increase of Blautia was found in these patients 46.
Figure 2.
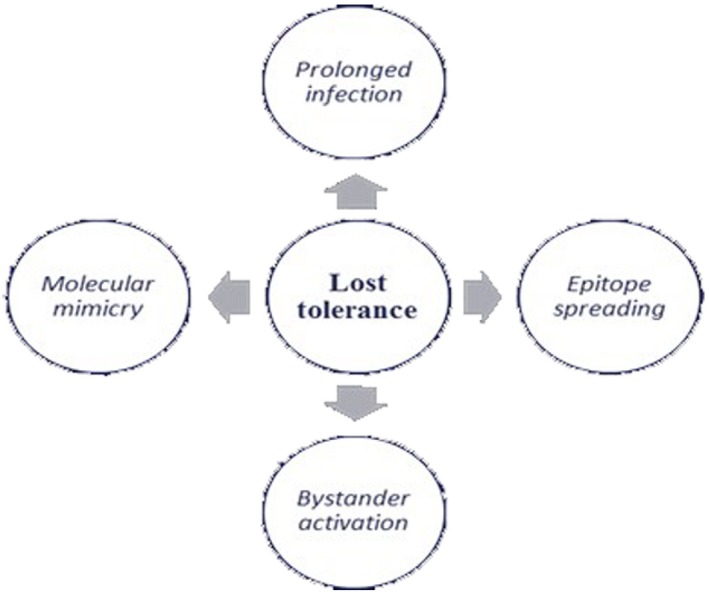
The loss of tolerance.
Rheumatoid arthritis (RA)
RA is a chronic autoimmune disease characterized by inflammation and pain of the joints with varying degrees of systemic involvement in the presence of rheumatoid factor (RF) and anti‐citrullinate peptide antibodies (ACPA). Although the genetic background plays an important role, other environmental risk factors, such as tobacco use and infection, have shown strong evidence for the pathogenesis of the disease. Recently, alteration of microbiota has attracted the attention of many researchers 47. Several studies have found an association between periodontitis and RA. The chronic oral inflammation caused by oral bacteria and leucocyte infiltration with progressive destruction of the alveolar bone seems to share the same pathogenetic mechanisms with RA: (i) accumulation of leucocyte infiltration; (ii) release of inflammatory cytokines and mediators such as prostaglandin E2 (PGE2), tumour necrosis factor (TNF)‐α, interleukin (IL)‐1b, IL‐6, IL‐12, IL‐17, IL‐18, IL‐33, granulocyte–macrophage colony‐stimulating factor (GM‐CSF), monocyte (CSF (M‐CSF), receptor activator of nuclear factor kappa‐Β ligand (RANKL), matrix metalloproteinases (MMPs) and nitric oxide (NO) 48. Furthermore, Porphyromonas gingivalis, a bacterium linked to the pathogenesis of the periodontitis, has the unique ability to convert the amino acid arginine to the amino acid citrulline: a process called citrullination, through the production of peptidylarginine deiminase. The protein, which contains the amino acid citrulline, is recognized by the autoantibodies ACPA that are highly specific for RA 49. Recently, Brusca et al. 50 found that there were more organisms than simply P. gingivalis which cause periodontal disease (i.e. Anaerglobus geminatus and Prevotella/Leptotrichia) and were related to the presence of anti‐citrullination antibodies. It was found that there is a positive correlation between antibodies to P. gingivalis and the presence of anti‐ cyclic citrullinated peptide (CCP) in patients with juvenile idiopathic arthritis. In the oral flora, bacteria such as P. intermedia/Tannerella forsythia were found and high titres of antibodies against these microorganisms have been detected in the serum and synovial fluids of patients with RA 51. Otherwise, other scientists 52 found that IgG antibodies to P. intermedia and C. ochracea were associated with a lower prevalence of RF 52, 53, 54. However, there is evidence that the treatment of periodontal disease may improve RA symptoms 55. Unlike these studies, analysis of a cohort of 292 patients with RA did not demonstrate any correlation between periodontal disease and RA 56. The oral cavity is not only the place in the human body that was colonized by bacteria. The majority of microorganisms present in our body are harboured in the human gut, thus affecting the balance between pro‐ and anti‐inflammatory immune responses 57. As mentioned above, the butyrate produced by intestinal bacteria could explain anti‐inflammatory properties through the differentiation of Treg lymphocytes. Also, polysaccharide A produced by B. fragilis binds TLR‐2 on the surface of lymphocytes and DCs. It thus promotes the maturation of CD4+ lymphocytes into Treg lymphocytes and the production of anti‐inflammatory cytokines such as IL‐10 54, 55, 56, 57, 58. Rogier et al. 59 reported a decrease of Bacteroidaceae and an increase of Firmicutes and Proteobacteria (i.e. Ruminococcaceae, Lachnospiraceae) and Desulfovibrinocaceae during the immune‐priming phase of arthritis in the collagen‐induced arthritis (CIA) mice model. The authors suggested particularly that an alteration of the intestinal microbiome during the immune‐priming phase could induce an inflammatory response in the joints. Conversely, administration of antibiotics decreased the severity of arthritis in mice models. This result is due to a decrease in the abundance of segmented filamentous bacteria, a commensal that influences the adaptive immunity and the innate immune system through an increase of IgA secretion and a development of Th17 lymphocytes, respectively 59, 60. Recently, other studies have demonstrated that the gut microbiome of a new‐onset RA was characterized by an increase of P. copri and lower numbers of Bifidobacteria, the Bacteroides–Porphyromonas group, the B. fragilis subgroup and Eubacterium rectal–Clostridium coccoides 52, 53, 61. Interestingly, Moreno et al. 62 suggested that P. copri might play a key role in the pathogenesis of RA in patients with a lower level of genetic susceptibility, where environmental factors are critical for the development of the disease. In accordance with Scher 63, P. copri was inversely proportional to the presence of shared‐epitope risk alleles and abundance of Bacteroides. Chen et al. 64 obtained different results on the basis of stool evaluations from 40 patients with RA and a control group. No relationship between the occurrence of P. copri with early RA was determined. In contrast, Eggerthella, Faecalibacterium and Colinsella were found in faecal samples of patients affected by RA. Larger counts of L. salivarius, L. iners and L. ruminis were found in patients in the control group and L. mucosae was found solely in patients with RA. Also, an increase of Clostridium bacteria was detected in the faecal samples of mice models of RA with the RA‐susceptible DRB1*0401 gene 65, 66. Other authors have hypothesized that the microbiome diversity could influence the response to therapy with methotrexate (MTX) in patients affected by RA. A microbiome rich in Prevotella has been shown to decrease tetrahydrofolate (THF) biosynthesis through a decrease of purine metabolic pathway. According to the author, this may have therapeutic implications, because methotrexate (MTX), a folate analogue, and a dihydrofolate (DHF) reductase inhibitor exhibits a pharmacological effect on the same metabolic pathway 63. Nevertheless, many other scientists have shown that the pharmacological activity of MTX occurs mainly through an increase of cyclic AMP concentration 67. Alterations of commensal bacteria have also been discovered in the low respiratory tract and in the urinary system. Notably, an association between erosive RA and Pseudonocardia colonization in the lung was suggested 68, and the number of Proteobacteriacea was increased in patients affected by RA 69. Despite previous speculations, other studies report a decrease in an abundance of Actinomyces, Prevotella and Porphyromonas in the bronchoalveolar lavage (BAL) of RA patients 68. Conversely, several studies found a strong correlation between urinary infections supported by Proteus mirabilis and rheumatic disease 70, given that patients affected by RA exhibit antibodies against P. mirabilis in the serum. Thus, the authors suggested that chronic infection triggers activation of the immune system and the development of RA in certain genetic backgrounds and in the presence of unknown environmental factors 71, 72.
Systemic lupus erythematosus (SLE)
SLE is a heterogeneous autoimmune disease with a wide range of clinical and serological manifestations. The disease course is marked by remissions and relapses and may vary from mild to severe. The prevalence ranges from 20 to 200 cases per 100 000 people and women are affected more often than men. The pathogenesis of SLE is not understood completely; it is thought to involve hormonal factors, environmental factors (infection, drugs, ultraviolet A light) and genetic causes 73. However, in previous years it has been suggested, as in other autoimmune diseases, that the gut microbiota could play an important role in the development of SLE (Table 1). In patients with SLE, a lower Firmicutes/Bacteroidetes ratio and the abundance of several genera have been reported: Rhodococcus, Eggerthella, Klebsiella, Prevotella, Eubacterium and Flavonifractor were enriched significantly, while Dialister and Pseudobutyrivibrio were decreased in SLE patients 74, 75. It is not known whether the alteration of commensal bacteria results as a consequence of the disease process or dysbiosis contributes to the lupus onset 76. According to Johnson et al. 77, dysbiosis is associated with local inflammatory responses (specifically the Th17 response) and high circulating levels of antibodies against ds‐DNA and histone. Moreover, immune responses generated by inflammatory commensals could be promoting the activation of the systemic lymphocyte and Treg–Th17 transdifferentiation. However, in patients with SLE, lower levels of Synergistetes (a microorganism associated positively with the Firmicutes to Bacteroidetes ratio) were found 78. Therefore, under physiological conditions, Synergistetes decrease serum levels of IL‐6 (a proinflammatory cytokine) and may stimulate B1 cells to secrete natural protective IgM anti‐phosphorylcholine. This can be achieved by down‐regulating inflammation in several ways [clearance of apoptotic cells and cellular debris, removal of oxidized lipids, blockade of mitogen‐activated protein kinase (MAPK) activation and other proinflammatory mediators] 79, 80. Reduction in the abundance of Lactobacillaceae and an increase of Lachnospiraceae were observed in patients with SLE 81, 82. Recently, it was demonstrated that Lactobacillis spp. and L. reuteri could have a positive effect on renal function in mice affected by lupus nephritis. In this study, treatment with Lactobacillus spp. improved the intestinal permeability (altered before the nephritis onset), decreased inflammatory cytokines (i.e. IL‐6 and IL‐18) and increased anti‐inflammatory cytokines (i.e. IL‐10, TGF‐β) and Tregs. It also demonstrated an improvement of renal disease through the decrease in IgG2a (one of the major immune deposits) and of the IFN‐γ level. However, such evidence was not present in male mice, suggesting that the influence of microbiome is sexually related, dependent and indicating a role for sex hormones in the regulatory function of the gut microbiota on lupus 83. In accordance with other findings, Bankole et al. 84 highlighted an increase of Protebacteria phyla and family of Lachnospiraceae and a decrease of Rikenellaceae, Odoribacteraceae, Christensenellaceae and Peptococcaceae families in samples from 21 patients with SLE.
Table 1.
Main alterations of microbiome in autoimmune diseases
| Bacteria |
S L E |
R A |
S S |
A P S |
S S c |
|---|---|---|---|---|---|
| Acinetobacter johnsonii | 94skin | ||||
| Akkermansia muciniphila | |||||
| Alistipes finegoldii | 97gut | ||||
| Anaerglobus geminatus | 50 oral | ||||
| Bacteroidetes | 59 gut | 94gut | |||
| Bacteroides fragilis | 100 gut | ||||
| Bifidobacterium | 53 gut | 97 gut | 100 gut | ||
| Campylobacter | |||||
| Capnocytophaga sputigena | 94oral | ||||
| Capnocytophaga ochracea | 94 oral | ||||
| Christensenellaceae | 74‐75 gut | ||||
| Clostridia‐like bacterium ( XIVa–IV) | 66 gut | 100gut | |||
| Clostridium coccoides | 53 gut | ||||
| Corynebacteriium amycolatum | 94 skin | ||||
| Dialister | 74 gut | ||||
| Eggerthella | 74 gut | 64 gut | |||
| Enterococcaceae | |||||
| Escherichia, Shigella, Enterobacter | 96 gut | ||||
| Eubacterium | 75gut | 53 gut | |||
| Eubacterium rectal | 61 gut | ||||
| Faecalibacterium prausnitzii [Link] | 64 gut | 96 gut | 101gut | ||
| Firmicutes [Link] | 74gut | 59 gut | 95 gut | ||
| Flavonifractor | |||||
| Fusobacterium | 96 gut | 100gut | |||
| Klebsiella | 74 gut | ||||
| Lachnospiraceae | 81 gut | 59 gut | |||
| Lactobacillaceae [Link] | 81,101gut | 64 gut | 101gut | ||
| Leptotrichia | 49 oral | 97 gut | |||
| Methanobrevibacter, | |||||
| Odoribacteraceae | 74 gut | ||||
| Peptococcaceae | |||||
| Porphyromonas gingivalis [Link] | 49 oral | ||||
| Prevotella disiens | 94 oral | ||||
| Prevotella Intermedia [Link] | 51oral | ||||
| Protebacteria | 69 lung | ||||
| Proteus mirabilis | 70 urinary t. | ||||
| Pseudobutyrivibrio | |||||
| Pseudocardia | 69 lung | ||||
| Rhodococcus | 74‐44 gut | ||||
| Rhodotorula glutinis | 102skin | ||||
| Rikenellaceae | 94‐94 gut | ||||
| Roseburia intestinalis | 89gut | ||||
| Ruminococcaceae | 59 gut | ||||
| Spirochetes | |||||
| Streptococcus | 95 oral | ||||
| Synergistetes | 78 gut | 95 oral | |||
| Tannerella forsythia | 51 oral | ||||
| Veillonella | 95oral |
A lower Firmicutes/Bacteroidetes ratio in systemic lupus erythematosus (SLE) patients. Conversely, Firmicutes are increased in rheumatoid arthritis (RA) and Sjögren’s syndrome (SS) patients.
Reduction in the abundance of Lactobacillaceae and an increase of Lachnospiraceae are observed in patients with SLE. In SSc patients Lactobacillaceae are increased.
High titres of antibodies against Prevotella intermedia have been detected in the serum and synovial fluids of patients with RA.
Porphyromonas gingivalis converts the amino acid arginine to the amino acid citrulline. The protein which contains the amino acid citrulline is recognized by the anti‐citrullinate peptide antibodies (ACPA).
Faecalibacterium prausnitzii is increased in patients with RA and decreased in SS and SSc patients.
Anti‐phospholipid syndrome (APS)
APS is an acquired autoimmune disorder that manifests clinically as recurrent venous or arterial thrombosis and/or fetal loss. Characteristic laboratory abnormalities in APS include persistently elevated levels of antibodies directed against membrane anionic phospholipids [i.e. anti‐cardiolipin (aCL) antibody, anti‐phosphatidylserine] or their associated plasma proteins, such as beta‐2 glycoprotein I (β2GPI) or evidence of a circulating anti‐coagulant. The incidence of APS is approximately five cases per 100 000 people per year, with a prevalence of approximately 40–50 cases per 100 000 people 85, 86. The pathogenesis is poorly understood: environmental factors and genetic background have been proposed and, in previous years, the role of the microbiome and infections have been suggested. Shoenfeld et al. 87 showed that mice immunized with proteins from Haemophilus influenzae, Neisseria gonorrhoeae or tetanus toxoid have developed antibodies that recognized cardiolipin, β2GPI and the amino acid sequences contained in the proteins. Conversely, naive mice infused with these antibodies developed significant thrombocytopenia, prolonged activated partial thromboplastin time and pregnancy loss similar to mice treated with pathogenic anti‐β2GPI. These studies suggested that molecular mimicry could be a potential explanation for the induction of pathogenic aPLs 88. Recently, it has been demonstrated that commensal, in particular segmented filamentous bacteria (SFB), influence T cell phenotypes and also both T‐dependent and T‐independent antibody production. If the microbiome homeostasis is disrupted (i.e. infections, drugs), proinflammatory interactions could occur with local and systemic effects on the immune system, including breaches of the mucosal barriers and generation of commensal‐specific memory T cells and autoantibodies. Therefore, it is likely that commensal bacteria may promote breaks in tolerance and the induction of persistent aPLs in genetically predisposed individuals 89. Notably, the authors suggested that Roseburia intestinalis, an anaerobic Gram‐positive bacterium extremely common in the gut of patients affected by APS, has many homologous sequences to both the major B and T cell epitopes and thus it could stimulate lymphocytes 89.
Sjögren’s syndrome (SS)
SS is a chronic autoimmune inflammatory disorder characterized by a reduction in the production of saliva, tears and pancreatic juice. Distinctive changes occur in the exocrine glands (salivary glands, tear glands, pancreas, glands of the alimentary and respiratory tracts). Also, lymphocytic (CD4+) T cells, DCs and B cells can infiltrate polyclonal B cell hyperactivity and autoantibody production (anti‐SSA/Ro60 antibodies). The incidence of SS is estimated at approximately seven per 100 000 people and the highest incidence rates are reported in studies from Europe and Asia 90. The pathogenesis of SS includes multiple genetic and non‐genetic interacting factors. There is an involvement of innate and adaptive immunity, as well as neuroendocrine and neuropathic processes. Biopsies of glandular and extra‐glandular sites are characterized by lymphocytic infiltration, with immune‐histological evidence for the involvement of numerous elements of innate and adaptive immune responses. Furthermore, cellular adhesive molecules, metalloproteinases and neural transmitters show alterations in the affected target organs 91. Several studies report an intimate association between SS and Epstein–Barr virus (EBV)/Coxsackie virus infections 92, 93. Conversely, commensal bacteria could have an important role in the pathogenesis of SS. Peptides derived from oral, gut and skin commensal bacteria (Table 1) may induce an immune response by activation of Ro60‐reactive T cells. Specifically, they are found in the oral flora (P. disiens, Capnocytophaga sputigena and C. ochracea) and in the gut flora (B. finegoldii, B. intestinalis, B. fragilis and Alistipes finegoldii) and two are found on the skin (Corynebacteriium amycolatum and Acinetobacter johnsonii) 94. Therefore, in SS patients, oral dysbiosis has been found with an increase of Firmicutes, specifically Streptococcus and Veillonella, and decreased in Synergistetes and Spirochaetes 95. Furthermore, de Paiva et al. 96 discovered that faecal samples from SS patients had an approximately 50% reduction in the genus Faecalibacterium, which includes F. prausnitzii, one of the predominant butyrate producers in the intestine. There was also a significant increase in the enteric pathogens, such as Escherichia/Shigella and Enterobacter. In addition, the researchers did not find any difference in the microbial composition between control and SS patients, except for an increase in Streptococcus and decrease in numbers of Leptotrichia and Fusobacterium. Moreover, patients with severe dysbiosis, meaning decreased levels of bacteria from the genera Bifidobacterium (38 versus 3%; P < 0·001) and Alistipes (19 versus 3%; P = 0·017) could have higher disease activity (evaluated by Sjögren’s syndrome Disease Activity Index), lower levels of complement component and higher levels of faecal calprotectin 97.
Systemic sclerosis (SSc)
SSc is a complex and heterogeneous disease, with clinical forms ranging from limited skin involvement (limited cutaneous systemic sclerosis) to forms with diffused skin sclerosis and severe and often progressive internal organ involvement (diffuse cutaneous systemic sclerosis). Moreover, immunological disturbances, such as positive anti‐nuclear antibody (ANA), anti‐topoisomerase I (anti‐Scl‐70) antibody, anti‐centromere antibody (ACA) and anti‐RNA polymerase III antibody (anti‐RNAPIII) were found 98, 99. It is well known that SSc patients have decreased commensal bacteria (Table 1), such as Faecalibacterium and Clostridium, and increased bacteria, such as Fusobacterium and γ‐Proteobacteria, compared with healthy controls. However, SSc patients also had increased Bifidobacterium and Lactobacillus, which are typically decreased during the inflammation state. Moreover, patients with moderate/severe gastrointestinal symptoms had decreased B. fragilis and increased Fusobacterium compared with SSc patients with no/mild symptoms 100. According to Andréasson et al. 101, dysbiosis (lower abundance of F. prausnitzii and Clostridiaceae and, at the same time, relatively high levels of Lactobacillus) was more pronounced among patients with pulmonary fibrosis, oesophageal dysfunction and malnutrition (Table 1). In another study, the researchers used ribosomal RNA sequencing of forearm skin biopsies taken from patients with early (< 6 months) diffused and limited SSc and healthy controls. They discovered increased expression of Rhodotorula glutinis sequences in the patient samples; it has been supposed that R. glutinis might activate the immune system and in this way induce skin fibrosis 102.
Discussion
The association between bacteria and autoimmune disease is well understood; alteration of microbiome ‘dysbiosis’ can induce autoimmune disease in people with certain genetic backgrounds and environmental factors. Dysbiosis can be categorized into three different types: (1) loss of beneficial organisms, (2) excessive growth of potentially harmful organisms and (3) loss of overall microbial diversity. Moreover, these three types are not mutually exclusive and can occur simultaneously 103. Besides, different commensal can increase or decrease in amount according to disease, i.e. in MS Prevotella decreases, while in RA it increases 44, 64. Recently, studies have focused upon reversing the negative effects mediated by the microbiota during the disease state. It is possible to restore the healthy flora through administration of (i) probiotics, Gram‐positive bacteria (i.e. Bifidobacteriaum spp., Lactobacillus spp., Lactococcus spp., Pediococcus spp. and other non‐pathogenic strains of E. coli) 104; and (ii) faecal microbiota transplantation (FMT), which consists of engrafting a healthy microbiota into patient recipients to reintroduce or re‐establish a stable environment that influences both the endogenous microbes and the host 105 (Fig. 3). The scientific literature is rich with studies concerning probiotic treatments in autoimmune disorder. Recently, it has been demonstrated that in non‐obese diabetic (NOD) mice the oral administration of a Lactobacillaceae protects mice from T1D by suppressing IL‐1b and promoting the differentiation of CD103+ tolerogenic DCs in the gut 106. Moreover, in a multi‐centre prospective cohort study, early probiotics supplementation has been shown to decrease the risk of islet autoimmunity in children with higher genetic risk of TDM1. 107. In 45 RA patients the administration of Bacillus coagulans has ameliorated pain and has improved disability, antagonizing microbes that may be contributing to an inflammatory response and producing short‐chain fatty acids such as butyric acid with anti‐inflammatory activities 108. In addition, the administration of L. casei reduces proinflammatory molecules (IL‐1β, IL‐2, IL‐6, IL‐12, IL‐17, IFN‐γ, TNF‐α and Cox‐2) in experimental arthritis 109. Similarly, Lactobacillus spp. improves lupus symptoms, diminishes inflammation and restores the intestinal barrier, thereby increasing the expression of adhesion molecules in the gut 83, 110. Despite pharmacological drugs and the use of antibiotics that produce a negative impact on gut microbiota, there is a great deal of evidence that autoimmune diseases can be treated with antibiotics. Mu et al. 111 showed that the administration of oral antibiotics in lupus‐prone MRL/lpr mice improve the disease by decreasing inflammatory cytokines (i.e. IL‐17) and increasing IL‐10, a cytokine with anti‐inflammatory activity. Moreover, the authors demonstrated that the administration of vancomycin reduced the levels of anti‐dsDNA IgG and proteinuria and improved intestinal permeability. In a recent review, Rosman et al. 112 described the usefulness of antibiotic therapy in autoimmune disorders through their anti‐inflammatory and immunomodulatory properties (Fig. 4). In conclusion, further studies will be required to investigate the relationship between mammalian and commensals in order to develop novel therapeutic targets. The intestine at birth is an aerobic environment and only facultative anaerobes, such as members of the Enterobacteriaceae family, can grow. After few days, the intestinal lumen turns anaerobic, allowing only anaerobes such as Bifidobacterium, Clostridium and Bacteroides to colonize 4. During the first few weeks, the microbiome of the infant gut is similar to the maternal skin and vaginal microbiome, with Enterococcaceae, Streptococcaceae, Lactobacillaceae, Clostridiaceae and Bifidobacteriaceae the predominant bacterial taxa. During the first months the diet is almost exclusively milk, favouring milk oligosaccharide fermenters such as Bifidobacterium. With the weaning and introduction of solids foods, another rapid and important shift in gut microbiota occurs. The introduction of a variety of nutrients, many of which are polysaccharides not digested by host enzymes, trigger an increase in the abundance of Bacteroides, Clostridium and Ruminococcus and a decrease in Bifidobacterium and Enterobacteriaceae. In the subsequent 12–30 months, the infant gut microbiome progresses into an adult‐like gut microbiota abundant in Ruminococcaceae, Lachnospiraceae, Bacteroidaceae and Prevotellaceae.
Figure 3.
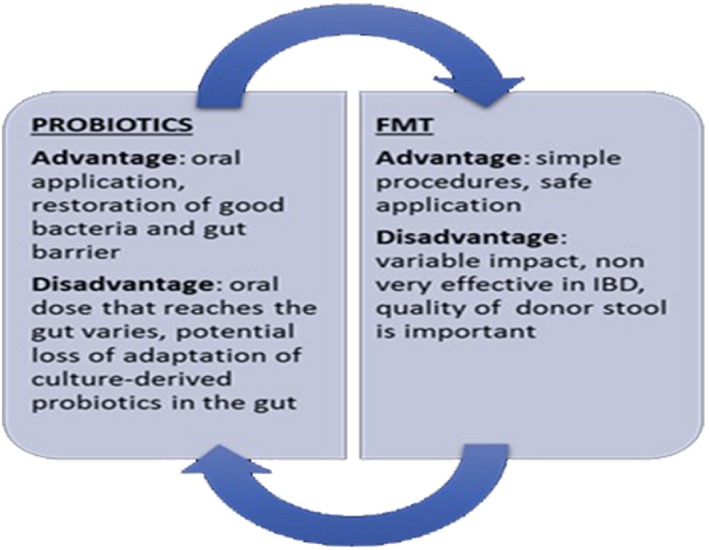
Advantages and disadvantages between probiotics and faecal transplantation (FMT).
Figure 4.
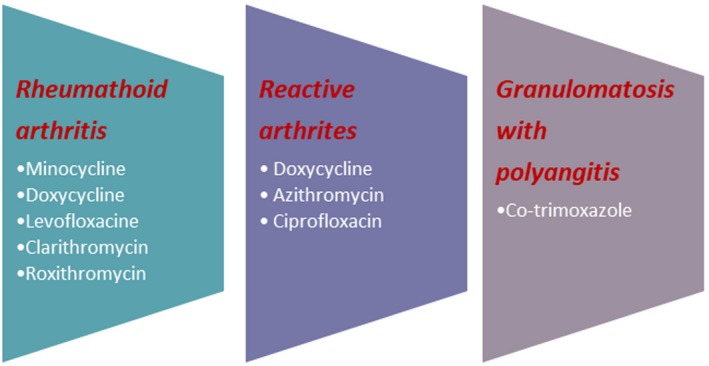
Utility of antibiotic in autoimmune diseases.
Microorganisms can elicit the immune response against the host if the mechanisms of tolerance fail in several ways: (i) epitope spreading consists of the development of autoimmune responses to endogenous epitopes following the release of self‐antigens during an inflammatory response, which is caused by a change in protein structure, i.e. changing of amino acid residue from arginine to citrulline. This may result in an immune reaction not only against the original protein or in its citrullinated form, but also against other citrullinated proteins 27; (ii) molecular mimicry is a mechanism by which infections can induce autoimmunity and occurs when foreign antigens share sequence or structural similarities with self‐antigens. The immune responses can be directed against peptides with similar charge distribution and overall shape 28. (iii) Bystander activation occurs when microbial infection stimulates Toll‐like receptors (TLRs) and other pattern recognition receptors on antigen‐presenting cells (APCs) with the production of proinflammatory mediators which, in turn, may lead to tissue damage 29; and (iv) prolonged infection with a virus, such as EBV or HCV, can induce constant activation and proliferation of T cells, resulting in the production of monoclonal and polyclonal antibodies as well as immune complexes, leading to loss of tolerance 30, 31.
Disclosure
The authors declare that they have no competing interests.
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
Pathogen infection and autoimmune disease. Clinical and Experimental Immunology 2019, 195:10–14.
Enterovirus infection and type 1 diabetes: unraveling the crime scene. Clinical and Experimental Immunology 2019, 195:15–24.
Pathogen infections and primary biliary cholangitis. Clinical and Experimental Immunology 2019, 195:25–34.
Pathogens and autoimmune hepatitis. Clinical and Experimental Immunology 2019, 195:35–51.
The potential role for infections in the pathogenesis of autoimmune Addison’s disease. Clinical and Experimental Immunology 2019, 195:52–63.
Mechanisms of lymphatic system‐specific viral replication and its potential role in autoimmune disease. Clinical and Experimental Immunology 2019, 195:64–73.
References
- 1. Belkaid Y, Hand T. Role of the microbiota in immunity and inflammation. Cell 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koren O, Goodrich JK, Cullender TC et al Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012;150:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen X, Liu S, Tan Q, Yehuda S, Zeng Y. Microbiome, autoimmunity, allergy, and helminth infection: the importance of the pregnancy period. Am J Reprod Immunol 2017;78:e12654. [DOI] [PubMed] [Google Scholar]
- 4. Arrieta M‐C, Stiemsma LT, Amenyogbe Ny, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol 2014;5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samriz O, Mizrahi H, Werbner M, Shoenfeld Y, Avni O, Koren O. Microbiota at the crossroads of autoimmunity. Autoimmun Revi 2016;15:859–69. [DOI] [PubMed] [Google Scholar]
- 6. Dominguez‐Bello MG, Costello EK, Contreras M et al Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010;107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pacheco AR, Barile D, Underwood MA, Mills DA. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci 2015;3:419–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect 2012;18:12–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 2012;22:1147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Srutkova D, Schwarzer M, Hudcovic T et al Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS‐induced colitis in strictly strain‐specific manner. PLOS ONE 2015;10:e0134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perez PF, Dore J, Leclerc M et al Bacterial imprinting of the neonatal immune system: lessons from matern cells? Pediatrics 2007;119:e724–32. [DOI] [PubMed] [Google Scholar]
- 12. PrabhuDas M, Adkins B, Gans H et al Challenges in infant immunity: implications for responses to infection and vaccines. Nat Immunol 2011;12:189–94. [DOI] [PubMed] [Google Scholar]
- 13. Siegrist CA. Neonatal and early life vaccinology. Vaccine 2001;19:3331–46. [DOI] [PubMed] [Google Scholar]
- 14. Valentini M, Piermattei A, Di Sante G, Migliara G, Delogu G, Ria F. Immunomodulation by gut microbiota: role of toll‐like receptor expressed by T cells. J Immunol Res 2014;2014:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guven‐Maiorov E, Tsai C‐J, Nussinov R. Structural host–microbiota interaction networks. PLOS Comput Biol 2017;13:e1005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kollmann TR, Levy O, Montgomery RR, Goriely S. Innate immune function by Toll‐like receptors: distinct responses in newborns and the elderly. Immunity 2012;37:771–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host–microbial relationships in the intestine. Science 2001;291:881–4. [DOI] [PubMed] [Google Scholar]
- 18. Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA 2002;99:15451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. An D, Oh SF, Olszak T et al Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014;156:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olszak T, An D, Zeissig S et al Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012;336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol 2008;8:523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shevach EM. Mechanisms of foxp3+ T regulatory cell‐mediated suppression. Immunity 2009;30:636–45. [DOI] [PubMed] [Google Scholar]
- 23. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA 2014;111:2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Furusawa Y, Obata Y, Fukuda S et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–50. [DOI] [PubMed] [Google Scholar]
- 25. Miller FW, Pollard KM, Parks CG et al Criteria for environmentally associated autoimmune diseases. J Autoimmun 2012;39:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramos‐Casals M, Brito‐Zerón P, Kostov B et al Google‐driven search for big data in autoimmune geoepidemiology: analysis of 394,827 patients with systemic autoimmune diseases. Autoimmun Rev 2015;14:670–9. [DOI] [PubMed] [Google Scholar]
- 27. Vanderlugt CJ, Miller SD. Epitope spreading. Curr Opin Immunol 1996;8:831–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guilherme L, Kalil J, Cunningham M. Molecular mimicry in the autoimmune pathogenesis of rheumatic heart disease. Autoimmunity 2006;39:31–9. [DOI] [PubMed] [Google Scholar]
- 29. Getts DR, Chastain EM, Terry RL, Miller SD. Virus infection, antiviral immunity, and autoimmunity. Immunol Rev 2013;255:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vojdani A. A potential link between environmental triggers and autoimmunity. Autoimmun Dis 2014;2014:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agmon‐Levin N, Ram M, Barzilai O et al Prevalence of hepatitis C serum antibody in autoimmune diseases. J Autoimmun 2009;32:261–6. [DOI] [PubMed] [Google Scholar]
- 32. Rinaldi M, Perricone R, Blank M, Perricone C, Shoenfeld Y. Anti‐Saccharomyces cerevisiae autoantibodies in autoimmune diseases: from bread baking to autoimmunity. Clin Rev Allerg Immunol 2013;45:152–61. [DOI] [PubMed] [Google Scholar]
- 33. Shoenfeld Y, Agmon‐Levin N. ‘ASIA’ – autoimmune/inflammatory syndrome induced by adjuvants. J Autoimmun 2011;36:4–8. [DOI] [PubMed] [Google Scholar]
- 34. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular–phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gevers D, Kugathasan S, Denson LA et al The treatment‐naive microbiome in new‐onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 2015;26:26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamps LW, Madhusudhan KT, Havens JM et al Pathogenic Yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn’s disease. Am J Surg Pathol 2003;27:220–7. [DOI] [PubMed] [Google Scholar]
- 38. Navaneethan U, Venkatesh PGK, Bo S. Clostridium difficile infection and inflammatory bowel disease: understanding the evolving relationship. World J Gastroenterol 2010;16:4892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chassaing B, Rolhion N, de Vallée A et al Crohn disease‐associated adherent‐invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. Clin Invest 2011;121:966–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang HX, Wang YP. Gut microbiota–brain axis. Chin Med J (Engl) 2016;129:2373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest 2015;125:926–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adamczyk Sowa M, Aldona M, Madej P, Michlicka W, Dobrakowski Hindawi P. Does the gut microbiota influence immunity and inflammation in multiple sclerosis pathophysiology? J Immunol Res 2017;2017:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miyake S, Kim S, Suda W et al Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLOS ONE 2015;10:e0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mete A, Garcia J, Ortega J, Lane M, Scholes S, Uzal FA. Brain lesions associated with Clostridium perfringens type D epsilon toxin in a Holstein Heifer calf. Vet Pathol 2013;50:765–8. [DOI] [PubMed] [Google Scholar]
- 45. Finnie JW, Blumbergs PC, Manavis J. Neuronal damage produced in rat brains by Clostridium perfringens type D epsilon toxin. J Comp Pathol 1999;120:415–20. [DOI] [PubMed] [Google Scholar]
- 46. Qi C‐J, Zhang Q, Yu M et al Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese children. Chin Med J 2016;129:1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tobon GJ, Youinou P, Saraux A. The environment, geo‐epidemiology, and autoimmune disease: Rheumatoid arthritis. J Autoimmun 2010;35:10–4. [DOI] [PubMed] [Google Scholar]
- 48. Adriano VM, Melo IM, Lima V. Relationship between periodontitis and rheumatoid arthritis: review of the literature. Mediat Inflamm 2015;2015:259074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Meulen TA, Harmsen HJM, Bootsma H, Spijkervet FKL, Kroese FGM, Vissink A. The microbiome–systemic diseases connection. Oral Dis 2016;22:719–34. [DOI] [PubMed] [Google Scholar]
- 50. Brusca SB, Abramson SB, Scher JU. Microbiome and mucosal inflammation as extra‐articular triggers for rheumatoid arthritis and autoimmunity. Curr Opin Rheumatol 2014;26:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caminer AC, Haberman R, Scher JU. Human microbiome, infections, and rheumatic disease. Clin Rheumatol 2017;36:2645–53. [DOI] [PubMed] [Google Scholar]
- 52. Goh CE, Kopp J, Papapanou PN, Molitor JA, Demmer RT. Association between serum antibodies to periodontal bacteria and rheumatoid factor in the third national health and nutrition examination survey. Arthritis Rheumatol 2016;68:2384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lange L, Thiele GM, McCracken C et al Symptoms of periodontitis and antibody responses to Porphyromonas gingivalis in juvenile idiopathic arthritis. Pediatr Rheumatol Online J 2016;14:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roszyk E, Puszczewicz M. Role of human microbiome and selected bacterial infections in the pathogenesis of rheumatoid arthritis. Reumatologia 2017;5:242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Monsarrat P, Vergnes JN, Cantagrel A et al Effect of periodontal treatment on the clinical parameters of patients with rheumatoid arthritis: Study protocol of the randomized, controlled ESPERA trial. Trials. 2013;14:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elizabeth V, Arkema Elizabeth W, Karlson Karen H, Costenbader A. Prospective study of periodontal disease and risk of rheumatoid arthritis. J Rheumatol 2010;37:1800–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhong D, Wu C, Zeng X, Wang Q. The role of gut microbiota in the pathogenesis of rheumatic diseases. Clin Rheumatol 2018;37:25–34. [DOI] [PubMed] [Google Scholar]
- 58. Rogier R, Koenders MI, Abdollahi‐Roodsaz S. Toll‐like receptor mediated modulation of T cell response by commensal intestinal microbiota as a trigger for autoimmune arthritis. J Immunol Res 2015;2015:527696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rogier R, Evans‐Marin H, Manasson J et al Alteration of the intestinal microbiome characterizes preclinical inflammatory arthritis in mice and its modulation attenuates established arthritis. Sci Rep 2017;7:15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research the immune. Comp Med 2014;64:90–8. [PMC free article] [PubMed] [Google Scholar]
- 61. Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol 2008;35:1500–5. [PubMed] [Google Scholar]
- 62. Moreno J. Prevotella copri and the microbial pathogenesis of rheumatoid arthritis. Reumatol Clin 2015;11:61–3. [DOI] [PubMed] [Google Scholar]
- 63. Scher JU, Sczesnak A, Longman RS et al Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen J, Wright K, Davis JM et al An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med 2016;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol 2013;67:170–6. [DOI] [PubMed] [Google Scholar]
- 66. Gomez A, Luckey D, Yeoman CJ et al Loss of sex and age driven differences in the gut microbiome characterize arthritis‐susceptible *0401 mice but not arthritis‐resistant *0402 mice. PLOS ONE 2012;7:e36095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cutolo M, Sulli A, Pizzorni C, Seriolo B, STRAUB R.. Anti‐inflammatory mechanisms of methotrexate in rheumatoid arthritis. Ann Rheum Dis 2001;60:729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Scher JU, Joshua V, Artacho A et al The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome 2016;4:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Willis VC, Demoruelle MK, Derber LA. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum 2013;65:2545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wilson C, Thakore A, Isenberg D, Ebringer A. Correlation between anti‐Proteus antibodies and isolation rates of P. mirabilis in rheumatoid arthritis. Rheumatol Int 1997;16:187–9. [DOI] [PubMed] [Google Scholar]
- 71. Ebringer A, Rashid T. Rheumatoid arthritis is caused by a Proteus urinary tract infection. APMIS 2014;122:363–8. [DOI] [PubMed] [Google Scholar]
- 72. Rashid T, Ebringer A. Rheumatoid arthritis is linked to Proteus – the evidence. Clin Rheumatol 2007;26:1036–104. [DOI] [PubMed] [Google Scholar]
- 73. Edworthy SM, Harris ED. Clinical manifestations of systemic lupus erythematosus In Harris ED.Jr, Budd RC, Genovese MC. et al., eds. Kelley's textbook of rheumatology, 7th edn. Philadelphia, PA: WB Saunders; 2005:1201–24. [Google Scholar]
- 74. Hevia A, Milani C, Lopez P et al Intestinal dysbiosis associated with systemic lupus erythematosus. MBio 2014;5:e01548–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. He Z, Shao T, Li H, Xie Z, Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathogens 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lo’pez P, Sa’nchez B, Margolles A, Sua’reza A. Intestinal dysbiosis in systemic lupus erythematosus: cause or consequence. Curr Opin Rheumatol 2016;28:515–22. [DOI] [PubMed] [Google Scholar]
- 77. Johnson BM, Gaudreau M‐C, Al‐Gadban MM, Gudi R, Vasu C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus‐prone SNF1 mice. Clin Exp Immunol 2015;181:323–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. López P, de Paz B, Rodríguez‐Carrio J et al Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep 2016;6:24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Grönwall C, Chen Y, Vas J. MAPK phosphatase‐1 is required for regulatory natural autoantibody‐mediated inhibition of TLR responses. Proc Natl Acad Sci USA 2012;109:19745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bao S, Husband AJ, Beagley KW. B1 B cell numbers and antibodies against phosphorylcholine and LPS are increased in IL‐6 gene knockout mice. Cell Immunol 1999;198:139–42. [DOI] [PubMed] [Google Scholar]
- 81. Neuman H, Koren O. The gut microbiota: a possible factor influencing systemic lupus erythematosus. Curr Opin Rheumatol 2017;29:374–7. [DOI] [PubMed] [Google Scholar]
- 82. Mu Q, Zhang H, Luo XM. SLE: another autoimmune disorder influenced by microbes and diet? Front Immunol 2015;6:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mu Q, Zhang H, Liao X et al Control of lupus nephritis by changes of gut microbiota. Microbiome 2017;5:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bankole A, Luo X. Husen Z. A comparative analysis of gut microbiota between systemic lupus erythematosus patients and non‐autoimmune controls: a single center cohort experience. Sci Med 2017;4 . 10.1136/lupus-2017-000215.354. [Google Scholar]
- 85. Negrini S, Pappalardo F, Murdaca G, Indiveri F, Puppo F. The antiphospholipid syndrome: from pathophysiology to treatment. Clin Exp Med 2017;17:257–267. [DOI] [PubMed] [Google Scholar]
- 86. Giannakopoulos B, Krilis SA. The pathogenesis of the antiphospholipid syndrome. N Engl J Med 2013;368:1033–44. [DOI] [PubMed] [Google Scholar]
- 87. Blank M, Krause I, Fridkin M et al Bacterial induction of autoantibodies to beta2‐glycoprotein‐I accounts for the infectious etiology of antiphospholipid syndrome. Clin Invest 2002;109:797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Blank M, Shoenfeld Y, Cabilly S, Heldman Y, Fridkin M, Katchalski‐Katzir E. Prevention of experimental antiphospholipid syndrome and endothelial cell activation by synthetic peptides. Proc Natl Acad Sci USA 1999;96:5164–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ruff WE, Vieira SM, Kriegel MA. The role of the gut microbiota in the pathogenesis of antiphospholipid syndrome. Curr Rheumatol Rep 2015;17:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ramos‐Casals M, Tzioufas AG, Font J. Primary Sjögren’s syndrome: new clinical and therapeutic concepts. So Ann Rheum Dis 2004;64:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mavragani CP, Nezos A, Moutsopoulos HM. New advances in the classification, pathogenesis and treatment of Sjogren’s syndrome. Curr Opin Rheumatol 2013;25:623–9. [DOI] [PubMed] [Google Scholar]
- 92. McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, Jame JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med 2005;11:85–9. [DOI] [PubMed] [Google Scholar]
- 93. Stathopoulou EA, Routsias JG, Stea EA, Moutsopoulos HM, Tzioufas AG. Cross‐reaction between antibodies to the major epitope of Ro60kD autoantigen and a homologous peptide of Coxsackie virus 2B protein. Clin Exp Immunol 2005;141:148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, Deshmukh US. T cell epitope mimicry between Sjögren’s syndrome antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin Immunol 2014;152:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Siddiqui H, Chen T, Aliko A, Mydel PM, Jonsson R, Olsen I. Microbiological and bioinformatics analysis of primary Sjögren’s syndrome patients with normal salivation. J Oral Microbiol 2016;8:31119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. de Paiva CS, Jones DB, Stern ME et al Altered mucosal microbiome diversity and disease severity in Sjögren Syndrome. Sci Rep 2016;6:23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mandl T, Marsal J, Olsson P, Ohlsson B, Andréasson K. Severe intestinal dysbiosis is prevalent in primary Sjögren’s syndrome and is associated with systemic disease activity. Arthritis Res Ther 2017;19:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A. classification criteria for systemic sclerosis: an American College of Rheumatology/European League against Rheumatism collaborative initiative. Arthritis Rheum 2013;65:2737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Joseph CG, Darrah E, Shah AA et al Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science 2014;343:152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Volkmann ER, Chang YL, Barroso N et al Association of systemic sclerosis with a unique colonic microbial consortium. Arthritis Rheumatol (Hoboken, NJ) 2016;68:1483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Andréasson K, Alrawi Z, Persson A, Jönsson G, Marsal J. Intestinal dysbiosis is common in systemic sclerosis and associated with gastrointestinal and extraintestinal features of disease. Arthritis Res Ther 2016;18:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Arron ST, Dimon MT, Li Z et al High Rhodotorula sequences in skin transcriptome of patients with diffuse systemic sclerosis. J Investig Dermatol 2014;134:2138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. De Gruttola AK, Low D, Mizoguchi A, Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis 2016;22:1137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Solis B, Samartín S, Gómez S, Nova E, de la Rosa B, Marcos A. Probiotics as a help in children suffering from malnutrition and diarrhea. Eur J Clin Nutr 2002;56:S57–9. [DOI] [PubMed] [Google Scholar]
- 105. Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 2011;53:994–1002. [DOI] [PubMed] [Google Scholar]
- 106. Dolpady J, Sorini C, Di Pietro C, Cosorich I, Ferrarese R, Saita D. Oral probiotic VSL3 prevents autoimmune diabetes by modulating microbiota and promoting indoleamine 2,3‐dioxygenase‐enriched tolerogenic intestinal environment. J Diabetes Res 2016;2016:7569431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Uusitalo U, Liu X, Yang J et al Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr 2016;170:20–8. [PMC]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mandel DR, Eichas K, Holmes J. Bacillus coagulans: a viable adjunct therapy for relieving symptoms of rheumatoid arthritis according to a randomized, controlled trial. BMC Complement Altern Med 2010;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. So JS, Kwon HK, Lee CG et al Lactobacillus casei suppresses experimental arthritis by down‐regulating T helper 1 effector functions. Mol Immunol 2008;45:2690–9. [DOI] [PubMed] [Google Scholar]
- 110. de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology 2017;152:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mu Q, Tavella VJ, Kirby JL et al Antibiotics ameliorate lupus‐like symptoms in mice. Sci Rep 2017;7:13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rosman Y, Lidar M, Shoenfeld Y. Antibiotic therapy in autoimmune disorders. Clin Pract 2014;11:91–103. [Google Scholar]


