Abstract
Despite high remission rates following CAR-T cell therapy in B-ALL, relapse due to loss of the targeted antigen is increasingly recognized as a mechanism of immune escape. We hypothesized that simultaneous targeting of CD19 and CD22 may reduce the likelihood of antigen loss, thus improving sustained remission rates. A systematic approach to the generation of CAR constructs incorporating two target-binding domains led to several novel CD19/CD22 bivalent CAR constructs. Importantly, we demonstrate the challenges associated with the construction of a bivalent CAR format that preserves bifunctionality against both CD19 and CD22. Using the most active bivalent CAR constructs, we found similar transduction efficiency compared to that of either CD19 or CD22 single CARs alone. When expressed on human T cells, the optimized CD19/CD22 CAR construct induced comparable interferon γ and interleukin-2 in vitro compared to single CARs against dual-antigen-expressing as well as single-antigen-expressing cell lines. Finally, the T cells expressing CD19/CD22 CAR eradicated ALL cell line xenografts and patient-derived xenografts (PDX), including a PDX generated from a patient with CD19− relapse following CD19-directed CAR therapy. The CD19/CD22 bivalent CAR provides an opportunity to test whether simultaneous targeting may reduce risk of antigen loss.
Keywords: chimeric antigen receptor, adoptive cell therapy, leukemia
Introduction
Genetically modified T cells expressing chimeric antigen receptors (CARs) targeting CD19 have demonstrated potent efficacy in children and young adults with relapsed and chemotherapy refractory B cell acute lymphoblastic leukemia (B-ALL).1, 2, 3 With complete remission rates approaching 65%–80%, US Food and Drug Administration (FDA) approval of CD19 CAR-T cells for pediatric ALL,4, 5 and ongoing investigation of other constructs, CAR-T cell therapy in ALL shows tremendous potential. However, not all patients respond, and relapse due to loss of CD19 can occur in over one third of cases.6 CD19 CAR-T cell therapy has also been approved for adult lymphoma treatment, with a lower remission rate (50%–70%),7 and CD19 antigen loss has also been observed in this setting.8 We recently developed and tested a highly active CAR targeting CD22,9 a pan-B cell antigen that has been successfully targeted using monoclonal-antibody-based strategies.10, 11 Despite a degree of clinical activity similar to that seen with CD19-directed CAR-T cells, relapse due to CD22 loss or diminution of surface expression was also seen.12
There is evidence in preclinical models that dual or multi-antigen targeting by CARs may result in synergistic responses in solid tumors compared to targeting a single antigen, optimizing response rates to therapy13, 14 and overcoming interpatient variability in antigen expression.15, 16 Indeed, the expression levels of CD19 and CD22 on B-ALL may be heterogeneous from patient to patient,2, 17 and immunophenotypic changes may occur with relapse, particularly in the era of targeted therapies. Thus, dual targeting by CAR-T cells may be a more broadly active therapy, which can account for baseline variability in antigenic expression and changes that occur over time. Lastly, simultaneous targeting of two antigens on the surface of B-ALL may reduce the likelihood of antigen-loss variants, as recently described in investigations simultaneously targeting both CD123 and CD19.6 We sought to develop and test various CAR strategies to simultaneously target both CD19 and CD22 based on two clinically validated single-CAR constructs.
Bispecific CAR strategies (any approach targeting two antigens) that we explored included co-infusion, co-transduction, and CAR constructs encoding a single protein with two targeting domains (which we refer to as a “bivalent” CAR). Through this exercise, which ultimately led to the development of an active bivalent CD19xCD22-targeted CAR now being tested in the clinic, we demonstrate the complexities encountered in designing and testing bivalent targeting constructs and how minor modifications in CAR assembly can have a major impact on CAR functionality, providing a framework for the optimization of bispecific CAR-targeting strategies.
Results
Heterogeneous Expression of CD19 and CD22 on B-ALL
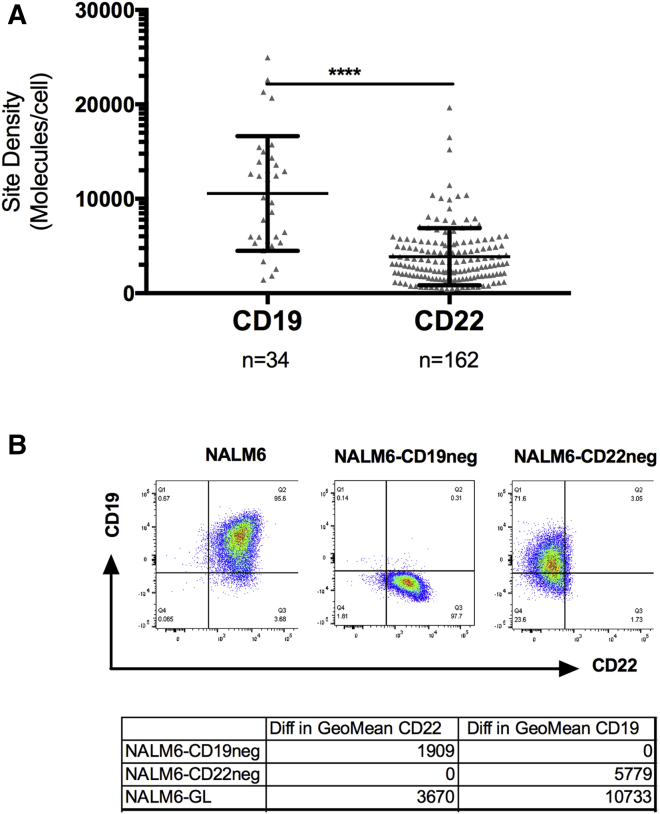
Patient samples, primarily derived from patients with multiply relapsed disease, were evaluated for CD19 and CD22 expression. As shown in Figure 1A, there was a broad range in expression of CD19 and CD22 prior to the administration of immunotherapy.
Figure 1.
Heterogeneous Expression of CD19 and CD22 on B-ALL
(A) CD19 and CD22 expression on primary patient samples was measured using a flow-cytometric quantitative assay prior to CAR T cell therapy. ****p < 0.0001. (B) CD19 and Cd22 expression on parental Nalm6 and following CRISPR-Cas9-mediated deletion of CD19 and CD22.
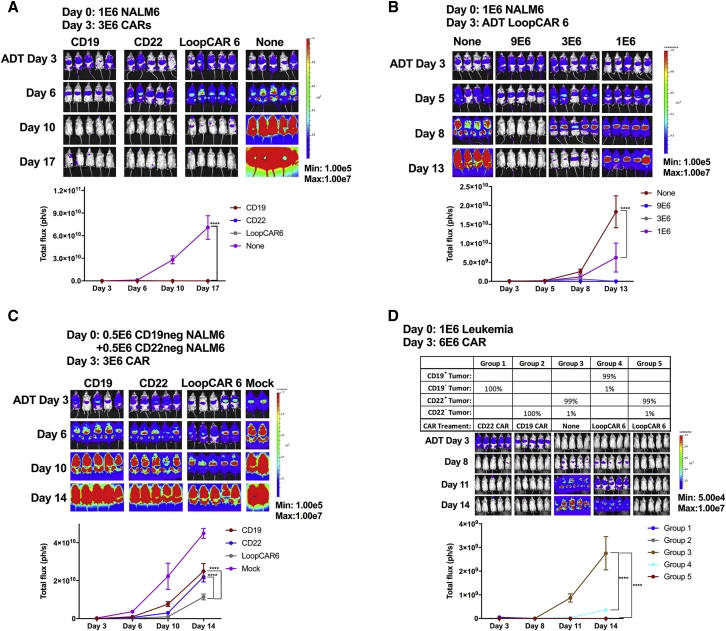
Simultaneous Targeting of Both CD19 and CD22 Is Superior to Sequential Treatment in Preventing Relapse or Disease Progression of Antigen Loss-Relapse Models
To model the CD19 and CD22 relapse phenomenon seen in clinical trials,1, 2, 3, 12 CRISPR-Cas9 gene editing was used to delete CD19 or CD22 from the B-ALL cell line NALM6 (Figure 1B). After single-cell cloning to ensure stability, both CRISPR-edited NALM6 lines and parental NALM6 demonstrated comparable disease progression when engrafted into NSG (NOD SCID gamma) mice, despite the deletion of CD19 or CD22 (Figure S1A).
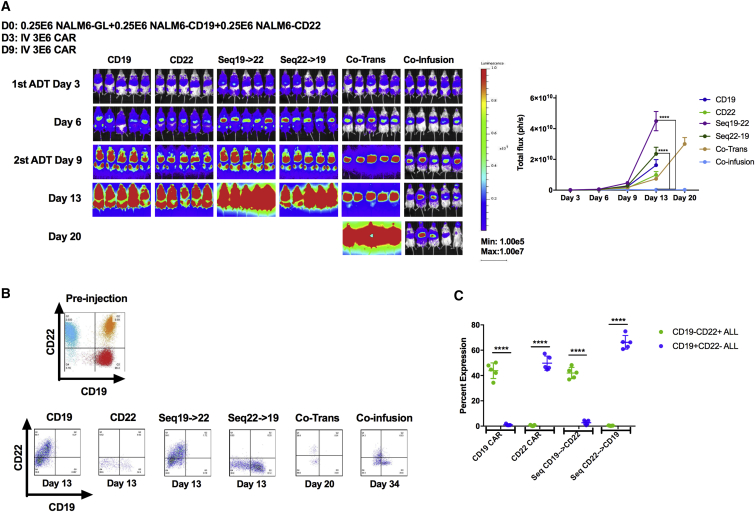
One approach to exert immunotherapeutic pressure on two antigens is via sequential infusion of CD19 CAR-T cells followed by CD22 CAR-T cells or vice versa, analogous to using the second CAR infusion as a salvage therapy. To test this strategy, mice were injected with a mixture of CD19−, CD22−, and parental NALM6 (CD19+/CD22+) ALL cells to simulate antigen-negative relapse (Figure 2A). As expected, single antigen-specific CAR-T cells resulted in the recurrence of leukemia not expressing the targeted antigen and validating the relapse model (Figures 2B and 2C). Surprisingly, sequential infusion of curative doses of CD19 and CD22 CAR-T cells separated by 6 days did not prevent ALL progression. Importantly, the relapse phenotype demonstrated a lack of efficacy of the second CAR infusion. Simultaneous administration (co-infusion) of both CD19- and CD22-targeted CAR-T cells was superior to sequential infusion but resulted in the progression of CD19− ALL still expressing CD22, suggesting that the CD19 CAR may dominate in functionality.
Figure 2.
Simultaneous Targeting on Both CD19 and CD22 Surface Antigens with Co-infusion of Two Mono-targeting CARs or Co-transduced T Cell Products Result in Slowing Down of Tumor Progression in the Relapse Leukemia Model
(A) NSG mice were challenged with a mixture of 2.5−E5 each of NALM6, NALM6-CD19neg, and NALM6-CD22neg leukemia lines on day 0. Mice in the sequential treatment group received 3E−6 CAR+ T cells on day 3 and 3E−6 CAR+ T cells on day 9. Mice in the co-injection group received a total of 6E−6 CAR+ T cells with 3E−6 CD19 CAR+ and 3E−6 CD22 CAR+ T cells on day 3. Mice in the co-infusion group received 3E−6 CD19 CAR+ and 3E−6 CD22 CAR+ T cells. Mice in the co-transduction group received 6E−6 CAR-expressing T cells (see Figure S1 for distribution of CAR-expressing cells). Mice in the CD19 or CD22 group received 3E−6 CD19 CAR+ or CD22 CAR+ T cells, respectively. Quantification of luminescence is shown on the right. (B). Representative flow-cytometric plots demonstrating the CD19 and CD22 surface expression of the leukemia before injection or at takedown at the times indicated post-CAR treatment. (C) Graphic plot presentation of the leukemia phenotype post-CAR treatment. ****p < 0.0001. ADT, adoptive transfer day (day at which CAR T cells were given).
See also Figure S1.
Based on the apparent dominance of the CD19 CAR when co-infused with the CD22 CAR, we next attempted to introduce both the CD19 and CD22 CARs into the same T cell through co-transduction, generating a pool of T cells containing dual-specific CAR-T cells. However, co-transduction efficiency was consistently low, yielding only a quarter of the total T cell product expressing both CD19 and CD22 CAR (Figure S1B). Furthermore, the relapse phenotype (CD22+CD19− and CD22−CD19−; Figure 2B) suggests that, again, the CD19 CAR-T cell may dominate when co-administered with T cells expressing both CARs or with the CD22 CAR. Thus, based on the inefficiency of gene transfer by two vectors, the technical challenges and costs associated with managing two vectors, and the possibility that inclusion of single-CAR-expressing T cells may impair expansion of the dual-specific T cell population, we pursued approaches to introduce dual specificity from the same vector.
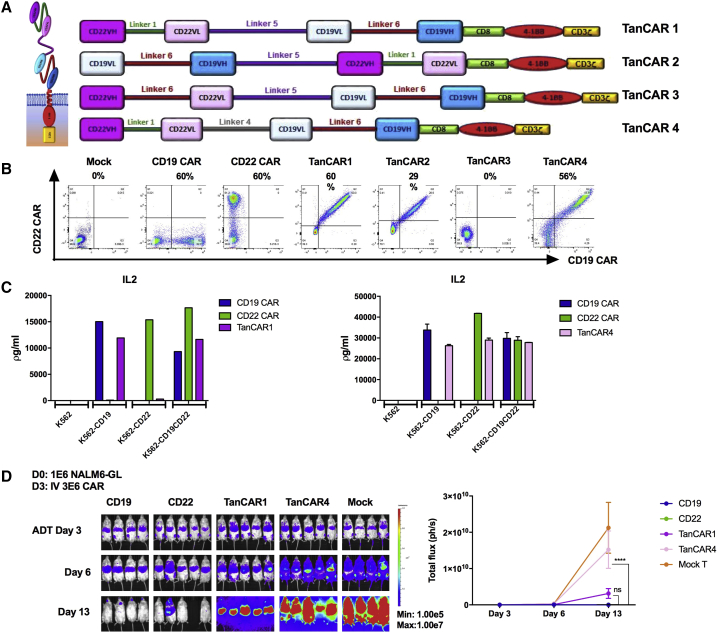
Development of the Bivalent CARs with Tandem Sequencing of scFv
Bivalent CARs were generated by coupling two different single-chain fragment variable (scFv) domains into a single-CAR construct (Figure S2).16 The approach we undertook in constructing CD19xCD22 CARs using the scFv regions validated in clinical trials (FM63 for CD19 and m971 for CD22) was to place the heavy and light chains (VH and VL, respectively) for each scFv in sequential order to make tandem CARs (TanCARs), as depicted in Figure 3A. For TanCAR1, we maintained the original linkers between the VH and VL from each single CAR and connected the two scFvs, using a flexible and protease-resistant (G4S)X5 linker,18 a format that could be detected at a level comparable to that of single-antigen-targeted CARs on the cell surface following transduction (Figure 3B). Importantly, all CAR-expressing T cells showed concordance between CD22 Fc fusion and anti-FMC63 idiotype binding. For TanCAR2, we reversed the order of anti-CD19 and anti-CD22 scFvs, which resulted in much lower detection on the surface. Despite good surface detection of TanCAR1 and comparable levels of interleukin-2 (IL2) production compared to monovalent CD19 CAR-T cells against CD19+/CD22− ALL, IL-2 production was extremely low when co-incubated with CD19−/CD22+ ALL (Figure 3C). Given the extremely short linker (G4S) between the anti-CD22 VH and VL in the parental CAR, we constructed TanCAR3 with increased linker length within the CD22 scFv, a format that abolished CD22 Fc and anti-idiotype binding (Figure 3A). For TanCAR4, we maintained the short parental linker for the CD22 scFv but reduced the length of the linker between CD19 and CD22 scFv. This resulted in CAR surface expression (CD22 Fc and anti-FMC63 idiotype binding, Figure 3B) and enhanced CD22-directed functionality compared to TanCAR1, as measured by the IL2 production against CD19−/CD22+ ALL (Figure 3C). Cytotoxicities of TanCAR1 and TanCAR4 were further evaluated, demonstrating activity comparable to that of CD19 and CD22 monovalent CARs (Figure S3). Surprisingly, despite in vitro activity, neither TanCAR1 nor TanCAR4 eradicated CD19+CD22+ ALL in vivo (Figure 3D). These results illustrate the challenges of generating bivalent CD19xCD22 CAR constructs that maintain bispecific activity (particularly against CD22) and highlight the importance of comprehensive testing of multivalent CAR formats, including in vivo experiments.
Figure 3.
Development of the Bivalent Tandem CARs
(A) Schematic of TanCAR structures. (B) Flow-cytometric plot demonstrating the surface binding of CD22Fc and CD19 idiotypes. (C) Cytokine production by CD19-CAR-, CD22-CAR-, TanCAR1-, and TanCAR4-expressing T cells co-incubated with K562, K562-CD19, K562-CD22, and K562-CD19CD22 cell lines. (D) Comparison of in vivo efficacy of TanCAR1 and TanCAR4 CAR T cells. NSG mice were challenged with 1E−6 luciferase-expressing NALM6 leukemia cells on day 0. On day 3, mice were i.v. injected with 3E−6 tandem-CAR-expressing T cells. Quantification of luminescence is shown on the right. ****p < 0.0001. ns, not significant.
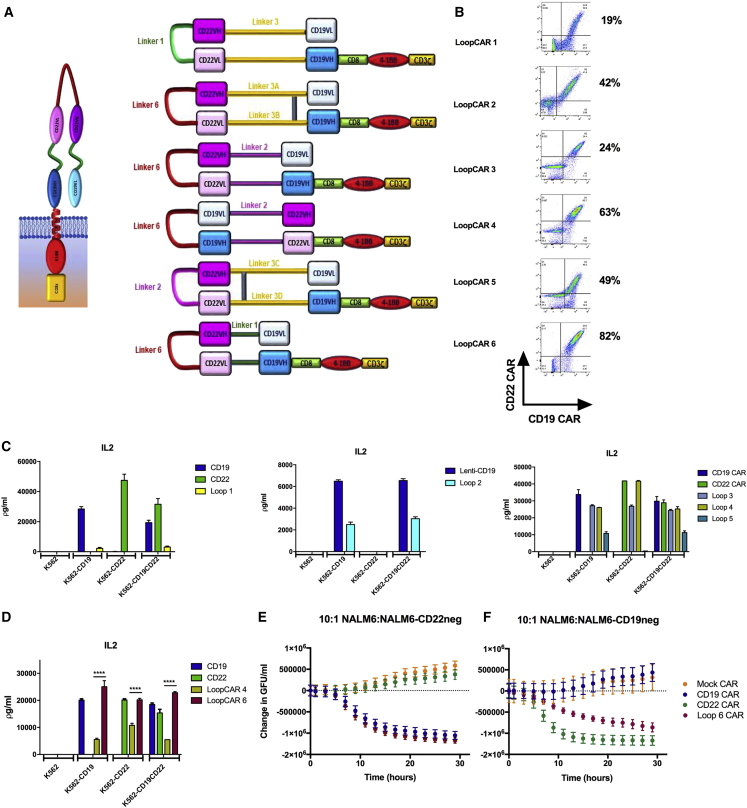
Development of the Bivalent CARs with Alternative Sequence of scFv Resulting in a Loop Structure
To optimize the CD19xCD22 bivalent CAR activity, we next built a series of CAR constructs (Figure 4A) based on previously described success generating bivalent antibodies using loop structures.19 LoopCAR1 was constructed with the CD22 scFv (maintaining the short linker) between the VH and VL of the CD19 ScFv, a format that could only be detected at low percentages on the cell surface (Figure 4B). For LoopCAR2, we increased the length of the linker between the heavy and light chain in the CD22 scFv in an attempt to facilitate folding of the loop structure and slightly modified the amino-acid structure of the linker between the CD19 variable chains and the CD22 scFv to facilitate disulfide bond formation. This improved CAR surface detection. As expected based on low surface detection, LoopCAR1 failed to generate IL-2 production against either CD19 or CD22 (Figure 4C). Despite improved surface detection and some IL-2 production against CD19, LoopCAR2 did not generate detectable IL-2 against CD22 antigen (Figure 4C). Thus, we further modified LoopCAR3 to reduce the length of the linker between the CD19 heavy chains and the CD22 scFv and maintained the slightly longer linker between the VH and VL introduced in LoopCAR2, resulting in improved IL-2 production against CD19−/CD22+ ALL (Figure 4C).
Figure 4.
Development of the Bivalent Loop CARs
(A) Schematic of Loop CAR structures. (B) Flow-cytometric plot demonstrating the surface binding of CD22Fc and CD19 idiotype. (C) Cytokine production of CD19 CAR, CD22 CAR, and LoopCAR1-5 with K562, K562-CD19, K562-CD22, and K562-CD19CD22 target cell lines. (D) Cytokine production of CD19 CAR, CD22 CAR, LoopCAR4, and LoopCAR6 with K562, K562-CD19, K562-CD22, and K562-CD19CD22 target cell lines. ****p < 0.0001. (E and F) Killing of a 10:1 ratio of NALM6:NALM6-CD19neg (E) and NALM6:NALM6-CD22neg (F) cells by CD19-CAR-, CD22-CAR-, and LoopCAR6-expressing T cells. GFU, green fluorescent units.
For the next series of constructs, we placed the CD19 scFv in a membrane-distal location and between the variable chains of the CD22 scFv. In LoopCAR4, we maintained the linker between CD19 scFv and the CD22 scFv variable chains introduced in LoopCAR3, resulting in high levels of CAR detection and superior IL2 production, compared to any of the previous formats (Figure 4C), suggesting that the CD22 scFv membrane-proximal location may be optimal in loop structure. Given that IL-2 production against CD19−/CD22+ ALL was still inferior to the CD22 monovalent CAR, we further modified LoopCAR5 to favor disulfide bond formation, a structure that did not improve cytokine production (Figure 4C). Finally, in LoopCAR6, we incorporated a short G4Sx1 linker between CD19 scFV and CD22 variable chains, which dramatically improved both CAR detection and IL-2 production against both CD19+/CD22− and CD19−/CD22+ ALL (Figures 4B and 4D), as well as in vitro killing of single-antigen-expressing ALL (Figures 4E and 4F). Of note, the kinetics of the killing of CD19− ALL by LoopCAR6-expressing T cells, compared to monovalent CD22-CAR-expressing T cells, suggested slightly less potency against CD22. LoopCAR6 produced multiple cytokines in response to both CD19 and CD22 (Figure S4), further confirming the potency and polyfunctionality of LoopCAR6-expressing T cells. Thus, we established that the loop design may be optimal for bivalent CARs incorporating CD19xCD22 specificity, likely due to challenges maintaining CD22 binding. Among the multiple constructs designed and tested, LoopCAR6 was identified as the most potent format and was further tested in in vivo models.
LoopCAR6 Effectively Eradicates CD19+/CD22+ and CD19− PDX
We next tested LoopCAR6 in NALM6 xenografts. LoopCAR6, at a dose of 3 × 106 cells, eradicated CD19+/CD22+ NALM6 (Figures 5A and 5B). However, at low doses, LoopCAR6 failed to completely eradicate a mixture of CD19− and CD22− ALL (Figure 5C), resulting in the progression of the CD19− NALM6 variant with lower expression of CD22 compared to parental NALM6 (Figure S1A). LoopCAR6 was further tested in a “spike-in” relapse model in which the engrafted ALL inocula contained 1% CD19− or CD22− ALL with 99% CD19+/CD22+ ALL, an assay that mimics relapse from a small, pre-existent clone as has been seen in trials.20 In this model, LoopCAR6 was comparable to CD19 CAR at clearing CD22− ALL, confirming the comparable potency of LoopCAR6 to the CD19 monovalent CARs against CD19. However, in contrast to the CD22 monovalent CAR, LoopCAR6 was unable to clear CD19−/CD22+ ALL with low CD22 site density (Figure 5D). Collectively, and as suggested by the kinetics of in-vitro-killing, CD22 single-expressing ALL cells (Figure 4E), the in vivo experiments suggest that LoopCAR6 has potency that is comparable to that of the CD19 monovalent CAR against CD19 but is slightly less potent than the CD22 monovalent CAR against CD22.
Figure 5.
LoopCAR6 Demonstrates Comparable Efficacy with CD19 CAR but Diminished Potency compared to CD22 CAR against CD19− NALM6 In Vivo
(A) NSG mice were challenged with 1E−6 luciferase-expressing NALM6 leukemia cells on day 0. On day 3, mice were i.v. injected with 3E−6 CAR-expressing T cells. (B) NSG mice were challenged with 1E−6 luciferase-expressing NALM6 leukemia cells on day 0. On day 3, mice were i.v. injected with titrated doses of 9E−6, 3E−6, and 1E−6 LoopCAR6-expressing T cells. (C) NSG mice were challenged with a mixture of 5E−5 NALM6-CD19neg and 5E−5 NALM6-CD22neg luciferase-expressing leukemia cells on day 0. On day 3, mice were treated with 3E−6 CAR-expressing T cells. (D) NSG mice were challenged with 1E−6 luciferase-expressing leukemia cells, as indicated in the table on day 0. On day 3, mice were treated with 6E−6 CAR-expressing T cells. Quantification of luminescence is shown below each image. ****p < 0.0001.
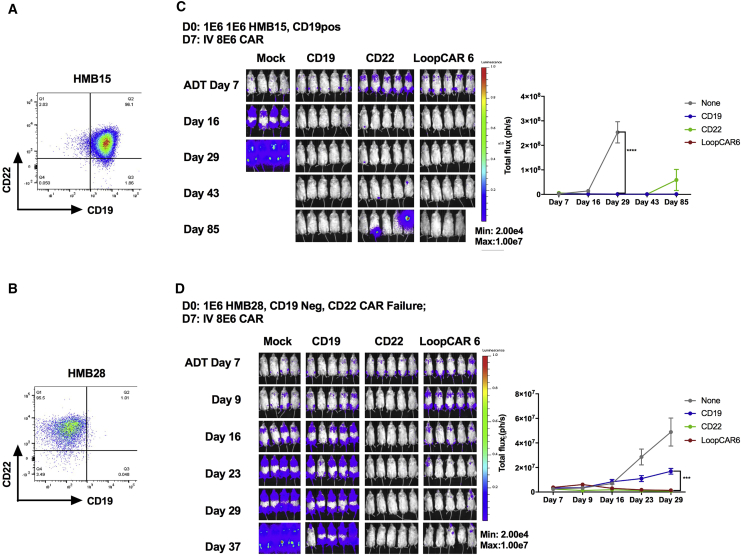
To further explore the in vivo activity of the LoopCAR6 in a clinically relevant model of CD19 CAR resistance, we utilized two different patient-derived xenografts (PDXs) generated from de novo relapse specimens: HMB15 (CD19+/CD22+) and HMB28 (CD19−/CD22+). Whole-exome and transcriptome sequencing were performed to characterize the two PDX model systems. HMB15 harbors a translocation that results in an in-frame fusion oncogene between the N terminus of MLL (exons 1–6) and the C terminus of MLLT10. The CD19 and CD22 genomic loci are intact in this model. The HMB28 PDX primary oncogenic driver is a point mutation of KRAS G12D. In addition, this model harbors a premature stop codon in the CD19 (Table 1). As expected, HMB15 was cleared by both monovalent CARs and LoopCAR6 (Figure 6). HMB28 was resistant to CD19 monovalent CAR and, therefore, a good model of CD19 CAR resistance. Encouragingly, LoopCAR6 prevented progression in HMB28, indicating that LoopCAR6 may be effective at preventing CD19 CAR resistance.
Table 1.
Genomics of Patient-Derived Xenografts
| PDX Model | Primary Oncogene | CD19 | CD22 |
|---|---|---|---|
| HMB15 | MLL-MLLT10 fusion | intact DNA and RNA | intact DNA and RNA |
| HMB28 | KRAS G12D | W214* | intact DNA and RNA |
Figure 6.
LoopCAR6 Eradicates CD19+CD22+ and CD19− Patient-Derived Xenografts
(A) Flow-cytometric plot showing the surface expression of CD19 and CD22 on HMB15 ALL PDXs. (B) NSG mice were challenged with 1E−6 of HMB15 cells on day 0. Mice received treatment with 8E−6 of CD19, CD22, or LoopCAR6 CAR+ T cells on day 7. (C) Flow-cytometric plot showing the surface expression of CD19 and CD22 on HMB28 ALL PDXs. (D) NSG mice were challenged with 1E−6 of HMB28 cells on day 0. Mice received treatment with 8E−6 CD19, CD22, or LoopCAR6 CAR+ T cells on day 7. Quantification of luminescence is shown on the right. ***p < 0.001; ****p < 0.0001.
No Evidence for Off Target Activity of the Optimized CD19xCD22 Bivalent Loop CAR
Given the possibility of mispairing of two different chains, VH and VL, resulting in potential off-tumor toxicity, we performed functional screening of T cells expressing the loop 6 bivalent CAR. LoopCAR6 T cells were co-incubated with human induced pluripotent stem cell (iPSC) lines representing multiple normal tissues, and interferon (IFN)γ production in culture supernatant was analyzed for IFNγ (as a sensitive measure of CAR activity), as described previously4 and in the Supplemental Information. NALM6 and REH-TSLPR, two separate ALL cell lines expressing both CD19 and CD22, were used as positive controls. As expected, in this assay, LoopCAR6 induced IFNγ in T cells against both NALM6 and REH-TSLPR. However, IFNγ production was not detected in supernatants of LoopCAR6-expressing T cells in the presence of any of the iPSC lines (Figure S6).
Discussion
Despite the great success of CAR-T cell therapy in leukemia, a number of obstacles have become apparent from the early clinical experience with CD19 CAR-T cells. One of these challenges is the frequency of CD19 antigen negative relapse, reducing the sustained remission rate.2, 5, 21, 22 Our institutional experience with an active CD22 CAR construct9 also demonstrated changes in antigen expression at relapse, typically associated with antigen diminution rather than complete loss, but sufficient for CAR evasion.12 Furthermore, with heterogeneity in the expression of target antigens, single-antigen targeting may lead to a predisposition to antigen-negative escape. We hypothesized that simultaneous targeting of multiple antigens may reduce the likelihood of relapse due to single antigen loss, similar to the well-established paradigm of multi-agent chemotherapeutic regimens.
Using clinically validated CD19 and CD22 CAR constructs, we tested mutliple bispecific targeting strategies in robust xenograft models of antigen escape. We initially tested the co-infusion of two separate T cell products, each with a single CAR. Interestingly, this approach appeared to favor expansion of the CD19-CAR-expressing T cells over those expressing CD22 CAR. We also tested co-transduction in which two viral vectors are introduced into a single T cell expansion. This approach resulted in low transduction efficiency of both CARs and preferential activity of the CD19 CAR. The mechanism for single-CAR dominance is unclear but under study. Whether this will be a general principle for CAR therapy targeting multiple tumor antigens is also not known.
We next attempted to design single-CAR constructs containing 2 different scFvs, thus generating a bivalent CAR with the goal of ensuring sustained bispecific antigenic activity. Bivalent CARs have been tested in a pre-clinical solid tumor model in which a second CAR target (CD19) is artificially introduced into the tumor cell. These studies provided proof of principle that multi-targeted CARs are active and that the scFv binding domains may act synergistically.13, 16 Thus, we systematically developed bivalent CARs to simultaneously target CD19 and CD22. Through the assembly of the bivalent CAR, we found that details of CAR design (scFv order, linker length, and CAR configuration; i.e., tandem versus loop) had a substantial impact on the potency of the bivalent CAR construct. Ultimately, we were able to generate a CAR (LoopCAR6) with activity both in vitro and in vivo against both CD19 and CD22, including the clearance of a CD19− PDX. This construct is being tested in ongoing clinical trials (NCT03241940, NCT03233854, and NCT03448393). Although prior preclinical studies suggested the possibility of synergistic anti-tumor activity,14, 16 this was not observed in our studies, suggesting that this effect may be dependent on the antigens targeted.
Particularly relevant to the assembly of the bivalent CAR constructs described here is that we based our bivalent design on clinically validated active monovalent CAR constructs. It is important to note that, during the preclinical development of the CD22 CAR, a high affinity scFv with therapeutic efficacy as a toxin conjugate was inactive, potentially due to membrane-distal binding to CD22 in contrast to the membrane-proximal binding, of the lower affinity scFv eventually validated in our clinical trial.9 The importance of membrane-binding location has previously been reported for CD22 CARs. Furthermore, we observed that the short linker in CD22ScFv in tandem-CAR format is optimal for CD22 CAR function. Changes in linker length between the VH and VL in the active monovalent CD22 CAR has also been reported to impair CAR activity.23 Thus, at least for certain antigens, spatial considerations appear to be important in CAR activity. Our studies presented here highlight that this will be even more complex when multiple scFvs are incorporated.
To our knowledge, the CD19/CD22 bivalent CAR reported here is the first demonstration of an active dual CAR targeting two naturally expressed antigens on B-ALL. Importantly, this construct demonstrated comparable efficacy for CD19 and/or CD22 CAR-T cells and provides the opportunity to target multiple antigens simultaneously, using a single construct with the potential to provide sustained pressure against two antigens expressed on leukemic cells. Results from ongoing clinical trials will be required to establish whether this approach reduces the incidence of relapse observed following monovalent CAR therapy.
Materials and Methods
Human Leukemia Samples
Patient samples were screened for antigen expression via a National Cancer Institute institutional review board (NCI-IRB)-approved screening protocol. Human ALL samples for xenograft generation were collected and stored after informed consent to an National Cancer Institute (NCI)-IRB-approved tissue acquisition protocol was obtained. All research specimens from human subjects were obtained with informed consent in accordance with the Declaration of Helsinki.
Cell Lines and Culture Conditions
The following leukemia cell lines were used: the erythroleukemia K562-CD22 (transduced with human CD22, GeneCopoeia, catalog no. EX-Z9364-Lv151); K562-CD19 (transduced with human CD19); K562-CD19CD22 (transduced with both human CD19 and CD22); non-transduced K562 as a negative control; and the B cell acute lymphoblastic leukemia lines NALM6, NALM6-GL (transduced with GFP and luciferase), and REH-TSLPR-GL.4 These cell lines were cultured in media supplemented with 10% heat-inactivated fetal bovine serum (FBS), 10 mM HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (Invitrogen). The Lenti-X 293T lentiviral packaging cell line (Clontech, catalog no. 632180) was cultured in DMEM (Invitrogen). Cell lines for CAR toxicity assay, including mesenchymal stem cell (MSC) (bone marrow-derived MSC [BMSC]), HUVEC (endothelial), cardio cell, iPSC (neuron), CCD19lu (fibroblast), MOVAS (fibroblast), SW403 (colon), 293T (kidney), H1299 (lung), and melanoma (skin), are from in-house stock. Endothelial, glutaneuron, cardiomyocyte, neuron, hepatocyte, and astrocyte cell lines were purchased from Cellular Dynamics International (Madison, WI), and all cell lines were cultured in recommended media according to recommended conditions.
Creation of the CD19− and CD22− Leukemia Relapse Model
CRISPR-Cas9 technology was used to edit NALM6 to generate NALM6-CD19−-GL (CRISPR CD19 on exon 3) and NALM6-CD22−-GL (CRISPR CD22 on exon 6). Lentiviral vectors for CRISPR-Cas9 gene editing of CD19 or CD22 on NALM6 was previously described.12 Simply, guide RNAs were optimized by http://crispor.tefor.net/ and cloned into LentiCRISPR v2 plasmid (Addgene, plasmid 52,961). Plasmids were then co-transfected with packaging plasmids and transformed into HEK293T cells. Two days later, supernatants were harvested, filtered, and concentrated. For viral transduction, 105 NALM6 cells were incubated with 10 mL concentrated viral supernatant for 2 days, followed by expansion in RPMI media. Cell phenotype was assessed by flow cytometry, followed by the sorting of cells with phenotypic alterations and single-cell cloning. Sequencing was performed on single-cell clones to confirm genotypic alterations.
CAR Lentiviral Vector Production and T Cell Transduction
Bivalent CAR constructs were designed and synthesized followed by cloning into lentiviral transfer plasmids. Bivalent CAR-encoding lentiviral vectors were produced by transient transfection of the Lenti-X 293T lentiviral packaging cell line modified from a previously described method. Briefly, Lenti-X 293T cells were plated into poly-D-lysine-coated 15-cm plates (BD Biosciences). The following day, Lenti-X 293T cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) with plasmids encoding the bivalent CAR along with packaging and envelope vectors (pMDLg/pRRE, pMD-2G, and pRSV-Rev). Lentiviral supernatants were harvested at 24 and 48 hr post-transfection, centrifuged at 3,000 rpm for 10 min to remove cell debris, frozen on dry ice, and stored at −80°C. Human peripheral blood mononuclear cells (PBMCs) from normal donors were obtained with an NIH-approved protocol and activated with CD3 and CD28 microbeads at a ratio of 1:3 (Dynabeads Human T-Expander CD3/CD28, Thermo Fisher Scientific, catalog no. 11141D) in AIM-V media containing 40 IU/mL recombinant IL-2 and 5% FBS for 24 hr. Activated T cells were resuspended at 2 million cells per 2 mL lentiviral supernatant plus 1 mL fresh AIM-V media with 10 μg/mL protamine sulfate and 100 IU/mL IL-2 in 6-well plates. Plates were centrifuged at 1,000 × g for 2 hr at 32°C and incubated overnight at 37°C. A second transduction was performed on the following day by repeating the same transduction procedure described earlier. The CD3:CD28 beads were removed on the third day following transduction, and the cells were cultured at 300,000 cells per milliliter in AIM-V medium containing 100 IU/mL IL-2, with fresh IL-2-containing media added every 2–3 days until harvest on day 8 or 9.
Flow Cytometry Analysis
Surface expression of CD22-CAR-transduced T cells was determined by flow cytometry, using a CD22-Fc chimera (R&D Systems), followed by incubation with phycoerythrin (PE)-F(ab)2 or antigen-presenting cell (APC)-F(ab)2 specific for human immunoglobulin G (IgG)-Fc (Jackson ImmunoResearch Laboratories). Surface expression of CD19-CAR-transduced T cells was detected with anti-CD19 Idiotype or Recombinant Human CD19 Fc Chimera Protein (R&D Systems) conjugated with APC by using the Lightning-Link APC Antibody Labeling Kit (Novus Biologicals). Expression of bivalent CARs was assessed using a combination of both detection reagents as indicated for individual figures. Expression of CD19 and CD22 on B-ALL lines was detected using the following anti-human antibodies: CD45-PerCP-Cy5.5 (eBioscience), CD19-Pacific Blue, CD19-APC-Cy7, CD10-PE-Cy7, and CD22-PE (BioLegend). T cells were characterized with the following antibodies: CD3-APC-Cy7, CD4-Pacific Blue, and CD8a-PE-Cy7 (BioLegend).
Cytotoxicity Assay
5E4 of target tumor cells in 100 μL RPMI media were loaded into a 96-well plate (Corning BioCoat Poly-L-Lysine 96-Well Clear TC-Treated Flat Bottom Assay Plate). An equal amount of CAR-T cells was added into the designated well on the following day. The plate was scanned for the GFP fluorescent expression to monitor apoptosis GFP+ cell disappearance using an IncuCyte ZOOM system every 30 min in a duration of 40 hr. The percentage of cell killing at each time point was determined relative to baseline.
Analysis of Cytokine Production
Target tumor cells and transduced CAR+ T cells were washed 3 times with PBS and resuspended in RPMI at 1−E6 cells per milliliter. 100 μL (1 × 105) tumor cell suspension and 100 μL CAR-T cell suspension was loaded into each well of a 96-well plate with T cell-only and tumor-cell-only controls in duplicates or triplicates. After 18 hr in a 37°C incubator, a culture supernatant was harvested for detection of the cytokines using either ELISA (R&D Systems) or a multiplex assay (Meso Scale Discovery).
In Vivo Studies
Animal experiments were carried out under protocols approved by the NCI Bethesda Animal Care and Use Committee. B-ALL cell lines and the xenografted human B-ALL specimens were intravenously (i.v.) injected into NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; Jackson Laboratories). For luciferase-expressing lines, leukemia was detected using the Xenogen IVIS Lumina (Caliper Life Sciences). Mice were injected intraperitoneally with 3 mg D-luciferin (Caliper Life Sciences) and were imaged 4 min later with an exposure time of 30 s for NALM6 and 2 min for PDXs. Living Image Version 4.1 software (Caliper Life Sciences) was used to analyze the bioluminescent signal flux for each mouse as photons per second per square centimeter per steradian. Leukemia burden in non-luciferase-expressing xenografts was measured by flow cytometry of peripheral blood, bone marrow, and spleen.
PDXs
The following primary samples were used for the generation of PDX models: CD19− ALL and the CD19+CD22dim de novo relapse specimens ALL_H0113_post22_r (HMB28) and ALL_H0090_post19_pd (HMB15). PDXs were created by injecting 1−E6 to 10−E6 of the patient ALL cells intravenously into NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson ImmunoResearch Laboratories). After 2 successful passages, PDX lines were transduced with lenti-GFP-Luc virus and sorted for high expression of GFP and luciferase after the first and second passages. GFP-transduced PDX leukemia in vivo burden was assessed by weekly fluorescence imaging, and animals were treated with CAR-T cells via tail vein injection once the human ALL was detectable by fluorescence imaging. Elutriated human lymphocytes from healthy donors were obtained from the Department of Transfusion Medicine at the NIH Clinical Center under an IRB-approved protocol. The human lymphocytes were cultured in AIM-V media.
Genomic Analysis of PDX Models
Nucleic acid extractions were performed on viably cryopreserved samples using QIAGEN AllPrep Micro Kits per the manufacturer’s protocol. DNA and RNA were quantified and assessed for quality using an Agilent 2100 BioAnalyzer. Poly-adenylated RNA libraries were generated and sequenced using TruSeq 4.0 chemistry on a HiSeq 2500 (Illumina) platform. Whole-exome data were generated using Agilent SureSelect XT Human All Exon V5 and TruSeq V4 chemistry and sequenced to a median of 100× coverage using the HiSeq 2500 (Illumina) platform.
Whole-exome and RNA-sequencing data were mapped and analyzed using the CCR Collaborative Bioinformatics Resource (CCBR) pipeline. Reads were aligned to reference genome Hg19. Somatic variant calling was performed using MuTect,24 and copy number alterations were analyzed using Nexus Copy Number 9.0, Discovery Edition (BioDiscovery). The integrity of the CD19 and CD22 genes was further interrogated by manual inspection using the Integrative Genome Viewer (IGV). RNA-sequencing reads for each sample were trimmed of their adapters and low-quality bases using Trimmomatic software and alignment with reference human Hg 38 and GENCODE V24 transcripts using STAR software.
Statistical Analysis
Statistics analyses were performed using Prism 7.0 software. Statistical significance was calculated using the Mann-Whitney test, two-way ANOVA, and unpaired Student’s t test. The significant p values were assigned as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 and designated on the plots.
Author Contributions
H.Q. designed CAR constructs and experiments, performed experiments, analyzed data, and wrote the manuscript. S.R., S.N., T.J.F., and A.P. performed experiments and analyzed data. J.F.S., analyzed RNAseq data and contributed in writing the manuscript. N.N.S. contributed in writing the manuscript and generated patient leukemia data. M.S.-S. and C.M.Y. generated antigen expression data on patient leukemia. T.J.F. conceived of the research, designed experiments, supervised research, analyzed data, and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We would like to thank John Buckley for his expert assistance in the animal experiments. We would also like to thank the Leidos Biomedical Research, Inc.’s Genomic Core at Frederick National Laboratory for Cancer Research for their work on RNA-seq. We would like to thank Haiyan Lei for helping on GEO data submission to a public database. This work was supported by the Intramural Research Program at the NIH.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.omto.2018.10.006.
Supplemental Information
References
- 1.Gardner R.A., Finney O., Annesley C., Brakke H., Summers C., Leger K., Bleakley M., Brown C., Mgebroff S., Kelly-Spratt K.S. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood. 2017;129:3322–3331. doi: 10.1182/blood-2017-02-769208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee D.W., Kochenderfer J.N., Stetler-Stevenson M., Cui Y.K., Delbrook C., Feldman S.A., Fry T.J., Orentas R., Sabatino M., Shah N.N. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin H., Cho M., Haso W., Zhang L., Tasian S.K., Oo H.Z., Negri G.L., Lin Y., Zou J., Mallon B.S. Eradication of B-ALL using chimeric antigen receptor-expressing T cells targeting the TSLPR oncoprotein. Blood. 2015;126:629–639. doi: 10.1182/blood-2014-11-612903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruella M., Barrett D.M., Kenderian S.S., Shestova O., Hofmann T.J., Perazzelli J., Klichinsky M., Aikawa V., Nazimuddin F., Kozlowski M. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J. Clin. Invest. 2016;126:3814–3826. doi: 10.1172/JCI87366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalabi H., Kraft I.L., Wang H.W., Yuan C.M., Yates B., Delbrook C., Zimbelman J.D., Giller R., Stetler-Stevenson M., Jaffe E.S. Sequential loss of tumor surface antigens following chimeric antigen receptor T-cell therapies in diffuse large B-cell lymphoma. Haematologica. 2018;103:e215–e218. doi: 10.3324/haematol.2017.183459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haso W., Lee D.W., Shah N.N., Stetler-Stevenson M., Yuan C.M., Pastan I.H., Dimitrov D.S., Morgan R.A., FitzGerald D.J., Barrett D.M. Anti-CD22-chimeric antigen receptors targeting B-cell precursor acute lymphoblastic leukemia. Blood. 2013;121:1165–1174. doi: 10.1182/blood-2012-06-438002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantarjian H.M., DeAngelo D.J., Stelljes M., Martinelli G., Liedtke M., Stock W., Gökbuget N., O’Brien S., Wang K., Wang T. Inotuzumab ozogamicin versus standard therapy for acute lymphoblastic leukemia. N. Engl. J. Med. 2016;375:740–753. doi: 10.1056/NEJMoa1509277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wayne A.S., Shah N.N., Bhojwani D., Silverman L.B., Whitlock J.A., Stetler-Stevenson M., Sun W., Liang M., Yang J., Kreitman R.J. Phase 1 study of the anti-CD22 immunotoxin moxetumomab pasudotox for childhood acute lymphoblastic leukemia. Blood. 2017;130:1620–1627. doi: 10.1182/blood-2017-02-749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry T.J., Shah N.N., Orentas R.J., Stetler-Stevenson M., Yuan C.M., Ramakrishna S., Wolters P., Martin S., Delbrook C., Yates B. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med. 2018;24:20–28. doi: 10.1038/nm.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grada Z., Hegde M., Byrd T., Shaffer D.R., Ghazi A., Brawley V.S., Corder A., Schönfeld K., Koch J., Dotti G. TanCAR: a novel bispecific chimeric antigen receptor for cancer immunotherapy. Mol. Ther. Nucleic Acids. 2013;2:e105. doi: 10.1038/mtna.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde M., Corder A., Chow K.K., Mukherjee M., Ashoori A., Kew Y., Zhang Y.J., Baskin D.S., Merchant F.A., Brawley V.S. Combinational targeting offsets antigen escape and enhances effector functions of adoptively transferred T cells in glioblastoma. Mol. Ther. 2013;21:2087–2101. doi: 10.1038/mt.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bielamowicz K., Fousek K., Byrd T.T., Samaha H., Mukherjee M., Aware N., Wu M.F., Orange J.S., Sumazin P., Man T.K. Trivalent CAR T cells overcome interpatient antigenic variability in glioblastoma. Neuro Oncol. 2018;20:506–518. doi: 10.1093/neuonc/nox182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegde M., Mukherjee M., Grada Z., Pignata A., Landi D., Navai S.A., Wakefield A., Fousek K., Bielamowicz K., Chow K.K. Tandem CAR T cells targeting HER2 and IL13Rα2 mitigate tumor antigen escape. J. Clin. Invest. 2016;126:3036–3052. doi: 10.1172/JCI83416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raponi S., De Propris M.S., Intoppa S., Milani M.L., Vitale A., Elia L., Perbellini O., Pizzolo G., Foá R., Guarini A. Flow cytometric study of potential target antigens (CD19, CD20, CD22, CD33) for antibody-based immunotherapy in acute lymphoblastic leukemia: analysis of 552 cases. Leuk. Lymphoma. 2011;52:1098–1107. doi: 10.3109/10428194.2011.559668. [DOI] [PubMed] [Google Scholar]
- 18.Wen D., Foley S.F., Hronowski X.L., Gu S., Meier W. Discovery and investigation of O-xylosylation in engineered proteins containing a (GGGGS)n linker. Anal. Chem. 2013;85:4805–4812. doi: 10.1021/ac400596g. [DOI] [PubMed] [Google Scholar]
- 19.Wu C. Diabodies: molecular engineering and therapeutic applications. Drug News Perspect. 2009;22:453–458. [PubMed] [Google Scholar]
- 20.Grupp S.A., Kalos M., Barrett D., Aplenc R., Porter D.L., Rheingold S.R., Teachey D.T., Chew A., Hauck B., Wright J.F. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sotillo E., Barrett D.M., Black K.L., Bagashev A., Oldridge D., Wu G., Sussman R., Lanauze C., Ruella M., Gazzara M.R. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015;5:1282–1295. doi: 10.1158/2159-8290.CD-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacoby E., Nguyen S.M., Fountaine T.J., Welp K., Gryder B., Qin H., Yang Y., Chien C.D., Seif A.E., Lei H. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat. Commun. 2016;7:12320. doi: 10.1038/ncomms12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruella M., Maude S.L., Engels B., Barrett D.M., Frey N., Marcucci K.T., Shestova O., Singh N., Perazzelli J., Christian D.A. Clinical efficacy of anti-CD22 chimeric antigen receptor T cells for B-cell acute lymphoblastic leukemia is correlated with the length of the Scfv linker and can be predicted using xenograft models. Blood. 2017;130:807. [Google Scholar]
- 24.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.