Abstract
Introduction
Celiac disease (CD) is a chronic inflammatory intestinal disorder. Different immunological factors, including inflammatory cytokines, may play an important role in disease susceptibility.
Aim
To investigate the relationship between -174G/C and -572G/C gene polymorphisms and the serum level of interleukin 6 (IL-6) and susceptibility to CD in the Iranian population.
Material and methods
In this case-control study blood samples were collected of 105 patients with CD and 106 healthy subjects randomly in 2016 and evaluated by polymerase chain reaction-restriction fragments length polymorphism (PCR-RFLP) method. A sequence was also used to confirm the results of both polymorphisms. The IL-6 concentration was measured using ELISA.
Results
The results showed a significant relationship between polymorphism -572G in CD patients when compared with control subjects by genotype (p = 0.001) and alleles (p = 0.022), respectively. There was no significant relationship between polymorphism 174G and frequency of genotype, but an association of this polymorphism with the frequency of alleles (p = 0.034), age (p = 0.001), and body mass index (p = 0.003) was seen. The serum level of interleukin-6 was significantly associated only with rs1800796 (p < 0.001).
Conclusions
The results confirm previous studies in different parts of the world and indicate that IL-6 (572G/C) polymorphism may play a role in susceptibility to CD in the Iranian population.
Keywords: celiac disease, polymerase chain reaction-restriction fragments length polymorphism, IL-6 gene polymorphism, autoimmune and inflammatory reaction
Introduction
Celiac disease (CD) is an immune disorder characterised by abnormalities in the small intestine caused by eating gluten, in susceptible individuals [1, 2]. Interleukin 6 (IL-6) stimulates the production of acute phase reactant proteins that cause inflammation or tissue injury [3, 4]. On the other hand, studies have shown that serum levels of IL-6 in CD patients increase after consumption of gluten-containing foods in untreated patients and decrease a year after commencement of a gluten-free diet [5, 6]. Studies in different populations have investigated the relationship between CD and gene polymorphisms of immunological parameters such as transforming growth factor β1 (TGF-β1), IL-10, IL-6, and tumor necrosis factor α (TNF-α) [7, 8] and shown that polymorphisms of -572G and -174G in the interleukin 6 promoter can effect the production and secretion of interleukin, which may play a role in the inflammation and pathogenesis of celiac disease [9–12].
Aim
The aim of this study was to investigate the serum level and role of IL-6 (-174) and (-572) G/C gene promoter regions in Iranian patients with CD.
Material and methods
A total of 105 patients with CD and 106 healthy individuals were recruited in this study during 2016. Diagnosis of CD was based on positive serology and confirmed by endoscopy and histopathological evaluation. This study was approved by the Local Ethics Committee of the Research Institute for Gastroenterology and Liver Diseases of Shahid Beheshti University of Medical Sciences (IR. SBMU.RIGLD.REC.1395.132). Demographic and clinical presentations were collected by questionnaire. The level of IL-6 in serum was determined using the ELISA method according to manufacturer’s instructions (eBioscience Human IL-6 ELISA Ready-SET-Go! Kit).
Blood collection
Blood samples (10 ml) were collected and serum and peripheral blood mononuclear cells (PBMC) were isolated and kept at –20°C for the subsequent steps.
DNA isolation
DNA extraction was performed in the following steps: (1) 70 μl of SDS (10%) was added to PBMC and mixed by pulse-vortexing, then incubated at 50°C for 10 min; (2) 20 μl of protein kinase was added and incubated at 50°C for 2 h; (3) 450 μl of NaCl2 (5 M) was added and mixed by pulse-vortexing, then it was centrifuged at 12,000 rpm for 10 min; (4) the supernatant was isolated, isopropanol (1 : 1 v/v %) was added and mixed slowly; (5) the sedimentation of DNA was transferred into new micro-tubes, 95% ethanol was added, and DNA was sedimented in the alcohol. Afterwards, it was centrifuged at 12,000 rpm for 10 min and washed with 70% ethanol. Finally, the DNA was diluted in 60 μl of TE (1×) and the DNA concentration was determined by spectroscopy (260 nm and 280 nm) using the following formula: double strand DNA (ng/μl) = (OD) × (50 ng/μl).
IL-6 polymorphism (rs1800796 and rs1800795 G/C)
IL-6 polymorphism was identified by polymerase chain reaction (PCR)-based restriction fragment length polymorphism (RFLP). For the IL-6 (-174 G/C) rs1800795 G/C polymorphism, the forward primer 5’-GAGCCAGAACACAGAAGAACT-3’ and reverse primer 5’-TGGGGCTGATTGGAAACCTT-3’ was designed. For the (-572 G/C) rs1800796 G/C polymorphism, the forward primer 5’-GTGCCCCAGTGAAACAGTG-3’ and reverse primer 5’-CAAGCCTGGGATTATGAAGA-3’ were designed, and the target areas were amplified by PCR-RFLP. The PCR products were visualised using 1% agarose gel electrophoresis and then digested by RFLP enzymes. For RFLP analysis, 10 μl of the PCR product was digested with SFANI and BSrBI restriction enzymes (New England Biolabs) for rs1800795 and rs1800796 polymorphisms, respectively, in a total volume of 25 μl. Afterwards, the samples were incubated in a bain-marie for 4 h for rs1800795 and 2 h for rs1800796 at 37°C. The products were visualised using 3% agarose gel electrophoresis using the 50 bp marker.
SFANI restriction enzyme cut points:
5’ … GCATC (N)5▾…3’
3’ … CGTAG (N)9▴…5’
BSrBI restriction enzyme cut points:
5’ … CCG▾CTC …3’
3’ … GGC▴GAG …5’
DNA sequencing
Approximately 10% of the PCR products were randomly selected for DNA sequencing and sent to Gene Fanavaran. DNA sequences were determined by Bioedit. The sequenced data were compared with sequences available at GenBank.
Statistical analysis
Statistical data analysis was done in SPSS software (version 21). Descriptive statistics were expressed as mean ± standard deviation and percentage for quantitative and qualitative variables, respectively. The differences were considered significant at p ≤ 0.05.
Results
Patient description
In the present study, of the 105 celiac patients (average age: 31.94 years; average body mass index (BMI): 23.37 kg/m2) 36 (34.3%) were male and 69 (65.7%) were female. Of the 106 (51%) healthy individuals (average age: 31.63 years; average BMI: 23.85 kg/m2) 51 (48.1%) were male and 55 (51.9%) were female. All were Iranian with an average age of 31.77 years (min. age: 1 year; max. age: 72 years). The average BMI was 23.6253 kg/m2 (min.: 2.21 kg/m2; max.: 35.91 kg/m2). There were 87 (41.2%) males and 124 (58.8%) females, 61.1% were married and 38.9% were single.
Polymerase chain reaction results
IL-6 gene polymorphism in positions -174 and -572 was analysed using PCR-RFLP. Briefly, the specific primer was designed for both polymorphisms, and the sequence of polymorphisms rs1800795 and rs1800796 were amplified by PCR reaction. The length of the PCR for rs1800795 was 442 and for rs18007956 was 358.
Enzymatic findings
For RFLP analysis of rs1800795 and rs1800796, the PCR product was digested and the cut fragments appeared on 3% agarose gel. The region of polymorphism of rs1800795 was identified using the SfaNI restriction enzyme (Table I). The RFLP-generated profile comprised three-fragment 442 for the CC genotype, (379 and 63) for the GG genotype and (442, 379, 63) for the CG genotype. The region of polymorphism of rs1800796 was identified using the BsrBI restriction enzyme (Table I). The presence of nucleotide G at the position of the polymorphism can be identified by restriction enzymes. The RFLP-generated profile comprised the three-fragment 358 for the CC genotype, (285 and73) for the GG genotype, and (358, 285, 73) for the CG genotype.
Table I.
Size of fragments produced by restriction enzyme
| Enzyme | Restriction site | Genotype | Fragment length [bp] |
|---|---|---|---|
| SfaNI | GCATC | CC | 442 |
| GG | 379, 63 | ||
| CG | 442, 379 and 63 | ||
| BsrBI | CCGCTC | CC | 358 |
| GG | 285 and 73 | ||
| CG | 358, 285, and 73 |
Direct sequencing
In order to confirm the results of PCR-RFLP, a number of samples were sequenced. Figures 1 and 2 show a fragment of the CG heterozygote genotype of rs1800795 and rs1800796, respectively.
Figure 1.
Sequence of rs1800795
Figure 2.
Sequence of rs1800796
Results of genotype determination of patients and healthy individuals
The genotypes and allele frequency distributions of the IL-6 rs1800795 and rs1800796 polymorphisms in CD patients and healthy controls are presented in Table II. The relationship between rs1800795 and body mass index (BMI) and age was significant (p = 0.001 and p = 0.003 respectively). The relationship between rs1800795 and sex was not statistically significant (p = 0.695). The relationships between rs1800796 and BMI, age, and sex were not significant (p = 0.102, p = 0.311 and p = 0.431, respectively).
Table II.
Genotypic and allelic frequency of IL-6 promoter polymorphism (-174) and (-572) in patients with CD and healthy controls
| Variable | Genotype and allele | Celiac patient (n = 105) | Healthy control (n = 106) | CI | OR | P-value |
|---|---|---|---|---|---|---|
| rs1800795 | GG | 47 (44.8%) | 61 (57.5%) | Ref. | Ref. | Ref. |
| CC | 11 (10.5%) | 5 (4.7%) | 0.372–1.157 | 0.656 | 0.145 | |
| CG | 47 (44.8%) | 40 (37.7%) | 0.114–1.077 | 0.35 | 0.06 | |
| G | 141 (67.1%) | 162 (76.4%) | Ref. | Ref. | Ref. | |
| C | 69 (32.9%) | 50 (23.6%) | 0.411–0.968 | 0.631 | 0.034 | |
| rs1800796 | GG | 38 (36.2%) | 12 (11.3%) | Ref. | Ref. | Ref. |
| CG | 60 (57.1%) | 90 (84.9%) | 0.451–7.260 | 1.810 | 0.403 | |
| CC | 7 (6.7%) | 4 (3.8%) | 2.297– 9.823 | 4.750 | < 0.001 | |
| G | 136 (64.8%) | 114 (53.8%) | Ref. | Ref. | Ref. | |
| C | 74 (35.2%) | 98 (46.2%) | 1.068–2.336 | 1.580 | 0.022 |
In the CD group, the frequency of the G allele (67.1%) of rs1800795 was much greater than that of the C allele (32.9%). The same was observed in the control group (p = 76.4% and p = 23.6%, respectively). In the CD group, the frequency of the G allele (64.8%) of rs1800796 was much greater than that of the C allele (35.2%). The same was observed in the control group (p = 53.8% and p = 46.2%, respectively).
Serum level of interleukin 6
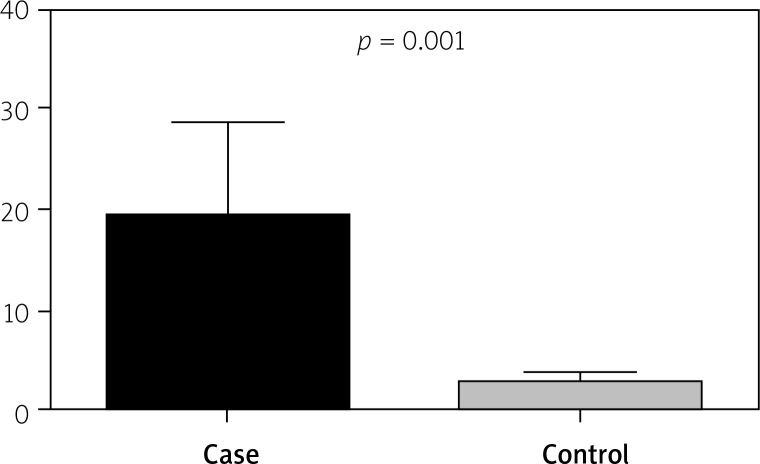
The mean serum IL-6 concentration in patients with CD was 19.11, and in the control group it was 2.78. Figure 3 shows that there was a significant difference between serum IL-6 levels in CD and healthy subjects (p = 0.0001). There was a significant relationship between rs1800796 polymorphism and serum IL-6 concentration (p < 0.001) but not for rs1800795 (p = 0.106).
Figure 3.
Relationship between serum levels in CD patients and healthy subjects
Discussion
Celiac disease is a T-cell-mediated an intestinal disorder that is caused by gluten in the diet of people who are genetically susceptible [13]. Genetic factors, including HLA class II genes, are the most important factors in the development of CD [14]. While 40–90% of patients are HLA-DQ2 positive, fewer are HLA-DQ8 positive. Other factors could be involved in the development of disease, such as genes encoded for several immune response molecules that are involved in CD sensitivity [14].
In CD, mucosal damage is caused by the activity of the immune system and acquired immunity. Previous studies have shown that inflammation of the intestines in CD is caused by different cytokines produced by T4 CD4 cells responsible for the pathogenesis of the disease [15, 16]. Cytokines are important intermediates of immunity. In the case of imbalance or deficiency, they can cause autoimmune diseases [17]. Studies have shown that the genes encoding inflammatory cytokines can be considered a predisposing factor in lesions caused by immune mediators in CD [18].
Interleukin 6 is an essential cytokine for adjustment of the immune system. Excessive production of this cytokine leads to inflammation and is associated with inflammatory autoimmune diseases [19]. Some polymorphism in the IL-6 promotor region has been studied, but polymorphism at positions -174 and -572 affects IL-6 expression [20]. This polymorphism has been investigated in rheumatoid arthritis, systemic lupus erythematous, and multiple sclerosis [21–23]. Several studies have examined the relationship between interleukin-6 polymorphism and interleukin-6 serum levels [24]. Compared with the CC genotype IL-6 (-572G/C), the GG genotype (G allele) gene is responsible for producing more IL-6 [25, 26].
Interleukin 6 plays an important role in the pathogenesis of CD. The current study evaluated genotypic distribution and allelic polymorphisms of IL-6 gene (rs1800796 G/C (-572 G/C) and rs1800795 G/C (-174 G/C)), and its serum level was evaluated among healthy and CD patients in the Iranian population.
In the current study, genotypic distributions for polymorphism at position -174 of the IL-6 gene promoter showed no significant difference between CD patients and the healthy control group. In this polymorphism, the relationship between allele prevalence, BMI, and age in the CD and healthy control groups was significant. In rs1800796, genotypic and allelic distributions were significant between the CD and healthy control groups, but in this polymorphism there was no significant relationship between BMI, age, and sex between the CD and healthy control group. The serum IL-6 levels were significantly higher in the CD group when compared with the healthy controls. There was no relationship between serum IL-6 and rs1800795, but there was a significant difference between the CD and healthy control groups in rs1800796.
In recent years, the association between rs1800795 and rs1800796 gene polymorphisms and susceptibility to some diseases has been studied. Sghaier et al. confirmed an association of IL-6, rs180795, and rs1800796 gene polymorphisms with hepatocellular carcinoma in HCV patients in Tunisia [27].
Note that similar studies in different populations can produce different results. Up to now, few studies have focused on the relationship between IL-6 polymorphisms and susceptibility to celiac disease. Woolley et al. studied the polymorphism of cytokines TGF-β1, IL-10, IL-6, interferon γ (IFN-γ), and TNF-α genetic in CD patients in Finland. They concluded that there was no association between CD and polymorphisms of cytokines TGF-β1, IL-10, IL-6 (rs1800795), and IFN-γ genes except for TNF-α gene [28]. In the current study, there was no association between CD susceptibility and IL-6 rs1800795 polymorphism, which confirms the results of Woolley’s study. Maranhão et al. showed that there is no association between CD and IL-6 (-174 G>C) polymorphism. The results of these studies indicate that gene polymorphism depends largely on geographic area [29]. In the current study, there was no association between CD susceptibility and IL-6 rs1800795 polymorphism, which confirms the results of Maranhão’s study.
Dema et al. examined the relationship between IL-6 -174G/C polymorphism and CD in Spanish females. This study was performed on 347 children with CD and 853 healthy children using TaqMan technology. The results showed that there is no relationship between polymorphism and subjects with CD; however, after segmentation of the genders, the 174C variant increased the risk of CD, especially in females [30]. In the current study, there was no association between CD and IL-6 rs1800795 polymorphism, which confirms the results; however, the relationships between alleles, BMI, and age with rs1800796 were found to be significant.
In a recent study, the association between IL-6 (-572G/C) and IL-17 (-197A/G) polymorphisms (rs2275913) and CD was investigated. This study was performed on 84 patients with CD and 83 healthy subjects as the control group. Polymorphisms were studied in the -572G/C region of interleukin-6 and -197A/G of the interleukin-17 gene using the PCR-RFLP method. The results were significant for the IL-6 gene, but not for the IL-17 gene polymorphism [7]. In the current study, there was a significant correlation between genotype prevalence and allele prevalence in rs1800796 for the CD and control group.
Several studies have been done to evaluate the serum concentration of IL-6 in some inflammatory diseases, including in CD. Romaldini et al. examined the serum concentration of the interleukin-2 receptor, IL-6 and tumour necrosis factor-α in children with CD. In accordance with the findings of the current study, serum levels of IL-6 in the CD group were significantly higher than in the control group [16]. Manavalan et al. confirmed the findings of the current study and showed that serum levels of pre-inflammatory cytokines (IL-1, IL-6, IL-8, TNF-α) in untreated CD patients and patients with positive antibodies in comparison with other groups showed a dramatic increase [10]. Garrote et al. reported that levels of IL-6 increased in the serum of CD patients [9]. Kapoor et al. also confirmed the results of the current study and showed that serum levels of interleukin-6 and interleukin-2 receptors have a good correlation with CD activity and can be used as reliable markers for detecting minimal transgression from GFD [15]. In the current study, the association between serum IL-6 concentration and rs1800795 and rs1800796 polymorphisms were investigated. The relationship between serum IL-6 and rs1800796 was significant.
Conclusions
The results of this study show a significant relation between IL-6 (-572G/C) (rs1800796) and CD. This suggests that the IL-6 (-572G/C) polymorphism should be evaluated as a risk factor in the development of CD. The rs1800796 polymorphism showed parallel links between serum IL-6 and the polymorphism of this cytokine in CD patients. The -572G/C polymorphism is a functional variant and promoter region directly responsible for serum levels of IL-6. Further studies with higher patient numbers and other interleukin-6 gene polymorphisms are needed to confirm the relationship between the IL-6 polymorphism and CD.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Dieterich W, Ehnis T, Bauer M, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 2.Fasano A, Berti I, Gerarduzzi T, et al. Prevalence of celiac disease in at-risk and not-at-risk groups in the united states: a large multicenter study. Arch Inter Med. 2003;163:286–92. doi: 10.1001/archinte.163.3.286. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rincon M. Interleukin-6: from an inflammatory marker to a target for inflammatory diseases. Trends Immunol. 2012;33:571–7. doi: 10.1016/j.it.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Clot F, Babron MC. Genetics of celiac disease. Mol Genet Metabol. 2000;71:76–80. doi: 10.1006/mgme.2000.3045. [DOI] [PubMed] [Google Scholar]
- 6.de la Concha EG, Fernández-Arquero M, Vigil P, et al. Celiac disease and TNF promoter polymorphisms. Hum Immunol. 2000;61:513–7. doi: 10.1016/s0198-8859(99)00187-1. [DOI] [PubMed] [Google Scholar]
- 7.Akbulut UE, Cebi AH, Sag E, et al. Interleukin-6 and interleukin-17 gene polymorphism association with celiac disease in children. Turk J Gastroenterol. 2017;28:471–5. doi: 10.5152/tjg.2017.17092. [DOI] [PubMed] [Google Scholar]
- 8.Singh PK, Chandra G, Bogra J, et al. Association of interleukin-6 genetic polymorphisms with risk of OSCC in indian population. Meta Gene. 2015;4:142–51. doi: 10.1016/j.mgene.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrote JA, Arranz E, Gomez-Gonzalez E, et al. IL6, IL10 and TGFB1 gene polymorphisms in coeliac disease: differences between DQ2 positive and negative patients. Allergol Immunopathol. 2005;33:245–9. doi: 10.1157/13080926. [DOI] [PubMed] [Google Scholar]
- 10.Manavalan JS, Hernandez L, Shah JG, et al. Serum cytokine elevations in celiac disease: association with disease presentation. Hum Immunol. 2010;71:50–7. doi: 10.1016/j.humimm.2009.09.351. [DOI] [PubMed] [Google Scholar]
- 11.Mazzarella G. Effector and suppressor T cells in celiac disease. World J Gastroenterol. 2015;21:7349–56. doi: 10.3748/wjg.v21.i24.7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amr K, El-Awady R, Raslan H. Assessment of the −174G/C (rs1800795) and −572G/C (rs1800796) interleukin 6 gene polymorphisms in Egyptian patients with rheumatoid arthritis. Open Access Maced J Med Sci. 2016;4:574–7. doi: 10.3889/oamjms.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuppan D. Current concepts of celiac disease pathogenesis. Gastroenterology. 2000;119:234–42. doi: 10.1053/gast.2000.8521. [DOI] [PubMed] [Google Scholar]
- 14.Trynka G, Wijmenga C, van Heel DA. A genetic perspective on coeliac disease. Trends Mol Med. 2010;16:537–50. doi: 10.1016/j.molmed.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Kapoor A, Patwari A, Kumar P, et al. Serum soluble interleukin-2 receptor, interleukin-6 and tumor necrosis factor alpha as markers of celiac disease activity. Indian J Pediatr. 2013;80:108–13. doi: 10.1007/s12098-012-0830-9. [DOI] [PubMed] [Google Scholar]
- 16.Romaldini CC, Barbieri D, Okay TS, et al. Serum soluble interleukin-2 receptor, interleukin-6, and tumor necrosis factor-alpha levels in children with celiac disease: response to treatment. J Pediatr Gastroenterol Nutrition. 2002;35:513–7. doi: 10.1097/00005176-200210000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Nemec P, Pavkova-Goldbergova M, Stouracova M, et al. Polymorphism in the tumor necrosis factor-alpha gene promoter is associated with severity of rheumatoid arthritis in the Czech population. Clin Rheumatol. 2008;27:59–65. doi: 10.1007/s10067-007-0653-7. [DOI] [PubMed] [Google Scholar]
- 18.Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–68. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 19.Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res Ther. 2006;8(Suppl 2):S2. doi: 10.1186/ar1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barisani D, Ceroni S, Meneveri R, et al. IL-10 polymorphisms are associated with early-onset celiac disease and severe mucosal damage in patients of Caucasian origin. Genet Med. 2006;8:169–74. doi: 10.1097/01.gim.0000204464.87540.39. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Chai W, Ni M, et al. The effects of gene polymorphisms in interleukin-4 and interleukin-6 on the susceptibility of rheumatoid arthritis in a Chinese population. BioMed Res Int. 2014;2014:265435. doi: 10.1155/2014/265435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godarzi EM, Sarvestani EK, Aflaki E, et al. Interleukin-6 gene polymorphism in Iranian patients with systemic lupus erythematosus. Clin Rheumatol. 2011;30:179–84. doi: 10.1007/s10067-010-1452-0. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, Liu J, Lin CY, et al. Interleukin-6 gene promoter-572 C allele may play a role in rate of disease progression in multiple sclerosis. Int J Mol Sci. 2012;13:13667–79. doi: 10.3390/ijms131013667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Tang RK, Yang X, et al. Lack of an association between interleukin-6 gene promoter polymorphisms (−174G/C,− 572G/C) and ischemic heart disease and/or ischemic stroke: a meta-analysis. Hum Immunol. 2011;72:641–51. doi: 10.1016/j.humimm.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 25.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Investig. 1998;102:1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerrard-Dunne P, Sitzer M, Risley P, et al. Inflammatory gene load is associated with enhanced inflammation and early carotid atherosclerosis in smokers. Stroke. 2004;35:2438–44. doi: 10.1161/01.STR.0000144681.46696.b3. [DOI] [PubMed] [Google Scholar]
- 27.Sghaier I, Mouelhi L, Rabia NA, et al. Genetic variants in IL-6 and IL-10 genes and susceptibility to hepatocellular carcinoma in HCV infected patients. Cytokine. 2017;89:62–67. doi: 10.1016/j.cyto.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Woolley N, Mustalahti K, Mäki M, et al. Cytokine gene polymorphisms and genetic association with coeliac disease in the finnish population. Scand J Immunol. 2005;61:51–6. doi: 10.1111/j.0300-9475.2005.01525.x. [DOI] [PubMed] [Google Scholar]
- 29.de Albuquerque Maranhão R, Martins Esteves F, Crovella S, et al. Tumor necrosis factor-alpha and interleukin-6 gene polymorphism association with susceptibility to celiac disease in italian patients. Genet Mol Res. 2015;14:16343–52. doi: 10.4238/2015.December.9.2. [DOI] [PubMed] [Google Scholar]
- 30.Dema B, Martinez A, Fernandez-Arquero M, et al. The IL6-174G/C polymorphism is associated with celiac disease susceptibility in girls. Hum Immunol. 2009;70:191–4. doi: 10.1016/j.humimm.2009.01.010. [DOI] [PubMed] [Google Scholar]