Abstract
Growth arrest and DNA damage-inducible beta (GADD45b) plays a pivotal role in many intracellular events in both cell survival- and cell death-related signaling. To date, the study of GADD35b has mainly focused on investigation of its function, as well as interacting molecules. However, studies of Gadd45b gene regulation are limited. In this study, we investigated the transcriptional regulation mechanism of Gadd45b. Since Gadd45b mRNA is highly induced by the PPARα agonist Wy-14,643 in the mouse liver, we analyzed the Gadd45b promoter using an in vivo reporter assay. Interestingly, the naked Gadd45b-luciferase construct strongly induced luciferase activity without any stimulant in our in vivo system. Therefore, we investigated the epigenetic changes in the Gadd45b promoter region using mouse liver genomic DNA, the methylation-specific restriction enzyme (HpaII), and disulfide conversion. Our results showed that two possible CpG methylation sites were methylated and demethylated by Wy-14,643 treatment. This study indicates that epigenetic change at the Gadd45b promoter is critical for Gadd45b induction.
Keywords: Gadd45b, PPARα, CpG methylation
1. Introduction
GADD45b is considered as a sensor molecule that is upregulated by internal and external stress. The role of GADD45b is primarily studied in cellular events, such as cell cycle arrest, DNA repair, cell survival, and apoptosis. GADD45b performs these many roles by interacting with other cellular molecules that are critical in the cell cycle and stress-related molecular machinery, such as p21, PCNA, cdc2/cyclinB, and MKK7/JNKK2, a kinase upstream of JNK [1,2]. However, the mechanisms involved in the transcriptional regulation of the Gadd45b gene are largely unknown. Previously, we reported that Gadd45b was highly induced by degradation of STAT3, a transcriptional repressor, following hepatic oxidative stress caused by PPARAα (peroxisome proliferator-activated receptor alpha) activation in vivo [3]. However, Gadd45b mRNA expression was not observed in vitro. Surprisingly, the Gadd45b promoter reporter assay showed that luciferase activity was highly increased in both in vivo and in vitro systems, and the levels of luciferase activity were similar even in the absence of drug treatment.
Peroxisome proliferator-activated receptor α (PPARα) is a member of the nuclear receptor superfamily and is activated by xenobiotic or endogenous ligands. PPARα plays an important role in lipid metabolism, inflammation, and hepatic carcinogenesis in the rodent [4,5]. PPARα activation is physiologically triggered in response to starvation, following the induction of genes involved in fatty acid transport, β-oxidation, and gluconeogenesis [6]. Additionally, phosphatidylcholine, an endogenous fatty acid metabolite, and fibrate, a synthetic ligand used clinically for hyperlipidemia in humans, are PPARα activators. In the presence of ligand, PPARα heterodimerizes with retinoid X receptor alpha (RXRα) and binds to specific DNA-response elements PPREs (peroxisome proliferator response elements) in the promoter of target genes [7,8]. In addition, unlike in humans, prolonged treatment of synthetic PPARα agonist in mice and rats leads to hepatocarcinogenesis [9–11].
DNA methylation, especially at CpG islands of gene promoter regions, has been widely known to affect the regulation of gene expression. CpG islands are located at the 5′-end gene promoters and found in the promoters of 60% of human gene [12]. Gene silencing by DNA methylation may directly inhibit the recruitment of DNA-binding proteins or transcription factors to their target sites, or indirectly facilitate the binding of repressors to the promoters of certain gene and allow histone modification [13].
Despite the drastic induction of Gadd45b mRNA expression by PPARα activation in the mouse liver, the promoter assay did not reflect the expression differences observed both in vivo and in vitro. Therefore, we assumed that epigenetic factors might be involved in Gadd45b mRNA regulation. If this was the case, the reporter plasmid DNA itself could induce the high level of luciferase activity observed in the in vivo reporter assay. The aim of this study is to investigate the possible involvement of CpG methylation in the regulation of Gadd45b mRNA induction in conjunction with PPARα activation.
2. Materials and methods
2.1. Mouse and in vivo reporter assay
Seven-to eight-week-old male WT mice (129 Sv background, five animals per group) were housed in a pathogen-free animal facility under standard 12 h light/dark cycle with water and chow ad libitum. For drug treatment, mice were fed with normal chow diet (control) or Wy-14,643 (0.1% w/w) diet (Bioserv, Frenchtown, NJ, USA) for 24 h. All experiments were carried out under the Association for Assessment and Accreditation of Laboratory Animal Care guidelines with approval from the NCI Animal Care and Use Committee. Ten micrograms of luciferase or eGFP reporter plasmid [3] containing the Gadd45b promoter region ( −2149 to +11, named as pLuc2K or peGFP2K) were suspended in 2.2 ml of TransIT-QR delivery solution and administered by mouse tail vein injection for 3–5 s. Two days later, the mice were fed with Wy-14,643 diet (0.1% w/w) for 24 h. For the in vivo luciferase reporter assay, the mice were dosed with 200 μl of D-luciferin (15 mg/ml DPBS, intraperitoneal) and imaging and bioluminescence were assessed using the IVIS Imaging 200 series (Xenogen, Hopkinton, MA).
2.2. Polymerase chain reaction (PCR)
For quantitative real-time PCR (qPCR), total RNA was isolated from mouse liver using Trizol (Invitrogen, Carlsbad, CA). Complementary DNA (cDNA) was synthesized from 1 μg total RNA using a SuperScript II reverse transcriptase kit (Invitrogen, Carlsbad, CA). qPCR was performed using an Applied Biosystems Prism 7900HT Sequence Detection System (Foster City, CA). All reactions were performed in a 10 μl volume consisting of 25 ng of cDNA, 300 nM of each primer and 5 μl of SYBR Green PCR Master Mix (Applied Biosystems) at 95 °C for 10 min and 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Expression levels of mRNA were normalized to β-actin. Primers for the qPCR were designed using qPrimer Depot (http://mouseprimerdepot.nci.nih.gov). For conventional PCR, harvested cells were processed using REDExtract-N-Amp™ Tissue PCR Kit (Sigma-Aldrich) and PCR reactions were followed at 95 °C for 2 min and 25 cycles of 95 °C for 33 s, 60 °C for 30 s and 72 °C for 45 s. The primers for eGFP amplification (265 bp) were: forward: 5′-GCA CGA CTT CTT CAA GTC CGC CAT GCC-3′, and reverse: 5′-GCG GAT CTT GAA GTT CAC CTT GAT GCC-3′.
2.3. Western blotting
Hepa1c1c7 cells were lysed with RIPA lysis buffer (150 mM NaCl, 0.5% Triton X-100, 50 mM Tris-HCl, pH 7.4, 25 mM NaF, 20 mM EGTA, 1 mM DTT, 1 mM Na3VO4, and protease inhibitor cocktail) for 30 min on ice, followed by centrifugation at 14,800×g for 15 min. Protein concentrations were measured with bicinchoninic acid (BCA) reagent. Protein (30 μg) was electrophoresed on a 4–15% gradient tris-HCl gel (Bio-Rad, Hercules, CA) and transferred onto a polyvinylidene difluoride membrane using Trans-Blot Turbo™ Transfer System (Bio-Rad, Hercules, CA). The membrane was blocked with 5% non-fat dry milk in phosphate buffered saline containing 0.1% Tween-20 (PBST) for 1 h. The membranes were probed with anti-GFP (Santa Cruz, California, USA) or anti-GAPDH (Abcam) primary antibodies and horseradish peroxidase-conjugated secondary antibodies using standard western blotting procedures. Proteins were visualized using the femto signal chemiluminescent substrate (Pierce) under the image analyzer (Alpha Innotech Corp., San Leandro, CA).
2.4. Epigenetics
Mouse liver genomic DNA was purified using Wizard® Genomic DNA Purification Kit (Promega, San Luis Obispo, CA, USA). To determine whether Gadd45b is regulated epigenetically by CpG methylation, genomic DNA (1 μg) was digested with HpaII, a restriction enzyme that recognizes and cuts the CCGG DNA sequence when CpG is unmethylated, overnight at 37 °C followed by PCR reaction with 20 ng of digested or undigested DNA. Purified liver genomic DNA was subjected to a bisulfite conversion experiment using EpiTect Bisulfite Kits (Qiagen, Valencia, CA, USA). Samples from two different experiments were subjected to PCR under the following conditions: initial activation step at 95 °C for 2 min, 34 cycles of 95 °C for 30 s, 60 °C for 30 s, 70 °C for 45 s, and a final extension at 70 °C for 5 min using the following primers: Region I, 5′- TCG GCC CTT TGT GCA TCT ACC AAT-3′ and 5′-AAG AAG ACA GAG ATG AAC GCA GAC CC-3′; Region II, 5′-AGC TCT CTC TCT AGA GCT CTC TGG CTT T-3′ and 5′-ATT GGT AGA TGC ACA AAG GGC CGA-3′; Region III (for DNA methylation specific), 5′-TTT CGT GTT GGC GAT GTT AGG GTG-3′ and 5′- TTC TCT CCT AAT TTC CCG ACA ACC GC-3′.
2.5. Statistical analysis
Experimental values are expressed as mean ± SD. Statistical analysis was performed by two-tailed Student’s t-test for unpaired data, with p < 0.05 considered statistically significant.
3. Results and discussion
3.1. Gadd45b-luciferase activity was strongly induced in the absence of Wy-14,643 in both in vitro and in vivo systems
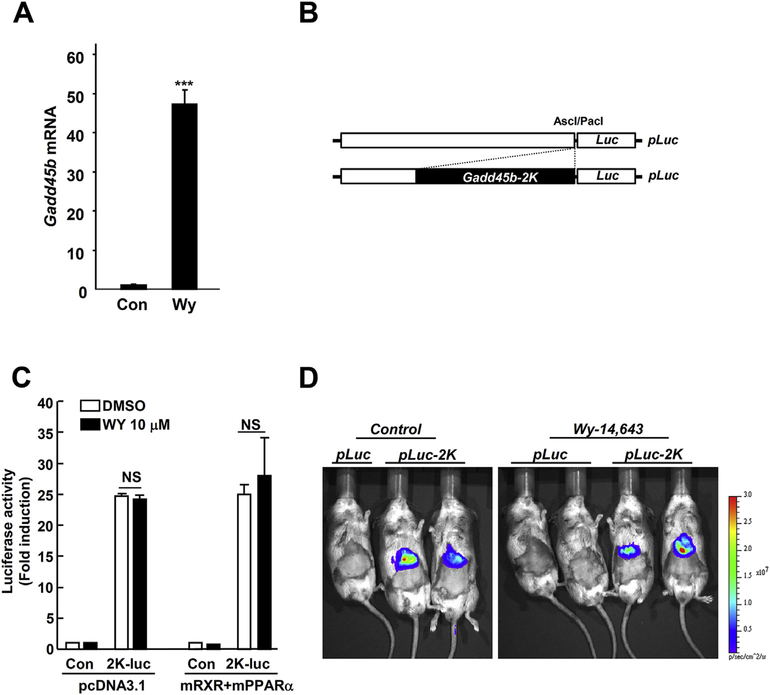
Gadd45b mRNA was strongly induced by Wy-14,643 in mouse liver (Fig 1A); therefore, we wanted to explore Gadd45b transcription factors in the mouse liver using the luciferase reporter assay. The 2 kb Gadd45b promoter was inserted in the luciferase construct (Fig. 1B) and transfected to the mouse hepatocarcinoma (Hepa1c1c7) cells or mouse liver. Surprisingly, Gadd45b-luciferase activity was strongly induced in both systems even in the absence of drug treatment. Furthermore, there were no significant changes in the luciferase activity between the control and Wy-14,643 treated groups (Fig. 1C and D). These results clearly show a discrepancy between Gadd45b mRNA expression and luciferase reporter-gene expression. Thus, we hypothesized that Gadd45b mRNA could be regulated through epigenetic modification. DNA hypermethylation of GpG islands in gene promoters is a major mechanism for epigenetic inactivation of tumor suppressor genes. Previously, Na et al. [14] reported that the methylation frequency of the Gadd45b promoter in human non-small cell lung cancer was significant, at 7.2%, and downregulated its mRNA expression levels. Thus, the downregulation of Gadd45b may lead to cancer proliferation. Inversely, it can be assumed that Gadd45b induction may inhibit cancer proliferation by enhancing the demethylation of CpG islands.
Fig. 1. Mouse hepatic Gadd45b mRNA induction by Wy-14,643 does not correlate with Gadd45b-promoter luciferase assay.
(A) Mice were fed with Wy-14,643 (0.1%, w/w) for 1 day and liver samples subjected to qPCR. (B) For in vivo Gadd45b-luciferase assay, Gadd45b promoter (2 Kb) was cloned into the modified pGL4.10 vector. (C) In vivo luciferase activity was measured using bioluminescent imaging system in the mice (Sv129 background). Mice were injected with 10 μg of pGL4.10-Gadd45b (2 Kbp inserted) plasmid for 2 days using the hydrodynamic tail vein injection method and followed by 24 h Wy-14,643 (0.1%, w/w) diet administration. Con, Control; pLuc, pGL4.10 vector; pLuc-2K, pGL4.10-Gadd45b-promoter (2 Kbp); Wy, Wy-14,643; ***, p < 0. 0001.
3.2. Wy-14,643 demethylated CpG islands of Gadd45b promoter
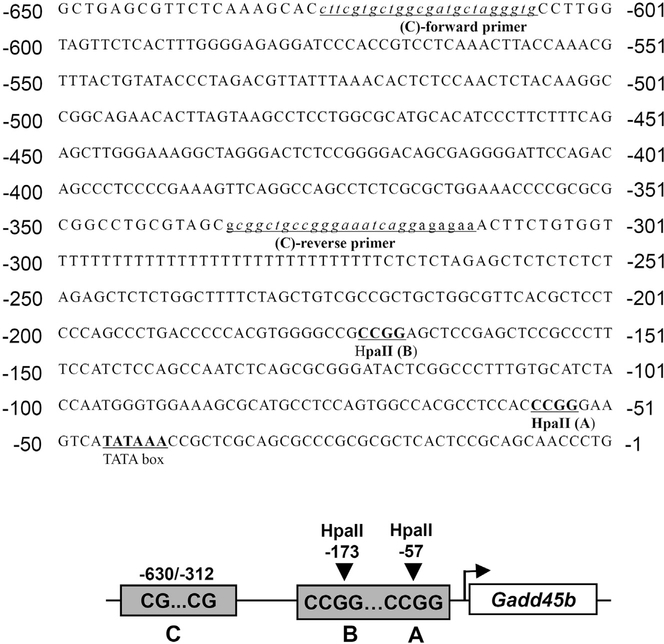
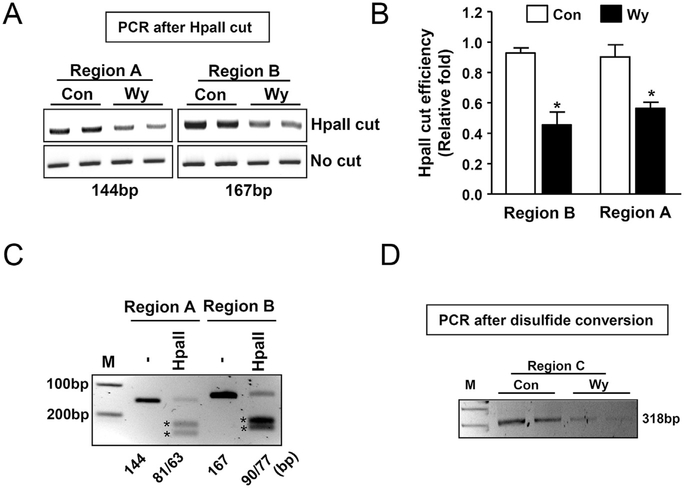
To determine whether Wy-14,643 could demethylate CpG islands of the Gadd45b promoter, mice were fed with control diet or Wy-14,643 (0.1% w/w) diet for one day and liver DNA was isolated for epigenetic analysis. We identified putative methylated CpG regions (Fig. 2) and designated them regions A, B, and C. Potential methylated cytosines were targeted with the HpaII restriction endonuclease, which recognizes and cleaves the CCGG sequence when the first, but not second, cytosine is methylated. Region A and B were identified as HpaII targeting sites whilst Region C was identified as suitable for disulfide conversion analysis. Epigenetic analysis showed that Wy-14,643 significantly reduced the quantity of PCR amplicons in both region A and B (Fig. 3A) and HpaII cut efficiency was indicated in Fig. 3B. PCR amplicons were digested with the HpaII enzyme to verify correct amplicon sizes (Fig. 3C). In addition, following disulfide conversion of liver genomic DNA, and amplification with specially designed PCR primers, the predicted PCR amplicon was significantly reduced by treatment with Wy-14,643 (Fig. 3D). Taken together, these epigenetic analyses indicate that the PPARα agonist Wy-14,643 could regulate Gadd45b mRNA expression by demethylating the CpG islands in the promoter.
Fig. 2. Gadd45b promoter sequence with possible CpG methylation sites.
Among the possible CpG methylation sites in the Gadd45b promoter, selected CpG methylation sequences (underlined) for the experiment are shown. Underlined italic lower case sequences were used as primers for the disulfide conversion experiment. Bold sequences in capital letters were targeted for the methylation-sensitive restriction enzyme (HpaII).
Fig. 3. Wy-14,643, a PPAR𝛂 agonist, decreases CpG methylation in the Gadd45b promoter region.
(A) To examine the cytosine methylation pattern in the selected CpG sequences, mice were fed with Wy-14,643 (0.1%, w/w) for 1 day and isolated liver genomic DNA was incubated with enzyme HpaII, a methylation sensitive restriction enzyme for epigenetic and cutting regions were amplified using conventional PCR reaction. (B) HpaII cut efficiency represented as a densitometry from (A). (C) To verify the correct PCR products from (A), PCR products were cut with the HpaII enzyme. The unmethylated PCR products were cut into two fragments to verify the correct sizes. Stars indicate the cutting sizes. (D) To demonstrate the epigenetic change of the Gadd45b promoter by Wy-14,643, the disulfide conversion method was applied. Unmethylated cytosine was delaminated to form uracil and samples were subjected to conventional PCR analysis with specifically designed primers. Con, control; Wy, Wy-14,643 (0.1%, w/w); *p < 0.05.
3.3. Wy-14,643 might trigger demethylation of methylated CpG islands of Gadd45b promoter-luciferase construct followed by the induction of luciferase activity
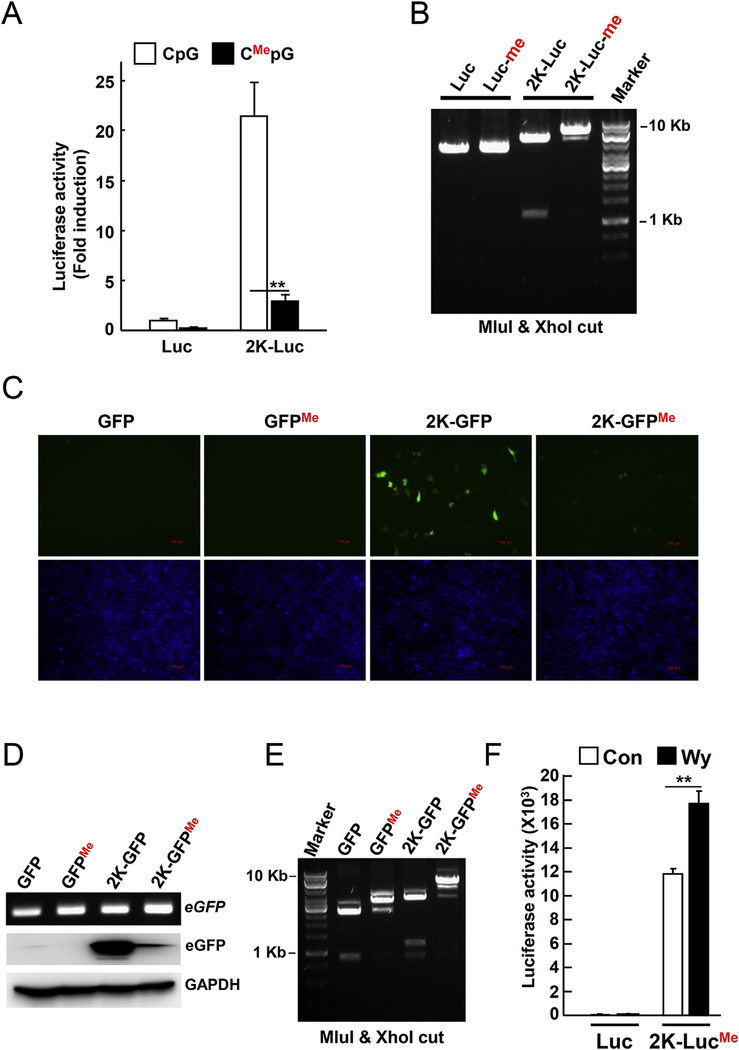
To determine if methylated CpG islands affect the expression of the Gadd45b gene, we used pGL4.10-Gadd45b-luciferase or pGL4.10-Gadd45b-eGFP reporter constructs. Reporter constructs were incubated with the CpG methyltransferase enzyme (M.SssI) to introduce a methyl group to the cytosines of CpG islands. The reporter assay revealed that CpG methylation of the pGL4.10-Gadd45b-luciferase or pGL4.10-Gadd45b-eGFP constructs resulted in significant suppression of luciferase activity or eGFP expression in the Hepac1c1c7 cells, respectively (Fig. 4A, C, and D). CpG methylation was confirmed by digestion with the CpG methylation sensitive restriction endonucleases, MluI and XhoI, and subsequent agarose gel electrophoresis (Fig. 4B and E). Furthermore, the CpG methylated pGL4.10-Gadd45b-luciferase construct analyzed in vivo in the mouse. The construct was injected into the mouse tail vein using the hydrodynamic injection method. After 1 day, the normal chow diet was replaced with the Wy-14,643 diet (0.1% w/w) for 1 day. Luciferase activity from the methylated Gadd45b-luciferase construct was significantly increased following Wy-14,643 treatment (Fig. 4D). Taken together, these results suggest that methylation on CpG islands of the Gadd45b promoter may result in the downregulation of Gadd45b expression. In addition, downregulation of Gadd45b expression can be reversed by Wy-14,643 via a CpG demethylation mechanism.
Fig. 4. Wy-14,643 increases the Gadd45b-luciferase activity by abolishing the CpG methylation.
(A) To demonstrate that CpG methylation could suppress gene expression, the Gadd45b-luciferase construct was incubated with CpG methyltransferase enzyme (M.SssI) to introduce methyl group in the cytosine of CpG and followed by reporter assay. Hepa1c1c7 cells were transfected with control or CpG methylated Gadd45b-luciferase construct (2K-luc) for 24 h (B) CpG methylation of the Gadd45b-luciferase construct was tested by electrophoresis following MluI and Xho restriction digestion. (C) Hepa1c1c7 cells were transfected with CpG methylated Gadd45b-eGFP reporter plasmid for 24 h. Green indicates the eGFP expression. Blue represents nuclei after Hoechst 33,342 staining. (D) PCR to confirm transfection by detecting GFP was performed on transfected samples from (C). eGFP expression was detected by Western blotting. GAPDH was used as a loading control. (E) CpG methylation of Gadd45b-eGFP constructs was tested by gel electrophoresis following MluI and Xho restriction digests. (F) To confirm if Wy-14,643 reversed the effect of CpG methylation of Gadd45b-luciferase in vivo, the constructs (10 μg) were transfected into mice by mouse-tail vein injection. 2K-luc, Gadd45b-luciferase construct (2 Kbp size); Me, methylated; **, p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Although long-term treatment with Wy-14,643 can induce liver cancer in murine models, it appears that mouse hepatic cells resist the cell proliferation phenotype by inducing anti-cell cycle inhibitors such as p21 and Gadd45b during short-term treatment with Wy-14,643 [3].
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF-2015R1A5A2008833), the Fund for New Professor Research Foundation Program, Gyeongsang National University (2014-04-007), the grant of Institute of Health Sciences (HIS GNU-2014-2), and the National Cancer Institute Intramural Research Program.
Abbreviations:
- PPARα
peroxisome proliferator-activated receptor alpha
- Gadd45b
growth arrest and DNA-damage-inducible beta
Footnotes
Conflict of interest
The authors have no financial conflicts of interest.
References
- [1].Abdollahi A, Lord KA, Hoffman-Liebermann B, Liebermann DA, Sequence and expression of a cDNA encoding MyD118: a novel myeloid differentiation primary response gene induced by multiple cytokines, Oncogene 6 (1991) 165–167. [PubMed] [Google Scholar]
- [2].Liebermann DA, Hoffman B, Gadd45 in stress signaling, J. Mol. Signal 3 (2008) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kim JH, Qu A, Reddy JK, Gao B, Gonzalez FJ, Hepatic oxidative stress activates the Gadd45b gene by way of degradation of the transcriptional repressor STAT3, Hepatology 59 (2014) 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Peters JM, Shah YM, Gonzalez FJ, The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention, Nature reviews, Cancer 12 (2012) 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pyper SR, Viswakarma N, Yu S, Reddy JK, PPARalpha: energy combustion, hypolipidemia, inflammation and cancer, Nucl. Recept. Signal 8 (2010) e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W, Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting, J. Clin. Investig 103 (1999) 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF, Identification of a physiologically relevant endogenous ligand for PPARalpha in liver, Cell 138 (2009) 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Desvergne B, Wahli W, Peroxisome proliferator-activated receptors: nuclear control of metabolism, Endocr. Rev 20 (1999) 649–688. [DOI] [PubMed] [Google Scholar]
- [9].Woods CG, Burns AM, Bradford BU, Ross PK, Kosyk O, Swenberg JA, Cunningham ML, Rusyn I, WY-14,643 induced cell proliferation and oxidative stress in mouse liver are independent of NADPH oxidase, Toxicol. Sci. Off. J. Soc. Toxicol 98 (2007) 366–374. [DOI] [PubMed] [Google Scholar]
- [10].Hays T, Rusyn I, Burns AM, Kennett MJ, Ward JM, Gonzalez FJ, Peters JM, Role of peroxisome proliferator-activated receptor-alpha (PPARalpha) in bezafibrate-induced hepatocarcinogenesis and cholestasis, Carcinogenesis 26 (2005) 219–227. [DOI] [PubMed] [Google Scholar]
- [11].Klaunig JE, Babich MA, Baetcke KP, Cook JC, Corton JC, David RM, DeLuca JG, Lai DY, McKee RH, Peters JM, Roberts RA, Fenner-Crisp PA, PPARalpha agonist-induced rodent tumors: modes of action and human relevance, Crit. Rev. Toxicol 33 (2003) 655–780. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Leung FC, An evaluation of new criteria for CpG islands in the human genome as gene markers, Bioinformatics 20 (2004) 1170–1177. [DOI] [PubMed] [Google Scholar]
- [13].Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A, Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex, Nature 393 (1998) 386–389. [DOI] [PubMed] [Google Scholar]
- [14].Na YK, Lee SM, Hong HS, Kim JB, Park JY, Kim DS, Hypermethylation of growth arrest DNA-damage-inducible gene 45 in non-small cell lung cancer and its relationship with clinicopathologic features, Mol. Cells 30 (2010) 89–92. [DOI] [PubMed] [Google Scholar]