Abstract
The transcription cycle can be roughly divided into three stages: initiation, elongation, and termination. Understanding the molecular events that regulate all these stages requires a dynamic view of the underlying processes. The development of techniques to visualize and quantify transcription in single living cells has been essential in revealing the transcription kinetics. They have revealed that (a) transcription is heterogeneous between cells and (b) transcription can be discontinuous within a cell. In this review, we discuss the progress in our quantitative understanding of transcription dynamics in living cells, focusing on all parts of the transcription cycle. We present the techniques allowing for single-cell transcription measurements, review evidence from different organisms, and discuss how these experiments have broadened our mechanistic understanding of transcription regulation.
Keywords: single-molecule, single-cell, kinetics, bursting, heterogeneity, imaging, fluorescence
INTRODUCTION
A quantitative understanding of gene regulation has been a biophysical pursuit since the original elucidation of the lac operon in Escherichia coli (59). Dynamics have always been at the heart of this pursuit. Looking back on Jacob and Monod’s description of the operon, one is reminded of how time-resolved measurements of gene expression and enzymatic activity played an essential role in formulating a mechanistic description of the metabolic shift to lactose metabolism. The same principle still holds: To understand mechanism is to understand kinetics. Our understanding of gene regulation has advanced tremendously in the last 50+ years, culminating in the genomic era and comprehensive measurement of virtually every aspect of gene regulation, from cis-acting regulatory sequences in DNA to trans-acting binding of transcription factors to the positions and posttranslational modifications of histones to the abundance of RNA. Yet time-resolved measurements—whether carried out in a test tube, in single cells, or on a genomic scale—are still the backbone of our understanding of gene regulation. In particular, observing the timescale of gene expression changes that occur on the order of milliseconds to seconds to hours to days allows one to elucidate the molecular machinery involved in RNA synthesis and processing. In this review, we focus on the molecular events responsible for generating mRNA. Broadly speaking, these molecular events are transcription initiation, elongation, termination, and 3′-end formation. Specifically, we aim to summarize the progress that has occurred in recent years in painting a kinetic picture of these processes in living cells.
Although kinetic measurements can be done in almost any context, we focus here on dynamic measurement in single cells for several reasons. First, there is a generic argument to be made for live-cell studies: The cell is the smallest unit of life and therefore the most amenable to reductionist approaches that still capture biological processes. Second, the molecular events involved in mRNA generation often occur at the same time and place in the nucleus, which is the active gene. There is consequently immense utility in the ability to visualize multiple processes at the same locus in the same cell. Third, the ability to follow cells over time provides a unique opportunity to connect the history of expression behavior to observed phenotypes. Finally, the time resolution of single-cell studies—ranging from milliseconds to days or longer—far surpasses that which is currently available in population measurements.
THE SHAPE OF DYNAMICS
The kinetics of gene expression can be broadly understood through the birth and death of mRNA. A simple view of this process, which can be considered the starting point for quantitative models of expression, is the following:
where N(t) is the number of RNA as a function of time t; c is a zeroth-order production rate of RNA, which is treated as constant and thus ignores the upstream regulatory processes; and k is the first-order degradation constant of the RNA. At steady state, the number of mRNAs in the cell is simply the ratio of synthesis and decay, c/k. This equation is the familiar ordinary differential equation description, which is based on deterministic behavior of a large population of molecules. However, as shown below, the number of mRNAs per cell is often quite low. For example, 80% of yeast mRNAs are present at six copies or fewer (56, 137). In this regime, the behavior of the system requires a mathematical description that is probabilistic and discrete. Therefore, instead of a single number of mRNAs per cell, there is a distribution.
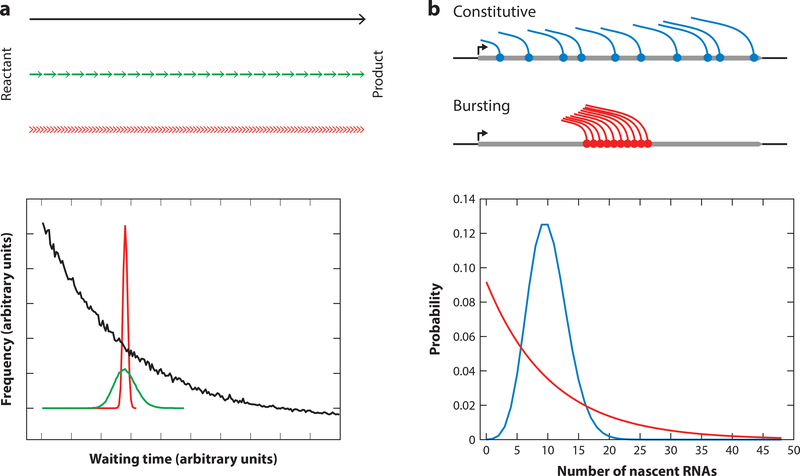
Perhaps the most straightforward expectation is that transcripts follow a Poisson distribution, which requires that both synthesis and decay are determined by single rate-limiting steps. Indeed, this behavior is observed for some genes in yeast (137). When a kinetic process is determined by a single rate-limiting step, the waiting time between so-called events (i.e., the interval at which RNA transcription is initiated from the gene, 1/c) follows an exponential distribution (125) (Figure 1a). In such cases, the most likely interval between events is 0, and there is a tail of longer waiting times. In the case in which many sequential kinetic steps are necessary and all have similar rates and therefore similar exponentially distributed waiting times, there is of course no particular rate-limiting step to reach the final product of the reaction. The aggregate rate for such a sequential process may be the same as the single-step case (i.e., c), but the waiting time between events is Gaussian distributed, which is a direct consequence of the central limit theorem. The classic case of this latter process is the time required during transcription elongation to synthesize a nascent RNA, which may be thousands of bases long (14). The important concept is the following: The average rate of a single-step process and multistep process might be identical, but the distribution of waiting times is exponential in the former and nonexponential in the latter (Figure 1a). For transcription, these different kinetic schemes can result in very different polymerase occupancy on the gene (Figure 1b). Such behavior can be best observed at the single-cell or single-molecule level and carries implications for the underlying mechanism and also the competition between processes.
Figure 1:
Theoretical concepts in transcription dynamics. (a) The distribution of waiting times between events for three kinetic processes that have the identical net rate going from reactant to product. If the process is a single-step process or has a rate-limiting step, the time between events is exponentially distributed (black line). If the process consists of sequential steps, the distribution is a peaked distribution, which resembles a Gaussian (green line). In the limit of many sequential steps, the time between events is sharply distributed around the mean value (red line). The distributions are from Monte Carlo simulations. Adapted from Reference 73 with permission. (b) Different transcription dynamics result in different distributions of the number of nascent RNAs per cell. For a gene that is transcribed with constitutive initiation, the number of RNAs at the transcription site follows a Poisson distribution (blue). When a gene is transcribed in bursts, the distribution follows a negative binomial distribution (red). In this example, burst size is 10, burst frequency is 1 burst/min, and the dwell time of the transcript at the transcription site is 1 min. The average RNA production of both transcription types in this example is the same.
DYNAMICS OF RNA SYNTHESIS
The transcription of pre-mRNA from the DNA template by RNA polymerase II (Pol II) can be broken down into multiple steps, each of which carries the potential for regulation (112). Initiation is the process whereby Pol II binds to the DNA, melts the DNA duplex to form single-stranded DNA, and begins to add nucleoside triphosphates to the nascent RNA strand. After initiation, Pol II breaks the contacts with the general transcription factors bound to the promoter and enters elongation. For many metazoan genes, there is an additional promoter-proximal pause within the first 50–100 nucleotides (nt) before Pol II begins productive elongation in the body of the gene. Finally, when Pol II encounters signals at the end of the gene, the nascent RNA is released, Pol II dissociates from the template, and the duplex reanneals.
The fundamental steps were mostly elucidated by population measurements. However, the average behavior measured in millions of cells often does not reflect the behavior in individual cells. Transcription in single cells is a stochastic process that arises from the random collision of molecules. Transcription factors diffuse through the nucleus with Brownian motion and associate nonspecifically with the DNA many times in search for specific binding sites (92). Even binding at the target motif represents short-lived dynamic interactions with the DNA (85). Often, many biochemical reactions have to take place before RNA polymerase can initiate transcription, such as recruitment of chromatin factors to remove or slide nucleosomes (1, 33). These random events can average out if molecule counts are high, but each gene is present in one or two copies per cell, making gene expression inherently stochastic.
Inferring Dynamics from Distributions
In the early 2000s, researchers realized that heterogeneity between genetically identical cells can be used to understand transcription dynamics (5, 13, 97, 124). Heterogeneity in gene expression had been recognized much earlier, but whether this heterogeneity was static or dynamic was unclear (70, 128). Even though initial measurements often did not have a time component, the steady-state distribution of protein expression in single cells yielded information on the relationship between the protein mean and its variance, from which different models of gene expression could be inferred through the comparison of the observed distribution to mathematical models. Thus, the models used to explain the data are intrinsically dynamic (5, 97, 124). These studies revealed that the variation in yeast for most proteins could be explained by random RNA synthesis and decay, consistent with single rate-limiting steps and exponentially distributed waiting times. Interestingly, some genes, such as stress genes or inducible genes, contained increased noise that could not simply be explained by a Poisson distribution. This noise was affected by mutations in TATA boxes, nucleosome-free regions, genome position, and transcription-factor concentration, supposedly through the regulation of the underlying transcription kinetics (7, 12, 25, 57).
In bacteria, a similar approach showed that almost all protein distributions follow a gamma distribution (124). In this study, the variation was explained not by stochasticity in mRNA synthesis, but mostly by discontinuous protein production and global differences between cells (extrinsic noise). The authors also showed that mRNA and protein levels are not correlated in individual cells. These results exemplify the danger of inferring transcription dynamics from steady-state protein levels. Protein distributions are affected by much more than transcription dynamics. For example, in both bacteria and human cells, mRNA has been shown to be transcribed more than once (21, 131, 134). The eventual protein distribution is a mix of the kinetics in transcription and translation synthesis, the dynamics of mRNA and protein decay, and cell-cycle or cell-division effects (58, 140). Distinguishing the relative contributions to heterogeneity of transcriptional versus posttranscription processes has proven to be a challenging task (36).
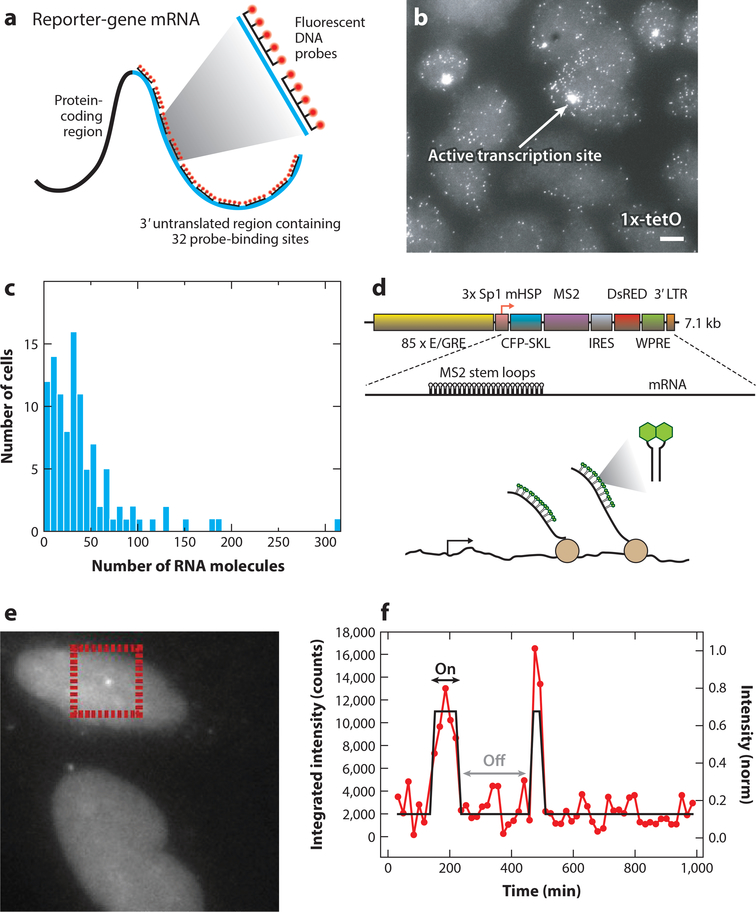
Instead of inferring RNA synthesis from protein distribution, several studies started measuring mRNA directly in single cells. Pioneered by the Singer lab (44), single-molecule fluorescence in situ hybridization (smFISH) emerged as a powerful technique to visualize single RNAs in fixed cells. Hybridization of several fluorescent probes along a gene gave single-molecule sensitivity, allowing for exact quantification of the number of a particular RNA in a cell (Figure 2a). Raj and coworkers (108) extended this approach to precisely quantify transcription in mammalian cells (Figure 2b). Using a reporter gene and an endogenous gene, the authors demonstrated that the variability in RNA copy number between cells is much higher than the variability of a Poisson distribution (Figure 2c). This variability could be explained through the assumption that transcription occurs in bursts, with periods of high activity followed by periods of inactivity. This mode of transcription had been theoretically predicted (69, 105) and is sometimes referred to as the random telegraph model. In this model, a promoter switches between two states, on and off, and transcription can take place only in the on state. The number of RNA produced in the on state is called the burst size, and the rate of switching to the on state is called burst frequency. In this two-state model, the on and off times are assumed to be regulated by single rate-limiting steps, with exponentially distributed time between bursts and geometrically distributed burst sizes. The resulting distribution of mRNAs per cell follows a negative binomial, which is a much wider distribution than a Poisson distribution (Figure 1b). Compared with constitutive transcription initiation, in which the probability of initiation is constant over time, bursting transcription thus results in more variability between cells. The distributions of mRNAs per cell from Raj et al. (108) suggested that mammalian genes are dominated by bursting transcription. Recent studies in embryonic stem cells and tissue still use wide RNA distributions as evidence for transcriptional bursting (3, 114).
Figure 2:
Measuring polymerase dynamics in fixed and living cells. (a) Schematic of the single-molecule fluorescence in situ hybridization (smFISH) procedure. Fluorescent probes are hybridized along the RNA, resulting in bright fluorescent spots. (b) Example of an smFISH experiment, showing mRNA of a reporter gene driven by a 1x-tetO-containing promoter in Chinese hamster ovary cells. (c) The distribution of the number of mRNAs per cell, obtained from an smFISH experiment. Panels a-c adapted from Reference 108 with permission. (d) Schematic of the reporter gene used to measure RNA in living U2-OS cells. When the MS2 sequences are transcribed, they form stem loops that are bound by fluorescent coat proteins. (e) Example of nascent RNA at an active transcription site of the reporter gene in living cells. (f) Quantification of the transcription site intensity, showing the on and off states of transcription. Panels d-f adapted from Reference 72 with permission. Abbreviations: E/GRE, ecdysone/glucocorticoid response element; Sp1, specificity protein one binding site; mHSP, minimal heat shock promoter; CFP-SKL, cyan fluorescent protein with a C-terminal SKL amino acid peroxisome-targeting motif; IRES, internal ribosome entry site; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; LTR, long terminal repeat.
This burst-like transcription agrees with studies from the late 1970s. Nuclear spreads of chromatin from Drosophila melanogaster embryos and amphibian oocytes were visualized by electron microscopy (also called Miller spreads) (84, 87). Although the familiar so-called Christmas tree images of transcribing genes in the nucleolus suggest that synthesis of rRNA by RNA polymerase I is continuous and nearly saturated, the same approach was also employed to look at synthesis of mRNA by Pol II. There, active transcription units showed internal fiber-free gaps in transcription, ranging from 1 to 5 kb in length. Around the fiber-free gaps, transcription initiation appeared uniform, leading the authors to conclude that polymerase-loading switches from high constant initiation frequency to negligibly low frequency (88), which is now referred to as bursting.
Direct Observations of Transcription in Living Cells
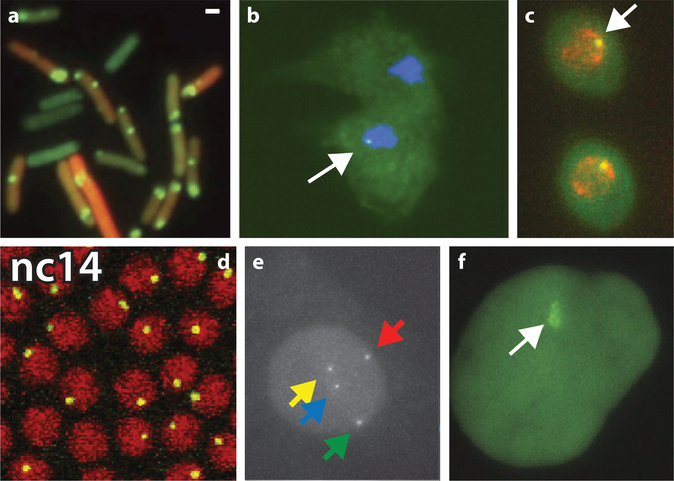
Measuring proteins and RNA distributions reveals the heterogeneity between cells but may be influenced by other factors such as cell size (100). Without a time component, static measurements are limited in revealing the kinetics of transcription itself, necessitating the development of dynamic approaches to measure transcription kinetics. In a landmark paper, Janicki et al. (60) visualized chromatin structure, RNA, and protein at the same locus to understand the kinetics of gene expression. To visualize transcription, the authors used the MS2 system, which was previously developed to follow the localization of single mRNAs (10). The MS2 system relies on the labeling of RNA by addition of multiple stem loops, each of which binds a dimer of the MS2 coat protein fused to a fluorescent protein instantaneously (Figure 2d-f). This technique was subsequently applied in many organisms to follow transcription in living cells (24, 29, 47, 48, 64, 72) and has even been used to label RNA of β-actin in a live mouse (77, 104) (Figure 3). Instead of inferring transcription kinetics from protein or RNA distributions, live-cell imaging of transcription now allowed for direct visualization of the fluctuations at the transcription site (Figure 2e,f). This method resulted in the first demonstration of bursting in bacteria in 2005 (Figure 3a), followed a year later by the eukaryote Dictyostelium discoideum (29, 48) (Figure 3b). Since these early studies, the regulation and properties of bursting transcription have been an intense area of study.
Figure 3:
Visualization of transcription in different organisms. (a) MS2-labeled RNA (green) from a reporter gene, driven by the P-lac/ara promoter. Panel a modified from Reference 48 with permission. (b) MS2-labeled RNA at a developmental gene in Dictyostelium. Panel b modified from Reference 29 with permission. (c) PP7-labeled RNA at the GLT1 gene with the POL1 promoter in Saccharomyces cerevisiae. Panel c modified from Reference 73 with permission. (d) MS2-labeled RNA at a reporter gene driven by the hunchback promoter and enhancer in a developing Drosophila embryo, nuclear cycle 14 (nc14). Panel d modified from Reference 47 with permission. (e) MS2-labeled Actb RNA after 13 min serum stimulation in immortalized mouse embryonic fibroblasts, obtained from an Actb-MBS knock-in mouse (77). (f) MS2-labeled RNA at a reporter gene in U2-OS cells. Panel f modified from Reference 60 with permission. Arrows in panels point to transcription sites.
Similar to the MS2 system, the PP7 bacteriophage can also be used for RNA labeling (26, 73). The MS2 and PP7 stem-loop sequences are orthogonal, allowing for visualization of two different RNAs or two parts of the same RNA in a single cell (35, 55, 74). Other techniques exist for visualizing RNA (17, 96, 101), but these have not yet been applied to measure the kinetics of transcription. Although the MS2 and PP7 systems so far yield the highest sensitivity and time resolution to follow transcription dynamics, the measurements are time consuming and not easily scalable to many genes. In addition, visualizing the RNA using the MS2 and PP7 system requires that repeat sequences be inserted into the gene of interest. Because precise genome editing has been difficult in mammalian cells, many investigators have used unstable RNA and short-lived luciferase or fluorescent protein reporter genes to follow gene expression changes at many different genes (38, 123).
Using RNA visualization or protein reporters, quantification of bursting parameters such as burst size and frequency was coupled with modulating input signals or regulatory factors in order to understand regulation of transcription dynamics. From these studies, we have learned that (a) transcriptional bursting is a conserved property of transcription, observed from bacteria to human, (b) the properties of transcriptional bursting appear to be organism dependent, and (c) not all genes show bursting. These observations suggest that bursting may have various origins and regulators, which may also differ per organism or per gene. Although the first observation of transcriptional bursting occurred almost 40 years ago (84), a mechanistic understanding of the biological steps that cause bursting is still lacking. However, the concept is pedagogically useful because it offers a more fine-grained view of transcription: Instead of saying transcription of a gene is upregulated, one can say that the firing is more frequent or the duration of the active state is longer, which potentially offers more mechanistic insight. In the following section, we review the current knowledge of transcription bursting dynamics in different organisms and discuss potential regulatory mechanisms underlying polymerase initiation in single cells.
Gene-Independent Bursting in Bacteria
In bacteria, bursting was first quantified at a reporter gene driven by P-lac/ara promoter (48) (Figure 3a). The average burst duration at this gene is 6 min, interspersed by average off periods of 37 min. Consistent with the two-state random telegraph model, burst size was found to be geometrically distributed, and the time between bursts followed an exponential distribution (48). At increasing expression levels, the burst duration becomes longer, but the burst frequency and initiation rate stay constant (48, 119). Interestingly, these properties are similar across 20 different promoters, indicating there may be a gene-independent mechanism to regulate bursting in bacteria.
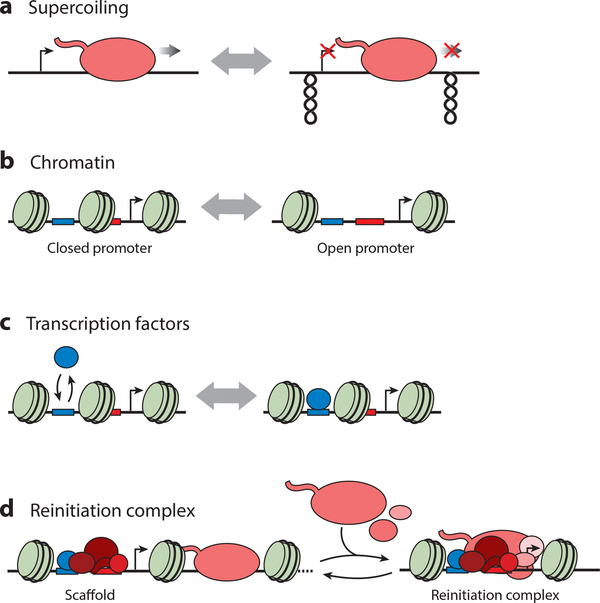
One explanation for this gene-independent bursting was recently suggested by Chong et al. (28). Using an in vitro single-molecule transcription assay as well as in vivo assays, the authors showed that positive and negative supercoiling in front of and behind the active polymerase inhibits transcription elongation and initiation (Figure 4a). The torsional constraints have to be released by the enzymatic activity of gyrase in order for transcription to resume. The model implies that if gyrase concentrations in the cell are limiting, highly expressed genes will show bursting in a gene-independent manner, agreeing with previous experimental results (119). Whether this model is sufficient to fully explain bursting in bacteria is unclear. For example, there is evidence that bacteria can express the same gene with both small and large bursts, suggesting additional layers of regulation (27). In this case, the different burst sizes were interpreted to result from partial or full dissociation of a repressor from the operator.
Figure 4:
Different hypotheses that have been proposed as a mechanism for transcriptional bursting. (a) Supercoiling buildup during transcription can result in inhibition of initiation and elongation and has to be released before transcription can resume. (b) Nucleosome positioning or binding in promoter regions may affect the accessibility of transcriptional regulators to DNA binding sites. (c) The binding dynamics of a specific regulatory factor, such as a gene-specific transcription factor, can determine bursting if initiation takes place as long as the factor is bound to DNA. (d) After initiation, several general transcription factors can remain bound to the promoter, forming a scaffold for rapid reinitiation and firing of multiple polymerases at the same time.
Gene-Specific Dynamics in Eukaryotes
In eukaryotes, bursting displays more gene-specific behavior than in bacteria. Different genes exhibit different bursting characteristics that are modulated at the level of burst size, burst frequency, or burst duration (38, 117, 123). Bursting properties are influenced by promoter architecture, genomic position, and transcription factors as well as chromatin. This gene-specific control of bursting is inconsistent with the supercoiling model as the (only) cause of bursting in eukaryotes and suggests the control via other mechanisms. Several models have been proposed (Figure 4), with varying degrees of support in different organisms, as is discussed below, but so far, the regulators of these bursts remain mysterious.
In yeast, transcription initiation dynamics appear to be divided into two classes, showing either random uncorrelated or bursting transcription initiation. As discussed above, for many yeast genes, the variability in protein abundance between cells is proportional to the mean, arising from stochastic transcription synthesis and decay (5, 97). Similarly, the cellular RNA distribution of three housekeeping genes follows a Poisson distribution, which is the expected distribution for constitutive uncorrelated transcription initiation (137).
Live-cell imaging confirmed that initiation from some promoters exhibits uncorrelated behavior (73). Random transcription initiation is likely the main mode of transcription for the majority of yeast genes and appears to be directly related to the promoter architecture (57). Low-noise genes generally show a nucleosome-free region upstream of the transcription start site, resulting from large poly(dA/dT) tracks that repel the formation of nucleosomes (22). Because nucleosomes do not have to be removed before activation, it seems plausible that transcription-factor binding can directly activate transcription for these genes. Accordingly, the measured initiation rate at the POL1 promoter correlates with the measured search time of the Mbp1 transcription factor, suggesting that the firing rate is determined by only one rate-limiting step of the transcription factor trying to find the gene (73).
Approximately 10% of yeast genes show higher expression variation. These genes are often inducible, generally contain a TATA box in their promoters, and are regulated by the transcriptional regulator SAGA (22). Their promoters typically do not contain nucleosome-free regions but instead display a more variable chromatin promoter architecture, making them more dependent on chromatin remodeling for their activation (22). Compared with housekeeping genes, smFISH of a SAGA-regulated gene showed a wider RNA distribution, which is more consistent with bursting than constitutive transcription (137). Yeast genes thus appear to be transcribed with multiple modes of transcription.
Direct observation of bursting in yeast was only recently shown (74). By direct imaging of GAL10 transcription, the measured burst size was small, with approximately two polymerases per burst, but bursts were very frequent, resulting in approximately eight polymerases at the gene at the same time. The promoter architecture of induced genes may be important for producing bursting transcription, because mutations in the TATA box were inferred to affect burst size, and mutations in different chromatin factors were inferred to change both burst size and frequency (13, 57, 109, 129). Although these latter measurements are based on protein distributions and may not reflect polymerase dynamics, they are suggestive of a model in which bursting may be regulated by chromatin (Figure 4b). The promoter on and off states may reflect different chromatin conformations, of which only some are permissive for transcription. Nucleosome binding might, for example, preclude the binding of transcription factors to the DNA, resulting in the off state. When nucleosomes are lost, the promoter is switched on, allowing the transcriptional machinery to assemble for multiple rounds of transcription.
In support of this model, mapping of the nucleosome conformations in single cells by electron microscopy or by methylation protection revealed eight different PHO5 promoter conformations (19, 118). The frequencies of the different conformations suggested stochastic transitions between conformations with assembly, disassembly, and position-specific sliding of nucleosomes (18, 19). Although nucleosome binding negatively correlated with gene expression levels (118), the nu- cleosome conformations that are permissive for transcription are unclear. smFISH distribution implied a promoter on frequency that was consistent with transcription activation when the middle of the three promoter nucleosomes was lost (19), but directly coupling nucleosome conformations with RNA production in the same cell to test this model further will be interesting.
Another model suggests that bursting is the direct result of the kinetics of transcription-factor binding (Figure 4c). In this model, transcription can take place as long as the transcription factor is bound to DNA. The inducible CUP1 array showed different cycles of transcription-factor binding upon induction, with a slow cycle correlating with transcription (64). Although bursting was not shown directly at the CUP1 array, cycles in transcription-factor binding appeared to reflect promoter accessibility, which might result in bursts of transcription.
Besides cycling on chromatin, some transcription factors have been reported to show pulses in nuclear localization (20, 51). Possibly, pulsing of transcription factors into the nucleus could generate bursts of transcription. This would also give multiple levels of control, as nuclear transcription-factor levels can be modulated by adjustment of the pulse frequency, duration, or amplitude. Combinatorial regulation can be achieved by regulating the temporal overlap between pulses of two transcription factors (76). For transcription factors that do not show regulated nuclear translocation, cyclic binding to DNA could be regulated by other means, such as degradation, or active removal by regulatory factors, creating bursts of transcription. Despite increasing efforts to measure transcription-factor binding and translocation dynamics (40, 50, 75, 98), how these kinetics relate to transcriptional bursting is still mostly unexplored.
In addition to chromatin or transcription factors, another theory suggests that the transcription initiation machinery itself may determine bursting (Figure 4d). Early in vitro transcription studies using purified yeast proteins or human nuclear extracts revealed the presence of a scaffold composed of part of the preinitiation complex (PIC) that remained bound to the promoter after polymerase release (53, 135). Because only a limited number of factors have to reassemble on the scaffold for transcription to reinitiate, this may potentially happen faster than de novo PIC assembly. Reinitiation of several polymerases may produce bursts of transcription, the size of which would be mainly determined by the stability of the reinitiation scaffold. Decreased scaffold stability has been used to explain why mutation in the TATA box can change variability between cells by changing the burst size (12). So far, there is no clear evidence for the reinitiation model in vivo, but a recent study reported an inhibitor of a metazoan TFIID (transcription factor IID) subunit that specifically inhibited the first round of transcription initiation, but not reinitiation (139).
The different proposed models are not necessarily mutually exclusive and may operate simultaneously on the same gene. For example, the reinitiation scaffold stability appears to be regulated by transcriptional activators (135), possibly linking transcription-factor binding dynamics to the stability of the preinitiation complex. Transcription-factor kinetics are, in turn, influenced by the chromatin environment. This interconnected regulatory strategy may give cells the ability to modulate different bursting properties independently, tuning not only the levels but also the variation between cells.
Transcriptional Bursting During Development
During embryonic development, cells undergo major changes in gene expression programs to allow differentiation from a single-cell zygote into a multicellular organism with many specialized cell types. Gene expression has to be tightly controlled to ensure that genes are expressed in the right place and at the right time. Any noise in transcription could potentially endanger robust expression, suggesting that cells may have developed ways to limit or control gene expression noise. As discussed above, transcriptional bursting results in significantly more gene expression variation than random transcription initiation. One might speculate that to reduce noise, cells may limit bursting transcription for developmentally regulated genes. Surprisingly, bursting seems to be the main mode of transcription for developmental genes. At several Drosophila genes, nascent transcript numbers from smFISH measurements show larger variations than can be explained by random noise, suggestive of bursting transcription (78, 103).
Live-cell measurement of eve transcription indicated burst durations of 4 to 10 min and large burst sizes of approximately 20 to 100 mRNAs (15). These bursts did not fit a simple two-state random telegraph model, suggesting more complicated regulation.
During Dictyostelium development, bursting is observed with different burst durations, frequencies, and intensities per gene (93). Bursting dynamics are also inherited in a chromatin-dependent manner from mother to daughter cells for many hours after cell division (94). During development, two strongly expressed housekeeping genes show decreasing burst duration, but surprisingly no change in bursting is observed for developmentally regulated genes (93, 122). Instead, developmental genes are regulated at the level of proportion of cells that showed expression (29, 122).
Binary regulation may be a common way to regulate transcription during development, as it is also seen during Drosophila development. A gradient of the transcription-factor Bicoid along the anterior posterior axis of the embryo influences the fraction of nuclei actively expressing the hunchback gene (47, 80). This regulatory strategy appears only in nuclear cycle 14 and is not present in earlier cycles. On top of this binary control, Bicoid also increases the rate of polymerase loading and the time window in which transcription can take place. Transcription initiation during development is thus regulated at multiple levels to ensure robust patterning. Moreover, precise mRNA distributions are obtained by averaging in time and space (78).
To summarize, there is no evidence that cells restrict bursting initiation during development. Instead, the level of expression is tuned by digital as well as analog control, and stochastic transcription at the individual gene is diluted by spatiotemporal averaging. Very likely, more mechanisms exist to buffer noisy transcription (4).
Bursting in Mammalian Cells
Mammalian gene transcription appears to be dominated by bursting (38). As in yeast, bursting properties seem to be modulated by chromatin and transcription factors. The main difference between bursting in mammalian cells compared with other organisms is the presence of a refractory period between bursts (2, 52, 90, 110, 123). A refractory period is a period of time when the template is no longer permissive for transcription. Because this refractory period usually occurs after a burst, it has been interpreted as a so-called reset time for the gene. The consequence of this refractory period is that for many genes in different systems, there is “memory” of previous transcription cycles. It has been speculated that in a cell population, a fraction of genes are poised for induction while another fraction are in a refractory state (52). This heterogeneity in cell state allows the population to have both an acute and a longer sustained response. Moreover, having a refractory state (or memory) indicates that multiple steps are required in order to reactivate a previously fired gene (34). Thus, the distribution of time between transcription on states is not exponentially distributed but rather shows a nonzero peak, consistent with the multistep process described at the beginning of this review. Studies describing a refractory period so far have used unstable luciferase/GFP reporter expression as a proxy for transcription, which may not reflect transcription itself. Live-cell imaging of the RNA should provide a more direct and precise view of these dynamics.
Refractory periods can also explain the existence of transcriptional oscillations. Several studies have identified oscillations in transcription-factor binding and Pol II recruitment that are distinct from the more traditional oscillations explained by negative feedback loops (such as in circadian transcription) (6, 62, 64, 67, 86,113). For example, during the estrogen response, several cofactors, polymerase, and histone modifying proteins are recruited in a cyclic manner to the proximal promoter of TFF1, a highly induced gene (86, 113). These oscillations vary in length with 40–60-min on times. If off times are distributed exponentially, as predicted for a single rate-limiting step in activation, there is no refractory period, and transcription can occur at any time, resulting in an absence of pronounced oscillations. The fact that such oscillations are visible in ensemble studies after hormone stimulation or inhibitor washout suggests uniformity in the process of transcription, which seems to contradict single-cell studies. More experiments are needed to resolve these differences.
The role of chromatin in shaping bursting patterns in mammalian cells is supported by the finding that expression of randomly integrated reporters from different genomic locations results in different burst sizes and frequencies (38). Because expression was driven by the same promoter sequence, the effect of genomic position can be interpreted to result from different chromatin environments, although bursting may also depend on local or distal contacts with enhancer elements. To increase activity, reporters at different positions modulated burst frequency at lower expression levels and burst size at higher expression levels. Similarly, a study using random HIV integration concluded that mostly burst size was regulated, although a weak trend of modulation of burst frequency can also be seen at low expression levels (117). An intriguing explanation might be that as frequency is increased for a gene, the refractory period sets an effective ceiling for RNA synthesis. The only way to increase expression for a gene at that point is to affect the burst size. Refractory periods could provide a regulatory threshold above which the gene switches to a different mode of regulation. However, this general mode of regulation contrasts with other studies that have reported gene-dependent or even stimulus-dependent regulation of bursting (89, 99, 123). For example, in mouse embryonic stem cells, endogenous Nanog expression showed changes in both burst frequency and burst size when the growth conditions were changed from serum to 2i conditions (99).
Interestingly, genomic integration appears to be crucial for bursting. When an MS2-labeled reporter gene was integrated into the genome, it displayed bursting transcription, but when expressed from a plasmid, it failed to show bursting (72). This finding is consistent with the model that chromatin determines bursting. To further question the role of chromatin, investigators repeatedly studied the effect of blocking histone deacetylation by the addition of trichostatin A (TSA). Different studies using TSA reported different results on the expression of protein reporters, with no changes in bursting on some genes but increased burst duration or burst size on other genes (52, 123). In chicken-cell lines with randomly integrated reporter genes, TSA mostly increased the burst frequency (126). The exact role of chromatin is therefore still an open question. More insight may be gained from direct perturbations of the chromatin machinery, targeted changes to histone modifications of certain genes (82), or imaging of chromatin modifications at a single locus (120).
Bursting properties are also influenced by gene-specific transcription factors. Increasing the affinity or the number of transcription-factor binding sites increases both burst size and frequency of protein reporters (123). Using a steroid-response exogenous reporter construct, Larson et al. (72) followed transcription with the MS2 technique in live cells after the addition of a hormone, allowing for direct measurement of on and off times (Figure 2d-f). The average burst duration at this reporter was approximately 40 min, with equally long average off times. An increase in the hormone level resulted in increased burst frequency, without affecting burst duration or burst size, suggesting that transcription-factor concentration appears to regulate the burst frequency (72). Similarly, detailed analysis of c-Fos bursting by smFISH and modeling suggests that during induction, increasing transcription-factor concentration regulates burst frequency (111). However, direct coupling between transcription-factor dynamics and gene bursting at the single-molecule level has not been performed, leaving the role of transcription factors in transcriptional bursting an open question.
In contrast to mammalian promoters, viral promoters integrated in mammalian cells show constant nonbursting expression. Visualizing expression of the cyclin D1 gene tagged with MS2, driven by either the cyclin D1 promoter or the viral CMV promoter, revealed that the endogenous promoter showed transcriptional bursting, whereas the CMV promoter had higher and constant expression (136). The difference in expression levels resulted from a higher rate of transcription initiation at the CMV promoter, with about twice as many polymerases loaded on the gene. The elongation rate of both constructs was similar. An HIV vector similarly showed no fluctuations in transcription (83). In contrast, another HIV reporter system reported variability consistent with bursting transcription, with burst sizes ranging from 2 to 10 mRNAs at different genomic positions (115). Integration of a retroviral transgene driven by the endogenous EF1α gene showed very rapid pulses, which lasted 144 s on average (79). Whether these pulses represent bursts of transcription or are single polymerases transcribing the gene is unclear. Transgenes or viral genes may thus have different kinetics than endogenous genes have, illustrating the limitations of using reporter genes as a model for endogenous gene control. With the recent development of more targeted genome-editing techniques, future studies will likely show increased focus on endogenous genes.
In summary, genes are transcribed with different kinetics, some displaying random initiation and others showing pulses or bursts. Transcriptional bursting is observed from bacteria to human cells, but whether the underlying mechanism of bursting is similar across organisms is still unclear. Live-cell studies have given valuable insight that points toward a role for chromatin and transcription factors in regulating bursting, but future studies directly measuring bursting parameters at endogenous genes will likely result in a more detailed view of the biochemical steps regulating Pol II dynamics.
TRANSCRIPTION ELONGATION DYNAMICS
Transcription regulation studies have traditionally focused on the control of initiation, but there is an emerging understanding that regulation can also occur at the level of elongation. Genomic approaches revealed that polymerase is not always evenly distributed along the gene, suggesting different elongation rates or pausing. At many genes in higher eukaryotes, polymerase shows a promoter-proximal pause that is highly regulated by positive and negative elongation factors (32). In addition, there is evidence for pausing at nucleosome dyads, or near splice sites (30, 61, 71). Pausing may have functional consequences, as changes in polymerase velocity have been suggested to affect the alternative splicing outcomes through kinetic competition (42). However, differences in polymerase densities detected by chromatin immunoprecipitation or nuclear run-on experiments do not always reflect changes in polymerase elongation rate (102), highlighting the importance of time-resolved measurements that directly measure elongation.
To measure elongation in living cells, many studies have used fluorescent recovery after pho-tobleaching (FRAP). With this technique, either a fluorescently labeled RNA polymerase subunit or RNA itself is bleached in a specific area, after which recovery of fluorescence is measured. A similar technique, called fluorescence loss of photobleaching (FLIP), or inverse FRAP, does the opposite and measures the reduction in fluorescence in one area when a different area is continuously bleached. The shapes of the recovery or bleaching curves give information about parameters such as the mobile fraction and the kinetics of the transcriptional process. In Chinese hamster ovary cells, FRAP and FLIP of a Pol II subunit at steady state revealed that approximately 25% of polymerases are transcriptionally engaged. The half-life of a typical transcription cycle was approximately 14–20 min. Because the recovery was measured at an arbitrary part of the nucleus and not at a specific locus, the exact transcription elongation rate could not be determined. Nonetheless, it was concluded that maximally half of the recovery was attributed to elongation (68). In contrast, photobleaching of a Pol II subunit at the MMTV (mouse mammary tumor virus) array during activation was interpreted as mostly reflecting elongation, but an early component in the bleaching curve also suggested the presence of abortive initiation events at the locus (8). In a study by Darzacq et al. (39), the complete transcription cycle was measured at approximately 200 repeats of a stably integrated gene cassette. The recovery curves of Pol II at this array revealed three kinetic components separated by two orders of magnitude, consisting of~5 s for transient promoter interaction,~50 s for initiation, and~500 s for elongation. Transition between steps was highly inefficient, and one of the oft-cited conclusions of this work is that only 1 in 90 polymerase interactions results in a complete RNA. A similar phenomenon of transient clustering of polymerases that do not enter into elongation was also observed at endogenous genes (31). Darzacq et al. (39) determined that polymerases move at 4.3 kb/min, but 4% of polymerases pause for approximately 4 min. The observed pausing is unlikely to represent promoter-proximal pausing but may instead be pausing in the gene body or at the polyadenylation site.
The polytene nuclei of salivary gland tissues in Drosophila allowed for the analysis of transcription dynamics at two native heat-shock genes (132). This system had previously been used to study the binding dynamics of the transcription factor HSF1, showing longer binding upon heat shock (133). Complete FRAP recovery of Pol II at these genes took approximately 100 s, resulting in an elongation rate of~1.5 kb/min. Because the shape of the curve could be explained solely by elongation, the conclusion was that recruitment, entry into elongation, and termination were too rapid to measure and must be extremely efficient (in contrast to conclusions from exogenous mammalian arrays). At later time points after heat shock, the FRAP curves showed incomplete recovery, suggesting local recycling of polymerase for multiple rounds of transcription (132).
Other studies of elongation using FRAP reported elongation rates that vary by two orders of magnitude. At the cyclin D1 gene, elongation rate was estimated at 0.31–0.78 kb/min, independent of the promoter (136). At a tandem array of an HIV construct, the elongation rate was 1.9 kb/min (14). Surprisingly, when the same gene was studied at a single integration site, the elongation rate increased to more than 50 kb/min (83). These large variations are challenged by population-based studies showing maximally fourfold differences in elongation rates across genes (37, 116). One potential explanation lies in the difficulty of FRAP curve interpretation (91). The recovery curves are a combination of the different transcriptional processes as well as nonspecific interactions and diffusion. Even if the curves would reflect only one transcriptional process, such as elongation, the measurements are an average of many polymerases that can have different kinetics or show pausing, complicating interpretation.
Other elongation measurements in single cells have used the MS2 or PP7 technique. Labeling the 5’ and 3’ ends of the same gene with the MS2 and PP7 approaches in yeast showed that elongation rates for the first round of transcription were highly variable between cells, ranging from 0.84 kb/min to 3.66 kb/min (55). In this study, the elongation rate did not depend on cell cycle. Another study, however, showed that the elongation rate during G1 was much slower than during the S/G2 phase, going from 1.2 kb/min to 2.8 kb/min (73). Variations in elongation rates inside a gene, between cells, or between cell-cycle stages exemplify the complexity of these measurements and illustrate the importance of single-cell measurements to elucidate regulatory control at the level of transcription elongation.
What do these numbers mean in the context of the cell? It is clear that elongation exists in an optimal range—faster and slower elongation rates are both deleterious in yeast (16, 63), and elongation rate has been shown to reflect metabolic state in human cells (41)—but such an optimum does not necessarily imply that processive Pol II elongation rate is a regulatory knob that the cell uses. Conversely, elongation provides a large dynamic range of potential responses. Because nucleic acid addition is a processive enzymatic process, the time to synthesize a nascent transcript is the sum of thousands of enzymatic addition cycles occurring in series, resulting in a nearly deterministic total time (Figure 1a). As such, elongation time is a unique kinetic quantity. In contrast, a regulatory pause might consist of only one or a few kinetic steps, resulting in a broader distribution of waiting times and outcomes (127). The missing biological component is an explanation of how a pause might act—one could imagine a kinetic delay acting through recruitment of some factor, RNA secondary structure, or something else entirely.
TERMINATION, CLEAVAGE, 3’ PROCESSING
The coordinated molecular events that occur at the 3’ ends of genes result in cleavage and release of the nascent RNA, and dismantling and removal of the elongation complex from the DNA template. The dynamics of this process are perhaps the most understudied in the transcription cycle. Furthermore, many studies have shown clear coupling between termination, cleavage, release, elongation, and splicing, although the mechanism and the implications remain elusive (9). There are several pathways through which termination is thought to take place in different organisms, but in this review, we focus on the poly(A)-dependent termination pathway found in eukaryotes; this pathway is the mechanism by which transcription of protein-coding genes, with the exception of histones, are thought to terminate.
The trigger for proper termination begins with transcription through the AAUAAApolyadeny-lation signal (PAS) and a GU-rich downstream sequence element (DSE) located approximately 40–60 nt further downstream (106). These sequences in the RNA are recognized and bound by complexes [in metazoans, called cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF)] that, in turn, recruit the full protein complex that cleaves the nascent RNA chain and adds a poly(A) tail to the 3’ end of the transcript. In the meantime, Pol II has continued elongation further downstream, before losing processivity and falling off at some point (95). ChIP-seq and GRO-seq studies show an accumulation of Pol II at the 3’ end of actively transcribing genes beyond the PAS with density extending a few kilobases downstream, supporting the idea of Pol II pausing or slowing down past the PAS (32, 107).
There have been a few efforts to measure the kinetics of termination and release in single living cells. Larson et al. (73) measured the dwell time of nascent transcripts at the site of transcription for an MDN1 reporter gene in yeast using PP7 labeling. By switching the position of the PP7 cassette within the gene, they were also able to measure the elongation rate of Pol II along the gene body. By subtracting the elongation time from the total dwell time of the transcript on the DNA template, they were able to determine that there is a window of 70 s between when Pol II elongates past the end of the gene to when the transcript is released from the chromatin. In another study using the MS2 labeling system, Boireau et al. (14) used a combination of FRAP and modeling to measure a time of 63 s for 3’-end processing and transcript release of an HIV-1 gene inserted into U2-OS cells. Coupling a time-resolved imaging approach with correlation analysis, Coulon et al. (35) measured a 3’-end dwell time of 116 s. Finally, using a gene array system, Darzacq et al. (39) measured a termination time of 8 min and a transcript release time of 30 s. However, at present, Pol II termination and RNA release have not been visualized simultaneously at the same locus.
There are two prevailing models for how Pol II is disassembled and transcription is terminated. In the conformational change model (sometimes also called the allosteric or anti-terminator model), transcription through the PAS is thought to cause a conformational change within the RNA Pol II complex, either by loss of elongation factors or by addition of termination factors. This rearrangement impairs the ability of Pol II to elongate efficiently and eventually disassembles the active complex. Candidates for such termination-enhancing factors include CstF-64, CstF-77, and Pcf11, which associate with the RNAPII elongating complex at the PAS, or CPSF-73 and Ssu72, which associate with the 3’ ends of genes (11, 46, 121). Either way, depletion of any of these factors results in read-through transcription. Conversely, factors such as the transcriptional coactivator PC4 disassociate from RNAPII at the PAS, allowing RNAPII to be termination competent (23). More importantly, although it has been observed only in vitro, termination can occur even in the absence of cleavage, providing further support for the conformational change model (138).
It has been proposed that in the torpedo model, cleavage of the nascent RNA provides an opening for a 5’-to-3’ exonuclease to latch onto the nascent chain still being synthesized by Pol II. The exonuclease chews back the nascent chain until it eventually catches up to Pol II and disassembles it. ChIP experiments show that the yeast 5’-to-3’ exonuclease Rat1 indeed localizes to the 3’ ends of genes (66). In wild-type cells, polymerase density is even throughout the gene body but decreases rapidly 200–400 nt downstream of the PAS, whereas in rat1 mutant cells, Pol II density did not decrease, thereby implicating Rat1 in the process of termination. Using nuclear run-on assays, researchers observed a similar result for Xrn2 (human homolog of Rat1) in HeLa cells (130). Furthermore, through the use of mutants of Pol II that transcribe at different rates, slow elongation has been shown to robustly shift termination upstream whereas fast elongation shifts it downstream (45). Similarly, increasing the rate of Pol II elongation in yeast results in significant read-through defects via altered kinetic competition with the helicase Sen1 (54). These results bolster the case for the torpedo model of termination in which Pol II and Xrn1 exist in kinetic competition with each other. The validity of the torpedo model is not clear-cut, as depletion of Rat1 did not completely abolish termination, nor did nuclease activity by itself lead to termination (49, 65). Thus, it appears that Rat1 by itself was not sufficient to initiate termination, and because Rat1 also functions in recruiting factors such as Pcf11 to the elongation complex, the actual model of termination might be a hybrid of both the conformational change and the torpedo models (43, 81). One prediction of the torpedo model is that the termination zone should be fairly narrow because both the helicase/exonuclease and the polymerase are processive enzymes that would catch up to one another in a narrowly distributed window of time. To our knowledge, exonuclease speeds have not been measured in vivo, but testing the various models of termination and cleavage are, in principle, excellent candidates for the live-cell dynamic approach.
CONCLUSIONS AND OUTLOOK
Single-cell studies provide a window into the heterogeneity of transcriptional responses in a population. In large part, this heterogeneity is due to the dynamics of RNA synthesis. These dynamics provide clues to the underlying molecular mechanisms and insight into the rate-limiting steps of transcription. So far, live-cell measurements of transcription have been performed mainly at reporter genes, and the field has likewise lacked the tools to do precise molecular perturbations. With the development of novel techniques to edit mammalian genomes, all tools are now in place to study transcription dynamics in higher eukaryotes at endogenous loci. Complementing live-cell techniques with emerging single-cell sequencing approaches will be interesting, and doing so may help advance the low-throughput live-cell measurements to a larger scale.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
This is a work of the US Government and is not subject to copyright protection in the United States.
LITERATURE CITED
- 1.Almer A, Rudolph H, Hinnen A, Hörz W. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5(10):2689–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong R, Wen W, Meinkoth J, Taylor S, Montminy M. 1995. A refractory phase in cyclic AMP-responsive transcription requires down regulation of protein kinase A. Mol. Cell. Biol 15(3):1826–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahar Halpern K, Tanami S, Landen S, Chapal M, Szlak L, et al. 2015. Bursty gene expression in the intact mammalian liver. Mol. Cell 58(1):147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balázsi G, van Oudenaarden A, Collins JJ. 2011. Cellular decision making and biological noise: from microbes to mammals. Cell 144(6):910–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, et al. 2006. Noise in protein expression scales with natural protein abundance. Nat. Genet. 38(6):636–43 [DOI] [PubMed] [Google Scholar]
- 6.Barrow J, Hay CW, Ferguson LA, Docherty HM, Docherty K. 2006. Transcription factor cycling on the insulin promoter. FEBS Lett. 580(2):711–15 [DOI] [PubMed] [Google Scholar]
- 7.Batenchuk C, St-Pierre S, Tepliakova L, Adiga S, Szuto A, et al. 2011. Chromosomal position effects are linked to Sir2-mediated variation in transcriptional burst size. Biophys. J 100(10):L56–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker M, Baumann C, John S, Walker DA, Vigneron M, et al. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3(12):1188–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet 15(3):163–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2(4):437–45 [DOI] [PubMed] [Google Scholar]
- 11.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280(5361):298–301 [DOI] [PubMed] [Google Scholar]
- 12.Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, et al. 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 24(6):853–65 [DOI] [PubMed] [Google Scholar]
- 13.Blake WJ, KÆrn M, Cantor CR, Collins JJ. 2003. Noise in eukaryotic gene expression. Nature 422(6932):633–37 [DOI] [PubMed] [Google Scholar]
- 14.Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, et al. 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol 179(2):291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. 2014. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. PNAS 111(29):10598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braberg H,Jin H,Moehle EA, Chan YA, Wang S, et al. 2013. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154(4):775–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bratu DP, Cha B-J, Mhlanga MM, Kramer FR, Tyagi S. 2003. Visualizing the distribution and transport of mRNAs in living cells. PNAS 100(23):13308–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown CR, Boeger H. 2014. Nucleosomal promoter variation generates gene expression noise. PNAS 111(50):17893–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown CR, Mao C, Falkovskaia E, Jurica MS, Boeger H. 2013. Linking stochastic fluctuations in chromatin structure and gene expression. PLOS Biol. 11(8):e1001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai L, Dalal CK, Elowitz MB. 2008. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455:485–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai L, Friedman N, Xie XS. 2006. Stochastic protein expression in individual cells at the single molecule level. Nature 440(7082):358–62 [DOI] [PubMed] [Google Scholar]
- 22.Cairns BR. 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461:193–98 [DOI] [PubMed] [Google Scholar]
- 23.Calvo O, Manley JL. 2001. Evolutionarily conserved interaction between Cstf-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7(5):1013–23 [DOI] [PubMed] [Google Scholar]
- 24.Campbell PD, Chao JA, Singer RH, Marlow FL. 2015. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development 142(7):1368–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey LB, van Dijk D, Sloot PMA, Kaandorp JA, Segal E. 2013. Promoter sequence determines the relationship between expression level and noise. PLOS Biol. 11(4):e1001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao JA, Patskovsky Y, Almo SC, Singer RH. 2008. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol 15(1):103–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi PJ, Cai L, Frieda K, Xie XS. 2008. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322(5900):442–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong S, Chen C, Ge H, Xie XS. 2014. Mechanism of transcriptional bursting in bacteria. Cell 158(2):314–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chubb JR, Trcek T, Shenoy SM, Singer RH. 2006. Transcriptional pulsing of a developmental gene. Curr. Biol 16(10):1018–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469(7330):368–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, et al. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341(6146):664–67 [DOI] [PubMed] [Google Scholar]
- 32.Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322(5909):1845–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosma MP, Tanaka T, Nasmyth K. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97(3):299–311 [DOI] [PubMed] [Google Scholar]
- 34.Coulon A, Chow CC, Singer RH, Larson DR. 2013. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat. Rev. Genet 14(8):572–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR. 2014. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife 3:e03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Csardi G, Franks A, Choi DS, Airoldi EM, Drummond DA. 2015. Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast. PLOS Genet. 11(5):e1005206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danko CG, Hah N, Luo X, Martins AL, Core L, et al. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50(2):212–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, et al. 2012. Transcriptional burst frequency and burst size are equally modulated across the human genome. PNAS 109(43):17454–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, et al. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol 14(9):796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, et al. 2009. Imaging transcription in living cells. Annu. Rev. Biophys 38:173–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.das Neves RP, Jones NS, Andreu L, Gupta R, Enver T, Iborra FJ. 2010. Connecting variability in global transcription rate to mitochondrial variability. PLOS Biol. 8(12):e1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, et al. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12(2):525–32 [DOI] [PubMed] [Google Scholar]
- 43.Dengl S, Cramer P. 2009. Torpedo nuclease Rat1 is insufficient to terminate RNA polymerase II in vitro. J. Biol. Chem 284(32):21270–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Femino AM, Fay FS, Fogarty K, Singer RH. 1998. Visualization of single RNA transcripts in situ. Science 280(5363):585–90 [DOI] [PubMed] [Google Scholar]
- 45.Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, et al. 2015. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol. Cell 60(2):256–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garas M, Dichtl B, Keller W. 2008. The role of the putative 3’ end processing endonuclease Ysh1p in mRNA and snoRNA synthesis. RNA 14(12):2671–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia HG, Tikhonov M, Lin A, Gregor T. 2013. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol 23(21):2140–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golding I, Paulsson J, Zawilski SM, Cox EC. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123(6):1025–36 [DOI] [PubMed] [Google Scholar]
- 49.Gu W, Wind M, Reines D. 1996. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. PNAS 93(14):6935–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hager GL, McNally JG, Misteli T. 2009. Transcription dynamics. Mol. Cell 35:741–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao N, O’Shea EK. 2012. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol 19(1):31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harper CV, Finkenstädt B, Woodcock DJ, Friedrichsen S, Semprini S, et al. 2011. Dynamic analysis of stochastic transcription cycles. PLOS Biol. 9(4):e1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hawley DK, Roeder RG. 1987. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem 262(8):3452–61 [PubMed] [Google Scholar]
- 54.Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. 2013. Kinetic competition between RNA polymerase II and Sen1-dependent transcription termination. Mol. Cell 49(1):55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH. 2013. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods 10(2):119–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, et al. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95(5):717–28 [DOI] [PubMed] [Google Scholar]
- 57.Hornung G, Bar-Ziv R, Rosin D, Tokuriki N, Tawfik DS, et al. 2012. Noise-mean relationship in mutated promoters. Genome Res. 22(12):2409–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh D, Paulsson J. 2011. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat. Genet. 43(2):95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol 3(3):318–56 [DOI] [PubMed] [Google Scholar]
- 60.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, et al. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116(5):683–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jonkers I, Kwak H, Lis JT. 2014. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3:e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang Z, Pirskanen A, Janne OA, Palvimo JJ. 2002. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem 277(50):48366–71 [DOI] [PubMed] [Google Scholar]
- 63.Kaplan CD, Jin H, Zhang IL, Belyanin A. 2012. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLOS Genet. 8(4):e1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, et al. 2008. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319(5862):466–69 [DOI] [PubMed] [Google Scholar]
- 65.Kim M, Ahn S-H, Krogan NJ, Greenblatt JF, Buratowski S. 2004. Transitions in RNA polymerase II elongation complexes at the 3; ends of genes. EMBOJ. 23(2):354–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, et al. 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432(7016):517–22 [DOI] [PubMed] [Google Scholar]
- 67.Kim S, Shevde NK, Pike JW. 2005. 1,25-dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid Xreceptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J. Bone Miner. Res 20(2):305–17 [DOI] [PubMed] [Google Scholar]
- 68.Kimura H, Sugaya K, Cook PR. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol 159(5):777–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ko MS. 1991. A stochastic model for gene induction. J. Theor. Biol 153(2):181–94 [DOI] [PubMed] [Google Scholar]
- 70.Ko MS, Nakauchi H, Takahashi N. 1990. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBOJ. 9(9):2835–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339(6122):950–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larson DR, Fritzsch C, Sun L,Meng X, Lawrence DS, Singer RH. 2013. Direct observation of frequency modulated transcription in single cells using light activation. eLife 2:e00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332(6028):475–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenstra TL, Coulon A, Chow CC, Larson DR. 2015. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol. Cell 60(4):597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levine JH, Lin Y, Elowitz MB. 2013. Functional roles of pulsing in genetic circuits. Science 342(6163):1193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Y, Sohn CH, Dalal CK, Cai L, Elowitz MB. 2015. Combinatorial gene regulation by modulation of relative pulse timing. Nature 527(7576):54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, et al. 2011. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8(2):165–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Little SC, Tikhonov M, Gregor T. 2013. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154(4):789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lo MYM, Rival-Gervier S, Pasceri P, Ellis J. 2012. Rapid transcriptional pulsing dynamics of high expressing retroviral transgenes in embryonic stem cells. PLOS ONE 7(5):e37130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lucas T, Ferraro T, Roelens B, De Las Heras Chanes J, Walczak AM, et al. 2013. Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr. Biol 23(21):2135–39 [DOI] [PubMed] [Google Scholar]
- 81.Luo W, Johnson AW, Bentley DL. 2006. The role of Rat1 in coupling mRNA 3’-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 20(8):954–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, et al. 2013. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 31(12):1137–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, et al. 2011. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 12(12):1280–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McKnight SL, Miller OL. 1977. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12(3):795–804 [DOI] [PubMed] [Google Scholar]
- 85.McNally JG, Müller WG, Walker D, Wolford R, Hager GL. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287(5456):1262–65 [DOI] [PubMed] [Google Scholar]
- 86.Métivier R, Penot G, Hübner MR, Reid G, Brand H, et al. 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115(6):751–63 [DOI] [PubMed] [Google Scholar]
- 87.Miller OL, Beatty BR. 1969. Visualization of nucleolar genes. Science 164(3882):955–57 [DOI] [PubMed] [Google Scholar]
- 88.Miller OL Jr., McKnight SL. 1979. Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell 17:551–63 [DOI] [PubMed] [Google Scholar]
- 89.Molina N, Suter DM, Cannavo R, Zoller B, Gotic I, Naef F. 2013. Stimulus-induced modulation of transcriptional bursting in a single mammalian gene. PNAS 110(51):20563–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morgan JI, Cohen DR, Hempstead JL, Curran T. 1987. Mapping patterns of c-fos expression in the central nervous system after seizure. Science 237(4811):192–97 [DOI] [PubMed] [Google Scholar]
- 91.Mueller F, Mazza D, Stasevich TJ, McNally JG. 2010. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: What do we really know? Curr. Opin. Cell Biol 22(3):403–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mueller F, Wach P, McNally JG. 2008. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys.J 94(8):3323–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muramoto T, Cannon D, Gierliński M, Corrigan A, Barton GJ, Chubb JR. 2012. Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. PNAS 109(19):7350–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muramoto T, Müller I, Thomas G, Melvin A, Chubb JR. 2010. Methylation of H3K4 is required for inheritance of active transcriptional states. Curr. Biol 20(5):397–406 [DOI] [PubMed] [Google Scholar]
- 95.Nag A, Narsinh K, Kazerouninia A, Martinson HG. 2006. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA 12(8):1534–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, et al. 2016. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165:488–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, et al. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441(7095):840–46 [DOI] [PubMed] [Google Scholar]
- 98.Normanno D, Dahan M, Darzacq X. 2012. Intra-nuclear mobility and target search mechanisms of transcription factors: a single-molecule perspective on gene expression. Biochim. Biophys. Acta 1819(6):482–93 [DOI] [PubMed] [Google Scholar]
- 99.Ochiai H, Sugawara T, Sakuma T, Yamamoto T. 2014. Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Sci. Rep 4:7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, et al. 2015. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol. Cell 58(2):339–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paige JS, Wu KY, Jaffrey SR. 2011. RNA mimics of green fluorescent protein. Science 333(6042):642–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palangat M, Larson DR. 2012. Complexity of RNA polymerase II elongation dynamics. Biochim. Biophys. Acta 1819(7):667–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paré A, Lemons D, Kosman D, Beaver W, Freund Y, McGinnis W. 2009. Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Curr. Biol 19(23):2037–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, et al. 2014. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343(6169):422–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peccoud J, Ycart B. 1995. Markovian modeling of gene-product synthesis. Theor. Popul. Biol 48(2):222–34 [Google Scholar]
- 106.Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev. 25(17):1770–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, et al. 2010. c-Myc regulates transcriptional pause release. Cell 141(3):432–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. 2006. Stochastic mRNA synthesis in mammalian cells. PLOS Biol. 4(10):e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raser JM, O’Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304(5678):1811–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sasaki K, Cripe TP, Koch SR, Andreone TL, Petersen DD, et al. 1984. Multihormonal regulation of phosphoenolpyruvate carboxykinase gene transcription. The dominant role of insulin. J. Biol. Chem 259(24):15242–51 [PubMed] [Google Scholar]
- 111.Senecal A, Munsky B, Proux F, Ly N, Braye FE, et al. 2014. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 8(1):75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shandilya J, Roberts SGE. 2012. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim. Biophys. Acta 1819(5):391–400 [DOI] [PubMed] [Google Scholar]
- 113.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103(6):843–52 [DOI] [PubMed] [Google Scholar]
- 114.Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, et al. 2014. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55(2):319–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. 2010. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys. J 98(8):L32–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singh J, Padgett RA. 2009. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol 16(11):1128–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Skupsky R, Burnett JC, Foley JE, Schaffer DV, Arkin AP. 2010. HIV promoter integration site primarily modulates transcriptional burst size rather than frequency. PLOS Comput. Biol. 6(9):e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Small EC, Xi L, Wang J-P, Widom J, Licht JD. 2014. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. PNAS 111(24):E2462–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.So L-H, Ghosh A,Zong C, Sepúlveda LA, Segev R, Golding I. 2011. General properties of transcriptional time series in Escherichia coli. Nat. Genet 43(6):554–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, et al. 2014. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516(7530):272–75 [DOI] [PubMed] [Google Scholar]
- 121.Steinmetz EJ, Brow DA. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol 23(18):6339–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stevense M, Muramoto T, Müller I, Chubb JR. 2010. Digital nature of the immediate-early transcriptional response. Development 137(4):579–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332(6028):472–74 [DOI] [PubMed] [Google Scholar]
- 124.Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, et al. 2010. Quantifying E. coli proteome and tran- scriptome with single-molecule sensitivity in single cells. Science 329(5991):533–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Van Kampen NG. 2011. Stochastic Processes in Physics and Chemistry. Amsterdam: Elsevier; 3rd ed. [Google Scholar]
- 126.Viñuelas J, Kaneko G, Coulon A, Vallin E, Morin V, et al. 2013. Quantifying the contribution of chromatin dynamics to stochastic gene expression reveals long, locus-dependent periods between transcriptional bursts. BMC Biol. 11:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Voliotis M, Cohen N, Molina-París C, Liverpool TB. 2008. Fluctuations, pauses, and backtracking in DNA transcription. Biophys. J. 94(2):334–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DI. 1995. Enhancers increase the probability but not the level of gene expression. PNAS 92(15):7125–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weinberger L, Voichek Y, Tirosh I, Hornung G, Amit I, Barkai N.2012. Expression noise and acetylation profiles distinguish HDAC functions. Mol. Cell 47(2):193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.West S, Gromak N, Proudfoot NJ. 2004. Human 5’→3’ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432(7016):522–25 [DOI] [PubMed] [Google Scholar]
- 131.Wu B, Buxbaum AR, Katz ZB, Yoon YJ, Singer RH. 2015. Quantifying protein-mRNA interactions in single live cells. Cell 162(1):211–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. 2007. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell 28(6):978–90 [DOI] [PubMed] [Google Scholar]
- 133.Yao J, Munson KM, Webb WW, Lis JT. 2006. Dynamics of heat shock factor association with native gene loci in living cells. Nature 442(7106):1050–53 [DOI] [PubMed] [Google Scholar]
- 134.Yu J, Xiao J, Ren X, Lao K, Xie XS. 2006. Probing gene expression in live cells, one protein molecule at a time. Science 311(5767):1600–3 [DOI] [PubMed] [Google Scholar]
- 135.Yudkovsky N, Ranish JA, Hahn S. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408(6809):225–29 [DOI] [PubMed] [Google Scholar]
- 136.Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. 2010. Single-allele analysis of transcription kinetics in living mammalian cells. Nat. Methods 7(8):631–33 [DOI] [PubMed] [Google Scholar]
- 137.Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol 15(12):1263–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang H, Rigo F, Martinson HG. 2015. Poly(A) signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Mol. Cell 59(3):437–48 [DOI] [PubMed] [Google Scholar]
- 139.Zhang Z, Boskovic Z, Hussain MM, Hu W, Inouye C, et al. 2015. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. eLife 4:e07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zopf CJ, Quinn K, Zeidman J, Maheshri N. 2013. Cell-cycle dependence of transcription dominates noise in gene expression. PLOS Comput. Biol 9(7):e1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]