Abstract
Icaritin is a natural flavonoid with anti-osteoporosis activity. This study aimed to characterize icaritin glucuronidation by pooled human liver microsomes (HLM) and pooled human intestine microsomes (HIM), and to determine the contribution of individual UDP-glucuronosyltrans-ferase (UGT) enzyme to icaritin glucuronidation.
Glucuronidation rates were determined by incubating icaritin with uridine diphosphate glucuronic acid (UDPGA)-supplemented microsomes. Kinetic parameters were derived by appropriate model fitting. Relative activity factors and activity correlation analysis were performed to identify main UGT isoforms.
UGT1A3, 1A7, 1A8, 1A9 and 2B7 were mainly responsible for catalyzing the formation of two glucuronides (G1 and G2). Icaritin 3-O-glucuronidation (G1) was significantly correlated with Chenodeoxycholic acid (CDCA) glucuronidation (r = 0.787, p = 0.002), propofol glucuronidation (r = 0.661, p = 0.019) and Zidovudine (AZT) glucuronidation (r = 0.805, p = 0.002). Similarly, icaritin 7-O-glucuronidation (G2) was also correlated with CDCA glucuronidation (r = 0.640, p = 0.025), propofol glucuronidation (r = 0.592, p = 0.043) and AZT glucuronidation (r = 0.661, p = 0.019). In addition, UGT1A3, 1A9 and 2B7 contributed 37.5, 33.8 and 21.3% for G1 in pooled HLM, respectively. Also, UGT1A3, 1A9 and 2B7 contributed 34.3, 20.0 and 8.6% for G2 in pooled HLM, respectively.
Icaritin was subjected to significant glucuronidation, wherein UGT1A3, 1A7, 1A8, 1A9 and 2B7 were main contributing enzymes.
Keywords: Activity correlation, glucuronidation, icaritin, reaction kinetics, UGTs
Introduction
Prenylflavonoids are reported to be a group of major active constituents present in genus Epimedium for several significant activities (Ma et al., 2011). Icaritin is the common flavonoid aglycone of these prenylflavonoids with many biological effects, such as anti-osteoporosis activities (Chen et al., 2013; Peng et al., 2013). Besides, icaritin could induce cell death in activated hepatic stellate cells through mitochondrial activated apoptosis and ameliorate the development of liver fibrosis in rats (Li et al., 2011). Meanwhile, icaritin has neuroprotective effects against MPP+-induced toxicity in MES23.5 cells. IGF-I receptor mediated activation of PI3K/ Akt and MEK/ERK1/2 pathways are involved in the neuroprotective effects of icaritin against MPP+-induced neuronal damage (Jiang et al., 2016). Additionally, icaritin is able to target androgen receptor and androgen receptor COOH-terminal truncated splice variants, to inhibit androgen receptor signaling and tumor growth with no apparent toxicity (Sun et al., 2015). Recently, icaritin had been shown as a potential agent for the treatment of systemic lupus erythematosus (Liao et al., 2016).
These biological activities had stimulated increasing interests in the in vivo metabolism of icaritin and its related prenylflavonoids. Poor bioavailability of prenylated flavonoids results from their poor intrinsic permeation across the human gastrointestinal tract (Chen et al., 2008). Meanwhile, it is shown that epimedium flavonoids could be hydrolyzed into secondary glycosides or aglycone by intestinal flora or enzymes, thereby enhancing their absorption and antiosteoporosis activity (Zhou et al., 2015). As so far, numerous researches of prenylflavonoids or individual flavonoid had been conducted in the fields of in vivo metabolites profiling (Chang et al., 2012; Cui et al., 2014; Jin et al., 2013; Qian et al., 2012; Zhao et al., 2010). Icaritin, as an intermediate aglycone, is easily metabolized and transformed into high polar conjugates to be preferentially excreted from the rat organism (Wang et al., 2015). Icaritin-3-O-glucuronide, icaritin-7-O-glucuronide and icaritin-5-O-glucuronide were the main Phase II conjugates, which indicated that glucuronidation is considered as the main clearance pathway for icaritin metabolism (Cui et al., 2014; Jin et al., 2013).
Glucuronidation is one of the most significant phase II conjugation reactions (Wu et al., 2011). Generally, it is a detoxification mechanism because the glucuronide is usually pharmacologically inactive and rapidly eliminated from the body due to its highly polar nature (Ritter, 2000). Meanwhile, it accounts for the clearance of ~35% drugs metabolized by phase II enzymes (Evans & Relling, 1999). The mechanism of glucuronidation reaction is that a glucuronic acid derived from the cofactor uridine diphosphoglucuronic acid (UDPGA) is transferred to the substrate, generating the glucuronidated metabolite (or glucuronide) via the action of the UDP-glucuronosyltransferase enzymes (UGTs) (Sun et al., 2015). Human UGT enzymes are systematically classified into four subfamilies, UGT1, UGT2, UGT3 and UGT8 (Mackenzie et al., 2005). Usually, the UGT1A and 2B enzymes contribute significantly to metabolism and detoxification of xenobiotics (Lu et al., 2016). The UGT1A family contains nine members (i.e. UGT1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9 and 1A10), whereas the UGT2B family contains seven members (i.e. UGT2B4, 2B7, 2B10, 2B11, 2B15, 2B17 and 2B28).
The UGT enzymes actively involved in glucuronidation of icaritin and their reaction kinetics still remained unknown. Therefore, the purpose of this study is to determine the metabolism of icaritin via the glucuronidation pathway, and to identify the main UGT enzymes involved in icaritin glucuronidation. The glucuronidation rates are determined by incubating icaritin with UDPGA-supplemented microsomes. Kinetic parameters are obtained by fitting an appropriate model to the data. Determination of the relative activity factors (RAF) and activity correlation analysis are performed to identify the main UGT enzymes contributing to the metabolism of icaritin. It is shown that UGT1A3, 1A7, 1A8, 1A9 and 2B7 are identified as the main contributors to glucuronidation of icaritin.
Materials and methods
Materials
Uridine diphosphate glucuronic acid (UDPGA), magnesium chloride (MgCl2), alamethicin and D-saccharic-1, 4-lactone were provided from Sigma-Aldrich (St Louis, MO). Pooled human liver microsomes (HLM), pooled human intestine microsomes (HIM) and recombinant expressed human UGT Supersomes™ (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 2B4, 2B7, 2B10, 2B15 and 2B17) were obtained from Corning Biosciences (New York, NY). Icaritin (purity>98%) were purchased from Jingzhu Medical Technology Co., Ltd (Nanjing, China). Chenodeoxycholic acid (CDCA), propofol and zidovudine (AZT) were purchased from Aladdin Chemicals (Shanghai, China). All other chemicals and reagents were analytical grade or the highest grade commercially available.
Glucuronidation assay
Icaritin was incubated with pooled HLM, HIM and expressed UGTs enzymes to determine the rates of glucuronidation as published references previously (Lu et al., 2016; Sun et al., 2015). Briefly, the incubation mixture mainly contained 50mM Tris-hydrochloric acid buffer (pH 7.4), 0.88mM MgCl2, 22mg/mL alamethicin, 4.4mM saccharolactone and 3.5mM UDPGA. The reaction was terminated by adding icecold acetonitrile (containing the internal standard quercetin). The samples were vortexed and centrifuged at 13 800g for 10 min. The supernatant was subjected to UPLC/q-tof-MS analysis. Incubation without UDPGA served as negative control to confirm the metabolites produced were UDPGA-dependent. All experiments were performed in triplicate.
Structural identification of icaritin glucuronides by UPLC/q-tof-MS
Metabolite screening of icaritin glucuronides were performed using a UPLC/q-tof-MS system (Waters Corporation, Manchester, UK). Chromatographic separation was performed on BEH C18 column (2.1 mm × 50 mm, 1.7 μm, Waters, Ireland, Part NO. 186002350) guarded with a column temperature at 35 C. The mobile phase was consisting of water (A) and acetonitrile (B) (both including 0.1% formic acid, V/V) at a flow rate of 0.4mL/min. The gradient elution program was 5% B from 0 to 0.5min, 5–40% B from 0.5 to 1.2min, keeping 40% B from 1.2 to 1.5min, 40% ~100% B from 1.5 to 2.8min, maintaining 100% B from 2.8 to 3.3min, 100%~5% B from 3.3 to 4.0min.
UPLC system was coupled to a hybrid quadrupole orthogonal time-of-flight (q-tof) tandem mass spectrometer (SYNAPT™ G2 HDMS, Waters, Manchester, UK) with electrospray ionization (ESI). The operating parameters were as follows: capillary voltage, 3kV (ESI+); sample cone voltage, 35V; extraction cone voltage, 4V; source temperature, 100 C; desolvation temperature, 300 C; cone gas flow, 50L/h and desolvation gas flow, 800L/h. The full scan mass range was 50–1500 Da. The method employed lock spray with leucine enkephalin (m/z 556.2771 in positive ion mode) to ensure mass accuracy.
Quantification of icaritin and its glucuronides
Due to lack of reference standard, quantification of icaritin glucuronide was based on the standard curve of the parent compound (icaritin) according to the assumption that parent compound and its glucuronide have closely similar UV absorbance maxima (Lu et al., 2016; Sun et al., 2014; Troberg et al., 2016). A series of working solutions of icaritin were determined by Acquity™ UPLC I-Class system equipped with a BEH C18 column (2.1 mm × 50 mm, 1.7μm, Waters, Ireland, Part NO. 186002350). The gradient elution program was same to the UPLC condition of structural identification of icaritin glucuronides above. The detection wavelength was set at 270 nm and the injection volume was 8μL.
The LOD and LOQ were calculated as 3-fold and 10-fold of the ratio of signal-to-noise (S/N), respectively. The LOD and LOQ for icaritin was 0.01 and 0.02μM, respectively. Calibration curves were constructed by plotting icaritin (analyte)/quercetin (internal standard) peak area ratios (Y) versus icaritin concentrations (X) using a 1/x2 weighting factor. Acceptable linear correlation (Y=1.7778X) was confirmed by correlation coefficients (r2) of 0.9991. The linear range was 0.02–20μM. The accuracy and precision of the intra-day and inter-day error were both less than 3.2%.
Enzymes kinetic evaluation
A preliminary experiment was performed to ensure that the rates of glucuronidation were determined under linear conditions with respect to the incubation time and protein concentration. As a result, the formation rate of icaritin glucuronides were linear over 120 min of incubation and 0.1–1.0mg/mL of microsomal protein. Meanwhile, less than 10% of icaritin (substrate) was metabolized in all incubations, the kinetic determinations were carried out using a microsomal protein concentration of 0.5mg/mL (HLM and UGTs) with 60-min incubation. Hence, the incubation time and protein concentration are 60min and 0.5mg/mL, respectively.
Serial concentrations of icaritin (0.4–20μM) were incubated with pooled HLM, HIM and expressed UGTs enzymes to determine icaritin glucuronidation rates. The kinetic models Michaelis–Menten equation and substrate inhibition equation were fitted to the data of metabolic rates versus substrate concentrations and displayed in Equations (1) and (2), respectively. Appropriate models were selected by visual inspection of the Eadie–Hofstee plot (Hutzler & Tracy, 2002). Model fitting and parameter estimation were performed by Graphpad Prism V5 software (San Diego, CA).
The parameters were as follow. V is the formation rate of product. Vmax is the maximal velocity. Km is the Michaelis constant and [S] is the substrate concentration. Ksi is the substrate inhibition constant. The intrinsic clearance (CLint) was derived by Vmax/Km for Michaelis–Menten and substrate inhibition models.
| (1) |
| (2) |
Activity correlation analysis
According to the glucuronidation assay protocol as described previously (Lu et al., 2016), the metabolic activities of individual HLMs (n=12) toward icaritin, CDCA (a probe substrate for UGT1A3), propofol (a probe substrate for UGT1A9) and AZT (a probe substrate for UGT2B7) were determined (Jiang et al., 2012; Li et al., 2016; Sun et al., 2014). Icaritin (2 μM) were incubated with UDPGA-supplemented individual HLM (1.0 mg/mL) for 60 min, whereas CDCA (250 μM), propofol (500 μM) and AZT (1.25mM) were separately treated with UDPGA-supplemented individual HLM (1.0mg/mL) for 120min. Correlation analysis were performed between icaritin glucuronidation (G1 and G2) and CDCA glucuronidation, between icaritin glucuronidation (G1 and G2) and propofol glucuronidation, and between icaritin glucuronidation (G1 and G2) and AZT glucuronidation. Correlation (Pearson) analysis was performed using GraphPad Prism V5 software.
Contribution of UGT isoforms
The contribution of individual expressed UGT enzymes to icaritin glucuronidation in pooled HLM was evaluated by the relative activity factor (RAF) approach as described in previous study (Liu et al., 2014; Sun et al., 2014). The RAF value was defined as the activity ratio of pooled HLM to an expressed UGT enzyme (Supersome) toward a probe substrate for this enzyme using Equation (3). The relative amount of icaritin glucuronidation in pooled HLM attributed to an expressed UGT enzyme was estimated by multiplying the glucuronidation activity (i.e. the intrinsic clearance) derived with this enzyme by the corresponding RAF. The RAFs were derived for UGT1A3, 1A9 and 2B7 using the well-recognized probe substrates CDCA, propofol and AZT, respectively. The contribution of individual UGT enzyme was calculated according to Equation (4).
| (3) |
| (4) |
Statistical analysis
Data are expressed as the mean ± SD (standard deviation). Mean differences between treatment and control groups were analyzed by two-tailed Student’s t-test. The level of significance was set at p<0.05 (*) or p<0.01 (**).
Results
Structural identification of icaritin metabolites
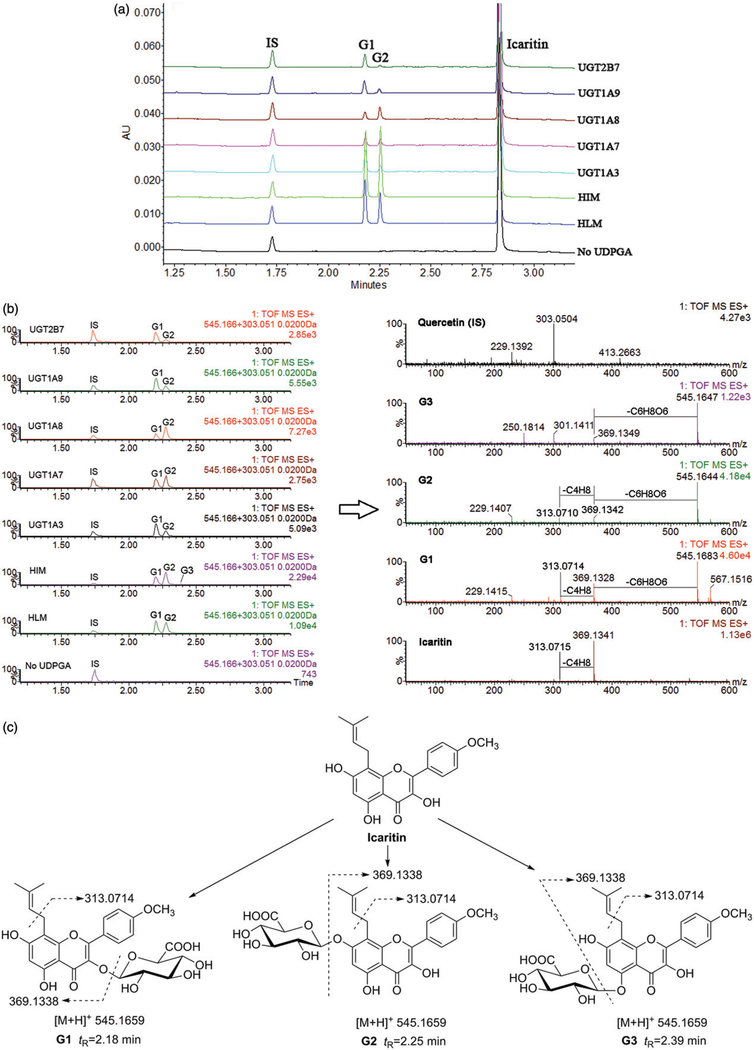
After incubation of icaritin with UDPGA-supplemented pooled HLM, three additional peaks which have a similar UV absorption profile in addition to icaritin were detected by UPLC-DAD analysis (Figure 1a). The retention times of icaritin and quercetin (IS) were 2.83 and 1.73 min, respectively. The peaks at 2.18, 2.25 and 2.39 min were referred to as metabolite G1, G2 and G3, respectively.
Figure 1.
Ultra high performance liquid chromatography analysis (a), EIC chromatograms and MS spectrum (b) and chemical structures (c) of icaritin and icaritin-3-O-glucuronide (G1), icaritin-7-O-glucuronide (G2) and icaritin-5-O-glucuronide (G3).
Icaritin exhibited a typical [M+H]+ ion at m/z 369.1336 (C21H21O6, _0.5 ppm), and a main daughter ion at m/z 313.0715 (C17H13O6) produced by losing a neutral fragment of C4H8 (56.0621 Da) (Figure 1b). The characteristic fragment was same as previous study (Zhao et al., 2010). Consistently, G1, G2 and G3 had the same adduct [M+Na]+ion at m/z 567.1462 (C27H28O12Na) and [M+H]+ ion at m/z 545.1655 (C27H29O12), which was 176.0325 Da higher (characteristic of the addition of glucuronic acid) than that of icaritin (Figure 1b). Based on these data, they were characterized as mono-glucuronide of icaritin. On the basis of previous study (Cui et al., 2014; Jin et al., 2013), icaritin-3-O-glucuronide, icaritin-7-O-glucuronide and icaritin-5-O-glucuronide were eluted successively on a reverse C18 chromatographic column. So, G1, G2 and G3 were characterized as icaritin-3-O-glucuronide, icaritin-7-O-glucuronide and icaritin-5-O-glucuronide, respectively (Figure 1c). Since the amount of icaritin-5-O-glucuronide (G3) was very few, and it could not be detected under the condition of 270nm. Hence, we did not perform the glucuronidation rate of icaritin-5-O-glucuronide (G3).
Glucuronidation of icaritin in human liver microsomes and human intestine microsomes
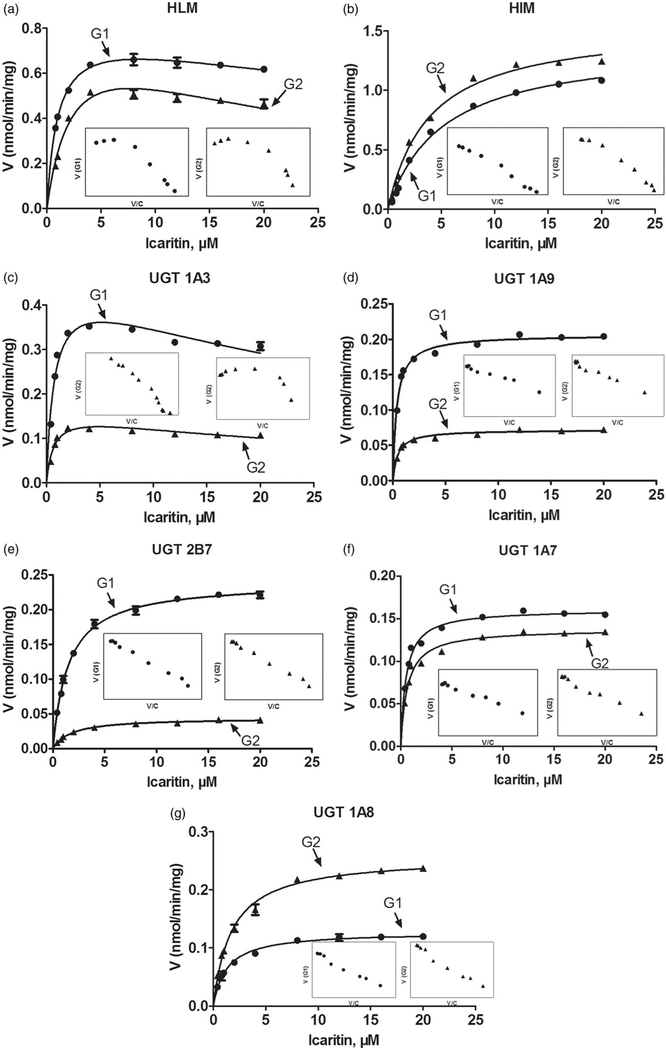
Kinetic profiling revealed that formation of icaritin-3-O-glucuronide (G1) and icaritin-7-O-glucuronide (G2) in pooled HLM well modeled by the substrate inhibition equation (Figure 2a), whereas they followed the classical Michaelis-Menten kinetics in HIM (Figure 2b). In contrast, the glucuronide formation of G1 (0.82 nmol/min/mg) and G2 (0.89 nmol/min/mg) in pooled HLM was similar as well as G1 (1.39 nmol/min/mg) and G2 (1.56 nmol/min/mg) in HIM. Glucuronidation G1 and G2 of icaritin in pooled HLM was efficient (CLint =0.80 and 0.35mL/min/mg, respectively), following the substrate inhibition kinetics with Km values of 1.03 and 2.55μM, respectively. Similarly, the CLint values of G1 and G2 in HIM were 0.27 and 0.40mL/min/mg, respectively, whereas the Km values of G1 and G2 in HIM in Michaelis–Menten model were 5.13 and 3.91 μM, respectively. In addition, Ki values of G1 and G2 in pooled HLM were 69.21 and 28.82 μM, respectively. The detailed parameters of G1 and G2 were listed in Table 1.
Figure 2.
Kinetic profiles for glucuronidation of icaritin by various types of microsomes. (a) Pooled human liver microsomes (HLM); (b) pooled human intestine microsomes (HIM). (c) Expressed UGT 1A3; (d) expressed UGT 1A9; (e) expressed UGT 2B7; (f) expressed UGT 1A7; (g) expressed UGT 1A8. In each panel, the insert figure showed the corresponding Eadie–Hofstee plot. All experiments were performed in triplicate.
Table 1.
Kinetic parameters of icaritin glucuronidation by pooled HLM, HIM and expressed UGT enzymes (Mean ± SD).
| Protein source | Metabolite | Vmax (nmol/min/mg) | Km (μM) | Ki (μM) | CLint (mL/min/mg) | Model |
|---|---|---|---|---|---|---|
| Pooled HLM | G1 | 0.82±0.02 | 1.03±0.06 | 69.21±8.32 | 0.80±0.05 | SI |
| G2 | 0.89±0.14 | 2.55±0.71 | 22.82±8.84 | 0.35±0.11 | SI | |
| HIM | G1 | 1.39±0.06 | 5.13±0.60 | N.A. | 0.27±0.03 | MM |
| G2 | 1.56±0.06 | 3.91±0.46 | N.A. | 0.40±0.05 | MM | |
| UGT1A3 | G1 | 0.47±0.04 | 0.74±0.17 | 35.93±11.85 | 0.63±0.15 | SI |
| G2 | 0.16±0.02 | 0.68±0.18 | 35.01±12.88 | 0.24±0.07 | SI | |
| UGT1A7 | G1 | 0.16±0.003 | 0.51±0.05 | N.A. | 0.31±0.03 | MM |
| G2 | 0.14±0.003 | 0.64±0.08 | N.A. | 0.21±0.03 | MM | |
| UGT1A8 | G1 | 0.13±0.002 | 1.28±0.10 | N.A. | 0.10±0.01 | MM |
| G2 | 0.26±0.005 | 1.75±0.13 | N.A. | 0.15±0.01 | MM | |
| UGT1A9 | G1 | 0.21±0.003 | 0.38±0.04 | N.A. | 0.55±0.05 | MM |
| G2 | 0.07±0.001 | 0.47±0.05 | N.A. | 0.15±0.02 | MM | |
| UGT2B7 | G1 | 0.24±0.002 | 1.49±0.06 | N.A. | 0.16±0.007 | MM |
| G2 | 0.04±0.001 | 1.77±0.15 | N.A. | 0.03±0.002 | MM |
All experiments were performed in triplicate.
Note: SI: substrate inhibition model; MM: Michael–Menten model; N.A.: not available.
Reaction phenotyping with expressed UGT enzymes
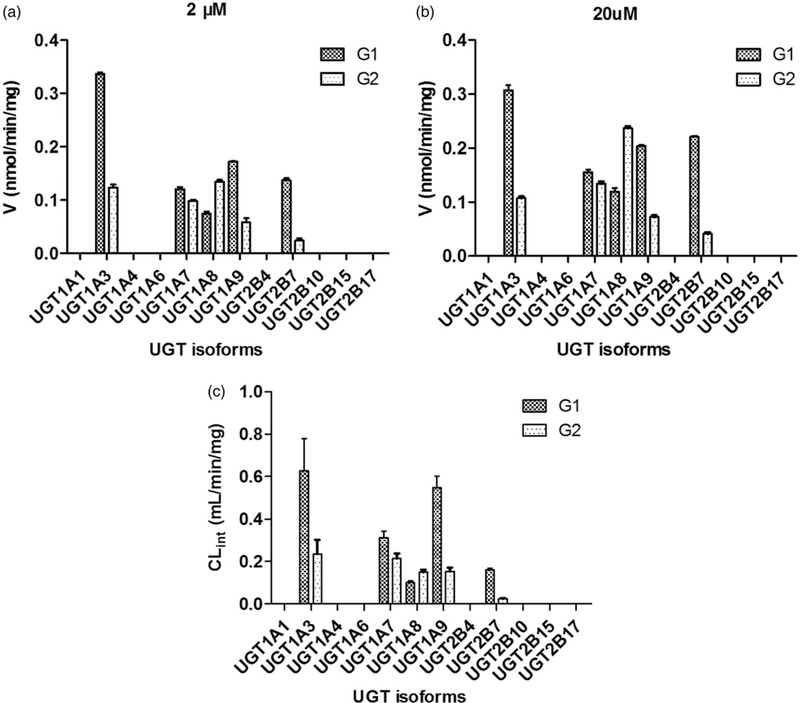
To identify the enzymes involving in the glucuronidation of icaritin, twelve expressed UGT enzymes were analyzed for their catalysis activities (expressed as nmol/min/mg protein) at the substrate concentrations of 2 μM (Figure 3a) and 20 μM (Figure 3b). The metabolic profiles for G1 and G2 were similar under two test substrate concentrations. All experiments were performed in triplicate. The formation of G1 and G2 mainly contributed to expressed UGT1A3, 1A7, 1A8, 1A9 and 2B7 enzymes. Other seven UGT enzymes were not capable of the production toward G1 and G2. In general, UGT1A3 showed the highest glucuronidation activities.
Figure 3.
Comparisons of glucuronidation rates (a: 2 μM; b: 20 μM) and the intrinsic values (CLint) (c) of icaritin by twelve expressed UGT enzymes. All experiments were performed in triplicate.
Glucuronidation kinetics by recombinant UGT enzymes
Based on reaction phenotyping results, glucuronidation kinetics of active recombinant UGT enzymes were analyzed using a series of substrate concentrations. Obviously, the kinetic profiles of G1 and G2 by UGT1A3 were both well modeled by substrate inhibition equation (Figure 2c), which was in line with its glucuronidation profiles in pooled HLM (Figure 2a), suggesting that UGT1A3 was indeed the most crucial enzyme responsible for glucuronidation of G1 and G2. In contrast, G1 and G2 glucuronidation of icaritin by UGT1A9 (Figure 2d) and 2B7 (Figure 2e) did not always followed the same kinetics as pooled HLM. For the metabolite G1, CLint values by UGT1A3, 1A9 and 2B7 were 0.63, 0.55 and 0.16 mL/min/mg, respectively. Meanwhile, CLint values of G2 by these three UGT enzymes were 0.27, 0.15 and 0.03 mL/min/mg, respectively. Hence, UGT1A3, 1A9 and 2B7 were the main UGT enzymes for glucuronidation (G1 and G2) of icaritin.
On the other hand, HIM-mediated glucuronidation of G1 and G2 followed the Michaelis–Menten kinetics (Figure 2b). Similarly, UGT1A7 and 1A8 enzymes were both described by the classical Michaelis–Menten kinetics (Figure 2f and g). The CLint values of G1 by UGT1A7 and 1A8 enzymes were 0.31 and 0.10 mL/min/mg, respectively. UGT1A7 and 1A8-mediated glucuronidation of G2 were with CLint values of 0.214 and 0.147 mL/min/mg, respectively. Taken together, UGT1A7 and 1A8 also played most important roles in the glucuronidation (G1 and G2) of icaritin. In general, kinetic profiles and calculated kinetic parameters of UGT1A3, 1A9, 2B7, 1A7 and 1A8, were shown in Figure 2c–g and Table 1, respectively, whereas their CLint comparison plot was shown in Figure 3c.
Contribution of UGT1A3, 1A9 and 2B7 to icaritin glucuronidation in pooled HLM
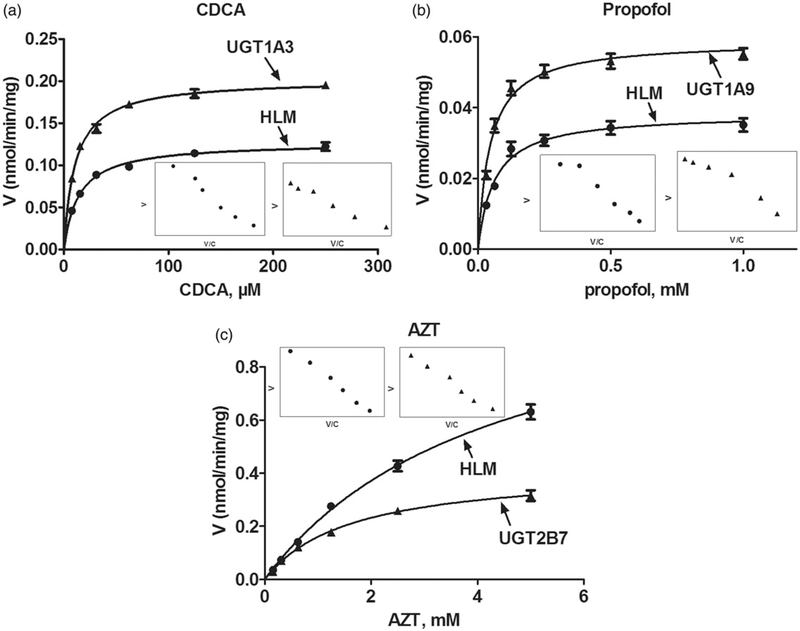
To estimate the exact contribution of UGT1A3, 1A9 and 2B7 to icaritin glucuronidation in pooled HLM, the RAF approach was calculated by CLint values of CDCA, propofol and AZT glucuronidation in pooled HLM and relative individual expressed UGT enzyme, respectively. The analytical conditions were adopted based on previous study (Li et al., 2016; Sun et al., 2014). As a result, the kinetic profiles of CDCA, propofol and AZT glucuronidation were modeled by the classical Michaelis–Menten kinetics (Figure 4). The derived RAFs for UGT1A3, 1A9 and 2B7 were 0.48, 0.49 and 1.04, respectively (Table 2). The scaled CLint values of G1 and G2 were 0.30 (=0.63 × 0.48) mL/min/mg and 0.12 (=0.24 × 0.48) mL/min/mg for UGT1A3 that represented 37.5% and 34.3% of the CLint values (0.80 and 0.35 mL/min/ mg) in pooled HLM. The scaled CLint values of G1 and G2 were 0.27 (=0.55 × 0.49) mL/min/mg and 0.07 (=0.15 × 0.49) mL/min/mg for UGT1A9, that was 33.8% and 20.0% of total glucuronidation activity in pooled HLM. The scaled CLint values of G1 and G2 were 0.17 (=0.16 × 1.04) mL/min/mg and 0.03 (=0.025 × 1.038) mL/ min/mg for UGT2B7, that was 21.3% and 8.6% of total glucuronidation activity in pooled HLM.
Figure 4.
Kinetic profiles for CDCA (a), propofol (b) and AZT (c) glucuronidation by pooled human liver microsomes (HLM) and individual UGTs enzymes; In each panel, the insert figure showed the corresponding Eadie–Hofstee plot. All experiments were performed in triplicate.
Table 2.
Kinetic parameters and RAF values of substrate glucuronidation by pooled HLM and individual expressed UGT enzyme (Mean±SD).
| Substrate | Protein source | Vmax (nmol/min/mg) | Km (μM) | CLint (μl/min/mg) | Model | RAF |
|---|---|---|---|---|---|---|
| CDCA | Pooled HLM | 0.13±0.003 | 14.32±1.16 | 8.86±0.74 | MM | 0.48 |
| UGT1A3 | 0.20±0.003 | 10.90±0.74 | 18.51±1.28 | MM | ||
| Propofol | Pooled HLM | 0.04±0.001 | 60.71±9.15 | 0.63±0.10 | MM | 0.49 |
| UGT1A9 | 0.06±0.002 | 45.91±5.91 | 1.28±0.17 | MM | ||
| AZT | Pooled HLM | 1.18±0.53 | 4359.0±341.4 | 0.27±0.02 | MM | 1.04 |
| UGT2B7 | 0.42±0.01 | 1604.0±121.0 | 0.26±0.02 | MM |
All experiments were performed in triplicate.
Note: MM: Michael–Menten model.
Activity correlation analysis
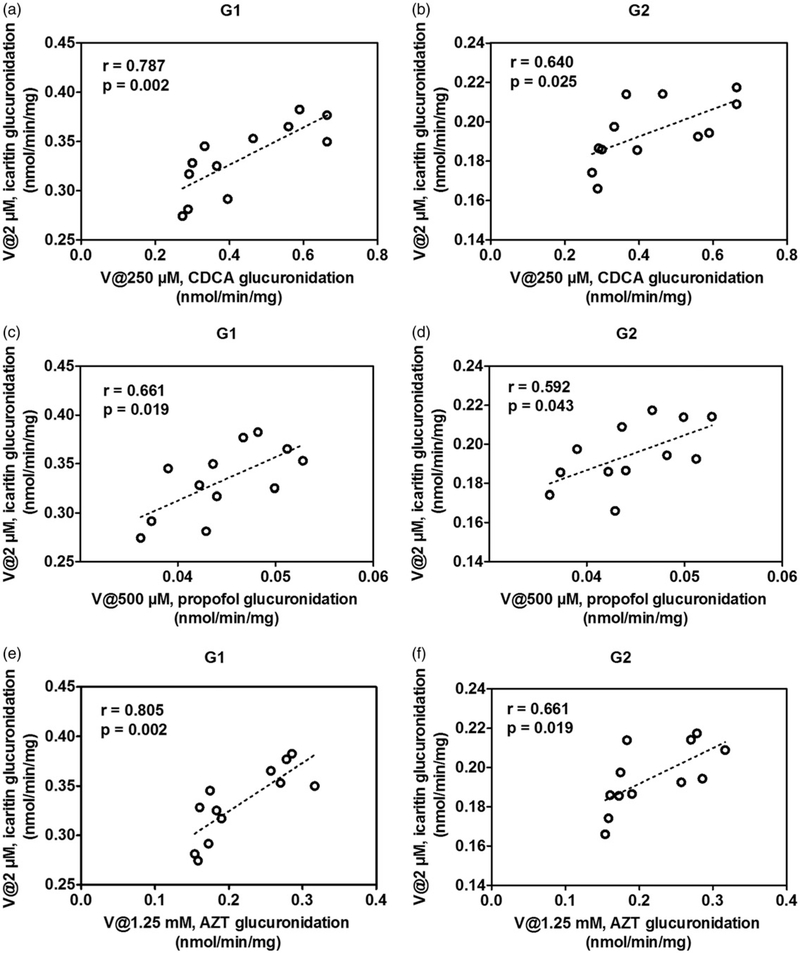
As mentioned, glucuronidation activity of CDCA in pooled HLM is a well-accepted functional marker for UGT1A3 (Li et al., 2016). Glucuronidation activities of individual HLMs (n=12) toward icaritin glucuronidation and CDCA glucuronidation were both determined. It was shown that icaritin 3-O-glucuronidation (G1) and 7-O-glucuronidation (G2) were strongly correlated with CDCA glucuronidation with correlation factors (r=0.787, p=0.002) and (r=0.640, p=0.025), respectively (Figure 5a and b). Similarly, icaritin 3-O-glucuronidation (G1) and 7-O-glucuronidation (G2) were significantly correlated with propofol glucuronidation (r=0.661, p=0.019) and (r=0.592, p=0.043), respectively (Figure 5c and d). In addition, icaritin glucuronidation (G1 and G2) was strongly correlated with AZT glucuronidation (r=0.805, p=0.002) and (r=0.661, p=0.019), respectively (Figure 5e and f). The result indicated that UGT1A3, 1A9 and 2B7 enzymes all played a critical role in icaritin glucuronidation and were the main expressed UGTs for icaritin glucuronidation.
Figure 5.
Correlation analysis between icaritin 3-O-glucuronidation and CDCA glucuronidation (a), icaritin 7-O-glucuronidation and CDCA glucuronidation (b) in a bank of individual human liver microsomes (n=12); Correlation analysis between icaritin 3-O-glucuronidation and propofol glucuronidation (c), icaritin 7-O-glucuronidation and propofol glucuronidation (d) in a bank of individual human liver microsomes (n=12); Correlation analysis between icaritin 3-O-glucuronidation and AZT glucuronidation (e), icaritin 7-O-glucuronidation and AZT glucuronidation (f) in a bank of individual human liver microsomes (n=12). All experiments were performed in triplicate.
Discussion
Icaritin is a natural flavonoid isolated from Herba Epimedii with multiple biological properties. It was shown that icaritin could be rapidly metabolized to phase II glucuronide conjugates and extensive glucuronidation conjugates were detected in rat urine samples after oral administration of individual prenylflavonoids or Herba Epimedii extracts (Chang et al., 2012; Cui et al., 2014; Jin et al., 2013; Zhao et al., 2010). Hence, it was speculated that glucuronidation reaction played an important role for icaritin clearance and determining its body exposure (bioavailability).
Our results demonstrated that icaritin could be rapidly glucuronidated in both pooled HLM and HIM in the presence of UDPGA, while three mono-glucuronides were formed (Figure 1b). The mono-glucuronides were characterized as 3-O-glucuronidation (G1), icaritin 7-O-glucuronidation (G2) and 5-O-glucuronidation (G3), respectively. Furthermore, strong evidence was provided that UGT1A3, 1A7, 1A8, 1A9 and 2B7 were the main contributors to glucuronidation of icaritin in the reaction phenotyping (Figure 3). Among all UGT enzymes, UGT1A3, 1A7, 1A8, 1A9 and 2B7 showed predominant activities towards icaritin (Figure 2). Icaritin 3-O-glucuronidation and icaritin 7-O-glucuronidation were both significantly correlated with CDCA glucuronidation (a functional marker of UGT1A3), propofol glucuronidation (a functional marker of UGT1A9) and AZT glucuronidation (a functional marker of UGT2B7) (Figure 5). Up to 92.6% of icaritin 3-O-glucuronidation in pooled HLM was attributed to UGT1A3, 1A9 and 2B7, basing on the RAF approach. Similarly, the contribution of UGT1A3, 1A9 and 2B7 for icaritin 7-O-glucuronidation in pooled HLM was over 60%. Although UGT1A7 and 1A8 also had high activities towards icaritin (Figure 3c), the authors did not determine their roles in glucuronidation because they are gastrointestinal enzymes and hardly found in the liver (Nakamura et al., 2008; Ohno & Nakajin, 2009).
The glucuronidation activity was obtained by kinetic profiling and modeling. Kinetic profiling required the determination of the rates of icaritin glucuronidation at a series of icaritin concentrations. The relative activities of different expressed UGT enzymes toward icaritin glucuronidation were evaluated by the derived CLint values. Use of CLint (=Vmax/Km) as an indicator of UGTs enzymes activity was advantageous, because (1) CLint represents the catalytic efficiency of the enzyme and is independent of the substrate concentration; (2) compared with other kinetic parameters such as Km and Vmax, CLint is more relevant in an attempt to predict clearance in vivo (Wu et al., 2013). Therefore, CLint values were used to determine icaritin glucuronidation activity of different expressed UGTs in this study.
It is also worthy to mention that the corresponding CLint value of G2 in HIM was 0.40 mL/min/mg, which is similar to that in pooled HLM with the CLint value of 0.35 mL/min/mg. These results demonstrated that human intestine also played important role in 7-O-glucuronidation (G2). This implied that icaritin may undergo a first-pass metabolism during the absorption process.
Characterization of icaritin glucuronidation assumed a great role in the understanding of its pharmacokinetics and bioavailability. Oral bioavailability is a major factor in determining the biological actions of icaritin in vivo following oral administration of the compound (Wu et al., 2011). This study suggested that the oral bioavailability of icaritin would be influenced by first-pass glucuronidation in the liver. Furthermore, UGT1A7 and 1A8 which were abundantly expressed in the intestine showed metabolic activities towards icaritin (Figure 3c). Thus, it was highly possible that intestinal glucuronidation had impact on the oral bioavailability. Moreover, the role of glucuronidation in determining the oral bioavailability of icaritin should not be underestimated.
Apart from the rapid glucuronidation of icaritin in human liver and intestine, kidney may also participate in icaritin glucuronidation in human body. Taking into account the fact that UGT1A9 displayed a high catalytic activity towards two icaritin glucuronides, and it also could be abundantly expressed in kidney (Knights et al., 2013). UGT1A9 may be the major contributor to the metabolic clearance of icaritin in human kidney. Considering that UGT1A9 and 2B7 are the main contributing UGT isoforms in icartin glucuronidation, it is readily conceivable that icaritin could be extensively glucuronidated after oral administration. Therefore, the activity and the excretion of these three icaritin glucuronides should be further investigated.
Taken together, icaritin could be metabolized by human UGTs and generates three icaritin glucuronides. Kinetic characterization and activity correlation analysis assays demonstrated that UGT1A3, 1A9 and 2B7 played important roles in glucuronidation of icaritin, while UGT1A7 and 1A8 are two major contributors to the formation of two icaritin glucuronides in HIM. In addition, the CLint values of icaritin glucuronidation generated in HIM indicated a first-pass metabolism in the intestinal border after oral administration, and the following glucuronidation should lead to a rapid elimination of icaritin from human body. Moreover, UGT1A3, 1A9 and 2B7 are highly polymorphic enzymes (Bhasker et al., 2010; Nagar & Blanchard, 2006). Since icaritin was predominantly metabolized by these three enzymes, a large pharmacokinetic variability was expected to exist among individuals with different UGT1A3, 1A9 and 2B7 genotypes.
In conclusion, icaritin 3-O-glucuronidation (G1) and icaritin 7-O-glucuronidation (G2) were efficient in pooled HLM with CLint (the intrinsic clearance) values of 0.80 and 0.35 mL/min/mg, respectively. Also, they were determined in HIM with CLint values of 0.27 and 0.40 mL/min/mg, respectively. In addition, UGT1A3, 1A7, 1A8, 1A9 and 2B7 showed significant metabolic activities toward icaritin. UGT1A3, 1A9 and 2B7 were the main enzymes with the highest activities. Furthermore, G1 and G2 were both significantly correlated with CDCA glucuronidation, propofol glucuronidation and AZT glucuronidation in a bank of individual HLMs (n=12). On the basis of RAF approach, it were estimated that UGT1A3, 1A9 and 2B7, respectively, contributed 37.5, 33.8 and 21.3% of icaritin 3-O-glucuronidation in pooled HLM. Similarly, UGT1A3, 1A9 and 2B7 contributed 34.3, 20.0 and 8.6% for icaritin 7-O-glucuronidation in pooled HLM, respectively. Overall, UGT1A3, 1A7, 1A8, 1A9 and 2B7 were the main contributors to the glucuronidation of icaritin.
Acknowledgments
Declaration of interest
This work was supported by Major Project for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 81220108028). All the authors report no declarations of interest.
References
- Bhasker CR, McKinnon W, Stone A, et al. (2010). Genetic polymorphism of UDP-glucuronosyltransferase 2B7 (UGT2B7) at amino acid 268: ethnic diversity of alleles and potential clinical significance. Pharmacogenetics 10:679–85. [DOI] [PubMed] [Google Scholar]
- Chang Q, Wang GN, Li Y, et al. (2012). Oral absorption and excretion of icaritin, an aglycone and also active metabolite of prenylflavonoids from the Chinese medicine Herba Epimedii in rats. Phytomedicine 19: 1024–8. [DOI] [PubMed] [Google Scholar]
- Chen SH, Lei M, Xie XH, et al. (2013). PLGA/TCP composite scaffold incorporating bioactive phytomolecule icaritin for enhancement of bone defect repair in rabbits. Acta Biomater 9:6711–22. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhao YH, Jia XB, et al. (2008). Intestinal absorption mechanisms of prenylated flavonoids present in the heat-processed Epimedium koreanum Nakai (Yin Yanghuo). Pharm Res-Dordr 25: 2190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Xu F, Jiang J, et al. (2014). Identification of metabolites of epimedin A in rats using UPLC/Q–TOF–MS. Chromatographia 77: 1223–34. [Google Scholar]
- Evans WE, Relling MV. (1999). Pharmacogenomics: translating functional genomics into rational therapeutics. Science 5439:487–91. [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Tracy TS. (2002). Atypical kinetic profiles in drug metabolism reactions. Drug Metab Dispos 30:355–62. [DOI] [PubMed] [Google Scholar]
- Jiang MC, Chen XH, Zhao X, et al. (2016). Involvement of IGF-1 receptor signaling pathway in the neuroprotective effects of Icaritin against MPP(+)-induced toxicity in MES23.5 cells. Eur J Pharmacol 786:53–9. [DOI] [PubMed] [Google Scholar]
- Jiang W, Xu B, Wu B, et al. (2012). UDP-glucuronosyltransferase (UGT) 1A9-overexpressing HeLa cells is an appropriate tool to delineate the kinetic interplay between breast cancer resistance protein (BRCP) and UGT and to rapidly identify the glucuronide substrates of BCRP. Drug Metab Dispos 40:336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wu CS, Zhang JL, et al. (2013). A new strategy for the discovery of epimedium metabolites using high-performance liquid chromatography with high resolution mass spectrometry. Anal Chim Acta 768: 111–17. [DOI] [PubMed] [Google Scholar]
- Knights KM, Rowland A, Miners JO. (2013). Renal drug metabolism in humans: the potential for drug–endobiotic interactions involving cytochrome P450 (CYP) and UDPglucuronosyltransferase (UGT). Br J Clin Pharmacol 76:587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Liu Y, Wu H, et al. (2016). The role of icaritin in regulating Foxp3/IL17a balance in systemic lupus erythematosus and its effects on the treatment of MRL/lpr mice. Clin Immunol 162:74–83. [DOI] [PubMed] [Google Scholar]
- Li F, Wang S, Lu D, et al. (2016). Identification of UDP-glucuronosyltransferases 1A1, 1A3 and 2B15 as the main contributors to glucuronidation of bakuchiol, a natural biologically active compound. Xenobiotica 47:365–79. [DOI] [PubMed] [Google Scholar]
- Li J, Liu P, Zhang R, et al. (2011). Icaritin induces cell death in activated hepatic stellate cells through mitochondrial activated apoptosis and ameliorates the development of liver fibrosis in rats. J Ethnopharmacol 137:714–23. [DOI] [PubMed] [Google Scholar]
- Liu H, Wu Z, Ma Z, et al. (2014). Glucuronidation of macelignan by human liver microsomes and expressed UGT enzymes: identification of UGT1A1 and 2B7 as the main contributing enzymes. Biopharm Drug Dispos 35:513–24. [DOI] [PubMed] [Google Scholar]
- Lu D, Ma Z, Zhang T, et al. (2016). Metabolism of the anthelmintic drug niclosamide by cytochrome P450 enzymes and UDP-glucuronosyltransferases: metabolite elucidation and main contributions from CYP1A2 and UGT1A1. Xenobiotica 46:1–13. [DOI] [PubMed] [Google Scholar]
- Lu D, Liu H, Ye W, et al. (2016). Structure- and isoformspecific glucuronidation of six curcumin analogs. Xenobiotica 47: 304–13. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, et al. (2005). Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–85. [DOI] [PubMed] [Google Scholar]
- Ma H, He X, Yang Y, et al. (2011). The genus Epimedium: an ethnopharmacological and phytochemical review. J Ethnopharmacol 134:519–41. [DOI] [PubMed] [Google Scholar]
- Nagar S, Blanchard RL. (2006). Pharmacogenetics of uridine diphosphoglucuronosyltransferase (UGT) 1A family members and its role in patient response to irinotecan. Drug Metab Rev 38:393–409. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nakajima M, Yamanaka H, et al. (2008). Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos 36:1461–4. [DOI] [PubMed] [Google Scholar]
- Ohno S, Nakajin S. (2009). Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos 37:32–40. [DOI] [PubMed] [Google Scholar]
- Peng S, Zhang G, Zhang BT, et al. (2013). The beneficial effect of icaritin on osteoporotic bone is dependent on the treatment initiation timing in adult ovariectomized rats. Bone 55:230–40. [DOI] [PubMed] [Google Scholar]
- Qian Q, Li SL, Sun E, et al. (2012). Metabolite profiles of icariin in rat plasma by ultra-fast liquid chromatography coupled to triple-quadrupole/time-of-flight mass spectrometry. J Pharm Biomed Anal 66:392–8. [DOI] [PubMed] [Google Scholar]
- Ritter JK. (2000). Roles of glucuronidation and UDP-glucuronosyltransferases in xenobiotic bioactivation reactions. Chem Biol Interact 129:171–93. [DOI] [PubMed] [Google Scholar]
- Sun F, Indran IR, Zhang ZW, et al. (2015). A novel prostate cancer therapeutic strategy using icaritin-activated arylhydrocarbon-receptor to co-target androgen receptor and its splice variants. Carcinogenesis 36:757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Wang H, Liu H, et al. (2014). Glucuronidation of capsaicin by liver microsomes and expressed UGT enzymes: reaction kinetics, contribution of individual enzymes and marked species differences. Expert Opin Drug Metab Toxicol 10:1325–36. [DOI] [PubMed] [Google Scholar]
- Sun H, Zhou X, Zhang X, et al. (2015). Decreased expression of multidrug resistance-associated protein 4 (MRP4/ABCC4) leads to reduced glucuronidation of flavonoids in UGT1A1-overexpressing HeLa cells: the role of futile recycling. J Agric Food Chem 63: 6001–8. [DOI] [PubMed] [Google Scholar]
- Troberg J, Jarvinen E, Ge GB, et al. (2016). UGT1A10 is a high activity and important extrahepatic enzyme: why has its role in intestinal glucuronidation been frequently underestimated? Mol Pharm. [Epub ahead of Print]. DOI: 10.1021/acs.molpharmaceut.6b00852. [DOI] [PubMed] [Google Scholar]
- Wang C, Wu C, Zhang J, et al. (2015). Systematic considerations for a multicomponent pharmacokinetic study of Epimedii wushanensis herba: From method establishment to pharmacokinetic marker selection. Phytomedicine 22:487–97. [DOI] [PubMed] [Google Scholar]
- Wu B, Kulkarni K, Basu S, et al. (2011). First-pass metabolism via UDPglucuronosyl transferase: a barrier to oral bioavailability of phenolics. J Pharm Sci 100:3655–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, et al. (2013). Quantitative prediction of glucuronidation in humans using the in vitro- in vivo extrapolation approach. Curr Top Med Chem 13:1343–52. [DOI] [PubMed] [Google Scholar]
- Zhao H, Fan M, Fan L, et al. (2010). Liquid chromatography-tandem mass spectrometry analysis of metabolites in rats after administration of prenylflavonoids from Epimediums. J Chromatogr B Analyt Technol Biomed Life Sci 878:1113–24. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ma YH, Zhou Z, et al. (2015). Intestinal absorption and metabolism of epimedium flavonoids in osteoporosis rats. Drug Metab Dispos 43:1590–600. [DOI] [PubMed] [Google Scholar]