Abstract
Xenobiotics are encountered by humans on a daily basis and include drugs, environmental pollutants, cosmetics, and even components of the diet. These chemicals undergo metabolism and detoxication to produce numerous metabolites, some of which have the potential to cause unintended effects such as toxicity. They can also block the action of enzymes or receptors used for endogenous metabolism or affect the efficacy and/or bioavailability of a coadministered drug. Therefore, it is essential to determine the full metabolic effects that these chemicals have on the body. Metabolomics, the comprehensive analysis of small molecules in a biofluid, can reveal biologically relevant perturbations that result from xenobiotic exposure. This review discusses the impact that genetic, environmental, and gut microflora variation has on the metabolome, and how these variables may interact, positively and negatively, with xenobiotic metabolism.
Keywords: pharmacometabolomics, gut microflora, interindividual variation, metabotype, UPLC, mass spectrometry
INTRODUCTION
Xenobiotics are foreign compounds that include not only drugs but also environmental pollutants, dietary supplements, and food additives. Human exposure to xenobiotics is pervasive; in a human lifetime, one might be exposed to 1–3 million xenobiotics (1). These compounds can be toxic or harmless, but nonetheless they are treated by the body as foreign. They are metabolized and ultimately eliminated through the urine, bile, and feces. Xenobiotics can be eliminated unchanged, but the vast majority utilize endogenous mechanisms such as enzymatic functionalization and/or conjugation reactions that facilitate their elimination, and they use processes that are also involved in the metabolism and transport of endogenous compounds such as bilirubin, lipids, and steroids. Thus, it is important to have a comprehensive knowledge of in vivo xenobiotic metabolism so that potential problems such as the generation of reactive metabolites or bioavailability issues when coadministering a drug can be ascertained. Metabolomics—the unbiased global survey of low-molecular-weight molecules or metabolites in a biofluid, cell, tissue, organ, or organism— represents an ideal solution for understanding and measuring the impact of xenobiotic exposure on a biological system. The term metabolome was first used in 1998 (2) and has been defined since as “the set of metabolites synthesized by a biological system” (3, p. 155); it encompasses all the small metabolites present in a particular biofluid (urine, blood, sebum, cerebral spinal fluid, saliva), cell, or tissue. As metabolites are the ultimate downstream products of genomic, transcriptomic, and/or proteomic perturbations, changes in metabolite concentration and/or flux can reveal biologically relevant changes to the system.
MANIPULATION OF THE METABOLOME
Genetic and Environmental Influences on the Metabolome
The metabolome can vary among individuals owing to numerous genetic and environmental factors. Environmental influences include diet, stress, medication, lifestyle, and disease. Genetic variation includes gender, epigenetics, and polymorphisms in genes encoding xenobiotic-metabolizing components such as Phase I and II enzymes, transporters, receptors, and ion channels. Age is also another host factor that can have physiological effects and thus affect xenobiotic metabolism and elimination. The gut microflora or microbiome of an individual represents yet another source of extragenomic variation. The combination of all these factors contributes to interindividual differences, but the interplay between genetic variation and environmental exposure can further confound results. For example, environmental exposures and disease can induce epigenetic changes (DNA methylation, histone modification) that potentially affect drug-metabolizing enzyme activity and capacity; these effects, in turn, can influence the efficacy and toxicity of a drug among individuals (4).
Genetic Variation
Genetic variation, although a major factor in defining the metabolomes of various populations, can also be masked by environmental influences. Recent large-scale human population studies have illustrated how genetic and environmental differences can impact the metabolome. Twenty-four-hour urine samples collected from 17 distinct populations in Japan, China, the United States, and the United Kingdom were analyzed by nuclear magnetic resonance (NMR) spectroscopy-based metabolomics, and the analysis revealed that geographic differences were a stronger influence than that of gender. Environmental pressure was also seen among the metabolic phenotypes of Japanese living in Japan and Japanese living in the United States. These two populations were well differentiated even though they were genetically similar, whereas the populations from the United States and the United Kingdom had similar metabolomes (5). Therefore, environmental factors such as lifestyle and diet have large effects on the metabolome and may even overshadow genetic inputs.
Genetic polymorphisms in key drug-metabolizing enzymes can influence the route of metabolism, and ultimately the bioavailability, efficacy (in the case of drugs), and toxicity of xenobiotics. Phase I metabolizing enzymes such as the cytochrome P450 (CYP) superfamily add functional groups that allow for direct excretion or the addition of conjugates that render the compounds more hydrophilic. CYPs are regulated by nuclear receptors, including pregnane X receptor (PXR), constitutive androstane receptor (CAR), peroxisome proliferator-activated receptor α (PPARα), and the aryl hydrocarbon receptor (AHR). There is a wide range of variation in the expression of the CYP enzymes and nuclear receptors among individuals; this is due not only to genetic polymorphisms but also to differences that result from age, gender (progesterone can induce CYP3A4 in women), body weight, and disease (liver diseases in particular can affect the capacity of a drug-metabolizing enzyme). The Phase II conjugating enzymes uridine 5’-diphospho-glucuronosyltransferases (UGTs), sulfotransferases, N-acetyltransferases, and glutathione 5-transferases are also subject to genetic polymorphisms, some of which cause debilitating diseases. For example, a UGT polymorphism involving the UGT1A1*28 allele has been linked to Gilbert’s syndrome (6), in which UGT1A1 has much lower activity, and subjects may develop hyperbilirubinemia owing to lack of conjugation and elimination of bilirubin. Xenobiotic metabolism can also be affected by other chemicals in tobacco smoke (7), alcohol (8), and industrial pollutants [2,3,7,8-tetrachlorodibenzo-p-dioxin activates AHR (9)], and, of course, by coadministration of pharmaceutical drugs. St. John’s Wort, a dietary supplement for the treatment of mild depression, is an agonist for PXR, which induces the expression of CYP3A4. Thus, when St. John’s Wort is coadministered with other drugs such as digoxin (10) and oral contraceptives (estrogen and progestin) (11), a marked decrease in the plasma concentrations of these drugs is seen, resulting in lower efficacy. Another interference can come from dietary grapefruit ingestion, which inhibits drug-metabolizing enzymes such as CYP3A4 and drug transporters (12). Grapefruit-drug interactions have been seen with antihypertensives, antimicrobials, benzodiazepines, antihistamines, statins, and chemotherapeutics (13). Therefore, it is important to establish the exact metabolic pathway and mechanisms of these xenobiotics to determine the metabolites produced and their effects on the metabolome.
The Microbiome and Metabolome
The metabolome of an organism is also influenced by the symbiotic gut microflora or microbiome. The metabolome of an individual can contain metabolites that are formed through the actions of the gut microbiota, and metabolism by these microbes may directly affect the metabolome of the host. Hippurate and phenylacetylglycine, for example, are seen in the urinary metabolome and are formed from the microbial breakdown of larger dietary phenols and phenylalanine, respectively. They generally reflect small disturbances to the host’s environmental conditions (14). The gut microflora have been associated with diseases such as inflammatory bowel disease (15), obesity (16), and diabetes (17). In humans, gut microflora influence immunity and anaerobic metabolism of peptides and proteins, are a defense against pathogens, and influence the development of intestinal microvilli of the organism (18).
Most importantly, with respect to metabolomics, gut microflora are involved in the metabolism of xenobiotics. Dehydroxylation, decarboxylation, dealkylation, dehalogenation, and deamination reactions have been reported as gut microflora–mediated reactions (19). They can influence the xenobiotic metabolite pool among individuals, and this influence may have major consequences for toxicity. Gut microflora stability itself can be affected by xenobiotics, in particular digoxin (20), which increases susceptibility to enteric infections (21). Antibiotic treatment in particular disturbs gut microflora equilibrium and affects many metabolic pathways, such as those involved in bile acid synthesis and steroid metabolism (22, 23). Other xenobiotics including those in dark chocolate (21), pomegranate by-products (24), and probiotics (25) have also been shown to modulate the gut microflora environment.
Phase I and II xenobiotic metabolism is influenced by the gut microflora. p-Cresol sulfate, phenyl sulfate, and indoxyl sulfate are bacterial metabolites of tyrosine and have been observed to be elevated by the action of gut microbes (26, 27). Given that sulfation is a key element of Phase II drug metabolism, this also has implications for xenobiotic elimination. Furthermore, some microbial species can produce xenobiotics, requiring further metabolism by the host by CYP enzymes (19). Lhoste et al. (28) reported an example of this in which germ-free and human microflora–inoculated rats had different levels of UGT, glutathione 5-transferase, and CYP2C11 enzyme induction when administered catechins. Xenobiotic metabolism in germ-free or conventionally raised mice also showed different metabolism of barbiturates owing to gut microflora-influenced liver expression of CAR and PXR (29). Considering the degree of contribution of the microbiome to the metabolome and the effects of genetic and environmental stimuli on both, gut flora metabolism adds a further dimension of complexity to the host’s overall metabolome and an extra source of interindividual variation.
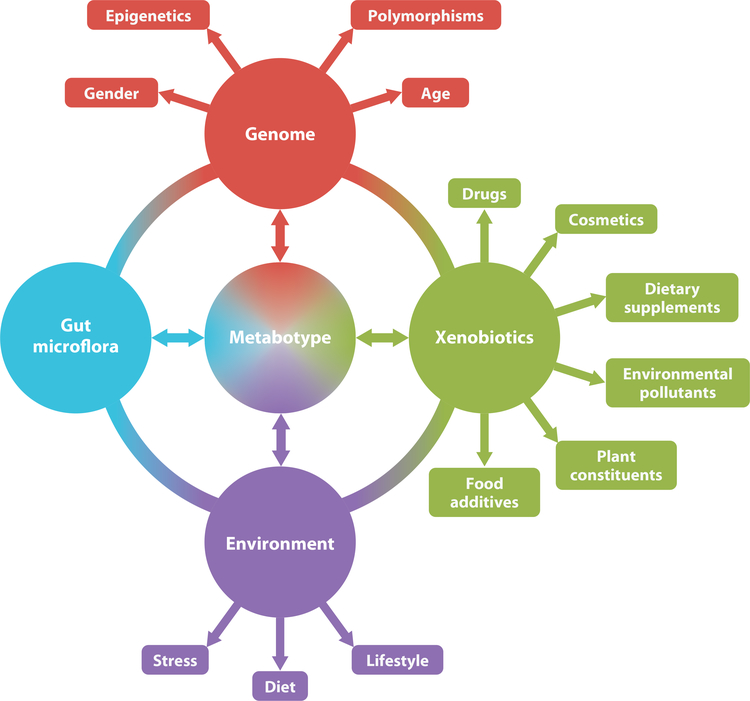
The concept of a metabotype encompasses all the genetic, environmental, and gut microflora modifications that are not necessarily readily observable, and it gives each individual a defining metabolomic fingerprint. The metabotype idea was first conceived and defined as “a probabilistic multiparametric description of an organism in a given physiological state based on analysis of its cell types, biofluids or tissues” (30, p. 173). As outlined in Figure 1, genetic and environmental factors can affect each other and give rise to interindividual variation and thus a unique metabotype. If one wishes to observe the effect of a specific intervention on an organism, the metabotype is an important consideration, in particular during drug development and in defining the drug’s metabolic fate. Patient stratification in clinical trials may start to rely more on metabotypes so that a population of responders/nonresponders can be defined; this definition could result in greater success in drug development by simultaneously considering environmental as well as genomic factors.
Figure 1.
Interindividual genomic, environmental, and gut microflora variation can contribute to an individual-specific metabotype or metabolomic fingerprint. Each of these factors can influence the others and determine the outcome of the metabotype. Conversely, the individual’s metabolome can affect each one of the factors.
Analysis of the Metabolome
Metabolomics was developed to identify and quantitate perturbations in the metabolome caused by genetic or environmental pressures. The analytical platforms used for metabolomics have been discussed extensively elsewhere (31–33). In brief, they include NMR spectroscopy and mass spectrometry (MS) coupled to chemometric or multivariate data analysis. No single platform can capture the whole metabolome owing to the different physical properties of metabolites, so the sample type and chemical constituents to be measured determine which system is optimal. Numerous NMR spectroscopic methods have been applied in metabolomics analysis, including magic-angle spinning (34); pulse sequences to optimize metabolite recovery, such as the Carr- Purcell-Meiboom-Gill spin echo sequence, which attenuates broad protein and lipoprotein signals (35); and the use of various nuclides such as 1H, 19F, 13C, and 31P. For sample introduction onto the mass spectrometer, the instrument can be connected to gas chromatography (GC), capillary electrophoresis (CE), or liquid chromatography (LC) systems. Ultraperformance liquid chromatography (UPLC) is the LC system of choice, preferred over the standard high-performance liquid chromatography (HPLC) system. When combined with orthogonal quadrupole time-of-flight (QTOF) MS, UPLC provides the advantage of high peak resolution with a lower limit of detection for ions and accurate mass determination (32). Recent advances in GC-MS technology for metabolomics analysis include the GCxGC-TOF-MS system, which allows for a much more complex sample analysis that can detect thousands more peaks. It uses two orthogonal separation phases, expanding the chromatographic plane and thus creating additional peak capacity in which peaks can be resolved. This setup enhances resolution and reduces the problem of coeluting peaks (36). Accurate quantitation can then be carried out by triple-quadrupole MS through multiple reaction monitoring to verify the concentration of the biomarker in each sample.
The most common chemometric techniques for data analysis include dimension-reduction methods such as principal components analysis, projection to latent structures discriminant analysis (PLS-DA), and orthogonal projection to latent structures. These methods are useful for revealing any systematic variation in the data and for finding patterns or groupings. As a complementary approach, the machine-learning algorithm Random Forests has been implemented in some metabolomics studies (37–40). This method is particularly superior for handling high-dimension data and provides a robust measurement of misclassification error (32). Another advancement in data analysis tools for metabolomics is the release of XCMS Online (https://xcmsonline.scripps.edu), which is a user-friendly program allowing the processing and analyzing of MS data. New innovative technologies and data processing techniques as well as enhancements to databases and data analysis methods are constantly under development to further optimize metabolomics as a powerful and essential analytical technique that can be applied in most academic settings. In addition, The Human Metabolome Database (http://www.hmdb.ca) and the METLIN Metabolite Database (http://metlin.scripps.edu) are of great value for interpretation of metabolomics data and metabolite identification.
APPLICATIONS OF METABOLOMICS IN XENOBIOTIC STUDIES
Knowing the metabolic fate of a xenobiotic will greatly aid in understanding its potential toxicity and also its mechanism of toxicity. Global metabolomics approaches can determine changes to metabolic pathways that may not be seen through normal, targeted biochemical assays or may not be present owing to the time delay from gene product to metabolic product. Multivariate data analysis and LC-MS were first combined for detection of xenobiotic metabolites by Plumb et al. (41). Since then, UPLC-MS-based metabolomics in particular have been applied successfully to numerous xenobiotic studies (see Table 1) and have revealed novel metabolites and pathways (42–52). Many of these novel metabolites were discovered for xenobiotics that are used by a large percentage of the population. Thus, metabolomics has expanded the knowledge surrounding these xenobiotics and led the way to understanding their metabolism, side effects, and possible health consequences. A good example of the power of using metabolomics for xenobiotic research involves acetaminophen (APAP) metabolism. Although this over-the-counter analgesic has been available for more than 50 years, three new metabolites of APAP—S-(5-acetylamino-2-hydroxyphenyl)mercaptopyruvic acid; 3,3′-biacetaminophen; and a benzothiazine compound—were recently discovered, which was surprising considering the wealth of knowledge surrounding APAP and its metabolism (46). Recently published studies discussed below further demonstrate the value and power of UPLC-MS-based metabolomics for xenobiotic and toxicology research.
Table 1.
Successful application of UPLC-MS-based metabolomics in xenobiotic studies
| Number of metabolites discovered |
|||||
|---|---|---|---|---|---|
| Xenobiotic | Animal model | Before | After | Key finding | Reference(s) |
| Arecoline, arecaidine (areca nut) | Wild-type mice | 4 | 11 | Novel pathways for arecoline and arecaidine metabolism | (47) |
| Arecoline 1-oxide | Wild-type mice | N/A | 13 | New metabolic pathways not previously recorded | (48) |
| PhIP | Wild-type,CYP1A2-humanized, and Cyp1a2-null mice | 9 | 17 | Importance of CYP1A2 in PhIP metabolism | (49, 84) |
| Fenofibrate | Sprague-Dawley rats, cynomolgus monkey | 4 | 9 | New metabolic pathways of fenofibrate | (45, 66) |
| APAP | (1) Cyp2e1-null and wild-type mice, (2) Wild-type mice, [acetyl-2 H3 ] APAP or 2,3,5,6-[2H4] APAP | 7 | 10 | Advantage of using a deuterated compound to identify and validate xenobiotic metabolites | (46) |
| Aminoflavone | Wild-type, Cyp1a2-null, and CYP1A2-humanized mice | 1 | 13 | Main metabolite is N 5-hydroxylated; 3-hydroxylation is preferable in humans | (50) |
| thioTEPA | Wild-type mice, liver microsome incubations | 3 | 9 | New metabolic pathways of thioTEPA | (52) |
| Melatonin | Wild-type, Cyp1a2-null, and CYP1A2-humanized mice | 7 | 14 | No interspecies difference with regard to CYP1A2-mediated metabolism | (42) |
| Cyclophosphamide,ifosfamide | Wild-type mice | 18 | 23 | Completion of metabolic map to S-carboxymethyl-cysteine and thiodiglycolic acid | (43) |
Abbreviations: APAP, acetaminophen; PhIP, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; thio TEPA, N,N′,N″-triethylenethiophowsphoramide; UPLC-MS, ultraperformance liquid chromatography-mass spectrometry.
Cyclophosphamide and Ifosfamide
Cyclophosphamide (CP) and ifosfamide (IF) are isomeric prodrugs used in cancer chemotherapy. Both drugs undergo complex Phase I and II metabolism to numerous metabolites. However, treatment with IF is known to cause nephrotoxicity and neurotoxicity, whereas CP treatment does not. Selective IF toxicity is thought to result from the production of 2-chloroacetaldehyde. The latter is converted to 2-chloroacetic acid (CAA), which can react with cellular thiols to produce S-carboxymethylcysteine (SCMC) and thiodiglycolic acid (TDGA). Although it is possible that SCMC and TDGA can induce encephalopathy and mitochondrial dysfunction via IF dosing, there are no reports to suggest CP toxicity from SCMC and TDGA production.
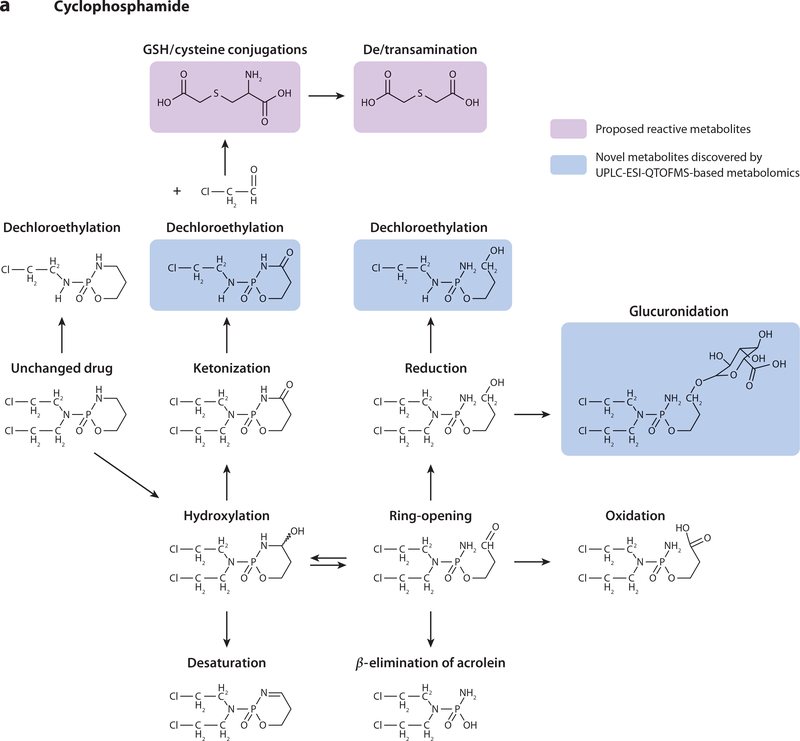
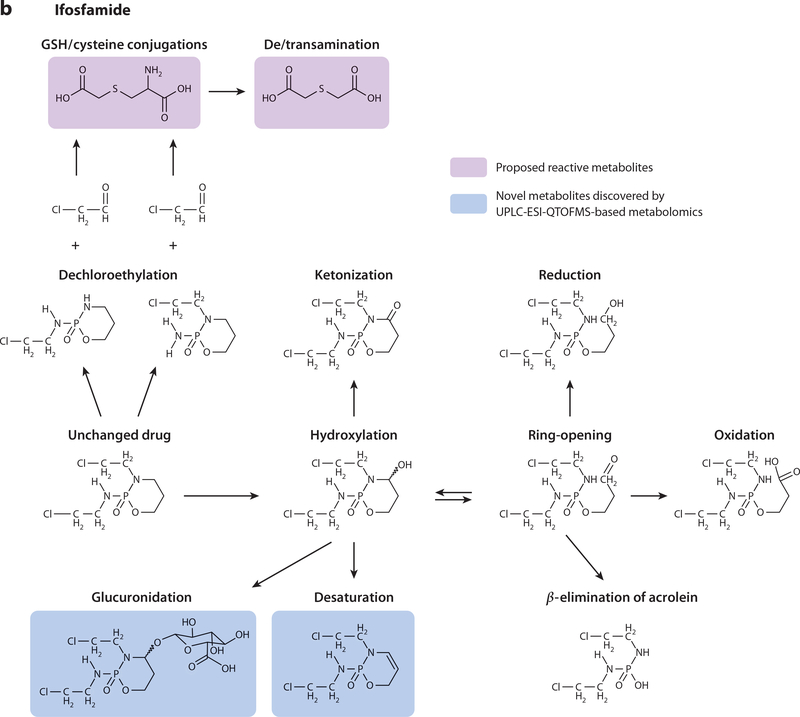
UPLC-ESI-QTOFMS-based metabolomics (i.e., metabolomics based on ultraperformance liquid chromatography–electrospray ionization–quadrupole time-of-flight mass spectrometry) was thus employed to perform a comprehensive comparative analysis of IF and CP metabolism. This was to determine whether IF or CP produced additional metabolites that could contribute to the observed pathologies. Twenty-four-hour urine samples were collected and analyzed from C57BL/6 mice dosed with IF (50 mg kg–1) or CP (50 mg kg–1) (43). Multivariate data analysis, specifically orthogonal projection to latent structures models, revealed 12 IF and 11 CP urinary metabolites, five of which were novel. A range of metabolic reactions produced the 23 metabolites, including dechloroethylation, hydroxylation, ketonization, dehydroxylation, alkylation, ring-opening, and conjugation reactions (Figure 2). Metabolomics revealed that one of the differences observed between the two prodrugs was increased excretion of CP ring-opened and ketonized metabolites compared with IF. In addition, the dechloroethylation reaction produced higher concentrations of IF metabolites than CP metabolites (twofold). CAA was also produced from the dechloroethylation reactions, which, in turn, produced SCMC and TDGA. SCMC and TDGA excretion was quantitated by triple-quadrupole MS, revealing that SCMC urinary excretion increased 32-fold and 44-fold above endogenous levels after administration of IF and CP. TDGA urinary excretion was increased by 14-fold and 17-fold after treatment with IF and CP, respectively. There were no significant differences in SCMC and TDGA excretion between the two prodrugs, which verified that the SCMC and TDGA metabolites did not confer toxicity with CP administration.
Figure 2.
Metabolic reactions of (a) cyclophosphamide and (b) ifosfamide in mouse urine. Boxed structures in pink represent proposed reactive metabolites; boxed structures in blue represent novel metabolites discovered by UPLC-ESI-QTOFMS-based metabolomics (i.e., metabolomics based on ultraperformance liquid chromatography–electrospray ionization–quadrupole time-of-flight mass spectrometry). Abbreviation: GSH, glutathione.
Hence, the results from this study (43) instead suggested that the toxic nature of IF could actually be derived from the CAA metabolite itself. The relative excretion of the dechloroethylated metabolites was much greater from IF dosing compared with CP dosing, signifying that CAA was produced in higher quantities upon IF administration, but because of the unstable nature of CAA, it was not quantified. As there was no significant difference between IF and CP with regard to SCMC and TDGA production, another mechanism of reaction may exist. Indeed, others observed a decrease in IF-induced nephropathy and glutathione depletion when IF was combined with N-acetylcysteine administration (53). In theory, this experiment would have produced N-acetyl SCMC, which would have blocked the production of TDGA, thus implying that TDGA is in fact the toxic metabolite. Another mechanism of IF toxicity could have resulted from favorable glutathione versus cysteine conjugation of CAA, which may have led to glutathione depletion. This then directs further studies on the differential metabolism and toxicity of the prodrugs to focus on the potential of CAA as a contributing toxicity factor.
Fenofibrate
Fibrate drugs are used for treatment of dyslipidemia resulting from increasing fatty acid β-oxidation; lower serum triglycerides result in reduced insulin resistance (54). Fenofibrate is well tolerated, but some adverse effects have been observed in rodent model systems, predominantly increased oxidative stress and myotoxicity (55–57). In humans, fenofibrate increases serum creatinine levels (58) and is associated with renal disorders (59); these scenarios are infrequent, but the toxicology of fenofibrate in humans is a concern. Fibrates are agonists of the nuclear receptor PPARα that control expression of genes involved in lipid oxidation, gluconeogenesis, and amino acid metabolism (60, 61). Chronic dosing of fibrates to rats, which activates PPARα, can result in hepatotoxicity and hepatocarcinogenesis, but the same is not seen in humans and nonhuman primates (61, 62). Fibrate metabolites may therefore contribute to the toxicity seen in rats. Three comprehensive UPLC-ESI-QTOFMS-based metabolomics studies were carried out to ascertain the full metabolic map of fenofibrate in different species. Previously, fenofibrate metabolites were reported in rats, guinea pigs, dogs, and humans. Fenofibric acid (FA) and reduced fenofibric acid (RFA) were seen in all species (63, 64), whereas fenofibric acid ester glucuronide (FAEG) and reduced fenofibric acid ester glucuronide (RFAEG) were seen in all species except dog (65). The metabolomics studies analyzed fenofibrate metabolism in cynomolgus monkeys (45), Sprague-Dawley rats (66), humans, and mice (39). The study in humans and mice aimed to specifically analyze the influence of fenofibrate on PPARα induction.
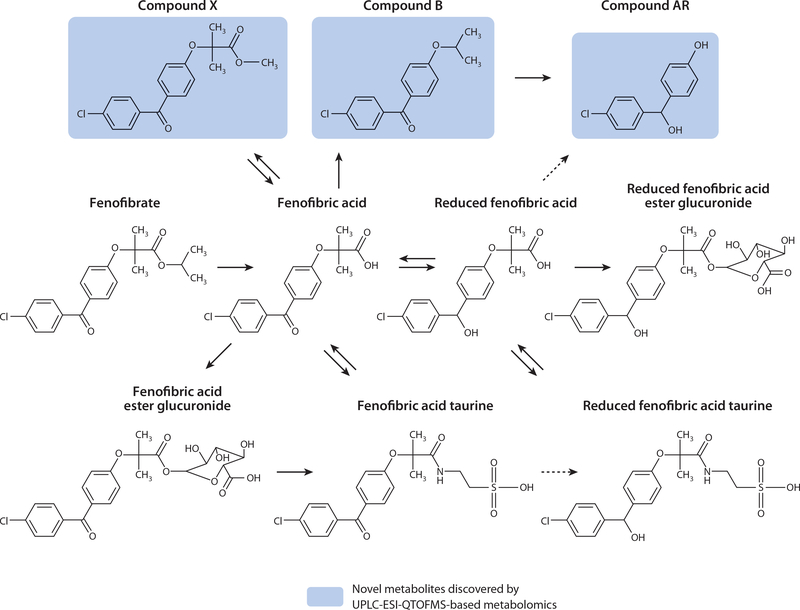
Sprague-Dawley rats were dosed twice daily for three days with 2,500 mg kg–1 fenofibrate (66). Predose urine and plasma samples were collected along with 12-h postdose samples. Sprague- Dawley rats were also used for the preparation of isolated hepatocytes. In brief, fenofibrate and eight fenofibrate metabolites were added to the hepatocyte cultures to determine the metabolic pathway of each metabolite. The cynomolgus monkeys were administered two different doses of fenofibrate, 30 mg kg–1 per day followed by 2,500 mg kg–1 per day, twice daily for 12 days, and urine was collected daily (45). A drug-free period of 4 weeks was allowed between the 30 mg kg–1 per day and the 2,500 mg kg–1 per day doses. Metabolomics analysis was carried out on the samples by supervised PLS-DA. The four previously identified fenofibrate metabolites were observed in monkey urine, and only three were seen in rat urine. Two novel taurine conjugates, fenofibric acid taurine (FAT) and reduced fenofibric acid taurine (RFAT), were observed in monkey urine, but only FAT was apparent in rat urine. Three further fenofibrate metabolites—named compounds AR, X, and B—were revealed through metabolomics analysis of the cynomolgus monkey urine but were not apparent in the rat data. Therefore, the metabolites not seen in rat urine (compounds AR, X, B, RFAT, and RFAEG) were considered to be possible metabolites, and LC-MS/MS was carried out on the ions of these specific metabolites from the rat plasma and hepatocyte incubation media. RFAT, RFAEG, and compounds X, B, and AR were seen in the hepatocyte media, and all of these except compound AR were seen in the plasma. The hepatocyte incubations also revealed that compound B was metabolized to compound AR. The data from this study yield a proposed metabolic map of fenofibrate (Figure 3).
Figure 3.
Proposed metabolism of fenofibrate after ultraperformance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOFMS)-based metabolomics analysis of dosed cynomolgus monkeys and rats. Items highlighted in blue are novel metabolites.
The PPARα-focused metabolomics study aimed to identify biomarkers of PPARα and increased fatty acid β-oxidation, and it involved a daily administration of 200 mg of fenofibrate to healthy human volunteers for 14 days (39). Twenty-four-hour urine samples and blood were collected at days 0, 7, and 14. Also, wild-type and Ppara-null mice were fed either a basal diet or a fenofibrate-enriched diet, and 24-h urines were collected after 7 days of dosing. The UPLC-ESI-QTOFMS analytical platform was used to analyze the human urine samples, and the data were analyzed by the machine-learning algorithm Random Forests. The most highly ranked urinary biomarkers that decreased after fenofibrate administration in humans included pantothenic acid, acetylcarnitine, propylcarnitine, isobutyrylcarnitine, (S)-(+)-2-methybutyrylcarnitine, and isovalerylcarnitine. Pantothenic acid and acetylcarnitine were quantitated from the wild-type mouse urine and were similarly depleted as in humans, but they were not depleted in Ppara-null mice. Therefore, metabolomics was able to identify biomarkers of PPARα-induced fatty acid β-oxidation and revealed how the fibrate drugs could affect lipid metabolism in both humans and mice. However, future studies are warranted in patient populations not restricted to healthy volunteers, in which fenofibrate is administered therapeutically. Studies in individuals with high levels of cholesterol and triglycerides will be important for identifying particular patients or metabotypes that, in particular, do not respond to the fenofibrate.
PHARMACOMETABOLOMICS
One of the more recent developments in metabolomics technology has been its use as a tool to predict drug efficacy and drug-induced side effects. This technique, termed pharmacometabolomics, has great potential to predict the response of an individual to drug therapy through knowledge of the metabotype. Through utilization of the metabotype, metabolic pathways that are disrupted in response to a treatment could aid significantly in drug development and result in greater success in clinical trials (67).
Pharmacometabolomics was first defined as “the prediction of the outcome of a drug or xenobiotic intervention in an individual based on a mathematical model of preintervention metabolite signatures” (68, p. 1073). The initial studies used NMR spectroscopy-based metabolomics to examine the toxic effects of galactosamine in rats. The results classified the rats into two distinct groups, responders and nonresponders, depending on the level of galactosamine-induced liver damage. The predose metabolomic profiles showed some discrimination between the outcomes of the two groups and suggested that information on individual responses to xenobiotics could be predicted from predose profiles.
A demonstration of the concept of pharmacometabolomics was undertaken on the basis of the hypothesis that drug-induced responses can be predicted from predose urinary metabolomic profiles without a priori knowledge of the genomic profile (68). Toxic-threshold levels of APAP were administered to rats, and the urinary metabolome was analyzed by NMR spectroscopy and PLS modeling. The incidence of drug-induced liver toxicity was also assessed. Clayton et al. (68) found that predose urinary metabolomic profiles could predict the ratio of postdose APAP glucuronide to APAP (unconjugated) in each urine sample. They also observed that most of the rats that had higher degrees of liver necrosis also had lower proportions of postdose urinary APAP sulfate. Predose and postdose profiles were obtained again for a similar follow-up study (69). Predose spectra were modeled in relation to drug metabolite excretion for detection of predose biomarkers of drug metabolites. The investigators observed that rats with high predose urinary levels of the gut-microbial metabolite p-cresol sulfate had low postdose urinary ratios of APAP sulfate to glucuronide, and vice versa. This was a consequence of competitive sulfation. These studies indicated that gut bacteria can influence the metabolome of the individual and that variation in gut microflora can directly influence drug-induced responses and the metabolic fate of drugs.
For proof of concept in humans, pharmacometabolomics was also used for prediction of APAP-induced liver injury from predose urine samples (70). Seventy-one volunteers were admitted as inpatients into a clinical research center and administered 1 g of APAP or placebo every 6 h for 7 days, and 24-h urine samples were collected. Some of the volunteers receiving APAP exhibited mild liver injury (2 × baseline elevation of alanine transaminase), but the predose urines could not be used to predict the alanine transaminase elevations. This is indicative of the obstacles that arise with analysis of human samples owing to large interindividual genetic and dietary variation. Perhaps larger study cohorts need to be carried out, or they need to restrict to distinct age, ethnicities, and/or genders.
Pharmacometabolomics has been adopted by many laboratories as part of their metabolomics and drug metabolism arsenal. For example, a novel strategy used pharmacometabolomics to “inform” a pharmacogenomics investigation (71). Pharmacogenomics examines the influence of genetic variation on drug response in patients, relating gene expression or single-nucleotide polymorphisms (SNPs) to drug efficacy or toxicity. This genetic variation could include polymorphisms in genes encoding CYP or UGT enzymes; these polymorphisms would also be evident by metabolomics as differences in drug metabolites formed. Therefore, with a combined pharmacometabolomics and pharmacogenomics approach, the metabolic and potential genetic causes for a differential response can be ascertained. Utilizing this strategy, Ji et al. (71) studied the efficacy of the selective serotonin reuptake inhibitors (SSRIs) citalopram and escitalopram in human patients. These drugs are used in the treatment of major depressive disorder, yet approximately 40% of the patients who are administered citalopram and escitalopram do not respond to treatment. The reason for this is unknown, and thus pharmacogenomics studies were carried out to identify potential polymorphisms in candidate genes. These studies, however, failed to produce biomarkers for the prediction of SSRI treatment outcome. To overcome these deficiencies, MS-based metabolomics was utilized to analyze urine from 40 individuals. Twenty of these were SSRI remitters [Quick Inventory of Depressive Symptomatology-Clinician Rated (QIDS-C) score ≤5 after 8 weeks of therapy], and 20 were nonremitters (QIDS-C score >5 after 8 weeks of therapy). There was no exclusion criteria set based on gender, race, or age, but these factors were not significantly associated with treatment outcome. Baseline metabolomic signatures showed that several metabolites in the nitrogen metabolism pathway, predominantly glycine, were negatively associated with treatment outcome (i.e., elevated glycine could be a marker for decreased SSRI response). Thus, glycine was focused upon as a target metabolite, and pharmacogenomics was employed to look for DNA sequence variations in genes that encoded enzymes involved in glycine synthesis and/or degradation. Only white non-Hispanics were included (~500 individuals), and a commonly occurring SNP (rs10975641) in the glycine dehydrogenase gene, associated with SSRI treatment outcomes, was identified. DNA samples from 1,926 patients were then genotyped for this SNP and were significantly associated with response in white non-Hispanics (1,245 individuals) and in all subjects when the data were adjusted for ethnicity. Thus, pharmacometabolomics and pharmacogenomics revealed a mechanism for different responses among individuals treated with these xenobiotics that could not previously be ascertained through pharmacogenomics alone. Therefore, the results of this study demonstrated a novel and innovative methodology for identifying biomarkers and could aid in designing individualized therapies for major depressive disorder. This study did show some variation among individuals, possibly caused by environmental or genetic interindividual variability. This finding suggests caution when designing studies to ensure that they are not underpowered (72) and to make sure that the combination of tools such as pharmacometabolomics and pharmacogenomics will confirm real changes in biological systems as observed here.
OVERCOMING THE COMPLICATIONS OF VARIATION IN HUMAN METABOLOMICS STUDIES
As mentioned previously, genetic variation can reveal the widest disparities between metabolomes, but the interplay of genetic variation and environmental intervention can confound results. Therefore, it is essential to control some of these factors in a metabolomics study to achieve optimal, interpretable results. The expression of Phase I and II drug-metabolizing enzymes is subject to considerable interindividual variation, resulting in altered pharmacokinetics, pharmacodynamics, and elimination. Many in vivo metabolomics experiments are carried out in mice; these experiments can be beneficial for a metabolomics study because breeding and cohousing of models can reduce differences in environmental bias. However, the genetic differences between mice and humans are large, and as such, they may not be useful as predictors of drug response (73). For the CYP enzymes, orthologs between mice and humans have been identified and have similar functions, but differences in activity have been seen in some studies (74–75). In addition, the regulation of the CYP enzymes occurs through the nuclear receptors AHR, PXR, CAR, and PPARα, and the enzymes’ expression and activation are also variable, which may contribute to confounding the results. A prime example in humans and mice involves species differences in the ligand specificity of human and mouse PXR: Rifampicin induces only human PXR, whereas pregnenolone-16a-carbonitrile induces only mouse PXR (76, 77).
To overcome this problem, transgenic humanized mouse models were generated to understand the human response to xenobiotics better. A number of these models were made: PXR-humanized mouse models were used to study black cohosh (74), rifaximin (78, 79), and all trans-retinoic acid (80), whereas PPAPα-humanized mice were used to study perfluorobutyrate (81), fenofibrate (73), and the PPARα ligand Wy-14,643 (82, 83). Aminoflavone (50), melatonin (42), and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) (49, 84) have been analyzed in a CYP1A2-humanized mouse model, and CYP2E1-humanized mice have been employed to study APAP toxicity (85). Double transgenic models were also generated and provide even better models for preclinical xenobiotic analysis. CYP3A4, one of the most important CYP enzymes in xenobiotic metabolism, is regulated by PXR. A PXR/Tg3A4 double-humanized mouse model was created to overcome the effects of species-selective ligand activation of PXR and hence activation of CYP3A4 (86). These mice have so far been used to study APAP (87) and black cohosh (74) and provide a comprehensive evaluation of xenobiotic metabolism that reflects metabolism in humans.
Other mechanisms and enzymes are, of course, involved in xenobiotic metabolism; UGT enzymes, for example, are involved in glucuronidation. Together with the CYPs, they account for more than 90% of xenobiotics cleared by the liver (6, 88). The UGTs encoded by the UGT1 locus are responsible for most xenobiotic clearance. A transgenic mouse model expressing a BAC-encoding human UGT1 locus has been generated (89). In addition, UGT-humanized mouse models have recently been produced; these carry commonly occurring human UGT1A1 polymorphisms, such as the UGT1A1*28 allele polymorphism (6).
The microbiome can also influence the metabolome and affect drug metabolism and toxicity. Gnotobiotic animals may be utilized to overcome this influence on the metabolome. These animals are germ-free at birth and can be colonized with different microbial species. Studies have used germ-free mice transplanted with human adult fecal microbiota (90) and microflora isolated from human baby feces (91). These systems essentially set up a human microbiome within a mouse and will provide insight into the role of human gut microflora in xenobiotic metabolism.
Aside from genetic or gut microflora variation, metabolomics studies carried out in human volunteers need to be designed in a way to reduce environmental variation. Some of the stipulations for these human studies are that they include healthy volunteers who are in the same age range, have normal body mass indices, and are nonsmokers with zero intake of pharmaceutical drugs or dietary supplements. These studies, however, are costly and are extremely difficult to control without admitting the volunteers to the clinic as inpatients for prolonged periods of time so that environmental factors such as alcohol, pharmaceuticals, and others can be restricted. This is well demonstrated in an aforementioned human APAP study (70) whereby volunteers were admitted into a clinic and placed on a standard whole-food diet for 14 days during APAP dosing. Thus, other external environmental factors could be controlled.
CONCLUSIONS
Metabolomics is a powerful analytical tool that can aid in understanding and revealing mechanisms that are involved in the metabolism of a xenobiotic. Through analysis of the metabolome, perturbations that occur as a result of the intervention can be ascertained. The metabolome consists of a pool of low-molecular-weight metabolites that are formed as a result of upstream genomic, transcriptomic, and proteomic processes. Interindividual variation influences this pool and can give rise to an individual metabotype. This metabotype can reveal the response of the individual to the xenobiotic and give clues to genetic and other influences. In the study of metabolomics in a human system, humanized and null-mouse models, either genetic or gnotobiotic, can overcome the complications of confounding environmental interactions and provide a stable model for analyzing a xenobiotic response.
SUMMARY POINTS.
The metabolome is subject to environmental, genomic, and gut microflora influences that affect xenobiotic metabolism. The metabotype encompasses all this variation and gives each organism a defining metabolic fingerprint. This will become an important tool for patient stratification in clinical trials, and the drug industry may find it useful for gaining better success in drug design and development.
UPLC-ESI-QTOFMS has been invaluable in the discovery of novel metabolites for numerous xenobiotics such as APAP, PhIP, and the areca nut alkaloids. This discovery of novel xenobiotic metabolites has contributed to the knowledge about the metabolic pathways of xenobiotics and the effects they may have on the biological system.
Pharmacometabolomics is a recent development in metabolomics technology and is highly effective for predicting drug efficacy and drug-induced side effects. This technique has been validated using human samples and developed further to lead pharmacogenomics investigations. It will be a valuable tool for future metabolomics research.
FUTURE ISSUES.
Do we have a good understanding of human metabolome variation to make metabolomics feasible for use in human studies? Large population studies such as the one carried out by Holmes et al. (4) can be valuable for assessing variation between groups and for designing patient stratification in clinical trials. More studies like these are needed to understand environmental and genomic influences on the metabolome.
One of the problems with moving from in vitro and in vivo models into human systems is overcoming the issue of genetic and environmental interindividual variation that can confound results. This difficulty can be partly resolved through the use of humanized mouse models for drug-metabolizing enzymes and gut microflora. These models provide insight into how an intervention such as a xenobiotic can affect the human gut microflora or a specific human enzyme/nuclear receptor and thus provide some clarity into the mechanisms and metabolic pathways involved.
The primary bottleneck in metabolomics research is identification of the metabolites shown to be biomarkers, especially from MS data. Identifying xenobiotic metabolites is relatively straightforward because they tend to be at concentrations much higher than concentrations of endogenous metabolites. However, identification of perturbations to the endogenous metabolome could be aided by better databases and a collaborative effort among metabolomics researchers. Unfortunately, owing to differences in instrumentation and methodologies among laboratories, this effort is somewhat restricted. Currently, identification using databases is through m/z and some fragmentation patterns, but these can also be different among laboratories when different collision energies for fragmentation are used. Therefore, standard operating procedures need to be set up and implemented to ensure homogeneity among laboratories. Reporting of data to a central database would also be of benefit to metabolomics researchers.
The development of new analytical tools for data analysis is also of importance. Innovative ways of maximizing recovery are welcomed; for example, the enhancement of GC-MS through the GCxGC-TOF-MS system increases data recovery immensely. The integration of platforms would be optimal for a comprehensive metabolomics study, allowing coverage of all metabolites.
Functionalization reaction: Phase I drug metabolism reaction that produces or uncovers functional groups that can be utilized for Phase II reactions
Conjugation reaction: Phase II drug metabolism reaction that conjugates a substrate to the functional group of a xenobiotic, producing a water-soluble metabolite that is excretable
Metabolome: the entire set of metabolites synthesized by a biological system, found in a biofluid
Microbiome: the gut microflora component of the host
NMR: nuclear magnetic resonance
CYP: cytochrome P450
PXR: pregnane X receptor
PPARα: peroxisome proliferator-activated receptor α
UGT: uridine 5′-diphospho-glucuronosyltransferase
Metabotype: the metabolic phenotype of the host that includes all the genetic, environmental, and gut microflora modifications in each individual
PLS-DA: projection to latent structures discriminant analysis
APAP: acetaminophen
CAA: 2-chloroacetic acid
SCMC: S-carboxymethylcysteine
TDGA: thiodiglycolic acid
UPLC-ESI-QTOFMS: Ultraperformance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry
Pharmacometabolomics: technique enabling the prediction of the outcome of a xenobiotic intervention based on knowledge of the predose metabolome
Humanized: describes an organism (typically a mouse) that expresses specific human genes or is transplanted with human cells/gut microflora in replacement for its own
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute Intramural Research Program.
Footnotes
DISCLOSURE STATEMENT
The authors have no affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Idle JR, Gonzalez FJ. 2007. Metabolomics. Cell Metab. 6:348–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver SG, Winson MK, Kell DB, Baganz F. 1998. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16:373–78 [DOI] [PubMed] [Google Scholar]
- 3.Fiehn O 2002. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 48:155–71 [PubMed] [Google Scholar]
- 4.Mathers JC, Strathdee G, Relton CL. 2010. Induction of epigenetic alterations by dietary and other environmental factors. Adv. Genet. 71:3–39 [DOI] [PubMed] [Google Scholar]
- 5.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, et al. 2008. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453:396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai H, Nguyen N, Peterkin V, Yang YS, Hotz K, et al. 2010. A humanized UGT1 mouse model expressing the UGT1A1*28 allele for assessing drug clearance by UGT1A1-dependent glucuronidation. Drug Metab. Dispos. 38:879–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantuck EJ, Kuntzman R, Conney AH. 1972. Decreased concentration of phenacetin in plasma of cigarette smokers. Science 175:1248–50 [DOI] [PubMed] [Google Scholar]
- 8.Ryan DE, Koop DR, Thomas PE, Coon MJ, Levin W. 1986. Evidence that isoniazid and ethanol induce the same microsomal cytochrome P-450 in rat liver, an isozyme homologous to rabbit liver cytochrome P-450 isozyme 3a. Arch. Biochem. Biophys. 246:633–44 [DOI] [PubMed] [Google Scholar]
- 9.Poland A, Knutson JC. 1982. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22:517–54 [DOI] [PubMed] [Google Scholar]
- 10.Johne A, Brockmoller J, Bauer S, Maurer A, Langheinrich M, Roots I. 1999. Pharmacokinetic interaction of digoxin with an herbal extract from St John’s Wort (Hypericum perforatum). Clin. Pharmacol. Ther. 66:338–45 [DOI] [PubMed] [Google Scholar]
- 11.Hall SD, Wang Z, Huang SM, Hamman MA, Vasavada N, et al. 2003. The interaction between St John’s Wort and an oral contraceptive. Clin. Pharmacol. Ther. 74:525–35 [DOI] [PubMed] [Google Scholar]
- 12.Hanley MJ, Cancalon P, Widmer WW, Greenblatt DJ. 2011. The effect of grapefruit juice on drug disposition. Expert Opin. Drug Metab. Toxicol. 7(3):267–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiani J, Imam SZ. 2007. Medicinal importance of grapefruit juice and its interaction with various drugs. Nutr. J. 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson CH, Patterson AD, Krausz KW, Lanz C, Kang DW, et al. 2011. Radiation metabolomics: 4. UPLC-ESI-QTOFMS-based metabolomics for urinary biomarker discovery in γ-irradiated rats. Radiat. Res. 175(4):473–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nell S, Suerbaum S, Josenhans C. 2010. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat. Rev. Microbiol. 8:564–77 [DOI] [PubMed] [Google Scholar]
- 16.Ley RE. 2010. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 26:5–11 [DOI] [PubMed] [Google Scholar]
- 17.Musso G, Gambino R, Cassader M. 2011. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 62:361–80 [DOI] [PubMed] [Google Scholar]
- 18.Nicholson JK, Holmes E, Wilson ID. 2005. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3:431–38 [DOI] [PubMed] [Google Scholar]
- 19.Wilson ID, Nicholson JK. 2009. The role of gut microbiota in drug response. Curr. Pharm. Des. 15:1519–23 [DOI] [PubMed] [Google Scholar]
- 20.Lindenbaum J, Rund DG, Butler VP Jr, Tse-Eng D, Saha JR. 1981. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N. Engl. J. Med. 305:789–94 [DOI] [PubMed] [Google Scholar]
- 21.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, et al. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antunes LC, Han J, Ferreira RB, Lolić P, Borchers CH, Finlay BB. 2011. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob. Agents Chemother. 55(4):1494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, et al. 2009. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril). Magn. Reson. Chem. 47(Suppl. 1):S36–46 [DOI] [PubMed] [Google Scholar]
- 24.Bialonska D, Ramnani P, Kasimsetty SG, Muntha KR, Gibson GR, Ferreira D. 2010. The influence of pomegranate by-product and punicalagins on selected groups ofhuman intestinal microbiota. Int. J. Food Microbiol. 140:175–82 [DOI] [PubMed] [Google Scholar]
- 25.Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, et al. 2008. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 4:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, et al. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 106:3698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. 2011. Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J. Proteome Res. 10:2842–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lhoste EF, Ouriet V, Bruel S, Flinois JP, Brézillon C, et al. 2003. The human colonic microflora influences the alterations of xenobiotic-metabolizing enzymes by catechins in male F344 rats. Food Chem. Toxicol. 41:695–702 [DOI] [PubMed] [Google Scholar]
- 29.Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. 2009. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE 4:e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavaghan CL, Holmes E, Lenz E, Wilson ID, Nicholson JK. 2000. An NMR-based metabonomic approach to investigate the biochemical consequences of genetic strain differences: application to the C57BL10J and Alpk:ApfCD mouse. FEBS Lett. 484:169–74 [DOI] [PubMed] [Google Scholar]
- 31.Lindon JC, Nicholson JK. 2008. Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Annu. Rev. Anal. Chem. 1:45–69 [DOI] [PubMed] [Google Scholar]
- 32.Patterson A, Lanz C, Gonzalez FJ, Idle JR. 2010. The role of mass spectrometry-based metabolomics in medical countermeasures against radiation. Mass Spectrom. Rev. 29:503–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. 2008. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 48:653–83 [DOI] [PubMed] [Google Scholar]
- 34.Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, et al. 2009. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 8:352–61 [DOI] [PubMed] [Google Scholar]
- 35.Waldram A, Holmes E, Wang YL, Rantalainen M, Wilson ID, et al. 2009. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res. 8:2361–75 [DOI] [PubMed] [Google Scholar]
- 36.Marriott P, Shellie R. 2002. Principles and applications of comprehensive two-dimensional gas chromatography. Trends Anal. Chem 21:573–83 [DOI] [PubMed] [Google Scholar]
- 37.Lanz C, Patterson A, Slavík J, Krausz K, Ledermann M, et al. 2009. Radiation metabolomics: 3. Biomarker discovery in the urine of γ-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat. Res. 172:198–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyburski JB, Patterson AD, Krausz KW, Slavík J, Fornace AJ Jr, et al. 2009. Radiation metabolomics: 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal γ-radiation exposure in mice. Radiat. Res. 172:42–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson AD, Slanar O, Krausz KW, Li F, Höfer CC, et al. 2009. Human urinary metabolomic profile of PPARα induced fatty acid β-oxidation. J. ProteomeRes. 8:4293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson AD, Bonzo JA, Li F, Krausz KW, Eichler GS, et al. 2011. Metabolomics reveals attenuation of the SLC6A20 kidney transporter in nonhuman primate and mouse models of type 2 diabetes mellitus. J. Biol. Chem. 286:19511–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plumb RS, Stumpf CL, Granger JH, Castro-Perez J, Haselden JN, Dear GJ. 2003. Use of liquid chromatography/time-of-flight mass spectrometry and multivariate statistical analysis shows promise for the detection of drug metabolites in biological fluids. Rapid Commun. Mass Spectrom. 17:2632–38 [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Chen C, Krausz KW, Idle JR, Gonzalez FJ. 2008. A metabolomic perspective of melatonin metabolism in the mouse. Endocrinology 149:1869–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F, Patterson AD, Höfer CC, Krausz KW, Gonzalez FJ, Idle JR. 2010. Comparative metabolism of cyclophosphamide and ifosfamide in the mouse using UPLC-ESI-QTOFMS-based metabolomics. Biochem. Pharmacol. 80:1063–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montoliu I, Martin FP, Collino S, Rezzi S, Kochhar S. 2009. Multivariate modeling strategy for inter-compartmental analysis of tissue and plasma JH NMR spectrotypes. J. Proteome Res. 8:2397–406 [DOI] [PubMed] [Google Scholar]
- 45.Liu A, Patterson AD, Yang Z, Zhang X, Liu W, et al. 2009. Fenofibrate metabolism in the cynomolgus monkey using ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry-based metabolomics. Drug Metab. Dispos. 37:1157–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, Krausz KW, Idle JR, Gonzalez FJ. 2008. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J. Biol. Chem. 283:4543–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giri S, Idle JR, Chen C, Zabriskie TM, Krausz KW, Gonzalez FJ. 2006. A metabolomic approach to the metabolism of the areca nut alkaloids arecoline and arecaidine in the mouse. Chem. Res. Toxicol. 19:818–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giri S, Krausz KW, Idle JR, Gonzalez FJ. 2007. The metabolomics of ( ± )-arecoline 1-oxidein the mouse and its formation by human flavin-containing monooxygenases. Biochem. Pharmacol. 73:561–73 [DOI] [PubMed] [Google Scholar]
- 49.Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, et al. 2007. A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) metabolism in the mouse using a multivariate data analysis approach. Chem. Res. Toxicol. 20:531–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Meng L, Ma X, Krausz KW, Pommier Y, et al. 2006. Urinary metabolite profiling reveals CYP1A2-mediated metabolism ofNSC686288 (aminoflavone). J. Pharmacol. Exp. Ther. 318:1330–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonzo JA, Patterson AD, Krausz KW, Gonzalez FJ. 2010. Metabolomics identifies novel Hnf1α-dependent physiological pathways in vivo. Mol. Endocrinol. 24:2343–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li F, Patterson AD, Höfer CC, Krausz KW, Gonzalez FJ, Idle JR. 2011. A comprehensive understanding of thioTEPA metabolism in the mouse using UPLC-ESI-QTOFMS-based metabolomics. Biochem. Pharmacol. 81:1043–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen N, Aleksa K, Woodland C, Rieder M, Koren G. 2008. N-Acetylcysteine prevents ifosfamide-induced nephrotoxicity in rats. Br. J. Pharmacol. 153:1364–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerre-Millo M, Gervois P, Raspé E, Madsen L, Poulain P, et al. 2000. Peroxisome proliferator-activated receptor α activators improve insulin sensitivity and reduce adiposity. J. Biol. Chem. 275:16638–42 [DOI] [PubMed] [Google Scholar]
- 55.Blane GF. 1987. Comparative toxicity and safety profile of fenofibrate and other fibric acid derivatives. Am. J. Med. 83:26–36 [DOI] [PubMed] [Google Scholar]
- 56.Clouâtre Y, Leblanc M, Ouimet D, Pichette V. 1999. Fenofibrate-induced rhabdomyolysis in two dialysis patients with hypothyroidism. Nephrol. Dial. Transplant. 14:1047–48 [DOI] [PubMed] [Google Scholar]
- 57.Nishimura J, Dewa Y, Muguruma M, Kuroiwa Y, Yasuno H, et al. 2007. Effect of fenofibrate on oxidative DNA damage and on gene expression related to cell proliferation and apoptosis in rats. Toxicol. Sci. 97:44–54 [DOI] [PubMed] [Google Scholar]
- 58.Davidson MH, Armani A, McKenney JM, Jacobson TA. 2007. Safety considerations with fibrate therapy. Am. J. Cardiol. 99(6):S3–18 [DOI] [PubMed] [Google Scholar]
- 59.Tahmaz M, Kumbasar B, Ergen K, Ure U, Karatemiz G, Kazancioglu R. 2007. Acute renal failure secondary to fenofibrate monotherapy-induced rhabdomyolysis. Ren. Fail. 29:927–30 [DOI] [PubMed] [Google Scholar]
- 60.Qi C, Zhu YJ, Reddy JK. 2000. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem. Biophys 32:187–204 [DOI] [PubMed] [Google Scholar]
- 61.Peters JM, Cheung C, Gonzalez FJ. 2005. Peroxisome proliferator-activated receptor-α and liver cancer: Where do we stand? J. Mol. Med 83:774–85 [DOI] [PubMed] [Google Scholar]
- 62.Gonzalez FJ, Shah YM. 2008. PPARα mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology 246:2–8 [DOI] [PubMed] [Google Scholar]
- 63.Weil A, Caldwell J, Strolinbenedetti M. 1990. The metabolism and disposition of 14C fenofibrate in human volunteers. Drug Metab. Dispos. 18:115–20 [PubMed] [Google Scholar]
- 64.Weil A, Caldwell J, Strolinbenedetti M. 1988. The metabolism and disposition of fenofibrate in rat, guinea-pig, and dog. Drug Metab. Dispos. 16:302–9 [PubMed] [Google Scholar]
- 65.Cornu-Chagnon MC, Dupont H, Edgar A. 1995. Fenofibrate: metabolism and species differences for peroxisome proliferation in cultured hepatocytes. Fundam. Appl. Toxicol. 26:63–74 [DOI] [PubMed] [Google Scholar]
- 66.Liu A, Chen Y, Yang Z, Feng Y, Rui W, et al. 2009. New metabolites of fenofibrate in Sprague-Dawley rats by UPLC-ESI-QTOF-MS-based metabolomics coupled with LC-MS/MS. Xenobiotica 39:345–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicholson JK, Lindon JC, Wilson ID. 2011. Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics 12:103–11 [DOI] [PubMed] [Google Scholar]
- 68.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C,et al. 2006. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 440:1073–77 [DOI] [PubMed] [Google Scholar]
- 69.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. 2009. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 106:14728–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winnike JH, Li Z, Wright FA, Macdonald JM, O’Connell TM, Watkins PB. 2010. Use of pharmacometabonomics for early prediction of acetaminophen-induced hepatotoxicity in humans. Clin. Pharmacol. Ther. 88:45–51 [DOI] [PubMed] [Google Scholar]
- 71.Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, et al. 2011. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 89:97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nebert DW, Zhang G, Vesell ES. 2008. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab. Rev. 40:187–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheung C, Gonzalez FJ. 2008. Humanized mouse lines and their application for prediction of human drug metabolism and toxicological risk assessment. J. Pharmacol. Exp. Ther. 327:288–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pang X, Cheng J, Krausz KW, Guo DA, Gonzalez FJ. 2011. Pregnane X receptor-mediated induction of Cyp3a by black cohosh. Xenobiotica 41:112–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Donato MT, Viitala P, Rodriguez-Antona C, Lindfors A, Castell JV, et al. 2000. CYP2A5/CYP2A6 expression in mouse and human hepatocytes treated with various in vivo inducers. Drug Metab. Dispos. 28:1321–26 [PubMed] [Google Scholar]
- 76.Cho JY, Kang DW, Ma X, Ahn SH, Krausz KW, et al. 2009. Metabolomics reveals a novel vitamin E metabolite and attenuated vitamin E metabolism upon PXR activation. J. Lipid Res. 50:924–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma X, Idle JR, Gonzalez FJ. 2008. The pregnane X receptor: from bench to bedside. Expert Opin. Drug Metab. Toxicol. 4:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng J, Shah YM, Ma X, Pang X, Tanaka T, et al. 2010. Therapeutic role of rifaximin in inflammatory bowel disease: clinical implication of human pregnane X receptor activation. J. Pharmacol. Exp. Ther. 335:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma X, Shah Y, Cheung C, Guo GL, Feigenbaum L, et al. 2007. The pregnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab. Dispos. 35:194–200 [DOI] [PubMed] [Google Scholar]
- 80.Wang T, Ma X, Krausz KW, Idle JR, Gonzalez FJ. 2008. Role of pregnane X receptor in control of all-trans retinoic acid (ATRA) metabolism and its potential contribution to ATRA resistance. J. Pharmacol. Exp. Ther. 324:674–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Foreman JE, Chang SC, Ehresman DJ, Butenhoff JL, Anderson CR, et al. 2009. Differential hepatic effects of perfluorobutyrate mediated by mouse and human PPAR-α. Toxicol. Sci. 110:204–11 [DOI] [PubMed] [Google Scholar]
- 82.Cheung C, Akiyama TE, Ward JM, Nicol CJ, Feigenbaum L, et al. 2004. Diminished hepatocellular proliferation in mice humanized for the nuclear receptor peroxisome proliferator-activated receptor α. Cancer Res 64:3849–54 [DOI] [PubMed] [Google Scholar]
- 83.Morimura K, Cheung C, Ward JM, Reddy JK, Gonzalez FJ. 2006. Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor α to Wy-14,643-induced liver tumorigenesis. Carcinogenesis 27:1074–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, et al. 2005. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem. Res. Toxicol. 18:1471–78 [DOI] [PubMed] [Google Scholar]
- 85.Cheung C, Yu AM, Ward JM, Krausz KW, Akiyama TE, et al. 2005The CYP2E1-humanized transgenic mouse: role of CYP2E1 in acetaminophen hepatotoxicity. Drug Metab. Dispos. 33:449–57 [DOI] [PubMed] [Google Scholar]
- 86.Ma X, Cheung C, Krausz KW, Shah YM, Wang T, et al. 2008. A double transgenic mouse model expressing human pregnane X receptor and cytochrome P450 3A4. Drug Metab. Dispos. 36:2506–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. 2009. Rifampicin-activated human pregnane X receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab. Dispos. 37:1611–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miners JO, Smith PA, Sorich MJ, McKinnon RA, Mackenzie PI. 2004. Predicting human drug glucuronidation parameters: application of in vitro and in silico modeling approaches. Annu. Rev. Pharmacol. Toxicol. 44:1–25 [DOI] [PubMed] [Google Scholar]
- 89.Chen S, Beaton D, Nguyen N, Senekeo-Effenberger K, Brace-Sinnokrak E, et al. 2005. Tissue-specific, inducible, and hormonal control of the human UDP-glucuronosyltransferase-1 (UGT1) locus. J. Biol. Chem. 280:37547–57 [DOI] [PubMed] [Google Scholar]
- 90.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, et al. 2007. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3:112. [DOI] [PMC free article] [PubMed] [Google Scholar]