Abstract
Although the aryl hydrocarbon receptor (AHR) and glucocorticoid receptor (GR) play essential roles in mammalian development, stress responses, and other physiological events, crosstalk between these receptors has been the subject of much debate. Metallothioneins are classic glucocorticoid-inducible genes that were reported to increase upon treatment with AHR agonists in rodent tissues and cultured human cells. In this study, the mechanism of human metallothionein 2A (MT2A) gene transcription activation by AHR was investigated. Cotreatment with 3-methylcholanthrene and dexamethasone, agonists of AHR and GR respectively, synergistically increased MT2A mRNA levels in HepG2 cells. MT2A induction was suppressed by RNA interference against AHR or GR. Coimmunoprecipitation experiments revealed a physical interaction between AHR and GR proteins. Moreover, chromatin immunoprecipitation assays indicated that AHR was recruited to the glucocorticoid response element in the MT2A promoter. Thus, we provide a novel mechanism whereby AHR modulates expression of human MT2A via the glucocorticoid response element and protein–protein interactions with GR.
Keywords: 3-Methylcholanthrene, Aryl hydrocarbon receptor, Dexamethasone, Glucocorticoid receptor, Metallothionein, Transcription
Introduction
The aryl hydrocarbon receptor (AHR), a member of the basic helix– loop–helix/Per–Arnt–Sim (bHLH/PAS) family, is a ligand-activated transcription factor responsible for the development and adaptive response to environmental changes elicited by toxic agents, hypoxia, and the light and dark cycle (Gu et al., 2000). AHR plays a key role in xenobiotic responses as a receptor for environmental contaminants such as dioxins, coplanar polychlorinated biphenyls, and benzo[ɑ]pyrene. In the absence of ligand, AHR exists in the cytoplasm in a complex with XAP2, two HSP90 chaperone proteins, and the co-chaperone protein p23. Upon binding with a ligand, AHR translocates to the nucleus, where it dimerizes with its heterodimer partner, AHR nuclear translocator (ARNT) (Denison et al., 2011). This AHR-ARNT heterodimer binds a DNA sequence termed the xenobiotic response element (XRE) and subsequently activates transcription of target genes represented by the phase I xenobiotic-metabolizing enzymes of cytochrome P450 (CYP) 1A1, CYP1A2, and CYP1B1 and by the phase II drug metabolizing enzymes of glutathione S-transferase and UDP-glucuronosyltransferase, as well as NAD(P)H quinone oxidoreductase 1 (Nebert et al., 2004; Omiecinski et al., 2011). Induction of these enzymes results in the metabolism and detoxification of xenobiotics, including some AHR ligands. However, increased metabolism can also cause adverse effects that result from oxidative stress and metabolic activation of certain xenobiotic chemicals. In addition to ligand-induced xenobiotic responses, AHR is related to various physiological events, such as cell proliferation and differentiation, development, reproduction, and immune system regulation.
While XRE-dependent mechanism is important for AHR transcriptional regulation, recent studies have indicated that AHR modulates gene expression without directly binding DNA. Instead, AHR interacts with other transcription factors, such as the RelA subunit of nuclear factor-kappa B (NFκB) (Tian et al., 1999; Kim et al., 2000) and estrogen receptor α (Beischlag and Perdew, 2005), and modulates the activity of these molecules in an XRE-independent manner. Thus, protein–protein interactions appear to be important for the various biological functions of AHR.
Glucocorticoids, including cortisol and corticosterone, are steroid hormones secreted from the adrenal glands that play a role in stress response through the regulation of carbohydrate metabolism and inflammation. The glucocorticoids are ligands of glucocorticoid receptor (GR), a member of the nuclear receptor superfamily. Glucocorticoid intracellular signaling is governed by GR. In the absence of ligand, GR exists in the cytoplasm in a complex with HSP90. After activation by ligand binding, GR enters the nucleus. GR forms a homodimer, and the GR dimer binds with a glucocorticoid response element (GRE) and regulates transcription of its target genes (Bamberger et al., 1996). On the other hand, protein–protein interactions between GR and other transcription factors are essential for GR-mediated transcriptional suppression. For example, synthetic glucocorticoids are widely used as therapeutic anti-inflammatory agents. In this mechanism, GR binds to NFκB and selectively represses transcription of proinflammatory cytokines such as IL-8 (Nissen and Yamamoto, 2000; Luecke and Yamamoto, 2005).
Both AHR and GR play essential roles in development, reproduction, immune system regulation, and stress responses, and there are apparent relationships between the activities of these two receptors. Cleft palate is a malformation that can be induced in the laboratory by excessive activation of AHR. Administration of dioxin to a pregnant mammal induces cleft palate in the fetus through activation of AHR (Mimura et al., 1997). Excess glucocorticoids also induce cleft palate. A synergistic effect was observed in embryonic cleft palate induced by cotreatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and hydrocortisol, AHR and GR agonists, respectively (Pratt, 1985; Abbott, 1995; Abbott et al., 1999). Using a reporter gene assay with a GRE-driven reporter construct, Dvořák1 et al. show that TCDD enhanced GR transactivation induced by dexamethasone (Dex), a synthetic GR agonist, in HepG2 cells (Dvořák et al., 2008). Wang et al. showed that AHR ligands such as polyaromatic hydrocarbons synergistically enhance Dex-induced transactivation of GR in ovarian granulosa HO23 cells (Vrzal et al., 2009). This synergistic effect was independent of the individual expression levels of AHR or GR. Others performed a comprehensive analysis to identify latent transcription factors associated with AHR-signaling (Frericks et al., 2008). A computational search for over-represented elements in the promoter region of TCDD-affected genes in a thymic epithelial cell line identified 37-transcriptional binding sites concluding with a GR binding sequence. These data imply that AHR and GR are closely associated with each other. However, the molecular mechanism of their interaction has not yet been clarified.
Mammalian metallothioneins (MTs) are classic glucocorticoid-inducible genes (Karin et al., 1980; Hager and Palmiter, 1981; Karin and Richards, 1982; Kelly et al., 1997). MTs are low molecular weight, cysteine-rich metal binding proteins. MTs are believed to play an important role in homeostasis of essential metals and biological protection against environmental toxicity represented by heavy metals or reactive oxygen species. In our previous study, DNA microarray analyses revealed that the mRNA levels of MT isoforms Mt1 and Mt2 were elevated in the liver of mice that were administered low doses of TCDD (Sato et al., 2008). Other groups also reported elevated mRNA or protein levels of MTs in rat (Nishimura et al., 2001; Fletcher et al., 2005) and mouse tissues (Kurachi et al., 2002; Boverhof et al., 2005) following acute exposure to high doses of TCDD, as well as in HepG2 cells treated with TCDD (Frueh et al., 2001). However, a functional XRE was not identified in the promoter region of MT genes, and the mechanism of transcriptional activation mediated by AHR is still unclear.
In this study, we identified human metallothionein 2A (MT2A) as an intrinsic gene whose transcription is regulated by AHR–GR interactions. In cultured cells, expression of human MT2A was cooperatively increased by cotreatment with Dex and 3-methylcholanthrene (3-MC), an AHR agonist, in a receptor-dependent manner. Our chromatin immunoprecipitation (ChIP) assay results indicate that AHR is recruited to the MT2A promoter. Moreover, coimmunoprecipitation experiments reveal a physical interaction between AHR and GR. Thus, we conclude that AHR modulates MT2A gene expression via the glucocorticoid response element and a novel protein–protein interaction with GR.
Materials and methods
Materials.
TCDD, 3-MC, Dex, and dimethyl sulfoxide (DMSO) were purchased from Sigma (St. Louis, MO). HepG2 cells were obtained from the Cell Resource Center for Biomedical Research, Tohoku University. HeLa and COS7 cells were kindly provided by Dr. Yoshida, Scientific University of Tokyo.
Animal experiments.
TCDD-treatment of mice and total RNA isolation from the liver were performed as described previously (Sato et al., 2008). Briefly, C57BL/6N wild-type and Ahr-null male mice, aged 6–7 weeks, were orally administered doses of 5, 50, or 500 ng TCDD/kg body weight (bw) in corn oil by gavage once a day for 18 days. Animals in the control groups were administered the same volume of corn oil without TCDD. Throughout the experimental period, the animals had free access to AIN93G standard diets (Reeves et al., 1993) and desalted water. On the 19th day, experimental animals were sacrificed and hepatic RNA was isolated using a phenol/guanidine-isothiocyanate-based reagent (Isogen; Nippon Gene Co., Tokyo, Japan).
The Animal Research-Animal Care Committee of the Graduate School of Agricultural Science, Tohoku University approved the experimental plan of the present study. All experiments were performed under the guidelines framed by this committee in accordance with Japanese governmental legislation (1980). The same committee supervised the care and use of mice in this study.
Northern hybridization analyses.
Total RNA (20 μg) from the liver was denatured in an RNA gel-loading buffer at 65 °C for 5 min and loaded onto 1.2% agarose gels containing formaldehyde (Sambrook et al., 1989). After electrophoresis, the RNA was transferred to Hybond N + nylon membranes (GE Healthcare, Tokyo, Japan) using the capillary method. DNA fragments encoding mouse metallothionein-1, rat metallothionein-2, and α-tubulin were labeled with [32P] dCTP (MP Biomedicals, Irvine, CA). The RNA-blotted filters and labeled cDNA were incubated at 68 °C for 1.5 h in ExpressHyb Hybridization Solution (Clontech Laboratories, Palo Alto, CA). After hybridization, the filters were washed twice with 2× SSC (150 mM sodium chloride, 150 mM sodium citrate) for 30 min and with 0.1× SSC for 40 min. The filters were then exposed on a Fuji imaging plate (Fuji Photo Film, Tokyo, Japan) for an adequate period of time and analyzed using a BioImage analyzer FLA-2000 (Fuji Photo Film). The relative mRNA expression levels were normalized to the amount of α-tubulin mRNA.
Cell culture.
HepG2, HeLa, and COS7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37 ˚C in a humidified atmosphere of 95% air and 5% CO2.
Quantitative reverse transcription-polymerase chain reaction (RTPCR).
HepG2 cells were plated and cultured for 16 h. The media was replaced with additive-free DMEM (without serum and antibiotics) and incubated for 24 h. Next, TCDD, 3-MC and/or Dex, or vehicle (DMSO) control was added at 0.1% v/v to the media and cells were incubated for 9 h. Cells were washed twice with phosphate-buffered saline (PBS), and total RNA was isolated. Total RNA (4 μg) was denatured at 65 ˚C for 5 min with 2.5 μM oligo-dT primer (GE Healthcare) and 0.5 mM dNTP (GE Healthcare). The RNA was incubated in 20 μl of RT buffer [50 mM Tris– HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol] containing 50 U SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and 20 U RNaseOUT RNase inhibitor (Invitrogen) for 60 min at 50 ˚C. An aliquot of synthesized cDNA was used as the template for quantitative PCR using an Applied Biosystems 7300 Real-Time PCR System (Foster City, CA). The target cDNAs were amplified using gene-specific primers (Table 1) and SYBR Premix Ex Taq (Takara Bio, Otsu, Japan) solution. The relative mRNA expression levels were normalized to the amount of eukaryotic translation elongation factor 1α1 (EEF1A1) mRNA.
Table 1.
Oligonucleotide sequences used in quantitative RT-PCR, chromatin immunoprecipitation assays, and RNA interference.
| Quantitative RT-PCR | ||
|---|---|---|
| MT2A mRNA | Forward | 5′-TGTACAACCCTGACCGTGACC-3′ |
| Reverse | 5′-TCACATTATTTCATAGAAAAAGGAATATAGC-3′ | |
| CYP1A1 mRNA | Forward | 5′-GATCAACCATGACCAGAAGCTATG-3′ |
| Reverse | 5′-CACCTTGTCGATAGCACCATC A-3′ | |
| EEF1A1 mRNA Forward | Forward | 5′-GATGGCCCCAAATTCTTGAAG-3′ |
| Reverse | 5′-GGACCATGTCAATGGCAG-3′ ays | |
| Chromation immunoprecipitation assays | ||
| MT2A GRE | Forward | 5′-TAACGGCTCAGGTTCGAGTACA-3′ |
| Reverse | 5′-TGCAGCGGGAGGACACA-3′ | |
| CYP1A1 XRE | Forward | 5′-GGCGCGAACCTCAGCTAGT-3′ |
| Reverse | 5′-ACGCTGGGCGTGCAGAT-3′ | |
| Imperfect XRE1 | Forward | 5’-GGAGCCGCAAGTGACTTCT-3′ |
| Reverse | 5’-CTAGAAAGAGCCGGGACGAG-3′ | |
| Imperfect XRE3 | Forward | 5’-ACAACACACTTCCTACAGTCCC-3′ |
| Reverse | 5’-TGGGGTGGTTGCTGATGAC-3′ | |
| RNA intrference | ||
| AHR siRNA | Sense | 5′-CCGAGUCCCAUAUCCGAAUGAUUAA-3′ |
| Anti-sense | 5′-UUAAUCAUUCGGAUAUGGGACUCGG-3′ | |
| GR siRNA | Sense | 5′-UAAUUGUGCUGUCCUUCCACUGCUC-3′ |
| Anti-sense | 5′-GAGCAGUGGAAGGACACAAUUA-3′ | |
| Control siRNA | Sense | 5′-CCGCCUUACCUAAGCGUAUAAGUAA-3′ |
| Anti-sense | 5′-UUACUUAUACGCUUAGGUAAGGCGG-3′ | |
Western blot analyses.
Cells were washed twice with PBS buffer and harvested with PBS. After removing PBS, cells were incubated on ice for 15 min in cell lysis buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40], followed by centrifugation at 12,000 ×g for 15 min. The protein concentration in the supernatant was measured using a protein assay reagent (Bio-Rad, Hercules, CA). Protein (15 μg) was denatured in sodium dodecyl sulfate (SDS) gelloading buffer, resolved by 10% or gradient SDS-polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking for 1 h with TBS-T [10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.1% Tween 20] containing 5% fat-free milk, the membrane was incubated with AHR (H-211; Santa Cruz Biotechnology, Santa Cruz, CA), GR (Perseus Proteomics, Tokyo, Japan), and β-actin (Thermo Fisher Scientific, Fremont, CA) anti-bodies. Proteins were detected using an Immobilon Western Chemiluminescent HRP Substrate (Millipore) and an LAS-4000 mini Lumino-image analyzer (Fuji Photo Film).
RNA interference.
HepG2 cells (0.5 × 106 cells) were plated in six-well plates 16 h before transfection. Stealth siRNA (20 pmol; Invitrogen) was used as control. AHR or GR was transfected using Lipofectamine 2000 with Opti-MEMI (Invitrogen) for 24 h according to the instruction manual. After transfection, the media was replaced with additive-free DMEM. HepG2 cells were cultured for 24 h, followed by incubation for 9 h with 3-MC and/or Dex, or DMSO alone. The specific siRNA sequences are described in Table 1.
Plasmid construction.
The wild-type MT2A promoter-driven luciferase reporter plasmid pMT2A-Luc was prepared using the following method. A HindIII–BamHI digested 840-bp insert of pMT-IIA was obtained from JCRB GENE Bank (Osaka, Japan) containing the human MT2A promoter region (Karin and Richards, 1982) and was cloned into the HindIII–BamHI site of pBluescript II SK+ (Stratagene, La Jolla, CA). The XhoI–BglII digested insert of this pBluescript II SK+/MT2A was cloned into the XhoI–BamHI site of the pGL3-basic luciferase vector (Promega, Madison, WI). The mutant GRE plasmid pGL3-mutGRE was prepared using the following method. PCR was performed with the 5′-phosphorylated oligonucleotide designed to amplify the linear pBluescript II SK+/MT2A altered GRE: Fwd, 5′GGCACCCAGCACCCGGacagCTGgacgCTCCCGCTGCACCCAGC-3′ and Rev, 5′-CACGCCGTGCGCCTCCGCCGTGT-3′ using KOD-Plus DNA Polymerase (Toyobo, Tokyo, Japan). This linear PCR product ligated itself after digestion of the template plasmid with DpnI. Next, the mutated promoter fragment was cloned into pGL3-basic as well as pMT2A-Luc. The mutant MT2A imperfect XRE2 reporter plasmid was prepared using the following method. PCR was performed with the oligonucleotide designed to amplify the linear pMT2A-Luc altered the imperfect XRE2: Fwd, 5′-CACGaaaccGGCACCCAGCACC-3′ and Rev, 5′CGCCTCCGCCGTGTGCACAG-3′ using Expand High Fidelity PCR System (Roche Applied Science, Mannheim, Germany). This linear PCR product was phosphorylated and ligated itself after digestion of the template plasmid with DpnI. AHR and GR expression plasmids were prepared as follows. Human AHR cDNA was amplified from pBluescriptR-human AHR (MHS1010–98075336, Open Biosystems) using the primer: Fwd, 5′-CTCGAGGGATGAACAGCAGCAGCGCC-3′ and Rev, 5′-CTCGAGTTACAGGAATCCACTGGATGTCAA-3′. Human GR cDNA was amplified from pCMV4-GR (kindly provided by Dr. Harata, Tohoku University) using the following primers: Fwd, 5′-CTCGAGGG ATGGACTCCAAAGAATC-3′ and Rev, 5′-CTCGAGCCAAGTCTTGGCCCTCT3′. The full-length AHR and GR cDNAs were cloned into the XhoI sites of pCMV-Myc and pCMV-HA, and the SalI sites of pM and pVP16 plasmid vectors (Clontech). The constructs pCMV-Myc-AHR, pCMV-Myc-GR, pCMV-HA-AHR, and pCMV-HA-GR were used in the immunoprecipitation assay, and pM-AHR and pVP16-GR were used in the mammalian two-hybrid assay.
Reporter gene assays.
HeLa cells at approximately 70% confluence were transfected with plasmid DNA (amounts are described in the figure legends) using Lipofectamine-PLUS reagent (Invitrogen) with additivefree DMEM in six-well plates. After 3 h, media was replaced with fresh additive-free DMEM containing 3-MC and/or Dex, or DMSO control, and cells were incubated for another 24 h. Cells were washed twice with PBS and lysed in 200 μl reporter lysis buffer (Promega). Lysates were centrifuged at 12,000 ×g for 2 min, and the luciferase activities in the supernatants were determined. Firefly luciferase activity of pGL3-MT2A, pGL3-mutGRE, or pGRE-Luc (Clontech) was determined using luciferase assay reagent [20 mM Tricine (pH 7.8), 1.07 mM (MgCO3)4Mg(OH)2, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 270 μM coenzyme A lithium salt, 470 μM luciferin, 530 μM ATP]. Renilla luciferase activity of the internal control pGL4-hRluc-TK (Promega) was determined using the Renilla luciferase assay system (Promega). Chemiluminescence was detected using a Luminescencer-MCA AB-2250 luminometer (Atto Co., Tokyo, Japan). Firefly luciferase activity was normalized to Renilla luciferase activity.
Mammalian two-hybrid assays.
COS7 cells were transfected with pM-AHR and pVP16-GR vectors (in concentrations specified in the figure legends) using Lipofectamine-PLUS reagent (Invitrogen) with additive-free DMEM in six-well plates. After 3 h, media was replaced with fresh additive-free DMEM, and cells were incubated for another 24 h. Firefly luciferase activity was normalized to Renilla luciferase activity from the internal control pGL4-hRluc-TK.
Immunoprecipitation.
HeLa cells at 60% confluence in a 10 cm dish were transfected with 2 μg of DNA (pCMV-HA-GR/pCMV-Myc-AHR or pCMV-Myc; pCMV-HA-AHR/pCMV-Myc-GR or pCMV-Myc) for 24 h. The cells were treated for 30 min with 1 μM 3-MC and 10 nM Dex. The cells were cross-linked with 1 mg/ml dithiobis (succinimidyl propionate) for 7 min at room temperature, and chased with Tris–HCl (pH 8.0; 100 mM final concentration). HepG2 cells were treated with 1 μM 3-MC and 100 nM Dex for 30 min. Cells were washed twice with PBS, lysed with 2.5 ml of IP buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, EDTA-free cOmplete Protease In-hibitor Cocktail (Roche Applied Science)], and centrifuged at 12,000 ×g for 15 min. Using 0.5 ml of the supernatant, immunoprecipitation was performed overnight at 4 ˚C using 20 μl of EZview red anti-c-Myc affinity gel (Sigma). Intrinsic proteins were immunoprecipitated as complexes of Dynabeads Protein G (Invitrogen) and antibodies against AHR (RPT9, Abcam, Tokyo Japan), GR, or TBP (QED Bioscience Inc., San Diego, CA). Next, the conjugate was washed three-times with 1 ml IP buffer and resuspended in 60 μl SDS gel-loading buffer. The boiled supernatant (20 μl) was used for Western blot analyses as described above.
Chromatin immunoprecipitation assays.
HepG2 cells were treated with 1 μM 3-MC and 100 nM Dex for 30 min. The cells were cross-linked with 1% formaldehyde for 15 min at room temperature, and chased with 125 mM glycine. The cells were washed twice with PBS. Cells were harvested with PBS and collected by centrifugation. Cell pellets were resuspended and incubated for 10 min in cell lysis buffer [10 mM Hepes–NaOH (pH 7.5), 0.5 mM EDTA, 2 mM MgCl2, cOmplete Proteinase Inhibitor (Roche Applied Science)], and nuclei were pelleted by centrifugation. Nuclei were resuspended in MNase buffer [10 mM Hepes–NaOH (pH 7.5), 5 mM CaCl2, cOmplete Proteinase Inhibitor (Roche Applied Science)] and treated with micrococcal nuclease (30 U/ml final, Takara Bio) to an average length of approximately 500 bp for 20 min. Fifteen micrograms of digested chromatin was incubated overnight at 4 ˚C with 1 μg of anti-AHR antibody (RPT9, Abcam) or control IgG (anti-β-actin antibody, Abcam). The immune complexes were collected using Dynabeads Protein G (Invitrogen) for 1 h at 4 ˚C. Next, the beads were washed twice with low salt wash buffer [10 mM Hepes–NaOH (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40], twice with high salt buffer [10 mM Hepes–NaOH (pH 7.5), 450 mM NaCl, 1 mM EDTA 1% NP-40], and suspended in decrosslink buffer (0.1 M NaHCO3, 20 mM NaCl, 1% SDS). The formaldehyde crosslink was reversed by incubation at 65 ˚C for 6 h. DNA fragments were purified using QIAquick PCR Purification Kit (Qiagen, Tokyo, Japan) after the treatment with RNase A and proteinase K. PCR was performed using MT2A imperfect XRE1 and XRE3, MT2A GRE, CYP1A1 XRE, and EEF1A1 gene-specific primers. Primer sequences are shown in Table 1.
Statistical analyses.
The data in Figs. 1, 2a, b, 7a, and b were analyzed using one-way analysis of variance (ANOVA) and multiple comparisons were made with Dunnett’s test. Data in Figs. 3a, b, c and e were analyzed using ANOVA and multiple comparisons were made with Scheffe’s tests. Data in Figs. 4b, c, and 5b were analyzed using Student’s-t test. Statistical analyses were performed using the StatView software (SAS Institute Inc., Cary, NC, USA).
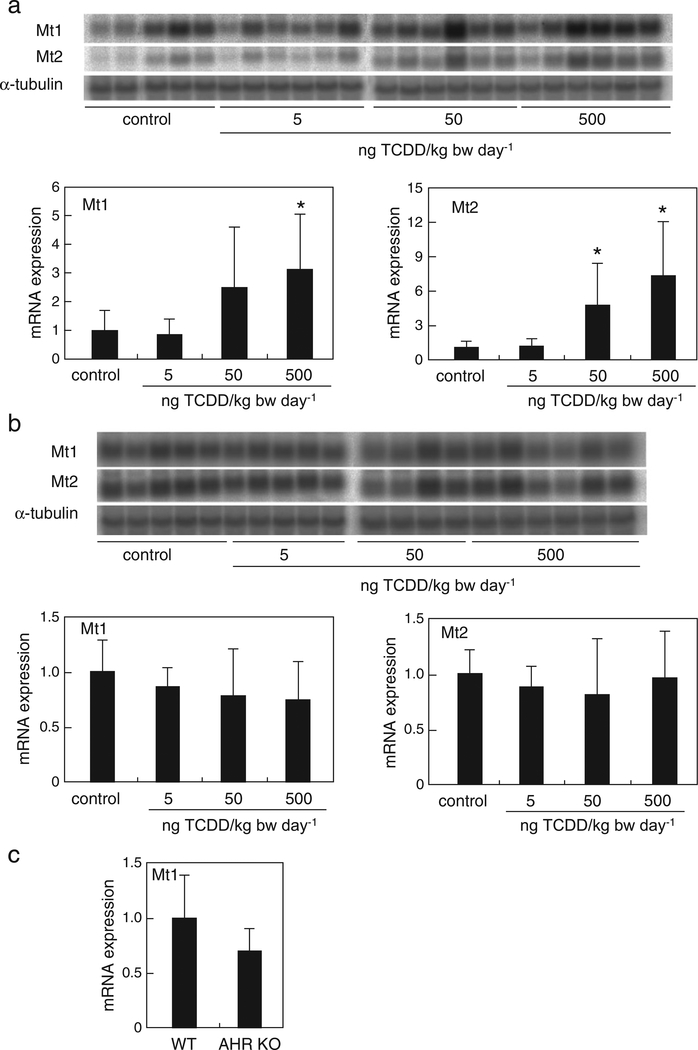
Fig. 1.
Expression of MT genes is induced by TCDD and mediated through the AHR signaling pathway in mouse liver. Mt1 and Mt2 mRNA expression levels in the liver of wild-type (a) or Ahr-null (b) mice following TCDD administration at 0, 5, 50, and 500 ng/kg bw·day−1 for 18 days were measured by northern hybridization. (c) Mt1 mRNA levels in the livers of wild-type and Ahr-null mice were determined by northern hybridization. The relative expression level of mRNA was normalized to the expression of α-tubulin mRNA. The values are the mean ± standard deviation (SD; n = 4–6). Values with an asterisk are significantly different from vehicle control at p < 0.05. Statistical analyses were conducted using one-way analysis of variance (ANOVA), followed by Dunnett’s tests.
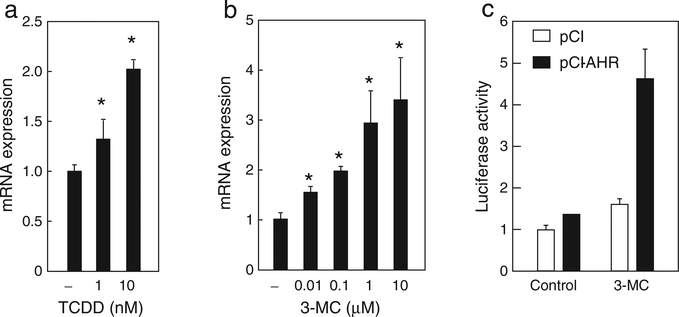
Fig. 2.
Human MT2A expression is induced by AHR agonists in human cultured cells. (a, b) HepG2 cells were treated with TCDD (a) or 3-MC (b) at the indicated concentrations for 9 h, and MT2A mRNA levels were determined using quantitative RT-PCR. Controls were treated with the vehicle (−). (c) HeLa cells were transfected with 0.25 μg of pMT2A-Luc and 0.15 μg of pGL4-hRluc-TK together with either 0.1 μg of AHR expression plasmid or control plasmid for 3 h. Cells were treated for 24 h with 1 μM 3-MC or vehicle, and relative luciferase activities were determined. The values are the mean ± SD of 3 (a, b) or 2 (c) independent experiments. Values with an asterisk are significantly different from vehicle control at p < 0.05. Statistical analyses were conducted using ANOVA, followed by Dunnett’s tests.
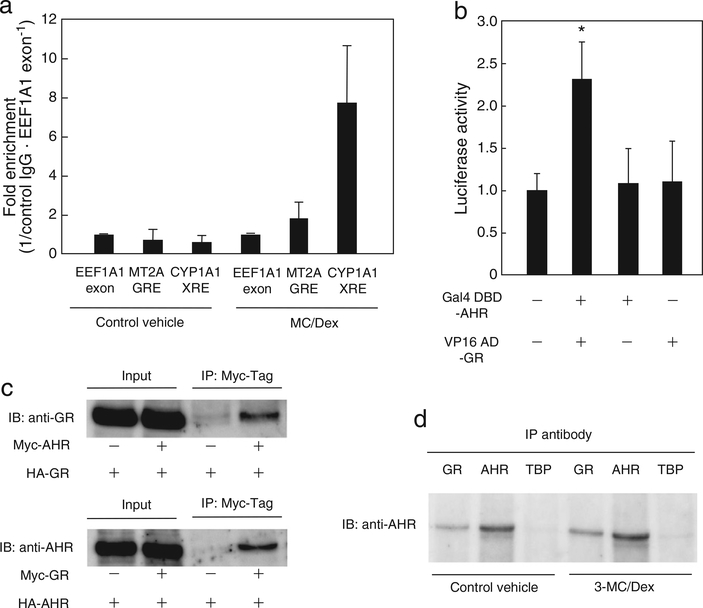
Fig. 7.
AHR interacts with GR. (a) HepG2 cells were treated for 30 min with 1 μM 3-MC and 100 nM Dex, or control vehicle, and ChIP assays were performed. The levels of the PCR products for the EEF1A1 exon, GRE and XRE were normalized to the control IgG. The fold enrichment of GRE or XRE is presented as a ratio to the EEF1A1 exon expression. Independent experiments were performed 3 times. (b) COS7 cells were transfected with 0.25 μg of the GAL4 response element-driven luciferase reporter plasmid and 0.15 μg of the pGL4-hRluc-TK construct together with either 0.1 μg of the expression plasmids containing the full-length receptor fusion sequence (pM-AHR and/or pVP16-GR) or the control plasmids (pM or pVP16) for 24 h. Cells were lysed and relative luciferase activities were determined. Values are the mean ± SD of 3 independent experiments. The value with asterisk is significantly different compared to control at p < 0.05. Statistical analyses were conducted using ANOVA, followed by Dunnett’s tests. (c) HeLa cells were transfected with 2 μg of DNA (upper panel, pCMV-HA-GR/pCMV-Myc-AHR or pCMV-Myc; lower panel, pCMV-HA-AHR/pCMV-Myc-GR or pCMV-Myc) for 24 h. The cells were treated for 30 min with 1 μM 3-MC and 10 nM Dex and cross-linked with DSP for immunoprecipitation assays. Independent experiments were performed 2 times. (d) HepG2 cells were treated for 30 min with 1 μM 3-MC and 100 nM Dex, or control vehicle, and immunoprecipitation assays were performed. Experiments were performed independently 2 times.
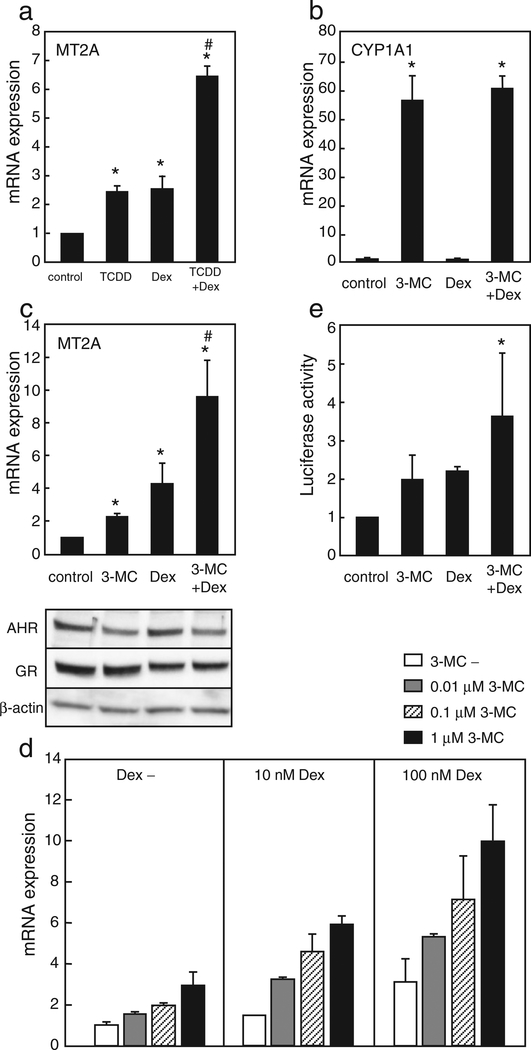
Fig. 3.
Dex and AHR agonists increase MT2A expression. HepG2 cells were treated for 9 h with vehicle, 10 nM TCDD and/or 100 nM Dex (a, b) or 1 μM 3-MC and/or 100 nM Dex (c). The mRNA levels were determined using quantitative RT-PCR. The protein levels of AHR and GR were determined by Western blotting (c, lower panel). (d) HepG2 cells were treated for 9 h with 3-MC and/or Dex, or vehicle at the indicated concentrations. MT2A mRNA levels were determined using quantitative RT-PCR. (e) HeLa cells were transfected with 0.25 μg of pMT2A-Luc and 0.15 μg of pGL4-hRluc-TK for 3 h. Cells were treated for 24 h with 1 μM 3-MC and/or 10 nM Dex, or vehicle, and relative luciferase activities were determined. The values are the mean ± SD of 3 (a, b, c, e) or 2 (d) independent experiments. Values with an asterisk are significantly different compared to vehicle control; the value with a sharp was significantly different compared to treatment with either 3-MC or Dex at p < 0.05. Statistical analyses were conducted using ANOVA, followed by Scheffe’s tests.
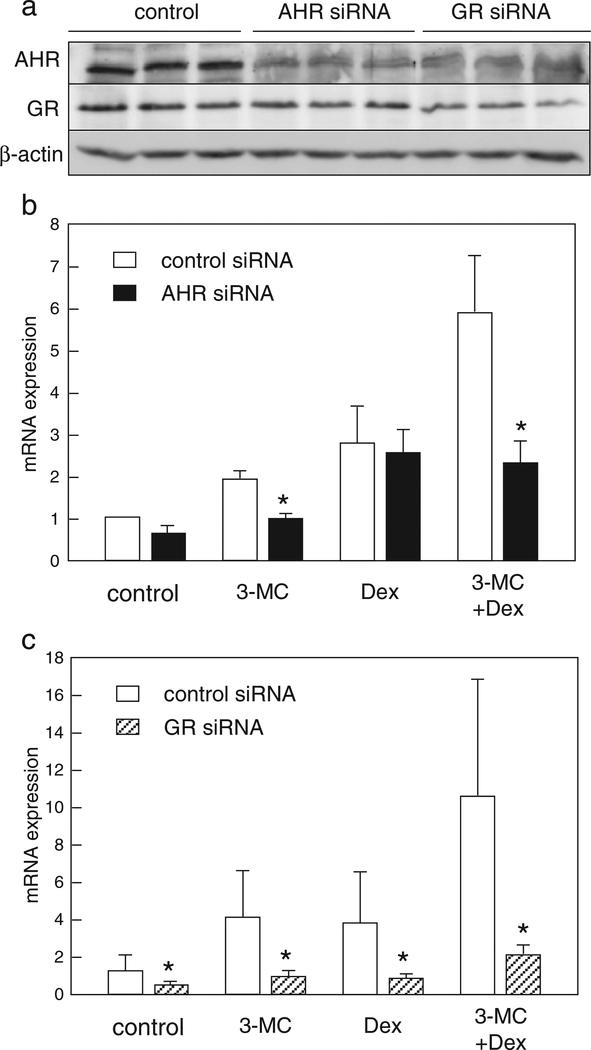
Fig. 4.
AHR- or GR-knockdown abolishes induced expression of MT2A mRNA upon treatment with the agonists. HepG2 cells were transfected for 48 h with siRNA for AHR, GR, or control. (a) After transfection, cells were lysed and AHR and GR protein levels were measured by Western blot analyses. (b, c) After transfection, cells were treated for 9 h with 1 μM 3-MC and/or 100 nM Dex, or vehicle, and MT2A mRNA levels were determined using quantitative RT-PCR. The values are the mean ± SDof 3 (b) or 4 (c) independent experiments. Values with an asterisk are significantly different from the control siRNA at p < 0.05. Statistical analyses were conducted using Student’s t-tests.
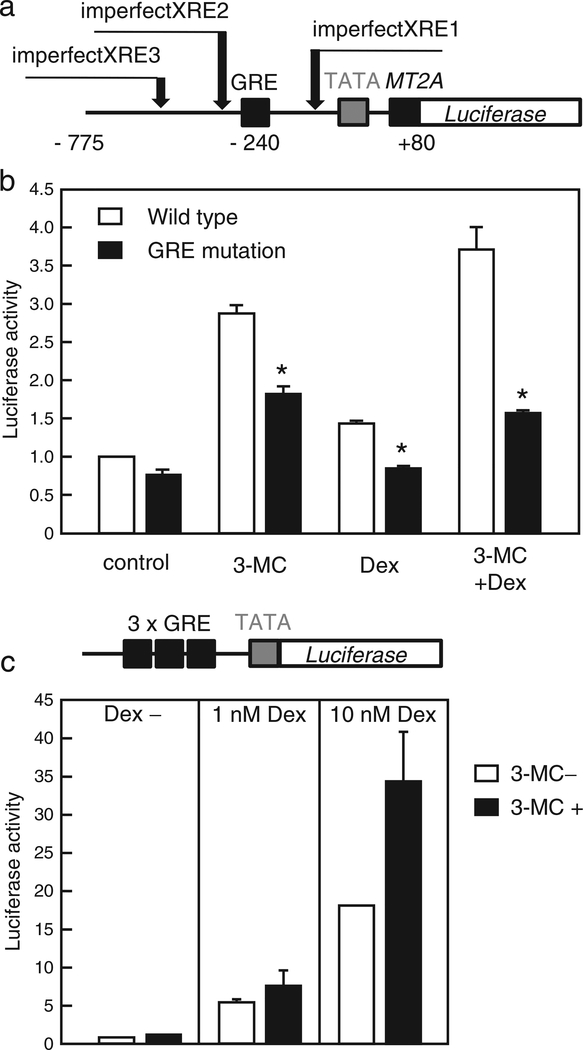
Fig. 5.
3-MC activates the GRE. (a) A schematic illustration of the pMT2A-Luc reporter construct showing 3 imperfect XRE in the MT2A promoter region. (b) HeLa cells were transfected with 0.25 μg of wild type (pMT2A-Luc) or GRE-mutant MT2A promoter-driven luciferase reporter plasmid together with 0.1 μg of AHR expression plasmid and 0.15 μg of pGL4-hRluc-TK for 3 h. Cells were treated for 24 h with 1 μM 3-MC and/or 10 nM Dex, or vehicle, and relative luciferase activities were determined. (c) HeLa cells were transfected with 0.25 μg of 3× GRE-driven luciferase reporter plasmid pGRE-Luc and 0.15 μg of pGL4hRluc-TK for 3 h. Cells were treated for 24 h with 100 nM 3-MC and/or 10 nM Dex, or vehicle, and luciferase activity was determined. Values are the mean ± SD of 3 (b) or 2 (c) independent experiments. Values with an asterisk were significantly different from the activity of the wild type MT2A promoter driven luciferase reporter at p < 0.05. Statistical analyses were conducted using Student’s t-tests.
Results
The AHR agonist induces metallothionein expression
We determined the mRNA levels of Mt1 and Mt2 in the liver from C57BL/6N wild-type and Ahr-null mice that were orally administered 5, 50, or 500 ng TCDD/kg bw·day−1 once a day for 18 days. In wild-type mice that were administered 50 and 500 ng TCDD/kg bw·day−1, Mt1 mRNA levels increased 2.5- and 3.2-fold, respectively, of that in vehicle-treated mice. Similarly, in these mice, Mt2 mRNA levels increased 4.6- and 7.1-fold, respectively, of that in control mice (Fig. 1a). In contrast, the mRNA levels of these genes were not altered in Ahr-null mice (Fig. 1b). In fact, the basal expression of Mt1 was slightly decreased in Ahr-null mice compared to the levels in wild-type mice (Fig. 1c). These data show that the increased mRNA levels of MTs induced by TCDD were mediated through the AHR signaling pathway in vivo.
Human MT2A is a homolog of mouse Mt1. Human hepatoma HepG2 cells were treated with AHR agonists for 9 h, and MT2A mRNA levels were measured. MT2A mRNA significantly increased in a dose-dependent manner following treatment with TCDD (Fig. 2a). It was also observed when the cells were treated with 3-MC (Fig. 2b). Similar results were obtained in mouse hepatoma Hepa-1c1c7 cells (data not shown). The 5′-flanking region of human MT2A gene has been well characterized as a transcriptional regulatory region by heavy metals and other extracellular stimuli. The transcriptional responsive elements, including multiple metal response elements, an anti-oxidant response element, and a glucocorticoid response element, are located in the 5′-flanking region of MT2A, whereas a sequence responsible for AHR-induced expression has not been found. To test whether MT2A transcription is activated by AHR, we constructed a luciferase reporter plasmid, pMT2A-Luc, containing the 5′-flanking region of MT2A (−775 to +80) (Karin and Richards, 1982). pMT2A-Luc was transfected into HeLa cells with or without an AHR expression plasmid, and cells were treated with 3-MC. Luciferase activity increased 1.6-fold upon addition of 3-MC. In AHR-overexpressing cells, luciferase activity increased 4.3-fold upon addition of 3-MC (Fig. 2c). These results indicate that MT2A transcription was upregulated by ligand-activated AHR.
Cotreatment with AHR agonist and Dex enhances human MT2A gene expression
Although MT2A transcription was activated by AHR, a typical XRE was not found in the MT2A promoter. Therefore, we presumed that AHR upregulates transcription of MT2A via interaction with a transcription factor that binds to the transcriptional regulatory elements of MT2A. We predicted that AHR might interact with GR because previous studies indicated that these two factors are potentially cooperative (Dvořák et al., 2008; Vrzal et al., 2009). First, to test this hypothesis, the effect of a GR agonist on TCDD-induced expression of MT2A was examined. HepG2 cells were treated with TCDD and/or Dex, a synthetic glucocorticoid. Following cotreatment with both TCDD and Dex, MT2A mRNA levels increased (2.6- and 2.5-fold) compared to the treatment with only TCDD or Dex, respectively (Fig. 3a), whereas CYP1A1 mRNA levels were not affected by Dex treatment (Fig. 3b). Similarly, the co-treatment with 3-MC and Dex resulted in increase of MT2A mRNA levels (4.2- and 2.3-fold) compared to the treatment with only 3-MC or Dex, respectively (Fig. 3c, upper panel). Western blot analyses demonstrated that the protein levels of AHR and GR were decreased by the respective ligands, and cotreatment of both ligands also did not increase their protein levels (Fig. 3c lower panel). In a dose-response study, a cooperative effect of 3-MC and Dex was observed in the MT2A mRNA expression (Fig. 3d). A statistical analysis of the data in Fig. 3c using two-factor ANOVA revealed a significant correlation (p < 0.0001) between the groups in the absence and presence of Dex, indicating that 3-MC and Dex affected the MT2A expression synergistically. Reporter gene assays showed that native MT2A reporter activity increased following treatment with 3-MC or Dex alone, and cotreatment with these reagents significantly potentiated the effect of each reagent in HeLa cells (Fig. 3e).
AHR and GR proteins are required for the cooperative effect of 3-MC and Dex
To confirm that AHR and GR are required for the increased expression of MT2A mediated by 3-MC and Dex, RNA interference was performed in HepG2 cells. The AHR protein levels in the AHR-specific siRNAtransfected cells were reduced to 34% of the levels in the control siRNAtransfected cells (Fig. 4a). In addition, AHR knockdown suppressed the increase in MT2A mRNA induced by 3-MC, but not by Dex (Fig. 4b). Importantly, the synergistic effect of 3-MC and Dex was abolished following AHR depletion (Fig. 4b). The protein level of GR in GRspecific siRNA transfected cells was reduced to approximately 40% of the level in control (Fig. 4a). GR knockdown suppressed the increase in MT2A mRNA mediated by Dex. Moreover, the effects of 3-MC disappeared in the absence or presence of Dex (Fig. 4c). Unexpectedly, depletion of GR expression could decrease AHR protein levels of that in control, whereas β-actin protein levels were not affected (Fig. 4a). This reduction in AHR protein level may partly influence the severe suppression of MT2A induction by 3-MC. These results indicate that both AHR and GR proteins are essential for the cooperative effect of 3-MC and Dex on MT2A induction.
GRE is a unique sequence affected by the AHR signaling pathway
A GRE consensus sequence is located in the MT2A transcriptional regulatory region (Karin and Richards, 1982). HeLa cells were transfected with the luciferase reporter plasmid, pMT2A-Luc, driven by the MT2A transcriptional regulatory region (Fig. 5a), together with the AHR expression plasmid. Luciferase activity increased upon 3-MC and/or Dex treatment (Fig. 5b). To examine whether the GRE is responsible for the induction of MT2A expression mediated by AHR, a point mutation was introduced into the GRE of pMT2A-Luc. As expected, the GRE mutation eliminated Dex-induced activation. The increased activity of pMT2A-Luc upon 3-MC addition was significantly suppressed by the GRE mutation, although the activity partially persisted. Furthermore, the additive effect of 3-MC and Dex on MT2A-Luc disappeared upon GRE mutation (Fig. 5b). These data suggest that the GRE contributes to cooperative transcriptional activation of MT2A by 3-MC and Dex, whereas 3-MC-induced activation partially depends on elements outside the GRE. Next, we tested whether 3-MC activated the 3× GRE-driven luciferase reporter plasmid. We observed a slight but insignificant enhancement of the luciferase activity in the 3× GRE-transfected cells treated with a low concentration of 3-MC (100 nM). However, in the presence of Dex, the 3× GRE construct was activated by 3-MC (Fig. 5c). Taken together, these results suggest that the GRE itself is activated by AHR along with the requirement of GR.
Although MT2A transcription was activated by AHR via GRE in the MT2A 5′-flanking region, no typical XRE motifs were found in the MT2A promoter. However, there are 3 imperfect XRE (GCGTG) motifs in the MT2A promoter region, so we examined whether these sequences affected the cooperative MT2A induction by AHR and GR (Fig. 5a). A ChIP assay for the imperfect XRE1 and XRE3 provided evidence that AHR did not bind to any of these sites and that these XRE core sequences were not involved in the MT2A induction (data not shown). Additionally, a reporter gene assay showed that a point mutation in the imperfect XRE2, i.e. in the vicinity of GRE, did not affect the MT2A promoter activation in the presence of 3-MC or Dex (1.4- and 1.5-fold compared to control, respectively, in HepG2 cells). Thus, we conclude that these GCGTG motifs are not responsible for the AHR-inducible MT2A expression.
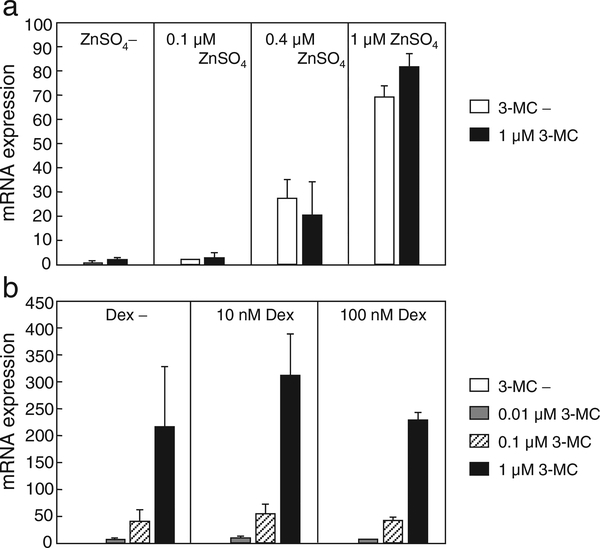
MTs, including human MT2A, are robustly induced by heavy metals via multiple metal response elements (MREs). To test whether the activation of the MRE is affected by AHR, HepG2 cells were treated with 3-MC and/or zinc, which is a typical activator of MREs. Levels of MT2A mRNA were increased by zinc in a dose-dependent manner, but were not influenced by 3-MC (Fig. 6a). To test whether XRE is affected by Dex, the expression level of CYP1A1, whose transcription is activated via multiple XREs, was measured. The 3-MC-induced CYP1A1 mRNA level was not influenced by Dex in HepG2 cells (Fig. 6b). Therefore, it was confirmed that the cooperative effect is specific for GRE.
Fig. 6.
The GRE is a unique sequence affected by the AHR signaling pathway. (a) HepG2 cells were treated for 9 h with 3-MC and/or ZnSO4 at the indicated concentrations, or vehicle, and MT2A mRNA levels were determined using quantitative RT-PCR. (b) HepG2 cells were treated for 9 h with 3-MC and/or Dex at the indicated concentrations, or vehicle, and the mRNA levels of CYP1A1 were determined using quantitative RT-PCR. Values are the mean ± SD of 2 independent experiments.
Protein–protein interaction between AHR and GR
To examine the recruitment of AHR to the MT2A gene regulatory region, ChIP assays were performed. In HepG2 cells co-treated with 3-MC and Dex the AHR antibody-precipitated DNA containing the GRE element in the MT2A promoter was enriched 2-fold compared to the AHR-independent control gene (EEF1A1), and 3-fold compared to that in the absence of the ligands (Fig. 7a). This result indicates that AHR is recruited to the MT2A gene regulatory region in the presence of ligands. Moreover, we examined the possibility of a physical interaction between AHR and GR. In mammalian two-hybrid assays using COS7 cells, when GAL4 DBD-fused AHR and VP16 activation domain-fused GR were co-expressed, reporter activity was clearly increased compared with the negative controls (Fig. 7b). Similar results were obtained in HeLa cells (data not shown). Moreover, coimmunoprecipitation assays demonstrated the complex of Myc-AHR and HA-GR (Fig. 7c, upper panel) and of Myc-GR and HA-AHR (Fig. 7c, lower panel). Importantly, the endogenous AHR–GR complex was detected by coimmunoprecipitation in HepG2 cells, and it appeared to form partially in a ligand-dependent manner (Fig. 7d). Thus, we conclude that AHR forms a complex with GR and modulates MT2A gene expression via the GRE, suggesting it could be a novel biological defense mechanism.
Discussion
AHR induces expression of xenobiotic-metabolizing enzymes such as CYP1A1, CYP1A2, and NAD(P)H quinone oxidoreductase 1 by binding to the XRE in a well-characterized mechanism. Here, we demonstrated a novel transcriptional mechanism in which AHR modulates GR-mediated transcriptional activation. We identified human MT2A as an intrinsic gene whose transcription is regulated by the AHR and GR interaction. In cultured cells, we showed that AHR forms a complex with GR that was recruited to the GRE in the MT2A regulatory region. Thus, expression of the human MT2A gene cooperatively increased with 3-MC and Dex treatment via GRE activation through the receptor-interaction mechanism.
Many groups have reported the possibility of an interaction between AHR and GR. In early studies, it was noted that glucocorticoids and TCDD work independently in the yolk sac and placenta, and affect palate formation via activation of each receptor in different cell types in the developing palate (Pratt, 1985). AHR regulates the expression pattern of several growth factors, such as TGFβ, in the embryonic palate (Puga et al., 2005). Abbott et al. showed that TCDD and hydrocortisone increased the expression of AHR and GR, and caused a synergetic effect in teratogenesis (Abbott, 1995, Abbott et al., 1999). However, the effects of AHR and GR agonists on the levels of these receptors and their target genes vary and appear to be regulated in a cell-specific manner and at multiple stages. Regarding AHR and its target genes, Dex reduced the AHR protein level, but increased the mRNA level of CYP1A2, but not CYP1A1, in human primary hepatocytes (Vrzal et al., 2009). On the other hand, Dex increased AHR mRNA and protein levels in a GR-dependent manner in mouse hepatoma Hepa-1 cells (Bielefeld et al., 2008). TCDD-induced Cyp1a1 mRNA expression was enhanced by Dex administration in rat hepatoma H4IIE cells but not in HepG2 cells (Sonneveld et al., 2007). Our data show that the basal protein level of AHR is dependent on GR level and that cotreatment of 3-MC and Dex did not increase either AHR or GR protein levels in HepG2 cells. Here, the mRNA level of CYP1A1 was not affected by Dex treatment in HepG2 cells which is consistent with previous studies (Sonneveld et al., 2007; Vrzal et al., 2009). Interestingly, in addition to the regulation accompanied by AHR and GR expression levels, the data suggest the possibility that the AHR and GR association affects other cellular functions. The increase in GRE-driven reporter activity by Dex was enhanced by TCDD in HepG2 cells (Dvořák et al., 2008), in agreement with our data. AHR ligands, such as polyaromatic hydrocarbons, synergistically enhanced Dex-induced activation of GRE- and MMTV promoter-driven reporter genes in ovarian granulosa HO23 cells (Wang et al., 2009). A combination study using an expression array and computational searches for over-represented elements showed frequent occurrences of GR binding sequences in the promoter region of TCDD-affected genes in a thymic epithelial cell line (Frueh et al., 2001). Our finding of a protein–protein interaction between AHR and GR can partially explain these reports.
Metallothionein proteins are small, cysteine-rich molecules containing free thiol residues that occupy one-third of their own amino acid residues. MTs play an important role in the homeostasis of essential metals (e.g., zinc) and biological protection against toxic metals (e.g., cadmium) via binding to these metals. Importantly, as large amounts of free thiol residues are able to scavenge oxygen free radicals effectively in vitro, MTs are believed to be anti-oxidative proteins in vivo (Kumari et al., 1998). We found that low-dose TCDD administration induced Mt1 and Mt2 in the mouse liver, which could be a biological defense against environmental toxicity.
MT genes are induced by various stimuli, including metals (zinc, cadmium, and copper), cytokines, physical stress, electrophiles, and anticancer drugs. Mammalian MTs are classic glucocorticoid-inducible genes (Karin et al., 1980; Hager and Palmiter, 1981; Karin and Richards, 1982; Kelly et al., 1997). For example, internal glucocorticoids increased MTs for restraint stress (Ghoshal et al., 1998). The gene regulatory regions of human MT2A and mouse Mt1 and Mt2 have been extensively analyzed and in addition to the functional GREs, multiple metal responsive elements and anti-oxidative response elements were identified in both species. Transcriptional activation via metal responsive elements is predominantly due to metal responsive transcription factor 1 (MTF1). MTF1 activity is regulated by reversible binding with zinc and is responsible for induction of MTs by heavy metals as well as by oxidative stress (Zhang et al., 2003). Thus, we tested an association between zinc and 3-MC and no cooperative effect was observed (Fig. 6a). However, the MT2A promoter luciferase construct was activated by 3-MC in AHR-overexpressing HeLa cells, and this activation was partly independent of the GRE (Fig. 5b). This partial activation might be related to an element in addition to the GRE.
In the last decade, it was reported that AHR mediates a variety of biological events by various mechanisms. In turn, AHR modulates gene expression without binding to the XRE. It was reported that the NFκB RelB subunit dimerizes with AHR, and the AHR/RelB dimer activates a novel DNA sequence distinct from XRE or NFκB binding site that exists in the regulatory region of chemokines such as IL-8 (Vogel et al., 2007). Estrogen receptor (ER) is also a member of the nuclear receptor superfamily and is reported to associate with the ligand-activated AHR. The estrogen-dependent activity of ER was inhibited by protein degradation, which occurred via direct binding with liganded AHR (Wormke et al., 2003). In contrast, AHR bound with ER in an estrogen-independent manner, and enhanced expression of ER-target genes related to reproduction via an estrogen response element (Beischlag and Perdew, 2005). Additionally, Hif1α, a member of the bHLH/PAS family as well as AHR, was reported to interact with GR (Kodama et al., 2003). Hif1α is activated by hypoxia-dependent stabilization and dimerization with ARNT, and the Hif1α/ARNT complex regulates target gene expression via a hypoxia response element (Epstein et al., 2001, Ivan et al., 2001). Hif1α forms a complex with GR, which enhances hypoxia-inducible gene expression (Kodama et al., 2003). However, Hif1α and GR seemed to form a complex indirectly through an interaction mediated by an unknown factor (Kodama et al., 2003). We could not determine the interaction domains using mammalian two-hybrid assays. Precise characterization of the protein interactions between the bHLH/PAS family and the nuclear receptor superfamily remains to be clarified in future studies. In this work, we identified a novel mechanism by which AHR and GR cooperatively regulate the human MT2A gene, which may be related to biological defense. AHR and GR proteins are expressed ubiquitously, and play essential roles in various biological events, such as biological clock or energy metabolism. Further studies are required to identify other target genes and to further evaluate the biological function of the AHR–GR interaction.
Acknowledgments
This work was partially supported by a grant from the Japan Food Industry Center. The authors gratefully acknowledge the technical assistance of Misato Maeda and Yoshie Higuchi in Tohoku University.
Abbreviations:
- 3-MC
3-methylcholanthrene
- AHR
aryl hydrocarbon receptor
- bHLH/PAS
basic helix–loop–helix/Per–Arnt–Sim
- ChIP
chromatin immunoprecipitation
- Dex
dexamethasone
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- MT
metallothionein
- MT2A
metallothionein 2A
- XRE
xenobiotic response element
Footnotes
Conflict of interest
There are no conflicts of interest.
References
- Abbott BD, 1995. Review of the interaction between TCDD and glucocorticoids in embryonic palate. Toxicology 105, 365–373. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Schmid JE, Brown JG, Wood CR, White RD, Buckalew AR, Heid GA, 1999. RT-PCR quantification of AHR, ARNT, GR, and CYP1A1 mRNA in craniofacialtissues of embryonic mice exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin and hydrocortisone. Toxicol. Sci 47, 76–85. [DOI] [PubMed] [Google Scholar]
- Bamberger CM, Schulte HM, Chrousos GP, 1996. Molecular determinants of glucocorticoid receptor function and tissue sensitivity to glucocorticoids. Endocr. Rev 17, 245–261. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Perdew GH, 2005. ERα-AHR-ARNT protein–protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription. J. Biol. Chem 280, 21607–21611. [DOI] [PubMed] [Google Scholar]
- Bielefeld KA, Lee C, Riddick DS, 2008. Regulation of aryl hydrocarbon receptor expression and function by glucocorticoids in mouse hepatoma cells. DrugMetab. Dispos 36,543–551. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR, 2005. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-mediated hepatotoxicity. Toxicol. Sci 85, 1048–1063. [DOI] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B, 2011. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci 124, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák Z, Vrzal R, Pávek P, Ulrichová J, 2008. An evidence for regulatory cross-talk between aryl hydrocarbon receptor and glucocorticoid receptor in HepG2 cells. Physiol. Res 57, 427–435. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ, 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- Fletcher N, Wahlström D, Lundberg R, Nilsson CB, Nilsson KC, Stockling K, Hellmold H, Håkansson H, 2005. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver, a microarray study. Toxicol. Appl. Pharmacol 207, 1–24. [DOI] [PubMed] [Google Scholar]
- Frericks M, Burgoon LD, Zacharewski TR, Esser C, 2008. Promoter analysis of TCDDinducible genes in a thymic epithelial cell line indicates the potential for cellspecific transcription factor crosstalk in the AhR response. Toxicol. Appl. Pharmacol 232, 268–279. [DOI] [PubMed] [Google Scholar]
- Frueh FW, Hayashibara KC, Brown PO,Whitlock JP Jr., 2001. Use of cDNAmicroarrays to analyze dioxin-induced changes in human liver gene expression. Toxicol. Lett 122, 189–203. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Wang Y, Sheridan JF, Jacob ST, 1998. Metallothionein induction in response to restraint stress. J. Biol. Chem 273, 27904–27910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YZ, Hogenesch JB, Bradfield CA, 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol 40, 519–561. [DOI] [PubMed] [Google Scholar]
- Hager LJ, Palmiter RD, 1981. Transcriptional regulation of mouse liver metallothionein-I gene by glucocorticoids. Nature 291, 340–342. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr., 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Karin M, Richards RI, 1982. Human metallothionein genes—primary structure of the metallothionein-II gene and related processed gene. Nature 299, 797–802. [DOI] [PubMed] [Google Scholar]
- Karin M, Andersen RD, Slater E, Smith K, Herschman HR, 1980. Metallothionein mRNA induction in HeLa cells in response to zinc or dexamethasone is a primary induction response. Nature 286, 295–297. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Sandgren EP, Brinster RL, Palmiter RD, 1997. A pair of adjacent glucocorticoid response elements regulate expression of two mouse metallothionein genes. Proc. Natl. Acad. Sci. U. S. A 94, 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Gazourian L, Quadri SA, Romieu-Mourez R, Sherr DH, Sonenshein GE, 2000. The RelA NF-κB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene 19, 5498–5506. [DOI] [PubMed] [Google Scholar]
- Kodama T, Shimizu N, Yoshikawa N, Makino Y, Ouchida R, Okamoto K, Hisada T, Nakamura H, Morimoto C, Tanaka H, 2003. Role of the glucocorticoidreceptor for regulation of hypoxia-dependent gene expression. J. Biol. Chem 278, 33384–33391. [DOI] [PubMed] [Google Scholar]
- Kumari MV, Hiramatsu M, Ebadi M, 1998. Free radical scavenging action of metallothionein isoforms I and II. Free Radic. Res 19, 93–101. [DOI] [PubMed] [Google Scholar]
- Kurachi M, Hashimoto S, Obata A, Nagai S, Nagahata T, Inadera H, Sone H, Tohyama C, Kaneko S, Kobayashi K, Matsushima K, 2002. Identification of 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive genes in mouse liver by serial analysis of gene expression. Biochem. Biophys. Res. Commun 292, 368–377. [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR, 2005. The glucocorticoid receptor blocks P-TEFb recruitment by NFκB to effect promoter-specific transcriptional repression. Genes Dev 19, 1116–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura J, Yamashita K, Nakamura K, Morita M, Takagi TN, Nakao K, Ema M, Sogawa K, Yasuda M, Katsuki M, Fujii-Kuriyama Y, 1997. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 2, 645–654. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ, 2004. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem 279, 23847–23850. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Miyabara Y, Suzuki JS, Sato M, Aoki Y, Satoh M, Yonemoto JTohyama, C., 2001. Induction of metallothionein in the livers of femaleSprague–Dawley rats treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Life Sci. 69, 1291–1303. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR, 2000. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14, 2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM, 2011. Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities. Toxicol. Sci 120, S49–S75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt RM, 1985. Receptor-dependent mechanisms of glucocorticoid and dioxin-induced cleft palate. Environ. Health Perspect 61, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga A, Tomlinson CR, Xia Y, 2005. Ah receptor signals cross-talk with multiple developmental pathways. Biochem. Pharmacol 69, 199–207. [DOI] [PubMed] [Google Scholar]
- Reeves PG, Nielsen FH, Fahey GC Jr., 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr 123, 1939–1951. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T, 1989. Molecular Cloning; A LaboratoryManual 2nd Ed. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- Sato S, Shirakawa H, Tomita S, Ohsaki Y, Haketa K, Tooi O, Santo N, Tohkin M, Furukawa Y, Gonzalez FJ, Komai M, 2008. Low-dose dioxins alter gene expression related to cholesterol biosynthesis, lipogenesis, and glucose metabolism through the aryl hydrocarbon receptor-mediated pathway in mouse liver. Toxicol. Appl. Pharmacol 229, 10–19. [DOI] [PubMed] [Google Scholar]
- Sonneveld E, Jonas A, Meijer OC, Brouwer A, van der Burg B, 2007. Glucocorticoidenhanced expression of dioxin target genes through regulation of the rat aryl hydrocarbon receptor. Toxicol. Sci 99, 455–469. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA, 1999. Ah receptor and NF-κB interactions, a potential mechanism for dioxin toxicity. J. Biol. Chem 274, 510–515. [DOI] [PubMed] [Google Scholar]
- Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F, 2007. RelB, a new partner of aryl hydrocarbon receptor mediated transcription. Mol. Endocrinol 21, 2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrzal R, Stejskalova L, Monostory K, Maurel P, Bachleda P, Pavek P, Dvorak Z, 2009. Dexamethasone controls aryl hydrocarbon receptor (AhR)-mediated CYP1A1 and CYP1A2 expression and activity in primary human hepatocytes. Chem. Biol. Interact 179, 288–296. [DOI] [PubMed] [Google Scholar]
- Wang SH, Liang CT, Liu YW, Huang MC, Huang SC, Hong WF, Su JG, 2009. Crosstalk between activated forms of the aryl hydrocarbon receptor and glucocorticoid receptor. Toxicology 262, 87–97. [DOI] [PubMed] [Google Scholar]
- Wormke M, Stone M.r, Saville B, Walker K, Abdelrahim M, Burghardt R, Safe S,2003. The aryl hydrocarbon receptor mediates degradation of estrogen receptor α through activation of proteasomes. Mol. Cell. Biol 23, 1843–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Georgiev O, Hagmann M, Günes Ç, Cramer M, Faller P, Vasák M,Schaffner W, 2003. Activity of metal-responsive transcription factor 1 by toxic heavy metals and H2O2 in vitro is modulated by metallothionein. Mol. Cell.Biol 23, 8471–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]