Abstract
Background:
Identifying genetic variation associated with plasma protein levels, and the mechanisms by which they act, could provide insight into alterable processes involved in regulation of protein levels. While protein levels can be affected by genetic variants, their estimation can also be biased by missense variants in coding exons causing technical artifacts. Integrating genome sequence genotype data with mass spectrometry-based protein level estimation could reduce bias, thereby improving detection of variation that affects RNA or protein metabolism.
Methods:
Here, we integrate the blood plasma protein levels of 664 proteins from 165 participants of the Tromsø Study, measured via TMT-mass spectrometry, with whole exome sequencing data to identify common and rare genetic variation associated with peptide and protein levels (pQTLs). We additionally use literature and database searches to prioritize putative functional variants for each pQTL.
Results:
We identify 109 independent associations (36 protein and 73 peptide), and use genotype data to exclude 49 (4 protein and 45 peptide) as technical artifacts. We describe two particular cases of rare variation: one associated with the complement pathway, and one with platelet degranulation. We identify putative functional variants and show that pQTLs act through diverse molecular mechanisms that affect both RNA and protein metabolism.
Conclusions:
We show that, while the majority of pQTLs exert their effects by modulating RNA metabolism, many affect protein levels directly. Our work demonstrates the extent by which pQTL studies are affected by technical artifacts, and highlights how prioritizing the functional variant in pQTL studies can lead to insights into the molecular steps by which a protein may be regulated.
Keywords: genetic association, genetics, proteonomics, proteomics, plasma
INTRODUCTION
Blood plasma is comprised of proteins generated from cells involved in diverse processes including thrombosis, hemostasis, immunity, and hematopoiesis. As it contains proteins from a wide variety of cells, blood plasma is a source for many potential biomarkers1 which may provide novel drug targets if they are causally related to disease2. Genetic variation which affects proteins can be used to assess the casual relationship between a particular biomarker and disease3; additionally, the molecular function of the variant can provide insight into processes important to the protein’s abundance. In particular, rare variation has proved to be an effective route for identifying drug targets4 as rare variants can have larger, wide reaching effects. By examining how variants that affect one protein are associated the levels of other proteins5, it may be possible to identify downstream targets of the initial protein. Additionally, delineating whether these genetic variants may act by modulating RNA or protein metabolism could provide insight(s) into alterable processes involved in the regulation of protein levels, thus elucidating insights into targeted therapeutics.
Recent advances in protein and genotype measurement have enabled the interrogation of genetic variants that affect protein levels (protein quantitative trait loci, pQTLs)6–9. While this advance has resulted in the identification of hundreds of plasma pQTLs in human samples, and is leading to insights into the proteomic consequences of risk for cardiovascular disease10, 11, it is currently unclear how often high throughput protein assays are affected by technical artifacts resulting from genetic variants. This is in part because the majority of previous pQTL studies6–10, 12 have utilized genotyping arrays that do not measure all variants that could disrupt the assay. In addition, these studies have utilized protein assays which sometimes rely on measurements from a single epitope (i.e., aptamer or antibody methods) and can therefore be less robust to genetic variation that causes an amino acid change which affects the assay’s quantification ability than assays that measure the total protein or multiple epitopes. Mass spectrometry, however, can measure multiple peptides per protein, with each peptide acting as a separate measure analogous to a separate epitope. DNA sequence information can then be used to identify specific peptides with missense variants that would result in artifactual associations due to inaccurate peptide quantification. Therefore, by integrating mass spectrometry peptide and protein level estimation with exome sequencing, it could be possible to identify true pQTLs and exclude those associated with artifactual associations, thereby improving the identification of variants associated with protein levels.
In this study, we utilized TMT-mass spectrometry to measure the blood plasma levels of 664 proteins across 165 participants from the Tromsø Study who have high depth exome sequence data available. We identified 109 independent, significant associations between common and rare genetic variation with peptide and protein levels. Our subsequent analyses determined that, while 60 of these (48 common cis, 3 rare cis, and 9 rare trans) were true associations, 49 (43 common cis and 6 rare cis) were likely due to a systematic, technical artifact driven by the presence of missense variants in coding exons. We examined common and rare associations for downstream effects on other proteins, and identified associations affecting the complement pathway and platelet degranulation. Using a combination of literature and database annotations, we prioritized and described putative functional variants for each locus. We show that approximately half of the pQTLs could be explained by variants with previous experimental evidence showing influence on the associated protein’s level. Furthermore, we identified many putative functional variants that affect protein metabolism and therefore would not have been detected in studies that solely examined gene expression. These results illustrate the potential for pQTL studies to characterize the effects of rare variation, and highlight a need for high throughput studies of protein levels to take into account technical artifacts caused by exonic genetic variation.
METHODS
TOP Guidelines statement
The whole exome sequencing data described in this study will not be made available, as the consent signed by the study participants does not allow the public release of these data. The proteome data has been made publically available through the MassIVE and proteomeXchange repositories and can be accessed at MSV000082489 and PXD010203, respectively. Full pQTL summary statistics are available from the corresponding author upon reasonable request.
IRB approval
This study was approved by an institutional review committee (The Regional Committee of Medical and Health Research Ethics in North Norway), and all subjects gave informed consent.
Methods are available as Supplemental Methods.
RESULTS
Data Generation
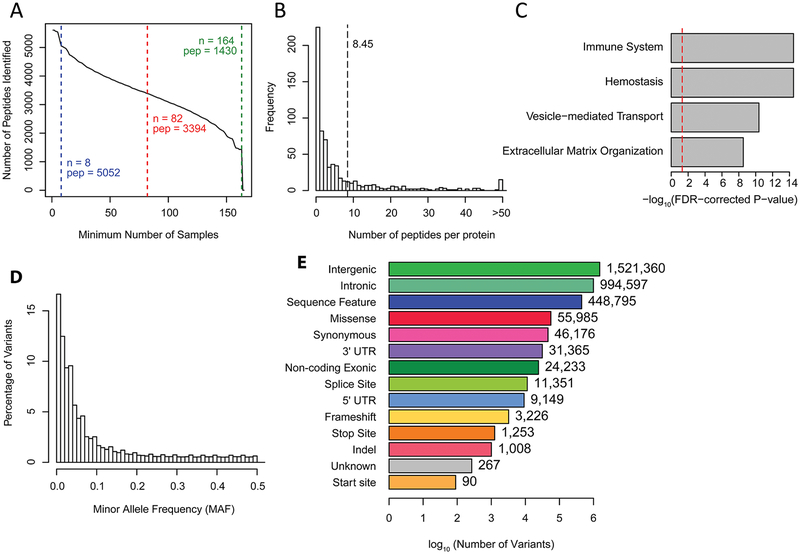
We examined peptide and protein levels from blood plasma, and genotype data from whole exome sequencing of blood DNA, from 165 individuals from the Tromsø Study (Figure 1A). These individuals consisted of 82 cases and 83 controls that were part of an effort to identify predictive biomarkers for venous thromboembolism13 and had genotype data available from whole exome sequencing generated as part of an ongoing study of the genetics of VTE14. To assess peptide and protein levels, we performed TMT-multiplexed mass spectrometry on blood plasma, identifying 5,608 peptides, corresponding to 664 proteins and 655 genes. Of the 5,608 peptides, 1,430 (25%) were present in all samples, 3,394 (61%) were identified in at least 50% (82 individuals), and 5,052 were identified in at least 5% (N=8) (Figure 2A). The identified peptides had an average length of 14.5 amino acids (range: 6 to 43) (Supplemental Figure 2A). We observed an average of 8.5 peptides mapping to each protein (range: 1 to 291) (Figure 2B); protein levels were calculated by summing these peptide measures. The functions of the proteins that were measured were consistent with their role in plasma, with the most enriched pathways (Reactome15 pathway analysis FDR q < 0.05) including the immune system and hemostasis (Figure 2C and Supplemental Figure 2B showing top level group associations, and Supplemental Table 2 showing all associations). From the whole exome sequencing data, we identified 501,682 genetic variants directly, and an additional 2,647,181 variants through imputation. Of the 3,148,863 total variants, 2,624,979 were evaluated in common variant analyses (minor allele frequency (MAF) ≥ 1%), and 1,690,437 were evaluated in rare variant analyses (MAF <5%) (Figure 2D). While most variants were in noncoding regions (intergenic or intronic), a total of 182,828 (5.8%) were located in UTR and exonic regions (Figure 2E). Overall, these analyses generated information on 664 proteins and 3,148,863 variants for genetic association analyses.
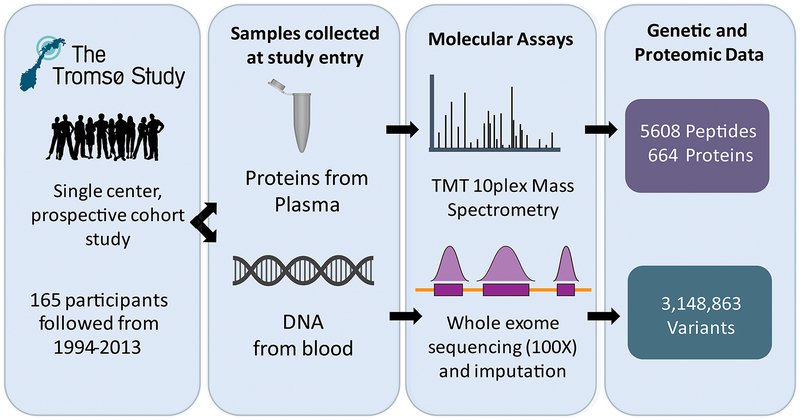
Figure 1.
Study overview: 165 individuals from The Tromsø Study were followed from 1994–2013. Between 1994 and 1995, blood plasma and whole blood were collected; blood plasma and whole blood were processed and subsequently used for protein quantification by mass spectrometry and whole exome sequencing, respectively. These analyses identified 5,608 peptides and 664 proteins from plasma, and 3,148,863 variants from whole blood, across all individuals.
Figure 2.
Description of protein and genotype data: (A) Cumulative distribution plot showing the number of peptides identified in at least N samples. 5,052 peptides were identified in at least 8 samples (blue), 3,394 peptides were identified in at least 82 samples (red), and 1,430 peptides were identified in all 165 samples (green). (B) Histogram showing the number of peptides identified for each of the 664 parent proteins. A mean of 8.45 peptides per parent protein were identified (dotted line). (C) Bar plot showing q-values from Reactome pathway analysis of the significantly enriched top level groups in the Reactome event hierarchy. The significance threshold of −log10(0.05) is shown by the red dotted line. (D) Histogram of the minor allele frequencies in this study for all 3,148,863 genetic variants identified across individuals. (E) Bar plot of the number of identified genetic variants within each SnpEff annotation. The number of variants with each annotation is also listed next to each bar.
Identification of peptide and protein cis pQTLs
We first identified cis pQTLs, i.e., those located near the gene encoding the plasma peptide and/or protein. We identified all variation within +/− 200 kb of the corresponding gene for each of the 5,608 peptides and 664 proteins. We tested for association between genetic variants and peptide or protein levels using EMMAX, a linear mixed effects model that includes a kinship matrix to account for population structure and family relatedness. Additionally, we modeled age, sex, BMI, smoking status, cancer status at the time of sample collection, VTE case-control status, and the TMT-multiplex experiment as covariates (see Methods). We identified 148 peptides and 31 proteins with significant associations (Bonferroni adjusted p < 0.05) with 80 and 31 cis genetic variants, respectively. Next, we identified additional independent significant pQTLs for each of the 148 peptides and 31 proteins by performing a step-wise analysis conditioned on the most significant variant, and found six peptides and two proteins that had a second cis genetic variant. In total, we identified 154 pQTLs associated with the levels of 148 peptides, and 33 pQTLs associated with the levels of 31 proteins (Supplemental Table 3).
Integration of peptide and protein pQTLs
As we expected that the peptide pQTLs would also be protein pQTLs for the parent protein, we investigated whether differences between peptide and protein pQTLs reflected technical artifacts introduced by genetic variants affecting the quantification process/pipeline. To examine the concordance between peptide and protein pQTLs, we determined the parent protein for all 154 peptide pQTLs and 33 protein pQTLs. We identified 67 unique parent proteins, of which 24 were associated with both a peptide pQTL and protein pQTL, 36 were only associated with peptide pQTL(s), and 7 were only associated with protein pQTL(s). For the 24 parent proteins with both peptide and protein pQTLs, we identified independent pQTL signals by examining whether the variants were different and not in linkage disequilibrium (LD; r2 < 0.2). We created three classifications for each independent pQTL: 1) those only associated with peptide levels (peptide-only pQTL), 2) those only associated with protein levels (protein-only pQTL), or 3) those associated with both peptide and protein levels (both pQTL). One of the peptide-only pQTLs (rs3742089) was associated with both C1L and its homologue C1LR and these were therefore considered the same parent protein. From this process, we obtained 91 independent pQTLs for 66 parent proteins, with some parent proteins being associated with multiple pQTL classes: 58 peptide-only pQTLs (42 parent proteins), 10 protein-only pQTLs, and 23 both pQTLs (22 parent proteins) (Supplemental Table 3). Using the exome sequencing data, we examined whether the peptides that were associated with the pQTLs (either directly or indirectly through LD with a polymorphic variant) affected the quantification process either by: 1) altering the sequence of the peptide, 2) altering the effectiveness of a trypsin digestion site, or 3) resulting in the association with a homologous protein, rather than the original parent protein. In total, 43 of the 91 independent pQTLs affected the quantification process by one of these three mechanisms and appeared to be technical artifacts (Supplementary Table 3). While the majority of artifact pQTLs were peptide-only pQTLs (39 of the 43), one protein-only pQTL and three both pQTLs appeared to be technical artifacts as well. After removing these 43 technical artifacts, the resulting data set had 48 independent associations with 37 proteins: 9 protein-only pQTLs, 19 peptide-only pQTLs (14 parent proteins), and 20 both pQTLs (19 parent proteins) with 5 parent proteins showing multiple types of independent pQTLs. Of note, 32 (8 protein-only, 15 peptide-only, and 9 both) of these independent associations were novel pQTLs that had not been identified in a previous study (Supplemental Table 3)6, 7, 9, 10, 16, 17.
Replication of common pQTLs
To replicate our findings, we compared our results to the recently published INTERVAL pQTL study18 which measured ~3000 plasma proteins using the SOMAscan aptamer approach. Of the 37 proteins that were associated with either a protein pQTL or a peptide pQTL in our final analysis, 7 were also measured in the INTERVAL study. For these 7 proteins, we found 9 pQTLs in our study. We obtained genome-wide association statistics from the INTERVAL study and observed that 7 of the 9 pQTLs (78%) were associated with the same protein at a nominal P < 0.05. As other pQTL studies have not made their full association statistics available, we investigated the overlap between our pQTLs and the genome-wide significant findings in other large-scale plasma pQTL studies6, 7, 9, 10, 16, and identified ten additional replications where the sentinel variant in our study was reported as genome-wide significant or was in LD (r2 > 0.8) with the reported variant (Supplementary Table 3). Additionally, the INTERVAL study raised concerns that protein coding variants could affect cis QTL results; therefore, we examined whether any of the proteins associated with the 43 pQTLs identified in our study due to technical artifacts were also reported in the INTERVAL study. We identified missense or digestion variants for 7 proteins that had been reported as cis pQTLs in the INTERVAL study, of which 6 were reported as non-significant after adjusting for protein coding variants. These results provide additional support that these variants bias protein measurements.
Collapsing variants to identify rare-variant cis pQTLs
To identify rare variants associated with protein levels, we tested the cumulative effects of sets of rare variants on peptide and protein levels. We collapsed rare variants using three different criteria: 1) MAF < 5%: all variants within the interval from 2kb upstream of the protein-coding gene to the transcription end of the gene with a minor allele frequency <5%; 2) Deleterious: all MAF <5% variants that were annotated using SNPEff19 as having an effect impact of high or moderate; and 3) CADD-score: all MAF <5% variants that have a PHRED-scaled CADD20 score >10. For each peptide or protein, after collapsing the rare cis variants for their corresponding gene, we identified associations between the peptide or protein, and rare variation using the optimal unified test SKAT-O21 which combines a kernel test with a burden test. We identified 16 rare cis pQTLs (12 associations with peptides and 4 associations with proteins), of which 10 were independent: 6 peptide-only, 2 protein-only, and 2 both pQTLs (Supplemental Table 4). As with common variation, we examined the associations for technical artifacts and found that all 6 of the peptide-only pQTLs overlapped a rare missense mutation; therefore, we excluded these 6 pQTLs from future analyses. As the threshold used for identifying common variation was MAF > 1%, some variants were included in both common and rare tests; we removed these associations, resulting in a total of 3 independent rare cis pQTLs, of which 2 were previously reported22, 23. Overall, while genetic variation was associated with substantial artifactual pQTLs in cis rare variant analysis, the significant non-artifactual associations were likely real as they were consistent with previous reports.
Trans rare-variant pQTLs
To identify downstream targets and pathways associated with pQTLs, and gain insight into the functional mechanism of the identified pQTLs, we tested for association in trans. We first tested all 2.6 million variants with MAF >1% genome wide for association (trans pQTLs) with each of the 5,608 peptides and 664 proteins; this method did not find any trans pQTLs at genome-wide significance (significance thresholds: peptide P < 8.91×10−12; protein P < 7.54×10−11). To increase our power, at each of the 655 loci encoding the measured proteins of this study, we performed association analyses using each of the three rare collapsing criteria to identify trans association with any of the peptides or proteins encoded at the other 654 loci. We identified 9 associations between rare variation and peptide levels (i.e., rare peptide-only trans-pQTLs) (Supplemental Table 5). One of the associations was a rare peptide-only trans-QTL between variation in FCN3, and levels of a peptide in the complement component C8 beta chain (C8B). FCN3 is an activator of the lectin complement pathway, and its pathway includes C8 in its final stages24. Notably, this variation was just below the significance threshold for being a rare peptide-only cis QTL for FCN3 (Figure 3A). We therefore examined the full established pathway of the lectin complement24. We observed that rare variation in FCN3 was associated with 8 other members of the lectin complement pathway at a nominal P < 0.05: C4a, C4b, C4BPa, C5, C6, C8b, C8a, and C8g (Supplemental Table 6), suggesting that the rare variation in FCN3 was broadly associated with the levels of proteins in the complement pathway. We next examined the other rare trans pQTLs, identifying five loci associated with levels of SERPINA1 (alpha-1-antitrypsin): CD109, CFL1, CLU, HYOU1, and RARRES2 (Figure 3B). Of the five genes, four encode proteins involved in platelet degranulation (Reactome15 enrichment FDR = 7.2×10−6). As alpha-1-antitrypsin is secreted into the plasma via platelet degranulation, these results suggest that rare variation in proteins associated with platelet degranulation could be important modulators of alpha-1-antitrypsin levels. The fifth gene, HYOU1, has not been implicated in platelet degranulation, but is upregulated in response to hypoxia25, an important risk factor for blood clotting26. Overall, these results suggest that rare variation in proteins can be associated with protein levels of downstream targets.
Figure 3.
Pathways identified from rare variation analyses: (A) An overview of the lectin complement pathway showing the relationship between FCN3 (Ficolin 3; teal) and the complement pathway. Nominal p-values are shown for the association between rare variation at the FCN3 locus and levels of the complement pathway proteins. C4, C3, C5, C8, and C6 were associated at a nominal P < 0.05 (purple), C2, C9, C7, or C5b were not associated (gray). (B) STRING database diagram of the five proteins associated with rare SERPINA1 variation (each labeled with their nominal association p-value). Connections between proteins are colored based on their evidence (see legend and STRING documentation).
Identifying putative common functional variants
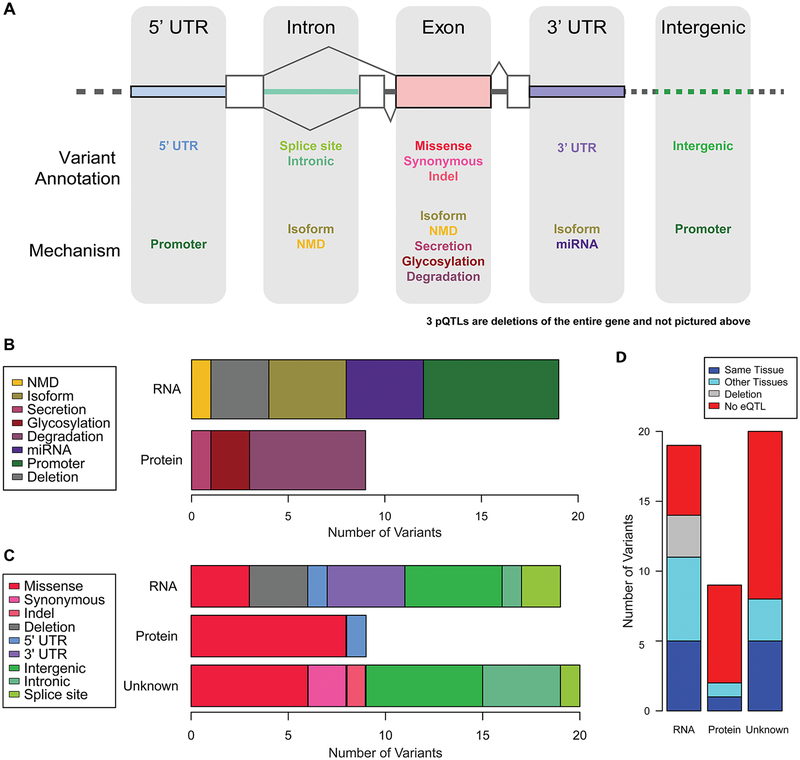
Due to LD, the most associated common variant (sentinel variant) may not be the causal variant. It is therefore necessary to examine variants in LD with the sentinel variant to prioritize causal variants that could be driving the association (putative functional variants: PFVs), and characterize the distribution of functional mechanisms underlying common pQTLs. Across the 48 common pQTLs, we observed an average of 151 variants in LD with each sentinel variant. We used a combination of database and literature searches to identify candidate variants at each pQTL locus (Figure 4A; see methods). For each locus, we categorized the strength of published evidence supporting a specific molecular mechanism (either proposed or validated) according to four categories ordered by strength: 1) known; 2) likely; 3) suggestive; or 4) unknown (see methods). Using these criteria, we selected the PFV at each locus as the variant with the strongest functional evidence (Supplemental Table 7). In total, we found 18 known, 5 likely, 5 suggestive, and 20 unknown PFVs; notably, 14 of the 23 PFVs with known or likely evidence were not the sentinel variant. Additionally, while a large proportion of the sentinel variants were intronic, the majority of PFV annotations were intergenic and coding annotations (Supplemental Figure 3A), suggesting that PFVs better capture causal variation than sentinel variants. Overall, approximately half of the 48 common pQTLs could be explained by variants previously experimentally shown to influence the associated protein’s level.
Figure 4.
Putative functional variant analyses: (A) Cartoon illustrating the genomic locations of variants with particular annotations and mechanisms, relative to the gene body of the pQTL. For example, indel annotated variants were only located within gene exons, but variants that have an underlying mechanism of “isoform” could be found in introns, exons, or the 3’ UTR. The three pQTLs where the PFV was a large genic deletion are not illustrated. (B) Stacked bar plot of the number of PFVs associated with each mechanism, subset by whether the mechanism affects the RNA molecule or the protein directly. (C) Stacked bar plot of the number of PFVs with each SnpEff annotation, subset by whether the PFVs’ mechanism affects the RNA molecule, the protein directly, or is unknown. (D) Stacked bar plot of the number of PFVs that were eQTLs in GETx, subset by whether the PFVs’ mechanism affects the RNA molecule, the protein directly, or is unknown.
Examining the functionality of PFVs
To examine the relative role of common genetic variation on different stages of protein level regulation – from gene expression to post-translational modifications – we classified the PFVs by their proposed molecular mechanism of action. We found the 28 PFVs with suggestive or better evidence to affect a wide range of processes: 19 (68%) were involved in RNA metabolism (7 affected the promoter, 4 affected isoform expression, 1 created a transcript that underwent nonsense-mediated decay, 3 resulted in large genic deletions, and 4 affected miRNA processing), and 9 (32%) were involved in protein metabolism (6 associated with protein degradation, 2 altered glycosylation, and 1 affected secretion) (Figure 4B; Supplemental Figure 3B). We next examined if PFV functional annotation varied by whether the PFV affected RNA metabolism or protein levels. We observed that PFVs associated with protein levels directly were more often missense variants, whereas PFVs that affected RNA levels were primarily located in non-coding regions (Figure 4C). The PFVs that did not have an established mechanism (i.e., unknown) were annotated as both missense and noncoding variants, suggesting that some of the unknown PFVs affect protein levels directly, whereas others affect RNA. As variants associated with RNA metabolism would also be expected to be associated with gene expression (e.g., an eQTL), we examined whether RNA metabolism PFVs were identified in GTEx more often than protein level PFVs. Excluding large deletions, we observed that PFVs which affected RNA metabolism (69%, 11/16) were more likely than protein PFVs (22%, 2/9) to be eQTLs (Figure 4D). The unknown PFVs were identified as eQTLs at an intermediate level (40%, 8/20), consistent with this group affecting both RNA and protein levels. These results suggest that, while common variants that affect protein levels often work through mechanisms associated with RNA, and therefore can be detected through eQTL analyses, many common variants affect protein levels without affecting RNA levels, and act through molecular mechanisms that are more challenging to measure with current high throughput methods.
Disease associations of common pQTLs
To determine whether the 48 common pQTLs were associated with human disease, we examined whether the sentinel variants (or variants in LD r2 >0.8) were associated with GWAS loci using PhenoScanner18. A total of seven pQTLs from six proteins (C4A, CFH, CFHR3, ECM1, LILRA3, and MST1) were associated with 10 diseases (Supplemental Table 8). In some cases, the relationship between the associated protein and the disease have been established, such as CFH for age-related macular degeneration27, or implicated, such as MST1 levels with inflammatory bowel disease28. For other GWAS loci, the relationships we identified were novel; for example, we observed that the pQTLs for C4A and ECM1 were associated with rheumatoid arthritis and atopic dermatitis, respectively. Overall, the identification of both known and novel GWAS association with 15% (7/48) of our pQTLs suggest that common cis pQTLs could aid in the identification of the causal genes underlying GWAS loci.
DISCUSSION
In this study, we leveraged TMT mass-spectrometry and deep whole exome sequencing data to identify 109 independent pQTLs, of which 60 (48 common cis, 3 rare cis, and 9 rare trans) were associated with 96 unique peptides and 30 proteins across the genome (Supplemental Table 9), while 49 were technical artifacts due to missense coding variants associated with specific peptide levels rather than the whole protein (43 common and 6 rare). We then utilized published papers and public databases to examine established molecular mechanisms underlying these pQTLs, and examined how often the mechanisms affected RNA or protein metabolism. We show that, while the majority of pQTLs exert their effects by modulating the gene’s RNA metabolism, many affect proteins directly through processes such as degradation, glycosylation, and translation. Our work thus not only shows the importance of identifying functional variation by directly assaying protein levels, but also highlights how identifying the causal variant in pQTL studies can lead to insights into the molecular steps by which the protein is regulated. Based on the types of protein mechanisms we describe, these results suggest that improved high throughput methods to assess variants that affect protein translation, modification, and degradation are needed.
It is currently unclear how often high throughput protein assays have technical artifacts resulting from genetic variants that affect the ability to correctly quantify peptide levels due to alterations of coding sequence through missense changes, isoform usage, or cleavage patterns. By integrating the individuals’ genotypes within coding sequences with standard TMT mass-spectrometry quantification techniques, we were able to identify pQTLs that were likely driven by genotype induced technical artifacts, and exclude them from our analyses. We observed the largest impact at the level of peptide-only associations, with the majority of independent peptide pQTLs (39/58; 67%) being driven by technical artifacts. The majority of independent associations at the protein level (87% of both pQTLs and 90% of protein-only pQTLs), however, were unaffected. These findings illustrate the importance of filtering variants that affect peptide quantification, and using quantification techniques that measure proteins at multiple locations and are therefore more resilient to peptide based quantification artifacts.
Rare variation is likely to be an important contributor to variation in protein levels. By focusing on the proteins that we measured, we identified trans associations between rare variation in FCN3 and the complement cascade. FCN3 has been established as a regulator of the lectin pathway of complement activation29. Additionally, an individual who was homozygous for a rare frameshift variant in FCN3 (FCN3+1637delC, one of the rare variants in our study) has been reported to have a lack of serum ficolin-3 and no complement activation via the ficolin‐3‐mediated pathway30. Our finding thus provides additional evidence that rare variation in FCN3, including FCN3+1637delC, is associated with variation in levels of the complement pathway proteins in the general population. Additionally, we identified five proteins with rare variation associated with levels of alpha-1 antitrypsin. Four of the proteins have been characterized as being involved in platelet degranulation, while the fifth, HYOU1, has been shown to act as an oxygen-inducible chaperone for proteins in the endoplasmic reticulum of macrophages31. Alpha-1 antitrypsin deficiency is a well-established genetic condition that predisposes an individual to chronic obstructive pulmonary disease, liver cirrhosis, and hepatocellular carcinoma32. While over 120 alleles of the SERPINA1 gene have been implicated in alpha-1 antitrypsin deficiency, variation in genes other than SERPINA1 have not yet been described32. While the individuals in this study have not been found to have alpha-1 antitrypsin deficiency, the finding that rare variation in many genes can contribute to alpha-1 antitrypsin plasma levels could have implications for the genetic architecture of the disorder. Additionally, seven of the common pQTL associations that we identified were associated with, or in LD with, human diseases. Thus, both common and rare pQTLs have the potential to provide insight into mechanisms underlying human disease.
Due to the fact that our analyses are based on high throughput data, the novel associations that we identified should be further validated by replication in an independent data set. As many of our findings were replicated in previous work, we expect that the majority of the novel associations will be replicated in future studies. It is also possible that we have missed associations due to incomplete coverage of the exome or imputation. While we had overall high sequencing coverage, exome capture methods are not fully complete, and therefore there may be coding and non-coding variants that were not captured and thus not tested. Additionally, the annotation of PFVs may have been biased for missense variants as we relied on published literature and databases, and past protein research may have focused on studying missense variation. However, as the majority of the PFVs that we identified were regulatory in nature, and the class of unknown variants showed annotations consistent with them affecting both RNA and protein metabolism, we believe that PFV annotations were likely not strongly biased for previously characterized missense variants.
Supplementary Material
ACKNOWLEDGMENTS:
The authors would like to thank Margaret K. R. Donovan for assistance with figure generation.
SOURCES OF FUNDING: This work was supported by an independent grant from Stiftelsen Kristian Gerhard Jebsen in Norway (J.B.H.) and by the Ray Thomas Edwards Foundation and the University of California Office of the President (D.J.G.). T.S. was supported by an institutional award to the UCSD Genetics Training Program from the National Institute for General Medical Sciences, T32 GM008666. W.W.G. was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number F31HL142151. J.D.L. is an IRACDA fellow supported by NIGMS/NIH (K12GM068524).
Footnotes
DISCLOSURES: None.
REFERENCES:
- 1.Jacobs JM, et al. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–85. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess S, et al. Mendelian randomization: where are we now and where are we going? Int J Epidemiol. 2015;44:379–88. [DOI] [PubMed] [Google Scholar]
- 4.Cohen JC, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. [DOI] [PubMed] [Google Scholar]
- 5.MacKeigan JP, et al. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Res. 2003;63:6928–34. [PubMed] [Google Scholar]
- 6.Johansson Å, et al. Identification of genetic variants influencing the human plasma proteome. Proc Natl Acad Sci U S A. 2013;110:4673–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 2008;4:e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, et al. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS One. 2013;8:e70269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folkersen L, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon T, et al. Associations Between Common and Rare Exonic Genetic Variants and Serum Levels of Twenty Cardiovascular-Related Proteins: The Tromsø Study. Circulation: Cardiovascular Genetics. 2016:CIRCGENETICS. 115.001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen SB, et al. Discovery of novel plasma biomarkers for future incident venous thromboembolism by untargeted synchronous precursor selection mass spectrometry proteomics. J Thromb Haemost. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carson AR, et al. Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinformatics. 2014;15:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabregat A, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lourdusamy A, et al. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet. 2012;21:3719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun BB, et al. Consequences Of Natural Perturbations In The Human Plasma Proteome. bioRxiv. 2017. [Google Scholar]
- 18.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, et al. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stengaard-Pedersen K, et al. Inherited deficiency of mannan-binding lectin-associated serine protease 2. N Engl J Med. 2003;349:554–60. [DOI] [PubMed] [Google Scholar]
- 23.Brantly M, et al. Repair of the secretion defect in the Z form of alpha 1-antitrypsin by addition of a second mutation. Science. 1988;242:1700–2. [DOI] [PubMed] [Google Scholar]
- 24.Garred P, et al. MBL2, FCN1, FCN2 and FCN3-The genes behind the initiation of the lectin pathway of complement. Mol Immunol. 2009;46:2737–44. [DOI] [PubMed] [Google Scholar]
- 25.Schofield CJ, et al. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–54. [DOI] [PubMed] [Google Scholar]
- 26.Reitsma PH, et al. Mechanistic view of risk factors for venous thromboembolism. Arterioscler Thromb Vasc Biol. 2012;32:563–8. [DOI] [PubMed] [Google Scholar]
- 27.Toomey CB, et al. Regulation of age-related macular degeneration-like pathology by complement factor H. Proc Natl Acad Sci U S A. 2015;112:E3040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Narzo AF, et al. High-Throughput Characterization of Blood Serum Proteomics of IBD Patients with Respect to Aging and Genetic Factors. PLoS Genet. 2017;13:e1006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garred P, et al. A journey through the lectin pathway of complement-MBL and beyond. Immunol Rev. 2016;274:74–97. [DOI] [PubMed] [Google Scholar]
- 30.Munthe-Fog L, et al. Immunodeficiency associated with FCN3 mutation and ficolin-3 deficiency. N Engl J Med. 2009;360:2637–44. [DOI] [PubMed] [Google Scholar]
- 31.Ozawa K, et al. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J Clin Invest. 2001;108:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoller JK, et al. A review of alpha1-antitrypsin deficiency. Am J Respir Crit Care Med. 2012;185:246–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.