Abstract
Among patients with asthma, heterogeneity exists regarding the pattern of airway inflammation and response to treatment, prompting the necessity of recognizing specific phenotypes. Based on the analysis of inflammatory cell count in induced sputum, patients with asthma can be classified in four unique phenotypes; eosinophilic, neutrophilic, mixed granulocytic, and paucigranulocytic asthma (PGA). PGA is an asthma phenotype with no evidence of elevated numbers of eosinophils or neutrophils in sputum or blood, and in which anti-inflammatory therapies are ineffective in controlling symptoms. While under-investigated, PGA is the most common asthma phenotype in patients with stable asthma. However, PGA is sometimes underestimated due to the exclusive reliance on induced sputum cell count which is variable among cohorts of studies prompting the necessity of developing improved biomarkers. Importantly, investigators have reported that inhaled corticosteroids had limited effect on airway inflammatory markers in patients with PGA defining, therefore, PGA as a potentially “steroidinsensitive” phenotype that requires exploration of alternative therapies. PGA manifests as an uncoupling of airway obstruction from airway inflammation that can be driven by structural changes within the airways such as airway smooth muscle (ASM) tissue hypertrophy. Animal models provide evidence that processes evoking airway hyperresponsiveness and ASM thickening occur independent from inflammation and may be a consequence of a loss of negative homeostatic processes. Collectively, further understanding of PGA with focus on the characterization, prevalence, clinical significance and pathobiology derived from animal studies will likely provide precision therapies that will improve PGA clinical outcomes.
Keywords: Irreversible airway obstruction, airway remodeling, structural cells, airway smooth muscle, steroid insensitivity, asthma phenotypes, asthma endotypes, precision medicine, biomarkers
Background.
Among patients with asthma, heterogeneity exists regarding the pattern of airway inflammation and response to treatment, prompting the necessity of recognizing specific phenotypes (1). Based on the analysis of inflammatory cell count in induced sputum, patients with asthma can be classified into four subtypes (2, 3) (Table 1). “Eosinophilic asthma (EoA)” describes patients with elevated sputum eosinophils (>3%) and whose symptoms are controlled by treatments aimed at suppressing eosinophils (4, 5). “Neutrophilic asthma (NeuA)” characterizes patients with elevated sputum neutrophils (≥61%5 or ≥64%6) (4, 6). When sputum eosinophils and neutrophils are both increased, patients can be classified as having a “mixed granulocytic asthma (MGA)”. Finally, patients can be determined to have “paucigranulocytic asthma (PGA)” if there is no evidence of elevated sputum eosinophils or neutrophils, and if treatments aimed at suppressing eosinophils and neutrophils are ineffective in controlling symptoms (2, 6). Occasionally, NeuA together with PGA are referred to as “non-eosinophilic asthma (NEoA)” (7). This review addresses current evidence on PGA, with focus on pathobiology derived from animal studies, characterization, prevalence, clinical significance and treatment strategies.
Table 1.
Asthma phenotypes.
| Asthma phenotypes | Eosinophilic | Non-Eosinophilic | ||
|---|---|---|---|---|
| Eosinophilic | Mixed Granulocytic | Neutrophilic | Paucigranulocytic | |
| Sputum Eosinophils* | >3% | >3% | <3% | <3% |
| Sputum Neutrophils* | <61% or <64% | >61% or >64% | >61% or >64% | <61% or <64% |
| Response to treatment** | + | +/− | - | - |
Commonly accepted thresholds;
treatment aimed at suppressing eosinophils.
Pathobiology of PGA: Insights from animal studies.
We and others have reported an uncoupling of airway obstruction (AO) from airway inflammation (AI) in several animal studies and demonstrated the role of neuronal factors, non-immunological mediators, signaling molecules and susceptible genes (Table 2). While most animal studies reported a different degree of overlap and interactions between AI and AO, we will focus mainly on animal studies reporting a significant uncoupling of AO from AI.
Table 2.
Potential mechanisms underlying the uncoupling of airway obstruction from airway inflammation in animal studies.
| Factors | Mechanisms mediating AHR increase | References | |
|---|---|---|---|
| Neuronal Factors | Nerve Growth factor | Alters ASM neuronal control | 9 |
| Non-lmmunological Mediators | Eicosanoids/Oxidized lipids: 20-HETE | Enhances ASM contractility | 10 |
| Critical Signaling Molecules | Regulator of GPCR: RGS5 | Its absence enhances calcium mobilization | 11 |
| Transmembrane proteins: Cav-1 | Its absence induces TGFβl and Orail expressions | 12 | |
| Growth transcriptional factors: Egr-1 | Its absence worsens TGFa-induced pulmonary pathology via EGFR-dependent pathways | 13 | |
| Sex hormone receptors: ERa | Its absence upregulates M2R | 14 | |
| Susceptible asthma genes present in chromosome 17q21 | GSDMB | Activates TGFβl and 5-LOX pathways | 16 |
| ORMDL3 | Induces TGFβl and ADAM8 dependent activation of ATF-6 and SERCA2b | 15,17 | |
ASM: Airway Smooth Muscle;
20-HETE: 20-Hydroxyeicosatetraenoic acid;
GPCR: G Protein Coupled Receptors;
RGS5: Regulators of G Protein Signaling-5;
Cav-1: Caveolin-1;
TGFβ1: Transforming Growth Factor Beta 1;
Orai1: Calcium release-activated calcium channel protein 1;
Egr-1: Early Growth Response- 1;
EGFR: Epidermal growth factor receptor;
ERα: Estrogen Receptor alpha;
GSDMB: Gasdermin B;
5-LOX: 5-Lipoxygenase;
M2R: Muscarinic receptor 2;
ORMLD3: Orosomucoid-like-3;
ADAM8: A Disintegrin And Metalloproteinase-8;
ATF-6: Activating Transcription Factor-6;
SERCA2b: Sarcoplasmic/endoplasmic reticulum calcium ATPase 2b.
We and others previously demonstrated that changes in airway smooth muscle (ASM) contractile properties play an important role in the development of AHR associated with chronic airway diseases such as asthma. Indeed, the shortening of ASM in response to G-proteincoupled receptor (GPCR) stimulation regulates airway caliber and bronchomotor tone. Such shortening is initiated by increases in cytosolic Ca2+ concentrations and the subsequent activation of the contractile apparatus, including the phosphorylation of myosin light chain (MLC)20. Interestingly, a variety of stimuli such as inflammatory cytokines, pollutants, mechanical strain and some therapeutic agents can prime the ASM to become ‘non-specifically’ hyperresponsive to contractile agonists (comprehensively reviewed in (8)). Collectively, this evidence suggests that ASM contractile function can mediate AHR in chronic airway inflammatory diseases.
Animal-based models suggested an altered neuronal control of ASM contractility as a possible mechanism promoting airway hyperresponsiveness (AHR) uncoupled from AI. For instance, mice treated with nerve growth factor promoted AHR to a similar extent as allergensensitized mice but without any AI (9) implicating a neurogenic mechanisms in the pathophysiology of AHR. Interestingly, the role of non-immunological mediators has also been proposed in the regulation of AHR uncoupled from AI. We previously showed that ozone acutely induced AHR to carbachol independently from AI, an induction that is insensitive to corticosteroid (CS) treatment (10). This was associated with the release of eicosanoids/oxidized lipids, e.g. 20-HETE directly modulating ASM contractile function (10).
Other studies demonstrated the role of critical signaling molecules in promoting AHR dissociated from AI. The loss of the Regulator of G protein signaling 5 (RGS5) using knockout mice dramatically enhanced constitutive AHR and calcium mobilization in ASM, and had little effect on AI suggesting that i) the dysregulation of pro-contractile G protein-coupled receptor (GPCR) signaling, expressed “selectively” in ASM, promotes AHR, ii) inflammation was not a prerequisite for AHR, and iii) that underlying abnormalities in ASM contribute to asthma diathesis (11). Other studies established the involvement of caveolin-1 (Cav-1), a transmembrane protein acting as a structural scaffold for organization of cytoplasmic signal complexes, in the modulation of AHR uncoupled from AI. Indeed, allergen-challenged Cav-1 knockout mice had an increase in AHR that was independent of inflammation, an effect that was more apparent as the mice aged (12). Further, the contribution of transcription factors involved in cell growth such as early growth response-1 (Egr-1) has been also suggested in the control of AHR uncoupled from AI. Using TGF-alpha-transgenic mice crossed with Egr-1 knockout mice, Kramer and colleagues showed that the lack of Egr-1 markedly worsened TGF-alpha-induced pulmonary disease, evoking AHR to methacholine and no AI (13). A role for sex hormones in inducing AHR without AI was also reported. Using a murine model of allergen-induced AHR, the absence of estrogen receptor-alpha increased AHR without affecting AI after allergen sensitization and challenge (14). Collectively, these studies provide evidence in animal models that processes evoking AHR and ASM thickening “without” inflammation may be due to the loss of negative homeostatic processes.
In other studies, mice overexpressing genes present in Chromosome 17q21, a chromosome linked with asthma susceptibility, developed spontaneous increased AHR without AI (15–17). Mice overexpressing Gasdermin B (GSDMB), a gene highly linked to asthma, showed spontaneous increases in AHR and airway remodeling, with increased ASM mass in the absence of AI (16). This was mainly due to GSDMB-induced activation of TGF-β1 and 5lipooxgenase pathways (16). Similarly, mice overexpressing human Orosomucoid-like (ORMDL3), a gene also highly linked to asthma, developed spontaneous increased AHR, ASM mass, subepithelial fibrosis and mucus in the absence of AI (17). This is potentially due to an increase in mediators promoting airway remodeling such as TGF-β1, and a disintegrin and metalloproteinase-8. These molecules selectively activate an unfolded protein response pathway that includes Activating Transcription Factor-6 and its target gene Sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA2b) (17). In support of these observations, precision cut lung slices (PCLSs) derived from hORMDL3Zp3-Cre mice manifested increased airway luminal narrowing through mechanisms involving ASM SERCA2b (15). Of note, hPCLS is an ex-vivo alternative to in vivo models for studying basic mechanisms affecting bronchoconstriction. This model provides a novel ex vivo system that retains in situ structural cell interactions, mast cells, and ciliary beat frequency providing a physiologically relevant platform for understanding how contractile signaling pathways are regulated in vivo. Further, studies using hPCLS recapitulate clinical observations of AHR in the absence of circulating immune cells. Using this model, we identified critical molecular signaling events that promote AHR (11, 18). Collectively, current evidence suggests that AHR can be uncoupled from AI by upregulating the in vivo expression of specific asthma susceptible genes.
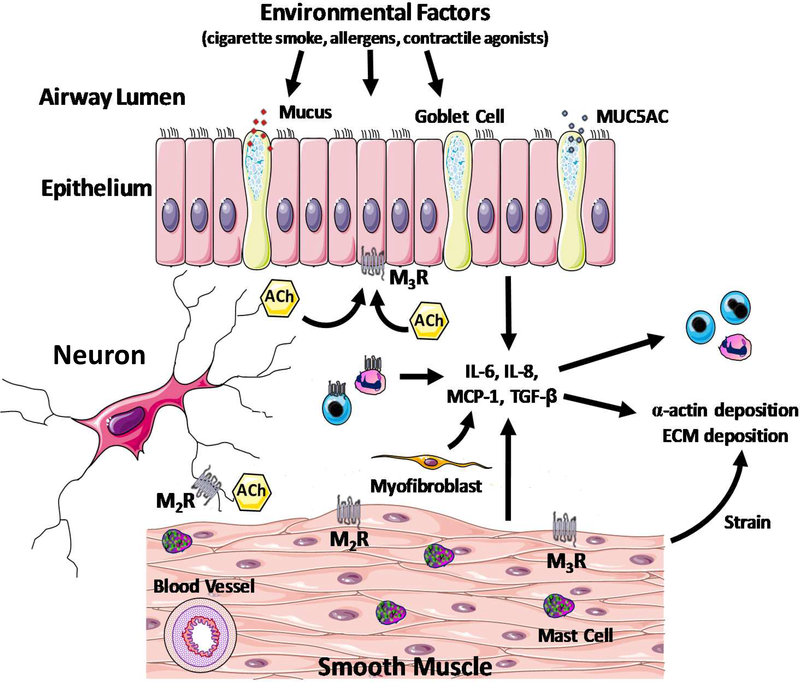
Together, these preclinical studies provide insight into the pathobiology of the uncoupling of AO from AI that will likely provide therapeutic targets that will improve PGA outcomes. However, since some discrepancies between animal and human systems exist, caution needs to be taken regarding the extrapolation of findings stemmed from animal studies to human asthma pathobiology. A conceptual model of the pathobiology of PGA that includes additional potential mechanisms is depicted in Figure 1.
Figure 1: A conceptual model of the pathobiology of paucigranulocytic asthma (PGA).
A variety of triggers induce alterations in structural cell function (myofibroblasts, epithelial, smooth muscle and mast cells) that engenders airway hyperresponsiveness. Typically, patients with PGA manifest insensitivity to corticosteroid therapy. In a corticosteroid-insensitive manner, structural cells also modulate airway inflammatory responses such as chemokine and cytokine secretion and leukocyte trafficking. MUC5AC: Mucin 5AC Oligomeric Mucus/Gel-Forming; Ach: Acetylcholine; M2R: Muscarinic Receptor 2; M3R: Muscarinic Receptor 3; IL: Interleukin; MCP-1: Monocyte Chemoattractant Protein 1; TGFb: Transforming growth factor beta; ECM: extracellular matrix.
Characterization/Identification of PGA.
Whereas the EoA and NeuA phenotypes likely reflect ongoing adaptive immune responses to allergens and innate immune responses to environmental triggers respectively, PGA remains poorly understood and its characterization is less well defined (19) (Table 3).
Table 3.
Potential markers discriminating asthma phenotypes.
| Asthma Phenotypes Potential Markers | EoA | NeuA | PGA | |
|---|---|---|---|---|
| Oxidative Stress enzymes* | PSSG | Reduced | Reduced | Unaltered** |
| Grx1 | Enhanced | Unaltered** | Enhanced | |
| Gal-3 & Gal-3BP | Gal-3 | Higher than NeuA | Lower than EoA and PGA | Higher than NeuA |
| Gal-3BP | Lower than NeuA | Higher than EoA and PGA | Lower than NeuA | |
| IL1RA & IL1 p | I LIRA | Higher than NeuA | Lower than EoA and PGA | Higher than NeuA |
| IL1β | Lower than NeuA | Higher than EoA and PGA | Lower than NeuA | |
| MMP9 & NE | MM9 | High | Low | Unaltered** |
| NE | Low | High | Unaltered** | |
| FeNO & AHR | FeNO | High | Low | Moderate |
| AHR | High degree | Low degree | Moderate degree | |
EoA: Eosinophilic Asthma;
NeuA: Neutrophilic Asthma;
PGA: Paucigranulocytic Asthma;
PSSG: S- glutathionylated proteins;
Grx1: glutaredoxins;
Gal-3: Galectin-3;
Gal-3BP: gal-3 binding protein;
IL-1RA: Interleukin-1 Receptor Antagonist;
MMP9: matrix metalloproteinase-9;
NE: neutrophil elastase;
FeNO: Fractional exhaled nitric oxide;
AHR: airway hyperresponsiveness to Mannitol;
Measured in sputum;
unaltered compared to healthy subject.
Induced sputum.
Analysis of induced sputum is commonly used to characterize asthma phenotypes. As mentioned earlier, PGA lacks elevation in eosinophils and neutrophils above certain thresholds. This needs to be appreciated within the context that variable cut-off points have been proposed to define neutrophilia. Threshold values commonly used for eosinophilia are ≥2% (20, 21), >2% (22) or ≥3% (4, 6, 23–25), of which ≥3% is the most sensitive biomarker of eosinophilic asthma (26). Importantly, eosinophil counts in sputum correlate with eosinophil numbers in bronchial tissue, suggesting that sputum eosinophils may serve as a surrogate for eosinophilic AI (27). Blood eosinophils can also provide a practical alternative to predict sputum eosinophilia, whereas blood neutrophils relate poorly to sputum numbers (23), (28). In contrast to eosinophil counts, to date there is little consensus regarding the cut-off value of sputuminduced neutrophils that defines NeuA as those proposed are extremely variable: >40% (29, 30), ≥50% (31), ≥61% (5), ≥64% (6), >65% (32), and ≥76% (23). Apparently, the higher the threshold value used to define NeuA, the greater the proportion of patients with PGA.
ASM phenotypic changes.
Considering the relative lack of increase in either eosinophils or neutrophils, PGA is considered to be non-inflammatory or at best a syndrome of low grade AI (28) that is associated with ASM dysfunction and AHR (19). In PGA patients, Zhang and colleagues proposed that the lack of increase in airway eosinophils and neutrophils was associated with airway remodeling, a possible consequence of “burnt-out” inflammation: past extensive inflammation having exhausted the pool of inflammatory cells, which ultimately manifests as a paucity of immunocytes (33). Such patients may have persistent airflow limitation and less variability in their disease (34). Undoubtedly, ASM mass increases, especially in those patients with severe disease (35–38). Thickening of the ASM layer is a pivotal component of airway remodeling that underpins exaggerated airway narrowing in asthma (39). However, the nature of this increased ASM mass remains unclear, but could involve myocyte hyperplasia, hypertrophy, and/or migration, driven by synergistic and additive effects of a number of micro-environmental cues (1). Interestingly, changes in ASM mass may exist independent of inflammation (35). Conceivably, in the absence of eosinophilia and neutrophilia, specific markers directly associated with increased ASM, such as thickening of the subepithelial basement membrane and elevated levels of TGFβ (33), could define PGA (40),(41).
Oxidative stress markers.
Studying disturbances in oxidative stress pathways may be an approach to phenotype PGA. Glutathione (GSH), the main pulmonary antioxidant, maintains the reduced state of protein thiols, mainly through covalent and reversible binding (42). The latter occurs under physiological conditions and is known as S-glutathionylated proteins (PSSG). GSH can be removed from proteins by glutaredoxins (Grx1) that restores the function of proteins targeted by PSSG (43). Evidence suggests that decreased lung function correlates with lower Grx1 and higher PSSG in induced sputum (44). Interestingly, such enzymes are altered in asthma (42). Indeed, sputum PSSG levels were significantly reduced in patients with EoA and NeuA but not in PGA patients (44). In addition, Grx1 protein levels were specifically enhanced in sputum supernatants of patients with EoA and PGA, but not in those with NeuA (44). Arguably, unaltered PSSG and increased Grx1 levels in sputum could specifically define PGA.
Galectin-3 and IL-1RA.
Analysis of sputum revealed reduced levels of galectin-3 (gal-3) that serve to recruit, activate, and remove neutrophils, in NeuA compared to EoA and PGA. In addition, while gal-3 binding protein (gal-3BP) and IL-1β levels were increased, the ratios of gal3/gal-3BP ratio and IL-1 receptor antagonist (RA)/IL-1β were significantly reduced in NeuA compared to EoA and PGA (45). Even though these markers do not discriminate between EoA and PGA, it does provide a provocative framework for exploring pathophysiological mechanisms characterizing unique asthma phenotypes.
MMP and neutrophilic elastase.
Whereas increased matrix metalloproteinase-9 (MMP-9) activity was found in EoA, higher neutrophil elastase activity and inactive MMP-9 levels were observed in NeuA. Within the specific asthma cohorts, an inverse relationship was observed between active MMP-9 (characteristic of EoA) and active neutrophil elastase (characteristic of NeuA) (3, 46). In PGA patients, however, levels of active MMP-9 and neutrophils elastase were unchanged compared to heathy controls (46).
Omics markers.
Analysis sputum transcriptomics may hold promise in identifying different asthma phenotypes. For instance, Baines and colleagues subjected whole-genome gene expression profiles from induced sputum of adults with stable asthma to unsupervised hierarchical cluster analysis (47). Interestingly, three distinct Transcriptional Asthma Phenotypes (TAPs) were identified that had similarities to previously defined sputum inflammatory phenotypes of EoA (TAP1), NeuA (TAP2), and PGA (TAP3) (47). In addition to transcriptomics, proteomics have recently been explored as a tool in phenotyping asthma. A large study by the Severe Asthma Research Program investigators focused on 18 cytokines detectable in bronchoalveolar lavage (BAL) and discriminated mild-to-moderate and severe asthmatic groups based on cytokine expression and its association with methacholine responsiveness (48). Whether PGA can similarly be identified using a specific protein profile in BAL remains, however, to be determined. Other studies employed protein microarray platforms using induced sputum and revealed differentially increased inflammatory protein markers among asthma phenotypes (49). However, the focus in this study was on phenotypes displaying either increased eosinophils and/or neutrophils and therefore, further analysis on markers relevant to PGA is warranted.
Fractional Exhaled nitric oxide (FeNO) and AHR.
FeNO levels can phenotype patients with asthma. Porsbjerg and colleagues showed that NeuA was associated with low levels of FeNO, whereas patients with PGA had levels of FeNO lower than those with EoA, but comparable to those with mixed granulocytic asthma (MGA) (50). Interestingly, when AHR response to mannitol was determined, similar results were observed: low degree of AHR in NeuA, moderate and comparable response in PGA and MGA, and highest degree of AHR was observed in EoA. These findings support that further sub-classification of asthma subtypes can be useful, as marked differences in FeNO and AHR were observed among these phenotypes (50).
Prevalence and Clinical Significance of PGA (Table 4).
Table 4.
Clinical characteristics associated with PGA patients in comparison to EoA patients:
| Clinical characteristics | PGA versus EoA |
|---|---|
| FEV1% | Similar (3) or higher (2, 23) |
| Atopy | Similar (3) or lower but still significantly present (2, 23) |
| Symptoms, emotions and activity | Similar (23) |
| Airway hyperresponsiveness (AHR) | Lower (23, 51) |
| Body Mass Index (BMI) | Lower (29) |
EoA: Eosinophilic Asthma;
PGA: Paucigranulocytic Asthma;
AHR: airway hyperresponsiveness;
BMI: Body Mass Index;
FEV1: Force Expiratory Volume during the first second.
Up to 50% of patients with asthma have a non-eosinophilic (NEoA) phenotype (7, 23, 29, 51). Different threshold values applied to determine sputum cell counts, as discussed earlier, may explain the variation in prevalence of different asthma subtypes. A large retrospective study conducted in 508 patients with asthma (cut-off values for eosinophils and neutrophils were ≥3% and ≥76%, respectively) revealed while 16% of patients were of the neutrophilic phenotype, the majority of asthma patients manifest an eosinophilic (42%) and paucigranulocytic (40%) phenotype (23). Similarly, in a study population of 93 patients with asthma (cut-off values for eosinophils and neutrophils were ≥1% and ≥61%, respectively), 31% of patients were identified as having PGA, 41% as having EoA, and 20% as having NeuA (3). An even higher proportion of NEoA phenotype (50%) was found in a multicenter study that enrolled 995 patients with asthma (21). More recently, a study conducted in 240 subjects revealed that 47.9% had PGA, while 40% manifested an eosinophilic subtype (2). Collectively, these studies suggest that PGA and EoA are the most common asthma phenotypes.
Wang and colleagues studied the relative frequency of asthma phenotypes (cut-off values for eosinophils and neutrophils were >3% and >61%, respectively) in stable and acute asthma in adults and children (52). In adults with stable asthma, the most frequent asthma phenotype was PGA (51.7%) followed by NeuA (27.6%). In children with stable asthma, the most frequent inflammatory phenotype was also PGA (49%) but was followed by EoA (28.6%). In contrast, acute exacerbations of asthma were predominantly eosinophilic in children (50%) and neutrophilic in adults (81.8%). Surprisingly, acute asthma was not associated with PGA in children or adults. Collectively, these findings suggest that PGA represents the most common asthma phenotype in stable asthma (52). Since these studies are a snapshot in the life cycle of asthma, it is possible that PGA, represents a cross sectional view related to disease activity rather than a stable phenotype. Further longitudinal studies, however, are needed to examine the stability of this phenotype.
Whether PGA is associated with asthma severity remains unclear. Evidence suggests that airway granulocytic inflammation is a common finding in severe asthma (53), (54). Since PGA manifests no sputum eosinophilia or neutrophilia, one may posit that PGA can manifest as a mild disease. Supporting this hypothesis, investigators showed that PGA patients had better lung function based on post-bronchodilation FEV1 (% predicted) and FEV1/FVC ratio as compared to patients with other asthma phenotypes (2). Further, PGA patients expressed lower levels of inflammatory markers in exhaled air and sputum, and severe refractory asthma occurred less frequently in PGA than in EoA and MGA (2). Similarly, others demonstrated that lung function was less altered in patients with PGA, while EoA phenotype patients exhibited higher FeNO levels, higher AHR and lower asthma control (23). Despite the fact that PGA was related to better lung function, a substantial proportion of patients with PGA (21.7%) was characterized as having severe refractory asthma and 14.8% of patients with PGA had an asthma control test score less than 19, suggesting that this subpopulation of PGA is not well controlled despite the absence of inflammatory cells in their sputum (2). Surprisingly, this latter cohort (i.e. PGA patients with severe refractory asthma), presented higher levels of FeNO, and sputum IL-13 and IL-8 compared to those with mild/moderate asthma (2). The presence of surrogate inflammatory markers in the absence of any granulocytic infiltration is intriguing and supports the need for a more comprehensive approach in the evaluation of AI in PGA patients with severe refractory asthma in order to develop adequate therapeutic strategies.
Importantly, airway remodeling is significantly more pronounced in patients with severe asthma than in patients with mild asthma (55). Evidence suggests that airway remodeling is relatively insensitive to ICS, as compared to allergen-induced airway inflammation (56, 57). Since airway remodeling represents a prominent histopathological finding of PGA, we speculate that PGA is more common in patients with severe asthma than patients with mild asthma and may contribute to inadequate ICS responsiveness seen in patients with severe asthma. Additional studies, however, are still needed to further explore the distinct difference of PGA in severe and mild asthma.
PGA: Current and Potential Future Therapeutic Strategies.
The identification of different asthma phenotypes based on sputum cell counts has supported the development of a variety of therapeutic strategies to control asthma symptoms.
Inhaled corticosteroids (ICS).
ICS are particularly effective against Th2-driven inflammation featuring mast cell and eosinophilic airway infiltration. Indeed, numerous studies in asthma revealed that ICS effectively reduces the percentage of sputum eosinophils (58, 59), represses the release of Th2 cytokines from lymphocytes (60) and eotaxin from epithelial cells (61). Unfortunately, ICS’ effects on innate immunity driven neutrophilic inflammation is rather poor (19).
Studies in a large asthma cohort (n=995) reported that in repeated measures analyses of patients with asthma not taking ICS, 22% of subjects had sputum eosinophilia on every occasion (persistent eosinophilia); 31% had eosinophilia on at least one occasion (intermittent eosinophilia); and 47% had no eosinophilia on every occasion (persistently non-eosinophilic) (21). Strikingly, anti-inflammatory therapy caused significant improvements in airflow obstruction in EoA, but not in persistently NEoA (21). Importantly, Ntontsi and colleagues provided strong evidence that such NEoA (which includes NeuA and PGA) represents a phenotype that does not benefit from CS treatment and that the CS dosing and adherence is not an explanation for poor CS responsiveness in this subgroup (2, 21). Accordingly, others compared the effects of ICS on sputum cell counts in PGA (28) and showed no significant differences between patients treated and untreated with ICS (28). Collectively, these studies suggest that ICS had limited effects on airway inflammatory markers in patients with PGA, thus defining PGA as a potentially “CS-insensitive” phenotype where the exploration of alternative therapeutic interventions is needed.
Therapies targeting airway remodeling.
Since granulocytic infiltration is not observed in PGA patients, symptoms in these patients may be primarily driven by ASM phenotypic changes or neuronal dysfunction. Thus, therapies directed towards ASM such as mast-cell directed therapies or in the most severe cases, bronchial thermoplasty, may benefit these patients. Although the mechanism of action is uncertain, bronchial thermoplasty aims to decrease ASM mass through the delivery of localized thermal energy (62, 63). Bronchial thermoplasty is indicated when AHR is severe (PC20 < 0.25) and when frequent exacerbations persist despite absent or controlled airway inflammation (62); this treatment may benefit the PGA subpopulation with severe refractory asthma. Since ICS-insensitive airway remodeling, the main driving component of PGA pathogenesis, is more prominent in patients with severe asthma than in patients with mild asthma, it is plausible that therapies such as bronchial thermoplasty aimed at targeting airway remodeling will be more beneficial in PGA patients with severe asthma than patients with mild asthma. Additional remodeling treatment options that could also be beneficial to this subpopulation were comprehensively reviewed elsewhere (64–66).
Future Perspectives.
The identification of PGA as a common phenotype in stable asthma both in adults and children has critical impact for future asthma research. Since surrogate inflammatory markers are present in the absence of any granulocytic infiltration, it is evident that in patients with PGA, other features than granulocytic infiltration should be investigated such as inflammatory mediators generated by airway structural cells such as ASM cells. Even though currently no drug convincingly reverses airway remodeling, non-pharmacological intervention by means of bronchial thermoplasty may benefit patients who suffer from severe refractory disease and are insensitive to current mainstay treatment options. Interestingly, previous studies identified a subgroup of patients with asthma insensitive to ICS treatment. Whether such CS insensitivity is caused by or a consequence of PGA, requires further investigation. While there are no compelling data to show that ICS are less effective in treating patients with PGA, we believe that ICS are less effective based on the pathogenesis of PGA where i) airway inflammation, the main target of ICS, is modest or absent in patients with PGA and ii) airway remodeling, which drives most of the pathogenesis of PGA, is relatively insensitive to ICS.
Since ICS increases airway neutrophils, we posit that ICS treatment of patients with PGA may increase airway neutrophils and switches asthma phenotype from PGA to NeuA, a possibility that also warrants further investigation. Since PGA represents a common phenotype in adults and children, studies aimed at identifying the molecular heterogeneity of PGA and its pathogenesis will likely provide precision therapies that will improve clinical outcomes of this disease.
Acknowledgement:
Funding sources:
This study was funded by NIH (HL 2P01HL114471–06, 7R01HL111541–06).
Abbreviations:
- 5-LOX
5-Lipoxygenase
- 20-HETE
20-Hydroxyeicosatetraenoic acid
- Ach
Acetylcholine
- ADAM8
A Disintegrin and metalloproteinase-8
- AHR
Airway hyperresponsiveness
- AI
Airway inflammation
- AO
Airway obstruction
- ASM
Airway Smooth Muscle
- ATF6
Activating Transcription Factor-6
- BAL
bronchoalveolar lavage
- Cav-1
Caveolin-1
- ECM
extracellular matrix
- EGFR
Epidermal growth factor receptor
- Egr-1
Early Growth Response-1
- EoA
Eosinophilic Asthma
- ER-alpha
Estrogen Receptor alpha
- FeNO
Fractional exhaled nitric oxide
- Gal-3
Galectin-3
- Gal-3BP
gal-3 binding protein
- GPCR
G Protein Coupled Receptors
- Grx1
glutaredoxins
- GSDMB
Gasdermin B
- GSH
Glutathione
- ICS
inhaled corticosteroid
- IL
Interleukin
- IL-1RA
Interleukin-1 Receptor Antagonist
- M2R
Muscarinic Receptor 2
- M3R
Muscarinic Receptor 3
- MCP-1
Monocyte Chemoattractant Protein 1
- MGA
mixed granulocytic asthma
- MMP9
matrix metalloproteinase-9
- MUC5AC
Mucin 5AC Oligomeric Mucus/Gel-Forming
- NE
neutrophil elastase
- NeuA
Neutrophilic Asthma
- NGF
nerve growth factor
- Orai1
Calcium release-activated calcium channel protein 1
- ORMLD3
Orosomucoid-like-3
- PCLS
precision cut lung slices
- PGA
Paucigranulocytic Asthma
- PSSG
S-glutathionylated proteins
- RGS5
Regulators of G Protein Signaling-5
- SERCA2b
Sarcoplasmic/endoplasmic reticulum calcium ATPase 2b
- TAPs
Transcriptional Asthma Phenotypes
- TGFβ
Transforming growth factor beta
Footnotes
Conflict of Interest:
The authors have nothing to disclose
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References.
- 1.Hirota JA, Nguyen TT, Schaafsma D, Sharma P, Tran T. Airway smooth muscle in asthma: phenotype plasticity and function. Pulm Pharmacol Ther. 2009;22(5):370–8. [DOI] [PubMed] [Google Scholar]
- 2.Ntontsi P, Loukides S, Bakakos P, Kostikas K, Papatheodorou G, Papathanassiou E, et al. Clinical, functional and inflammatory characteristics in patients with paucigranulocytic stable asthma: Comparison with different sputum phenotypes. Allergy. 2017;72(11):1761–7. [DOI] [PubMed] [Google Scholar]
- 3.Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology. 2006;11(1):54–61. [DOI] [PubMed] [Google Scholar]
- 4.Aleman F, Lim HF, Nair P. Eosinophilic Endotype of Asthma. Immunol Allergy Clin North Am. 2016;36(3):559–68. [DOI] [PubMed] [Google Scholar]
- 5.Taylor SL, Leong LEX, Choo JM, Wesselingh S, Yang IA, Upham JW, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018;141(1):94–103 e15. [DOI] [PubMed] [Google Scholar]
- 6.Svenningsen S, Nair P. Asthma Endotypes and an Overview of Targeted Therapy for Asthma. Front Med (Lausanne). 2017;4:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson NC. Novel approaches to the management of noneosinophilic asthma. Ther Adv Respir Dis. 2016;10(3):211–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amrani Y, Tliba O, Deshpande DA, Walseth TF, Kannan MS, Panettieri RA Jr. Bronchial hyperresponsiveness: insights into new signaling molecules. Curr Opin Pharmacol. 2004;4(3):230–4. [DOI] [PubMed] [Google Scholar]
- 9.Braun A, Quarcoo D, Schulte-Herbruggen O, Lommatzsch M, Hoyle G, Renz H. Nerve growth factor induces airway hyperresponsiveness in mice. Int Arch Allergy Immunol. 2001;124(1–3):205–7. [DOI] [PubMed] [Google Scholar]
- 10.Cooper PR, Mesaros AC, Zhang J, Christmas P, Stark CM, Douaidy K, et al. 20-HETE mediates ozone-induced, neutrophil-independent airway hyper-responsiveness in mice. PLoS One. 2010;5(4):e10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balenga NA, Jester W, Jiang M, Panettieri RA, Jr., Druey KM. Loss of regulator of G protein signaling 5 promotes airway hyperresponsiveness in the absence of allergic inflammation. J Allergy Clin Immunol. 2014;134(2):451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabehart KE, Royce SG, Maselli DJ, Miyasato SK, Davis EC, Tang ML, et al. Airway hyperresponsiveness is associated with airway remodeling but not inflammation in aging Cav1−/− mice. Respir Res. 2013;14:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer EL, Mushaben EM, Pastura PA, Acciani TH, Deutsch GH, Khurana Hershey GK, et al. Early growth response-1 suppresses epidermal growth factor receptor-mediated airway hyperresponsiveness and lung remodeling in mice. Am J Respir Cell Mol Biol. 2009;41(4):415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey MA, Card JW, Bradbury JA, Moorman MP, Haykal-Coates N, Gavett SH, et al. Spontaneous airway hyperresponsiveness in estrogen receptor-alpha-deficient mice. Am J Respir Crit Care Med. 2007;175(2):126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Miller M, Unno H, Rosenthal P, Sanderson MJ, Broide DH. Orosomucoid-like 3 (ORMDL3) upregulates airway smooth muscle proliferation, contraction, and Ca(2+) oscillations in asthma. J Allergy Clin Immunol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S, Miller M, Beppu AK, Mueller J, McGeough MD, Vuong C, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc Natl Acad Sci U S A. 2016;113(46):13132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koziol-White CJ, Yoo EJ, Cao G, Zhang J, Papanikolaou E, Pushkarsky I, et al. Inhibition of PI3K promotes dilation of human small airways in a rho kinase-dependent manner. Br J Pharmacol. 2016;173(18):2726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleich F, Demarche S, Louis R. Biomarkers in the Management of Difficult Asthma. Curr Top Med Chem. 2016;16(14):1561–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastie AT, Moore WC, Li H, Rector BM, Ortega VE, Pascual RM, et al. Biomarker surrogates do not accurately predict sputum eosinophil and neutrophil percentages in asthmatic subjects. J Allergy Clin Immunol. 2013;132(1):72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrath KW, Icitovic N, Boushey HA, Lazarus SC, Sutherland ER, Chinchilli VM, et al. A large subgroup of mild-to-moderate asthma is persistently noneosinophilic. Am J Respir Crit Care Med. 2012;185(6):612–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133(2):388–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schleich FN, Manise M, Sele J, Henket M, Seidel L, Louis R. Distribution of sputum cellular phenotype in a large asthma cohort: predicting factors for eosinophilic vs neutrophilic inflammation. BMC Pulm Med. 2013;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagener AH, de Nijs SB, Lutter R, Sousa AR, Weersink EJ, Bel EH, et al. External validation of blood eosinophils, FE(NO) and serum periostin as surrogates for sputum eosinophils in asthma. Thorax. 2015;70(2):115–20. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XY, Simpson JL, Powell H, Yang IA, Upham JW, Reynolds PN, et al. Full blood count parameters for the detection of asthma inflammatory phenotypes. Clin Exp Allergy. 2014;44(9):1137–45. [DOI] [PubMed] [Google Scholar]
- 26.Simpson JL, McElduff P, Gibson PG. Assessment and reproducibility of non-eosinophilic asthma using induced sputum. Respiration. 2010;79(2):147–51. [DOI] [PubMed] [Google Scholar]
- 27.Arron JR, Choy DF, Laviolette M, Kelsen SG, Hatab A, Leigh R, et al. Disconnect between sputum neutrophils and other measures of airway inflammation in asthma. Eur Respir J. 2014;43(2):627–9. [DOI] [PubMed] [Google Scholar]
- 28.Demarche S, Schleich F, Henket M, Paulus V, Van Hees T, Louis R. Detailed analysis of sputum and systemic inflammation in asthma phenotypes: are paucigranulocytic asthmatics really noninflammatory? BMC Pulm Med. 2016;16:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore WC, Hastie AT, Li X, Li H, Busse WW, Jarjour NN, et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol. 2014;133(6):1557–63 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair P, Gaga M, Zervas E, Alagha K, Hargreave FE, O’Byrne PM, et al. Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: a randomized, placebocontrolled clinical trial. Clin Exp Allergy. 2012;42(7):1097–103. [DOI] [PubMed] [Google Scholar]
- 31.Chaudhuri R, Norris V, Kelly K, Zhu CQ, Ambery C, Lafferty J, et al. Effects of a FLAP inhibitor, GSK2190915, in asthmatics with high sputum neutrophils. Pulm Pharmacol Ther. 2014;27(1):62–9. [DOI] [PubMed] [Google Scholar]
- 32.Nair P, Aziz-Ur-Rehman A, Radford K. Therapeutic implications of ‘neutrophilic asthma’. Curr Opin Pulm Med. 2015;21(1):33–8. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JY, Wenzel SE. Tissue and BAL based biomarkers in asthma. Immunol Allergy Clin North Am. 2007;27(4):623–32; vi. [DOI] [PubMed] [Google Scholar]
- 34.Haldar P, Pavord ID. Noneosinophilic asthma: a distinct clinical and pathologic phenotype. J Allergy Clin Immunol. 2007;119(5):1043–52; quiz 53–4. [DOI] [PubMed] [Google Scholar]
- 35.Benayoun L, Druilhe A, Dombret MC, Aubier M, Pretolani M. Airway structural alterations selectively associated with severe asthma. Am J Respir Crit Care Med. 2003;167(10):1360–8. [DOI] [PubMed] [Google Scholar]
- 36.Carroll N, Elliot J, Morton A, James A. The structure of large and small airways in nonfatal and fatal asthma. Am Rev Respir Dis. 1993;147(2):405–10. [DOI] [PubMed] [Google Scholar]
- 37.Ebina M, Yaegashi H, Chiba R, Takahashi T, Motomiya M, Tanemura M. Hyperreactive site in the airway tree of asthmatic patients revealed by thickening of bronchial muscles. A morphometric study. Am Rev Respir Dis. 1990;141(5 Pt 1):1327–32. [DOI] [PubMed] [Google Scholar]
- 38.Woodruff PG, Dolganov GM, Ferrando RE, Donnelly S, Hays SR, Solberg OD, et al. Hyperplasia of smooth muscle in mild to moderate asthma without changes in cell size or gene expression. Am J Respir Crit Care Med. 2004;169(9):1001–6. [DOI] [PubMed] [Google Scholar]
- 39.Lambert RK, Wiggs BR, Kuwano K, Hogg JC, Pare PD. Functional significance of increased airway smooth muscle in asthma and COPD. J Appl Physiol (1985). 1993;74(6):2771–81. [DOI] [PubMed] [Google Scholar]
- 40.Slats AM, Janssen K, van Schadewijk A, van der Plas DT, Schot R, van den Aardweg JG, et al. Bronchial inflammation and airway responses to deep inspiration in asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;176(2):121–8. [DOI] [PubMed] [Google Scholar]
- 41.Bradding P, Walls AF, Holgate ST. The role of the mast cell in the pathophysiology of asthma. J Allergy Clin Immunol. 2006;117(6):1277–84. [DOI] [PubMed] [Google Scholar]
- 42.Reynaert NL. Glutathione biochemistry in asthma. Biochim Biophys Acta. 2011;1810(11):1045–51. [DOI] [PubMed] [Google Scholar]
- 43.Xiong Y, Uys JD, Tew KD, Townsend DM. S-glutathionylation: from molecular mechanisms to health outcomes. Antioxid Redox Signal. 2011;15(1):233–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuipers I, Louis R, Manise M, Dentener MA, Irvin CG, Janssen-Heininger YM, et al. Increased glutaredoxin-1 and decreased protein S-glutathionylation in sputum of asthmatics. Eur Respir J. 2013;41(2):469–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao P, Gibson PG, Baines KJ, Yang IA, Upham JW, Reynolds PN, et al. Anti-inflammatory deficiencies in neutrophilic asthma: reduced galectin-3 and IL-1RA/IL-1beta. Respir Res. 2015;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson JL, Scott RJ, Boyle MJ, Gibson PG. Differential proteolytic enzyme activity in eosinophilic and neutrophilic asthma. Am J Respir Crit Care Med. 2005;172(5):559–65. [DOI] [PubMed] [Google Scholar]
- 47.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127(1):153–60, 60 e1–9. [DOI] [PubMed] [Google Scholar]
- 48.Brasier AR, Victor S, Boetticher G, Ju H, Lee C, Bleecker ER, et al. Molecular phenotyping of severe asthma using pattern recognition of bronchoalveolar lavage-derived cytokines. J Allergy Clin Immunol. 2008;121(1):30–7 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125(5):1028–36 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porsbjerg C, Lund TK, Pedersen L, Backer V. Inflammatory subtypes in asthma are related to airway hyperresponsiveness to mannitol and exhaled NO. J Asthma. 2009;46(6):606–12. [DOI] [PubMed] [Google Scholar]
- 51.Brooks CR, van Dalen CJ, Zacharasiewicz A, Simpson JL, Harper JL, Le Gros G, et al. Absence of airway inflammation in a large proportion of adolescents with asthma. Respirology. 2016;21(3):460–6. [DOI] [PubMed] [Google Scholar]
- 52.Wang F, He XY, Baines KJ, Gunawardhana LP, Simpson JL, Li F, et al. Different inflammatory phenotypes in adults and children with acute asthma. Eur Respir J. 2011;38(3):567–74. [DOI] [PubMed] [Google Scholar]
- 53.Louis R, Lau LC, Bron AO, Roldaan AC, Radermecker M, Djukanovic R. The relationship between airways inflammation and asthma severity. Am J Respir Crit Care Med. 2000;161(1):9–16. [DOI] [PubMed] [Google Scholar]
- 54.Duncan CJ, Lawrie A, Blaylock MG, Douglas JG, Walsh GM. Reduced eosinophil apoptosis in induced sputum correlates with asthma severity. Eur Respir J. 2003;22(3):484–90. [DOI] [PubMed] [Google Scholar]
- 55.Woodruff PG, Fahy JV. Airway remodeling in asthma. Semin Respir Crit Care Med. 2002;23(4):361–7. [DOI] [PubMed] [Google Scholar]
- 56.Chung KF. Defining phenotypes in asthma: a step towards personalized medicine. Drugs. 2014;74(7):719–28. [DOI] [PubMed] [Google Scholar]
- 57.Perry MM, Baker JE, Gibeon DS, Adcock IM, Chung KF. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am J Respir Cell Mol Biol. 2014;50(1):7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fahy JV, Boushey HA. Effect of low-dose beclomethasone dipropionate on asthma control and airway inflammation. Eur Respir J. 1998;11(6):1240–7. [DOI] [PubMed] [Google Scholar]
- 59.van Rensen EL, Straathof KC, Veselic-Charvat MA, Zwinderman AH, Bel EH, Sterk PJ. Effect of inhaled steroids on airway hyperresponsiveness, sputum eosinophils, and exhaled nitric oxide levels in patients with asthma. Thorax. 1999;54(5):403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corrigan CJ, Haczku A, Gemou-Engesaeth V, Doi S, Kikuchi Y, Takatsu K, et al. CD4 T-lymphocyte activation in asthma is accompanied by increased serum concentrations of interleukin-5. Effect of glucocorticoid therapy. Am Rev Respir Dis. 1993;147(3):540–7. [DOI] [PubMed] [Google Scholar]
- 61.Lilly CM, Nakamura H, Kesselman H, Nagler-Anderson C, Asano K, Garcia-Zepeda EA, et al. Expression of eotaxin by human lung epithelial cells: induction by cytokines and inhibition by glucocorticoids. J Clin Invest. 1997;99(7):1767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cox C, Kjarsgaard M, Surette MG, Cox PG, Nair P. A multidimensional approach to the management of severe asthma: Inflammometry, molecular microbiology and bronchial thermoplasty. Can Respir J. 2015;22(4):221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cox G, Thomson NC, Rubin AS, Niven RM, Corris PA, Siersted HC, et al. Asthma control during the year after bronchial thermoplasty. N Engl J Med. 2007;356(13):1327–37. [DOI] [PubMed] [Google Scholar]
- 64.Berair R, Brightling CE. Asthma therapy and its effect on airway remodelling. Drugs. 2014;74(12):1345–69. [DOI] [PubMed] [Google Scholar]
- 65.Hall C, Nici L, Sood S, ZuWallack R, Castro M. Nonpharmacologic Therapy for Severe Persistent Asthma. J Allergy Clin Immunol Pract. 2017;5(4):928–35. [DOI] [PubMed] [Google Scholar]
- 66.Sandstrom T Effects of pharmacological and non-pharmacological interventions. Clin Respir J. 2010;4 Suppl 1:41–8. [DOI] [PubMed] [Google Scholar]