Abstract
Background
We aim to establish a multicenter registry collecting clinical, imaging, and follow-up data for patients who undergo myocardial perfusion imaging (MPI) with the latest generation SPECT scanners.
Methods
REFINE SPECT - REgistry of Fast Myocardial Perfusion Imaging with NExt generation SPECT, uses a collaborative design with multicenter contribution of clinical data and images into a comprehensive clinical-imaging database. All images are processed by quantitative software. Over 290 individual imaging variables are automatically extracted from each image dataset and merged with clinical variables. In the prognostic cohort, patient follow-up is performed for major adverse cardiac events. In the diagnostic cohort (patients with correlating invasive angiography), angiography and revascularization results within 6 months are obtained.
Results
To date, collected prognostic data includes scans from 20,418 patients in 5 centers (57% male, 64.0 ± 12.1 years) who underwent exercise (48%) or pharmacologic stress (52%). Diagnostic data includes 2,079 patients in 9 centers (67% male, 64.7 ± 11.2 years) who underwent exercise (39%) or pharmacologic stress (61%).
Conclusion
The REFINE SPECT registry will provide a resource for collaborative projects related to latest generation SPECT-MPI. It will aid in the development of new artificial intelligence tools for automated diagnosis and prediction of prognostic outcomes.
Keywords: SPECT, high-efficiency SPECT, myocardial perfusion imaging, coronary artery disease, machine learning, artificial intelligence, quantitative analysis
INTRODUCTION
Coronary artery disease (CAD) is the leading cause of death in the United States in both men and women and remains a significant public health problem worldwide (1). Single photon emission computed tomography (SPECT) myocardial perfusion imaging (MPI) has long been a valuable diagnostic tool to evaluate patients with diagnosed or suspected CAD. Recently, SPECT-MPI has undergone major technological advances. These advances include high-efficiency scanners incorporating cadmium zinc telluride (CZT) solid-state detectors, specialized collimators, and software-based resolution recovery. The new systems have shown much higher imaging efficiency than traditional Anger SPECT cameras, by dramatically improved count sensitivity and image quality. Further advances have been recently achieved with CT-based attenuation correction.
We aim to develop a general imaging research resource for these next-generation SPECT scanners, and in doing so, we have developed the REgistry of Fast Myocardial Perfusion Imaging with NExt generation SPECT (REFINE SPECT). This registry uses a novel collaborative design with the contribution of clinical data and image datasets from nine (to date) investigative centers around the world into a comprehensive clinical-imaging database established at the central core laboratory at Cedars-Sinai Medical Center (CSMC) for automatic quantitative analysis. The registry will allow researchers to perform various collaborative projects, including the development of novel machine learning methods tailored for the analysis of next-generation SPECT MPI and subsequent integration of quantitative image data variables with “pre-scan” clinical information. This report describes the rationale and design features of the REFINE SPECT registry.
METHODS
Study Objective
The study objective is to build an imaging registry of patient-level new generation SPECT MPI data – the REFINE SPECT registry – to include structured data in the form of rich clinical data, stress testing and imaging data variables, and image datasets, with correlating follow-up information (in the prognostic cohort) or invasive coronary angiography information (in the diagnostic cohort).
Overall Study Design
We seek to establish a multicenter, international, registry collecting clinical, procedural, imaging, and follow-up data of patients undergoing latest generation SPECT MPI. The registry reflects the clinical routine of the investigative centers and uses a novel collaborative design with contribution and merger of clinical data and image datasets (Figure 1). Data are used to create a comprehensive clinical-imaging database at the central core laboratory for automatic analysis of images with advanced techniques, including machine learning methods.
Figure 1. Study Design for the REFINE SPECT Registry.
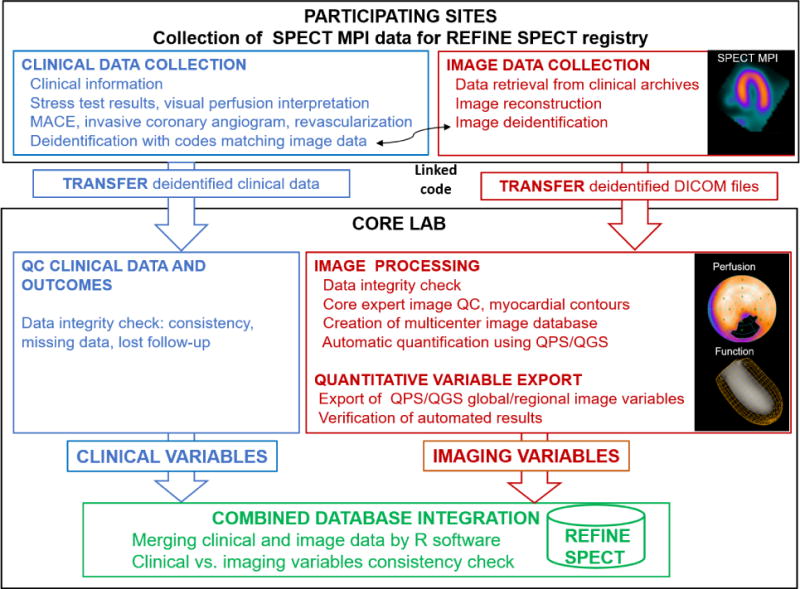
Overview of the data acquisition and processing scheme for REFINE SPECT registry. The activities include clinical data and image collection and de-identification at participating sites, data transfer to core laboratory (core lab) for further data processing (quality control, quantification in QPS/QGS software) and combined database integration. Blue: clinical data collection and analysis, red: imaging data collection and analysis, green: integration of clinical and imaging databases.
DICOM: Digital Imaging and Communications in Medicine; MACE: major adverse cardiac event; QC: quality control; QPS/QGS: Quantitative Perfusion SPECT/Quantitative Gated SPECT software.
Imaging datasets are collected for each patient in Digital Imaging and Communications in Medicine (DICOM) format. Clinical information is collected for each patient, including patient demographics, risk factors and history of cardiovascular events. For patients in the prognostic dataset, follow-up information for major adverse cardiac events (MACE) is also collected. For patients in the diagnostic dataset, invasive coronary angiography findings are collected. All imaging data is quality control checked by experienced core laboratory technologists and images are quantified with conventional nuclear cardiology software. Data management of the imaging and clinical data is conducted through local checks at the investigative center as well as at the core laboratory. Once the data is checked for completion (including the verification of the code match and the check of clinical data against image data), the imaging results and clinical data are merged using R software version 3.4.0 (Vienna, Austria) (2).
The investigators consulted the institutional ethics committee at each center to evaluate and approve the study protocol, before data collection and transfer. In addition, the investigational review board at Cedars-Sinai approved the overall collection of the data for the registry.
Target Population
The overall population includes consecutive patients at each center referred for SPECT imaging (overall time frame from 2009 until 2014) for suspected or known CAD who had invasive coronary angiography within 6 months or prognostic follow-up. If there were multiple scans for the same patients obtained on different dates, the first scan of the given patient was considered for the registry. The diagnostic cohort includes patients with no known prior CAD, myocardial infarction (MI), or coronary revascularization (percutaneous coronary intervention [PCI] or coronary artery bypass graft surgery [CABG]) who underwent clinically indicated invasive coronary angiography within 6 months of MPI. The prognostic cohort enrolls patients who have follow-up MACE data available. Participating centers enroll consecutive patients, collect data, de-identify clinical and image data and transfer data to the central core laboratory.
Study Endpoints (Prognostic Cohort)
The primary endpoint for the prognostic cohort is MACE, which comprises all-cause mortality, non-fatal MI, unstable angina (UA), or late (>3 months after MPI) coronary revascularization (PCI or CABG).
Study Endpoints (Diagnostic Cohort)
Outcomes for the diagnostic data of the registry include findings from invasive coronary angiography (ICA) performed within 6 months of the MPI and information regarding revascularization during this time.
Site Patient Follow-Up (Prognostic Cohort)
Follow-up for MACE is performed locally at each participating center. Non-fatal MI and UA are defined based on hospital admission for recent onset chest pain and admission diagnostics and MI by elevated cardiac enzyme levels and ischemic ECG changes (3, 4). All-cause-mortality determination methods differ by country. At the United States centers, all-cause mortality is retrieved from the Social Security Death Index and is combined with MACE obtained from hospital electronic medical records or patient contact. In Canada, all-cause mortality is determined by chart review at the institute and physician’s office through the OACIS Clinical Information System. In Israel, all-cause mortality is determined by the Ministry of Health Database.
Invasive coronary angiogram results, revascularization details and data regarding hospitalization due to unstable angina or MI are collected from email questionnaires, telephone contact, or medical records (including all clinics, cardiology groups, and hospital visits). For each patient considered to have a MACE, medical records regarding non-fatal events, are reviewed and verified for correctness by site physicians. After medical record and physician review, all data and all events are entered in the site spreadsheet. The first event for each patient is used as the MACE outcome.
Invasive coronary angiography and revascularization information (Diagnostic Cohort)
All coronary angiograms are visually interpreted by an on-site cardiologist. Luminal diameter narrowing of ≥ 70% in the left anterior descending artery (LAD), left circumflex artery (LCx), or right coronary artery (RCA)), or of ≥50% in the left main (LM) coronary artery is considered significant. Coronary artery dominance is also recorded. Dates of revascularization (if performed), revascularization type (PCI or CABG) and location (which coronary artery) are also collected.
Imaging Acquisition and Reconstruction
Data is acquired using 2 imaging positions (supine/prone or upright/supine), when applicable or one position with attenuation correction, when available. Reconstructed images are generated from the list mode data by vendor-recommended iterative reconstruction optimized for each scanner at each participating site. If CT is obtained for attenuation correction, CT images are collected, and attenuation corrected data are included (5).
Clinical MPI Reporting
Visual analysis is performed during clinical reporting. At each center, expert clinicians score SPECT perfusion images either by segmental scoring or by perfusion abnormality scoring according to site-specific protocols. If visual segmental scoring with a 17-segment American Heart Association model (6) is performed clinically, summed segmental scores are collected from centers. The overall interpretations are performed by expert readers during routine clinical reporting based on all available data, including stress perfusion data, raw projection data, gated functional data and clinical information. For consistent aggregate analysis of segmental and categorical results for all sites, clinical perfusion findings in the registry are re-categorized using a four-point scoring system (0: normal; 1: probably normal; 2: equivocal; 3: abnormal).
Clinical Data
A master template spreadsheet is provided by the core laboratory to all sites, to ensure a homogenized clinical data standard for the registry. The collected clinical variables are shown in Table 1. Investigative centers perform HIPPA (Health Insurance Portability and Accountability Act)-compliant de-identification of the data; the central core laboratory does not receive any information that would directly or indirectly identify the study patients.
Table 1.
Clinical Variables.
| Type | Variable |
|---|---|
| General Clinical Variables | Patient Location (In-patient, Out-patient, Emergency Department) |
| Age, Gender | |
| Height (cm), Weight (kg), Body Mass Index (kg/m2) | |
| Clinical Indications for Test | |
| Pre-Test Likelihood of Coronary Artery Disease | |
| Family History of Coronary Artery Disease | |
| Hypertension | |
| Diabetes Mellitus | |
| Dyslipidemia | |
| Currently Smoking | |
| Current Medications | |
| Anginal Presenting Symptoms | |
| Imaging Protocol | |
| Quality of MPI Study | |
| Visual Perfusion Assessment | |
| Summed Stress Score, Summed Rest Score, Summed Difference Score | |
| History of Heart Disease | Past Myocardial Infarction |
| Past PCI/CABG | |
| Past Transcatheter Aortic Valve Replacement | |
| Past Cardiac Transplant | |
| Past Other Open-Heart Surgery | |
| Peripheral Artery Disease | |
| Left Ventricular Hypertrophy | |
| Conduction Disease | |
| Stress variables | Pharmacologic Stress Agent |
| Stress Test Type Agent | |
| Exercise Protocol | |
| Resting Heart Rate (bpm) | |
| Peak Heart Rate at Stress (Pharmacologic or Exercise Stress) (bpm) | |
| Resting Blood Pressure (BP): Systolic, Diastolic (mmHg) | |
| Peak Blood Pressure at Stress: Systolic, Diastolic (mmHg) | |
| % of Maximal Predicted Heart Rate | |
| Exercise Duration (min) | |
| Reason for Termination | |
| ECG Response to Stress | |
| Clinical Response to Stress | |
| ECG ST Deviation (mm) Direction, and Slope | |
| Invasive Coronary Angiography and Revascularization | Invasive Coronary Angiography: Time Interval to MPI |
| Severity of Coronary Stenosis Per Coronary Artery Territory | |
| Revascularization: Time Interval to MPI, Type (PCI or CABG), vessel | |
| MACE Variables | Type MACE (All-Cause Death, Myocardial Infarction, Unstable Angina, Revascularization), MACE Interval from MPI, Follow-up Interval. |
ECG: Electrocardiogram; CABG: Coronary artery bypass grafting; PCI: Percutaneous coronary intervention, MACE: major adverse cardiac events.
SITE PROCESSES
Image Database
Image datasets are reconstructed and deidentified on-site. Study patient identifiers such as medical record number, names or dates of imaging tests are removed using dedicated image deidentification software. The images (in DICOM format) are anonymized, and data are transferred securely to the core laboratory at CSMC for quantification and analysis using HIPPA compliant storage. Each center retains a cross-referenced list containing patient code and link to the original image data; however, the REFINE SPECT registry does not contain any link to identifiable patient information, and core laboratory does not have any access to any identifiable information in the images.
All centers transfer anonymized DICOM images via Box web server (cloud service) or physical CDs with anonymized DICOM files. Once received, deidentified image data is checked for completeness and loaded into the REFINE SPECT imaging database.
CORE LABORATORY PROCESSES
Quality Control
The de-identified image files are quality control checked by experienced core laboratory technologists without knowledge of the clinical data. Automatically processed Quantitative Perfusion SPECT (QPS)/ Quantitative Gated SPECT (QGS) software (Cedars-Sinai Medical Center, Los Angeles, CA) is used to generate myocardial contours. When needed, image contours are adjusted to correspond to the myocardial boundaries (7). Any processing errors or technical problems with images are noted by the technologists.
Quantification with standard software
After quality control, images are quantified in QPS/QGS using batch-processing mode. This mode quantifies all image datasets for all patients in an automated mode while optimizing the computational resources required to process the image registry. The software outputs a single row of the image data variables quantified per image dataset, with multiple rows per patient –corresponding to static and gated studies for stress and rest acquisitions in different positions, with combined quantification if applicable (8), and with/without attenuation correction if applicable). Subsequently, the image data is reshaped to list one unique row per-patient, consolidating the patient’s perfusion and functional variables–with any missing variables set to blanks. If applicable, combined perfusion parameters, (computed from two positions) and ischemic perfusion deficit (difference of stress and rest) are also included (9).
In total, 32 imaging variable categories including general imaging, perfusion, and functional parameters are automatically quantified (Table 2). These variable categories include separate values for stress/rest and static/gated scans, values by three regional (left anterior descending, left circumflex, right coronary artery) and 17-segment myocardial models (over 290 variables if standard per vessel regions are used and over 3,500 if 17-segment variables are included).
Table 2.
Imaging Variables.
| Type | Variable | Stress and rest | Regional/segmental* |
|---|---|---|---|
| General Imaging Variables | Myocardial Counts (kCounts) | ✓ | |
| LV Segmentation QC Metrics | ✓ | ||
| Patient Position (Supine, Upright, Prone), Attenuation Correction | ✓ | ||
| LV Dimensions (mm) | ✓ | ||
| Myocardial Mass (g) | ✓ | ||
| LV Shape Index, Eccentricity | ✓ | ||
| Injected Dose (MBq) | ✓ | ||
| Perfusion Variables | Total Perfusion Deficit (%) | ✓ | ✓ |
| Perfusion Severity | ✓ | ✓ | |
| Perfusion Defect Extent (%) | ✓ | ✓ | |
| Stress-Rest Change | ✓ | ||
| Segmental Scores | ✓ | ✓ | |
| Normalized Raw Perfusion Uptake | ✓ | ✓ | |
| Functional Variables | Ejection Fraction (%) | ✓ | |
| Volumes: EDV, ESV (ml) | ✓ | ||
| Ventricle Length: Diastolic, Systolic (mm) | ✓ | ||
| Motion (mm) | ✓ | ✓ | |
| Motion Defect Extent (%) | ✓ | ✓ | |
| Motion Score | ✓ | ✓ | |
| Thickening (mm) | ✓ | ✓ | |
| Thickening Defect Extent (%) | ✓ | ✓ | |
| Thickening Score | ✓ | ✓ | |
| Phase SD | ✓ | ✓ | |
| Phase Bandwidth | ✓ | ✓ | |
| Phase Dyssynchrony | ✓ | ✓ | |
| Phase Entropy | ✓ | ✓ | |
| Diastolic Parameters: PER (EDV/s), PFR (EDV/s), MFR, (EDV/s), TTPF (ms) | ✓ | ||
| Average RR Interval in ECG (ms) | ✓ | ||
| Transient Ischemic Dilation |
Variables also computed for the 17-segment AHA subdivisions, coronary artery territories (left anterior descending, left circumflex, and right coronary artery) and for myocardial walls (apical, lateral, inferior, septal, and anterior). LV: left ventricular; EDV: end-diastolic volume; ESV: end-systolic volume; PER: peak ejection rate; PFR: peak filling rate; MFR: mean filling rate; MBq: megaBecquerel; SD: standard deviation; TTPF: time to peak filling; QC: automatic quality control; AHA: American Heart Association.
Management and quality control
The data received from the centers are verified to assure that all required information has been provided. Additional efforts are made to standardize all possible mismatches between centers regarding the coding of the clinical information. Codes used to report invasive coronary angiography findings, definitions of categorical variables, and encoding of missing data elements are verified. Additionally, the clinical data is checked for duplicated entries per patient and against quantified image data (one record for every image). Any inconsistencies are resolved with the investigative centers.
Combined database integration
After quantification, the reshaped imaging data are identified by the unique anonymized patient code (assigned at the participant center) and by the site (center name). The patient code matches the code in the corresponding clinical data. The site and patient codes are used to merge the reshaped imaging data information with the clinical data using R software.
Verification of the combined database integrity
Combined data integrity is subsequently verified at the core lab. For the imaging data, intra-patient studies are verified for mismatches in gender and mismatches in patient age. Although the date information is anonymized at the site, the core team checks the difference between relative study times which should be consistent with MPI protocols performed (one-day, two-day or stress-only). Patients with mismatches in DICOM gender and age are followed up with the center and any discrepancies resolved.
RESULTS TO DATE
Participating Investigative Centers
To date, the REFINE SPECT registry includes nine participating centers with solid-state cardiac SPECT MPI scanner systems (D-SPECT, GE Discovery NM530c, and NM/CT570c), as shown in Table 3. Five centers have provided both prognostic and diagnostic data; four centers have provided only diagnostic data.
Table 3.
Imaging Protocols.
| Center | Location | N | Radiotracer | System | Imaging Position | Stress Test Type (Frequency, %) |
|---|---|---|---|---|---|---|
| Centers Providing Prognostic and Diagnostic Data | ||||||
| Cedars-Sinai Medical Center | Los Angeles, CA, USA | Prognostic: 3,674 | Tc-99m sestamibi | D-SPECT | Supine, Upright | Exercise (39%), Pharmacologic (61%) |
| Diagnostic: 199 | Exercise (32%), Pharmacologic (68%) | |||||
| Brigham and Women’s Hospital | Boston, MA, USA | Prognostic: 2,426 | Tc-99m sestamibi | D-SPECT | Supine, Upright | Exercise (71%), Pharmacologic (29%) |
| Diagnostic: 356 | Exercise (69%), Pharmacologic (31%) | |||||
| Ottawa Heart Institute | Ottawa, Ontario, Canada | Prognostic: 3,480 | Tc-99m tetrofosmin | GE Discovery NM530c | Supine, Prone | Exercise (34%), Pharmacologic (66%) |
| Diagnostic: 378 | Exercise (35%), Pharmacologic (65%) | |||||
| Oregon Heart and Vascular Institute | Springfield, OR, USA | Prognostic: 2,691 | Tc-99m sestamibi | D-SPECT | Supine, Upright | Exercise (38%), Pharmacologic (62%) |
| Diagnostic: 336 | Exercise (31%), Pharmacologic (69%) | |||||
| Assuta Medical Center | Tel Aviv, Israel | Prognostic: 8,147 | Tc-99m sestamibi | GE Discovery NM530c | Supine, Prone | Exercise (54%), Pharmacologic (46%) |
| Diagnostic: 314 | Exercise (23%), Pharmacologic (77%) | |||||
| Centers Providing Diagnostic Data only | ||||||
| Columbia University | New York, NY, USA | Diagnostic: 116 | Tc-99m sestamibi | GE Discovery NM530c | Supine, Prone | Exercise (47%) Pharmacologic (53%) |
| Aspire Foundation (CVIT) | Kansas City, MO, USA | Diagnostic: 279 | Tc-99m sestamibi | D-SPECT | Supine, Upright | Exercise (38%) Pharmacologic (62%) |
| Yale University | New Haven, CT, USA | Diagnostic: 65 | Tc-99m tetrofosmin | GE Discovery NM/CT570c | Supine NC and AC | Exercise (38%) Pharmacologic (62%) |
| University of Zurich | Zurich, Switzerland | Diagnostic: 36 | Tc-99m tetrofosmin | GE Discovery NM/CT570c | Supine NC and AC | Exercise (8%) Pharmacologic (92%) |
AC: attenuation corrected; NC: non-attenuation corrected.
Patient Characteristics
20,489 consecutive patients have been registered for the prognostic dataset to date. 71 patients have been excluded from the registry due to lost follow up, missing clinical information, or non-diagnostic MPI (no clinical perfusion read possible). Datasets with imaging artifacts which could be interpreted were not excluded. For the diagnostic dataset, 2,082 consecutive patients have been registered. Three patients have been excluded from the registry due to missing clinical information or non-diagnostic MPI (no clinical diagnosis possible). Patient characteristics of the prognostic (n=20,418) and diagnostic (n=2,079) data of the REFINE SPECT registry collected to date with complete and verified information are shown in Table 4.
Table 4. Patient Characteristics.
Continuous variables reported as mean ± SD; categorical variables reported as n (%).
| Patient Characteristics | Prognostic Data (n= 20,418) | Diagnostic Data (n= 2,079) |
|---|---|---|
|
| ||
| Age (Years) | 64.0 ± 12.1 | 64.7 ± 11.2 |
|
| ||
| Male | 11,642 (57) | 1,385 (67) |
|
| ||
| Body Mass Index (kg/m2) | 28.4 ± 6.2 | 28.6 ± 5.8 |
|
| ||
| Diabetes | 5,212 (26) | 610 (29) |
|
| ||
| Hypertension | 12,920 (63) | 1,422 (68) |
|
| ||
| Dyslipidemia | 12,903 (63) | 1,297 (62) |
|
| ||
| Family History of CAD | 5,642 (28) | 694 (33) |
|
| ||
| Current Smoker | 3,875 (19) | 521 (25) |
|
| ||
| History of PAD | 2,420 (12) | 123 (6) |
|
| ||
| Chest Pain Typicality | ||
| Asymptomatic | 9,576 (47) | 638 (31) |
| Nonanginal | 5,025 (25) | 243 (12) |
| Atypical | 4,592 (22) | 649 (31) |
| Typical | 1,224 (6) | 427 (21) |
|
| ||
| Prior CAD | 5,796 (28) | 0 (0) |
SD: standard deviation, CAD: coronary artery disease, PAD: peripheral artery disease.
Image Acquisition Protocols (Prognostic Cohort)
D-SPECT (Spectrum-Dynamics, Haifa, Israel) (10) scanners have been used at three centers (n=8,791; 43%) and GE Discovery NM 530c scanners (GE Healthcare, Haifa, Israel) (11) at two centers (n=11,627; 57%) (Table 3). Image acquisition protocols of the registry include rest-stress or stress-rest 1-day, rest-stress 2-day, or stress-only protocols (Table 5) (12). Patients have undergone either symptom-limited Bruce/modified Bruce protocol treadmill exercise testing (9,732; 48%) or pharmacologic stress testing combined with low-level exercise whenever possible (10,686; 52%).
Table 5.
Imaging Acquisition. Variables reported as n (%).
| Prognostic Data (n= 20,418) | Diagnostic Data (n= 2,079) | |
|---|---|---|
|
| ||
| Imaging Protocol (Stress) | ||
| 2-Position NC | 18,222 (89.2) | 1,759 (84.6) |
| 1-Position NC | 2,196 (10.8) | 219 (10.5) |
| Supine AC | – | 101 (4.9) |
|
| ||
| Imaging Protocol (Rest) | ||
| 2-Position | 3,460 (16.9) | 422 (20.2) |
| 1-Position NC | 13,910 (68.1) | 1,483 (71.3) |
| Supine AC | – | 95 (4.6) |
|
| ||
| Stress Gated | 20,395 (99.9) | 2,077 (99.9) |
|
| ||
| Rest Gated | 17,323 (84.8) | 1,996 (96.0) |
|
| ||
| Imaging Acquisition | ||
| Rest-Stress on Same Day | 11,858 (58.1) | 1,221 (58.8) |
| Stress-Rest on Same Day | 5,580 (27.3) | 717 (34.5) |
| Stress and Rest on Separate Days | 217 (1.1) | 66 (3.2) |
| Stress Only | 2,763 (13.5) | 74 (3.6) |
2-position (supine/upright or supine/prone); AC: attenuation corrected; NC: non-attenuation corrected.
Image Acquisition Protocols (Diagnostic Cohort)
D-SPECT scanners have been used at four centers (n=1,170; 56%), GE Discovery NM 530c at three centers (n=808; 39%), and GE Discovery NM/CT570c with attenuation correction at two centers (n=101; 5%) (Table 3). Image acquisition protocols are shown in Table 5. Patients have undergone either symptom-limited Bruce/modified Bruce protocol treadmill exercise testing without adjuvant pharmacologic stress (805; 39%) or pharmacologic stress testing (1,274; 61%).
General SPECT Findings
Tables 6 and 7 show an overview of main SPECT findings of the REFINE SPECT registry for the prognostic and diagnostic datasets collected to date.
Table 6.
General SPECT Findings (Prognostic). Variables reported as mean ± SD.
| Visual Perfusion Assessment | Stress TPD (%) | Ischemic TPD (%) | Stress EF (%) | Rest EF (%) |
|---|---|---|---|---|
|
Normal n=12,249 (60%) |
1.9 ± 2.4 | 1.4 ± 1.8 | 64.7 ± 10.0 | 64.8 ± 11.2 |
|
Probably
Normal n=3,386 (17%) |
3.6 ± 2.9 | 2.8 ± 2.4 | 61.2 ± 10.3 | 63.2 ± 10.9 |
|
Equivocal n=681 (3%) |
4.9 ± 3.5 | 3.5 ± 2.6 | 59.2 ± 10.7 | 61.2 ± 11.7 |
|
Abnormal n=4,102 (20%) |
14.6 ± 11.0 | 7.1 ± 5.1 | 50.9 ± 13.9 | 53.2 ± 14.4 |
|
Total Population n = 20,418 |
4.9 ± 7.4 | 2.9 ± 3.6 | 61.1 ± 12.2 | 61.7 ± 12.9 |
TPD: total perfusion deficit, EF: ejection fraction.
Table 7.
General SPECT Findings (Diagnostic). Variables reported as mean ± SD.
| Visual Perfusion Assessment | Stress TPD (%) | Ischemic TPD (%) | Stress EF (%) | Rest EF (%) |
|---|---|---|---|---|
|
Normal n= 482 (23%) |
3.6 ± 4.7 | 2.5 ± 3.3 | 61.7 ± 13.1 | 62.5 ± 14.6 |
|
Probably
Normal n= 70 (3%) |
5.1 ± 3.9 | 3.3 ± 3.9 | 57.7 ± 15.0 | 60.7 ± 14.5 |
|
Equivocal n= 85 (4%) |
6.6 ± 5.0 | 4.3 ± 4.0 | 58.6 ± 13.5 | 62.0 ± 15.0 |
|
Abnormal n= 1,442 (69%) |
13.9 ± 10.2 | 9.5 ± 7.3 | 55.7 ± 14.2 | 57.9 ± 14.4 |
|
Total
Population n= 2,079 |
10.9 ± 10.0 | 7.5 ± 7.0 | 57.3 ± 14.2 | 59.2 ± 14.6 |
TPD: total perfusion deficit, EF: ejection fraction.
DISCUSSION
In summary, we have established a large international imaging registry, derived rich set of 28 clinical data variables, 17 stress test variables, and 32 imaging data variable categories (comprising of over 290 individual imaging variables including regional variables per patient), along with the images for each study patient (20,418 for the prognostic dataset and 2,079 for the diagnostic dataset). All imaging variables were automatically quantified and merged with the clinical data. Patient follow-up for images collected to date was completed for revascularization and invasive coronary angiography in diagnostic study patients, and for MACE events in prognostic study patients. All image variables were derived automatically at the REFINE SPECT core laboratory. Additional image variables could be derived automatically from collected and verified MPI images without any human intervention.
A variety of collaborative projects can result from the establishment of the REFINE SPECT registry. Since this registry includes all the imaging datasets, direct analysis of images from the registry in new projects is possible. In one very recent report, Betancur et al. developed a deep learning artificial intelligence model for automatic prediction of obstructive disease utilizing data directly from the registry (13). Further analyses can be conducted aimed to describe end-points and disease per subpopulations (e.g., gender and obesity). Any additional enhancements to automatic quantitation by emerging machine learning techniques (e.g., deep learning) can be explored for automatic quality control, image analysis, diagnosis and prognosis for MPI. Since all the imaging data variables are quantified automatically, the MPI performance for disease diagnosis or patient prognosis from stress-only protocols can be assessed, and the incremental value of rest imaging can be determined. The automated analysis may facilitate adaptation of stress-only protocols (by allowing automated decisions to cancel rest scans). We plan to further expand the registry including data from additional centers with other types of latest generation MPI, including latest generation SPECT-CT systems.
There are limitations of the REFINE SPECT registry. Coronary stenosis on invasive coronary angiography is assessed by visual assessment at each site, which is known to overestimate the prevalence of functionally significant disease when compared with fractional flow reserve (FFR) (14). Fractional flow reserve measurements are not available in this population, as these are not commonly performed clinically. The accuracy of stenosis interpretation may also differ between centers. The collected MACE events include all-cause death since the definition of cardiac death can be very difficult, particularly among elderly patients with multiple diseases (15). As with other observational registries, there are selection biases for patients referred for coronary angiography after SPECT imaging. There is also potential heterogeneity between centers, and inter-observer and inter-center variability in visual SPECT interpretation. However, this limitation is mitigated by the availability of image data and objective quantitative analysis of all the imaging data.
CONCLUSION
The information collected from the REFINE SPECT registry will provide a valuable resource for researchers to perform collaborative projects related to the diagnostic and prognostic efficacy of the new generation MPI, including the development of novel, automatic analysis with latest machine learning approaches.
Supplementary Material
New Knowledge Gained.
A novel extensive imaging registry has been created for next-generation SPECT MPI. This resource will allow objectifying automated diagnosis of coronary artery disease and prediction of future cardiac events, employing both standard quantitation and machine learning.
Acknowledgments
Conflict of Interest:
This research was supported in part by grant R01HL089765 from the National Heart, Lung, and Blood Institute/National Institutes of Health (NHLBI/NIH) (PI: Piotr Slomka). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures
Drs. Germano, Berman, and Slomka participate in software royalties for QPS software at Cedars-Sinai Medical Center. Dr. Slomka has received research grant support from Siemens Medical Systems. Drs. Berman, Dorbala, Einstein, and Miller have served as consultants for GE Healthcare. Dr. Dorbala has served as a consultant to Bracco Diagnostics; her institution has received grant support from Astellas. Dr. DiCarli has received research grant support from Spectrum Dynamics and consulting honoraria from Sanofi and GE Healthcare. Dr. Ruddy has received research grant support from GE Healthcare and Advanced Accelerator Applications. Dr. Einstein and his institution has received research support from GE Healthcare, Philips Healthcare and Toshiba America Medical Systems. Dr. Miller has served as a consultant for Bracco Inc; and he and his institution has received grant support from Bracco Inc. Dr. Berman’s institution has received grant support from HeartFlow. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
References
- 1.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, et al. Heart disease and stroke statistics–2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:480–6. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Team RC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2017. [Google Scholar]
- 3.Amsterdam EA, Wenger NK, Brindis RG, Casey DE, Ganiats TG, Holmes DR, et al. 2014 AHA/ACC Guideline for the Management of Patients With Non–ST-Elevation Acute Coronary Syndromes. A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. 2014;130:e344–e426. doi: 10.1161/CIR.0000000000000134. [DOI] [PubMed] [Google Scholar]
- 4.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD. Third Universal Definition of Myocardial Infarction. Circulation. 2012;126:2020–35. doi: 10.1016/j.gheart.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 5.van Dijk JD, Mouden M, Ottervanger JP, van Dalen JA, Knollema S, Slump CH, et al. Value of attenuation correction in stress-only myocardial perfusion imaging using CZT-SPECT. Journal of nuclear cardiology : official publication of the American Society of Nuclear Cardiology. 2017;24:395–401. doi: 10.1007/s12350-015-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–42. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Kavanagh P, Fish M, Gerlach J, Ramesh A, Lemley M, et al. Automated quality control for segmentation of myocardial perfusion SPECT. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2009;50:1418–26. doi: 10.2967/jnumed.108.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakazato R, Tamarappoo BK, Kang X, Wolak A, Kite F, Hayes SW, et al. Quantitative upright-supine high-speed SPECT myocardial perfusion imaging for detection of coronary artery disease: correlation with invasive coronary angiography. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2010;51:1724–31. doi: 10.2967/jnumed.110.078782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slomka PJ, Nishina H, Berman DS, Akincioglu C, Abidov A, Friedman JD, et al. Automated quantification of myocardial perfusion SPECT using simplified normal limits. Journal of nuclear cardiology : official publication of the American Society of Nuclear Cardiology. 2005;12:66–77. doi: 10.1016/j.nuclcard.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Gambhir SS, Berman DS, Ziffer J, Nagler M, Sandler M, Patton J, et al. A novel high-sensitivity rapid-acquisition single-photon cardiac imaging camera. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2009;50:635–43. doi: 10.2967/jnumed.108.060020. [DOI] [PubMed] [Google Scholar]
- 11.Herzog BA, Buechel RR, Katz R, Brueckner M, Husmann L, Burger IA, et al. Nuclear myocardial perfusion imaging with a cadmium-zinc-telluride detector technique: optimized protocol for scan time reduction. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2010;51:46–51. doi: 10.2967/jnumed.109.065532. [DOI] [PubMed] [Google Scholar]
- 12.Henzlova MJ, Duvall WL, Einstein AJ, Travin MI, Verberne HJ. ASNC imaging guidelines for SPECT nuclear cardiology procedures: Stress, protocols, and tracers. Journal of nuclear cardiology : official publication of the American Society of Nuclear Cardiology. 2016;23:606–39. doi: 10.1007/s12350-015-0387-x. [DOI] [PubMed] [Google Scholar]
- 13.Betancur J, Commandeur F, Motlagh M, Sharir T, Einstein AJ, Bokhari S, et al. Deep Learning for Prediction of Obstructive Disease From Fast Myocardial Perfusion SPECT: A Multicenter Study. JACC Cardiovascular imaging. 2018 doi: 10.1016/j.jcmg.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonino PA, Fearon WF, De Bruyne B, Oldroyd KG, Leesar MA, Ver Lee PN, et al. Angiographic versus functional severity of coronary artery stenoses in the FAME study fractional flow reserve versus angiography in multivessel evaluation. Journal of the American College of Cardiology. 2010;55:2816–21. doi: 10.1016/j.jacc.2009.11.096. [DOI] [PubMed] [Google Scholar]
- 15.Lauer MS, Blackstone EH, Young JB, Topol EJ. Cause of death in clinical research: time for a reassessment? Journal of the American College of Cardiology. 1999;34:618–20. doi: 10.1016/s0735-1097(99)00250-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


